Abstract
Active transport of materials across the cellular membrane is one the most fundamental processes in biology. In order to accomplish this task, membrane transporters rely on a wide range of conformational changes spanning multiple time and size scales. These molecular events govern key functional aspects in membrane transporters, namely, coordinated gating motions underlying the alternating access mode of operation, and coupling of uphill transport of substrate to various sources of energy, e.g., transmembrane electrochemical gradients and ATP binding and hydrolysis. Computational techniques such as molecular dynamics simulations and free energy calculations have equipped us with a powerful repertoire of biophysical tools offering unparalleled spatial and temporal resolutions that can effectively complement experimental methodologies, and therefore help fill the gap of knowledge in understanding the molecular basis of function in membrane transporters.
Keywords: membrane transporters, molecular dynamics simulation, large-scale conformational change, active transport, alternating access mechanism, free energy calculations
Introduction
Exchange of molecules across the cellular membrane is one of the most essential biological processes in all living organisms. This fundamental task is carried out by a structurally and mechanistically diverse class of membrane proteins known as membrane transporters, which provide highly efficient transport machinery for a wide range of molecular species. They couple the translocation of their (transport) substrates, which often takes place against the electrochemical gradient across the membrane, to various source of chemical energy in the cell. Despite diversity in structure and mechanistic details, active membrane transporters all operate via the alternating access model (1, 2), during which substrate accessibility is switched between the two sides of the membrane. This process relies on a highly coordinated set of complex structural transitions (at different scales and involving various structural elements in the transporter protein) that give rise to interconversion between two major functional states, referred to as the inward-facing (IF) and outward-facing (OF) states respectively, through transition pathways visiting multiple intermediate states involving at least one occluded state which prevents free diffusion of the substrate along the gradient. The involvement of such complex conformational changes in the mechanism and how they are fueled by a particular energy-providing mechanism and at the same time coupled to vectorial translocation of the transported species, highlight the indispensable role of protein dynamics in transporter function. Given technical challenges in describing the dynamics of membrane transporters, in spite of the rapid pace of their structural studies, critical mechanistic aspects remain largely unknown.
High spatial and temporal resolutions offered by molecular dynamics (MD) simulations can be exploited to trace motions of both the transported species, and the entire membrane transporter, thus elucidating the transport mechanism at an atomic level. The rapid growth of powerful computational resources and the development of more efficient computational sampling algorithms in the past decade have significantly expanded the scope of MD simulations in biophysical studies of membrane transporters. Here we provide a brief survey of recent simulation studies performed on membrane transporters where detailed molecular events relevant to function have been successfully described.
The transport cycles of membrane transporters involve highly diverse events of structural changes, ranging from local rearrangements at the binding sites and their gating elements, to global conformational transitions furnishing the shift of their accessibility between the two sides of membrane. Depending on the caliber and/or timescale of these events, it may be covered with conventional MD simulations or requires different methods of model simplification or sampling enhancement.
This review is structured based on the nature and magnitude of such molecular events. We will first describe more localized events, e.g., gating motions or binding and unbinding of the substrate and cotransported ions, as well as the molecular events arising from close coupling of the transporter with its environment of water and lipids. Then in the final section we focus on most recent studies aiming at describing large-scale structural transitions in transporters and novel methodological approaches required.
Dissecting Functionally Relevant Chemical Details
MD simulations have been widely employed to study localized molecular events involved in the transport cycle, providing atomic-level dynamical description of the membrane protein, transported ligands, and parts of the environment essential to the transport function. Combined with free energy calculations and enhanced sampling methods such as metadynamics, umbrella sampling (US), replica exchange (RE), thermodynamic integration (TI), and free energy perturbation (FEP), MD simulations can describe some critical events, on timescales ranging from picoseconds to tens of microseconds, in detail with sufficient statistics.
Molecular driving forces fueling the transport
The coupling between the driving force and driven species is at the heart of active transport, thus characterizing chemical details of interactions between protein and species providing driving forces, e.g., ions or ATP, is key to our understanding of the transport mechanism. MD simulations have been successfully employed to identify ion-binding sites, the binding sequence of different ions, and the structural effects coupled to ions in secondary active transporters. Similarly, the method has been employed to model the impact of ATP binding and hydrolysis in ATP-driven transporters such as ABC transporters.
As the most prominent ion to provide the driving force in secondary transporters, Na+ binding and ensuing conformational response of transporters have been extensively studied in several Na+-coupled transporter families (3–16). A novel putative Na+-binding site was predicted by MD simulations and structural symmetry analysis of betaine transporter (BetP). The predicted site was validated by experimentally observed changes in apparent Km, Kd, and Na+ transient current, in response to side chain mutations (4). In Na+/Ca2+ exchangers (NCXs), a combined MD and FEP study identified optimal occupancy and specificity for four putative ion binding sites (3). The processes of Na+ binding and unbinding and associated energetics have been characterized by extended equilibrium simulations complemented by metadynamics or US in a number of transporters (5, 8–12, 14–16). Some of these studies also reported on Na+-induced protein conformational changes, e.g., in BetP (5), leucine transporter (LeuT) (8, 9), glutamate transporter homologue (GltPh) (12), and a multidrug transporter (NorM) (14). Interestingly, the involvement of intermediate (quasi-stable) Na+-binding sites has been inferred for several LeuT-fold transporters (a superfamily sharing structural architecture of LeuT) (8–11). In a recent concerted experimental and computational study, restrained ensemble simulations guided by EPR measurements were used to probe the effect of Na+ binding on different conformational states of LeuT-fold transporters (6, 7).
H+ is another important ion in secondary active transporters. In order to probe the effect of H+ binding (protonation) on protein dynamics, conventional MD simulations can be performed on and compared between protonated and unprotonated states. Several studies exploiting this strategy have demonstrated H+-induced conformational changes for H+- coupled symporters from the major facilitator superfamily (MFS), namely, lactose permease (LacY) (17), oligopeptide transporter (POT) (18), and xylose permease (XylE) (19), as well as in two H+-coupled antiporters, i.e., LeuT (20) and Na+/H+ antiporter NhaA (16). With the goal of more quantitatively describe the protonation state of titratable residues in H+-coupled transporters, an integrated approach based on FEP and TI has been employed to identify H+-binding sites, and the binding order of ions and substrate in an excitatory amino acid transporter (EAAT3) (21).
Active transport in ABC transporters is powered by their ATPase domains, which are considered almost always nucleotide-bound due to their low Km. Given an apo ABC transporter structure, the enzymatic substrate MgATP can be accurately modeled in its binding site computationally by using high resolution structures of nucleotide-bound ATPase domains as a template (22). The free energy of ATPase dimerization (induced by ATP binding) has been computed and decomposed into enthalpic and entropic terms, showing that water entropy might play an important role in driving the process (23). The chemical reaction of ATP hydrolysis within the ATPase domains has been examined with QM/MM simulations supporting a general acid catalysis mechanism (24). The conformational changes of the ATPase domains after the hydrolysis have been simulated using an isolated ATPase dimer bound to the hydrolysis products (ADP and Pi), showing that the homodimeric ATPase domains consistently exhibit asymmetric conformations (25).
Substrate binding, unbinding, and translocation
Substrate transport consists of three major components, including (using an example of an importer) binding to the OF state, translocation, and release (unbinding) from the IF state. These components have been best studied computationally for the superfamily of LeuT-fold transporters, with numerous studies providing critical mechanistic details (5, 26–32). Combining MD simulations with crystallography and other experimental measurements, substrate-induced progressive reshaping and dehydration of the central binding site (5), and the mechanism by which specific hydrogen bonds guide the motion of the substrate during its binding (26) are demonstrated for two LeuT-fold transporters. The process of substrate release has been also captured computationally, either in microsecond-long equilibrium simulations as performed for carnitine/gamma-butyrobetaine antiporter (CaiT) (29), or using steered MD (27) or metadynamics (28) in a bacterial Na+-coupled glucose transporter (vS-GLT). The dynamical perspective offered by these simulation studies allows a more informed assessment of the functional state of a particular conformation. For example, the above studies (27, 29) suggest an open (IF) state for the crystallographically captured structure of the transporter. Substrate translocation in LeuT has been also described by combining the results of a comprehensive set of equilibrium, accelerated, and targeted MD simulations (30, 31). In a recent computational study, a new information theory-based analysis method was employed to characterize functionally important coupling between substrate binding and the intracellular gate in LeuT (32).
For glutamate transporter homolog GltPh, metadynamics simulations were used to study substrate uptake, release, and its coupling to cotransported Na+ ions (12). Free energy profiles were calculated at various stages, proposing a sequence of events for extracellular uptake and cytoplasmic release of the substrate and Na+ ions, and elucidating their coupling to gate opening and closure events. A subsequent study also investigating the process of ligand release in GltPh, supplemented an important element missing in the previous study, i.e., the Na+ ion in the Na3 site (13). Another study taking advantage of multiple simulations, each on the order of several microseconds, provides a detailed view on gating elements and their coupling in GltPh, concluding that helical hairpin 2 (HP2) acts as the main cytoplasmic gate in this transporter (33).
Drug binding to multidrug exporter Pgp, which can be viewed as the most biomedically relevant ABC transporter, likely represents the most computationally studied binding process in primary transporters. Most studies, however, have employed a conventional docking approach based on single structures obtained crystallographically for this transporter. Since Pgp exhibits a high degree of structural flexibility (22), a more meaningful docking would take advantage of conformation ensembles generated by MD simulations (34, 35). A more rigorous approach has been utilized to model the bound substrate in a different ABC transporter, where the protein-substrate interactions were exhaustively sampled using replica-exchange simulations (36).
Active participation of water and lipids in transporter function
Membrane transporters perform their function while intimately interacting with lipid molecules and water. Explicit representation of these molecular species in all-atom MD simulations enables us to closely probe many possible mechanisms by which they can affect transporter function.
Monitoring the dynamics of water molecules has brought novel insights into the critical aspects of transport mechanism. Direct involvement of water molecules in coordinating bound ions has been observed in MD simulations, e.g., in BetP (4). More interestingly, simulation studies have established a strong correlation between the level of hydration and binding/unbinding events, both for ions and the substrate, in diverse secondary transporters (5, 9, 14, 27, 28, 31).
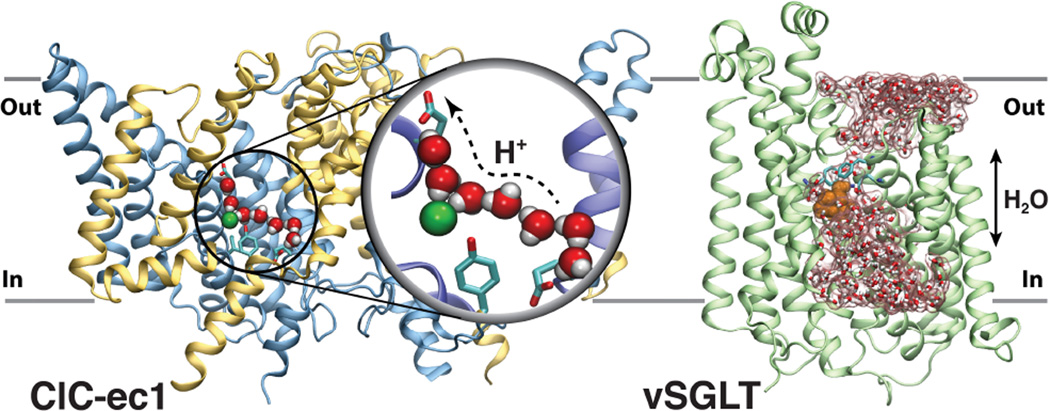
Water molecules are commonly present in H-bonded networks within biomolecules. Owing to their ability of both accepting and donating H-bonds, water can act as an effective H+ transfer mediator. In the H+/Cl− exchangers of the Cl− channel (CLC) family, MD simulations and free energy calculations demonstrated a coupling between Cl− binding and luminal hydration (37, 38). Repeated formation of transient water wires within a 15-Å hydrophobic region connecting the two known H+ binding sites (Fig. 2) was found to depend on the presence of Cl−, thereby providing a novel mechanism of coupling between Cl− and H+ transport in CLC antiporters (37).
Figure 2.
Monitoring water dynamics within proteins brings novel insights into transport mechanism. (Left) The continuous water wire spanning the ClCec1 lumen depend on the presence of Cl− (green sphere), as the H+ transport pathway. See (37) for further details. (B) Transient water-conducting state observed in vSGLT, which is permeable to water but occluded to the substrate (orange van der Waals). See (39) for further details.
A large set of extended equilibrium MD simulations performed on several membrane transporters representing different classes and distinct functional states characterized spontaneous formation of transient water-conducting (channel-like) states (Fig. 2) as a general phenomenon in transporters (39). The results led to a revised structural framework for the alternating access mechanism in transporters that does not require a tight sealing of gating elements during the transport cycle, thereby allowing for conduction of small molecules such as water (39). Extensive sampling provided by nearly 10 µs of MD simulations was used to characterize structural elements responsible for differential water permeability in bacterial and mammalian SGLT homologs (40).
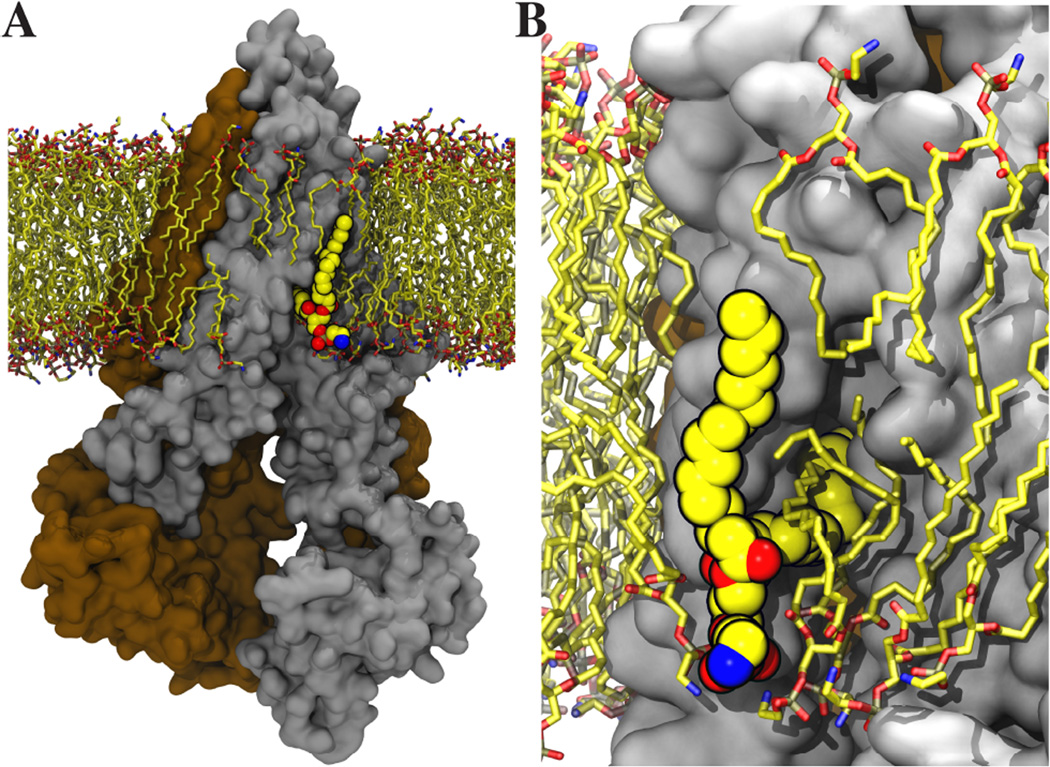
Lipid molecules can influence the activity of membrane transporters through various mechanisms. Although this role has not been extensively investigated computationally, a number of recent simulation studies have probed strong lipid dependence of transporter structure and dynamics. Examples include the demonstration of differential dynamics of LacY in bilayers with different lipid compositions (17), and quantification of energetic cost associated with hydrophobic mismatch of LeuT in mixed lipid bilayers (41). A special case of functionally relevant lipid-protein interaction captured in MD simulations is the protrusion of an annular lipid into the lumen of Pgp, in line with the known flippase activity of the transporter (Fig. 3) (22).
Figure 3.
Example of protein-lipid interactions captured in MD simulations. (A) The protrusion of an annular lipid into the substrate binding site of Pgp, which is a known lipid flippase. (B) Close-up view of the protrusion site along the crevice opening. See (22) for further details.
The majority of membrane protein structures are derived from detergent-solubilized samples, raising the concern about artefactual effect of detergents on the structure and its physiological relevance. Indeed, several computational studies have demonstrated the substantial changes in structural and functional properties of transporters due to detergent (42–44). For instance, combining activity measurements and molecular simulations it was demonstrated that the structure and function of the mitochondrial anion carrier UCP2 can be severely altered by a zwitterionic detergent (42). The illustration that the secondary substrate-binding site of LeuT (45, 46) can be occupied by a neutral detergent, as captured in MD simulations (43) represents a remarkable example of direct functional impact of detergents. Similarly, simulating Pgp under its crystallization conditions has also shown that anionic detergent molecules not only aggregate around the transmembrane domains, they also nonspecifically bind to the soluble regions (44), suggesting that the effect of detergents on the protein may be more extensive than expected.
Probing Large-Scale Structural Transitions
Most membrane transporters rely on complex, large-scale conformational changes for their function. however, we have a limited understanding of structural dynamics and conformational transitions between the major functional states of these proteins. From a computational perspective, the most daunting challenge is to overcome the sampling limitations. Global conformational changes in transporters, e.g., the transitions between OF and IF states, usually occur on timescales ranging from tens of milliseconds to seconds, which are still far beyond the reach of the conventional equilibrium MD simulations. Recent significant technological advances have already extended all-atom MD simulations for membrane proteins to hundreds of microseconds, as reported, e.g., for a voltage-gated K+ channel (47), and for a G protein-coupled receptor (48). These massive simulations, however, require specific computational resources accessible only to limited research groups. Currently the MD simulations for membrane transporters may reach up to tens of microseconds (33, 40). Due to the sampling limitations, the scope of full-atomic simulation studies is often limited to the local conformational changes. The global conformational change between the IF and OF states, on the other hand, is often studied either using qualitative techniques or simplified representations. State-of-the-art supercomputing and novel enhanced sampling techniques, however, have begun to open new opportunities for the quantitative study of large-scale conformational changes at atomic resolutions as discussed below.
Accelerating the dynamics by reduced representation
A common strategy to overcome the sampling problem and describe large-scale conformational changes of transporters is to employ simplified representations of the protein and/or environment. For instance, implicit membrane simulations (49) and coarse-grained models such as mixed elastic network models (mENM) (50), two-state anisotropic network models (51) and GŌ-like models (52) have been used to probe the OF-IF transition of transporters, often accompanied with the use of an enhanced sampling method such as dynamic importance sampling (DIMS) (53, 54), weighted ensemble (WE) path-sampling (52), and self-guided Langevin dynamics (SGLD) (49). The predicted transition pathways obtained from simplified models are sometimes converted into full-atomic representations and analyzed using all-atom unbiased MD simulations launched from the intermediates (50, 51, 53).
DIMS simulations of hydantoin transporter (Mhp1) suggest that IF–OF transition is primarily achieved by a rigid body movement of four transmembrane helices relative to the rest of the protein (54). Using WE path-sampling and a coarse-grained representation, two transport modes for Mhp1 have been suggested: one consistent with a strict alternating access model, and another decoupling the inner and outer gates (52). The IF–OF transition of LacY has been probed by SGLD and implicit/explicit membrane simulations, in which protonation of a glutamate residue and sugar binding are found to trigger the transition toward the OF state, and sugar undergoes an orientational change during the transition (49). Employing DIMS and implicit solvent models, simulations suggest an intricate coupling between two flexible gates during the transition, rather than a simple rigid-body motion for LacY (53). The two-state anisotropic network model has been employed to construct the transition pathway of GltPh (51). Only using a few low frequency normal modes, this study was able to describe the entire transition as a rigid-body motion of the transport domain relative to the trimerization domain, followed by intra-domain rearrangements (51). Another study combining motion planning and mENM concluded that the transition of GltPh mainly involves movement of the transport and trimerization domains in opposite directions along the membrane normal (50).
Towards complete description of the transport cycle
OF-IF conformational transition is beyond the time scales allowed by conventional all-atom MD; however, extended conventional equilibrium simulations may sample the conformational ensemble within the same state which could also involve large-scale conformational fluctuations. Such studies have been especially prominent in the case of the IF state of ABC exporters. MD simulations of bacterial and mammalian ABC exporters have revealed their distinct flexibility in the IF state (22, 55, 56), the finding strongly supported by experimental approaches such as hydrogen/deuterium exchange (55) and EPR-based measurements (22, 56).
Qualitative study of global OF-IF transition without compromising the atomic representation is often accomplished by using targeted MD (TMD), a conventional method to speed up the transition towards a final “target” structure in a nonequilibrium setting. This approach has been widely used to describe structural transitions (19, 30, 31), to explore unknown functional states (34, 35, 51, 57), and to generate an initial pathway connecting two end states (11) in membrane transporters. However conventional TMD may not always be suitable for OF-IF transition in transporters. For instance, it is shown that TMD simulations of MsbA results in a pore-like structure of the transmembrane domains permeable for substrate to both sides of the membrane (58, 59), clearly indicating the functional irrelevance of the emerging pathway. In order to overcome such problems, a novel approach has been developed which makes use of nonequilibrium simulations in a more systematic way by taking advantage of an extensive initial search for system-specific reaction coordinates combined with nonequilibrium work relations (58, 59). TMD simulations can be also relaxed using methods such as accelerated MD along with unbiased simulations, as was used in the study of LeuT to reconstruct the transport cycle (31). Along with the already known conformations, two substrate-bound and substrate-free intermediates were characterized which were both minimally hydrated and appeared during the OF→IF and IF→OF transitions, respectively (31).
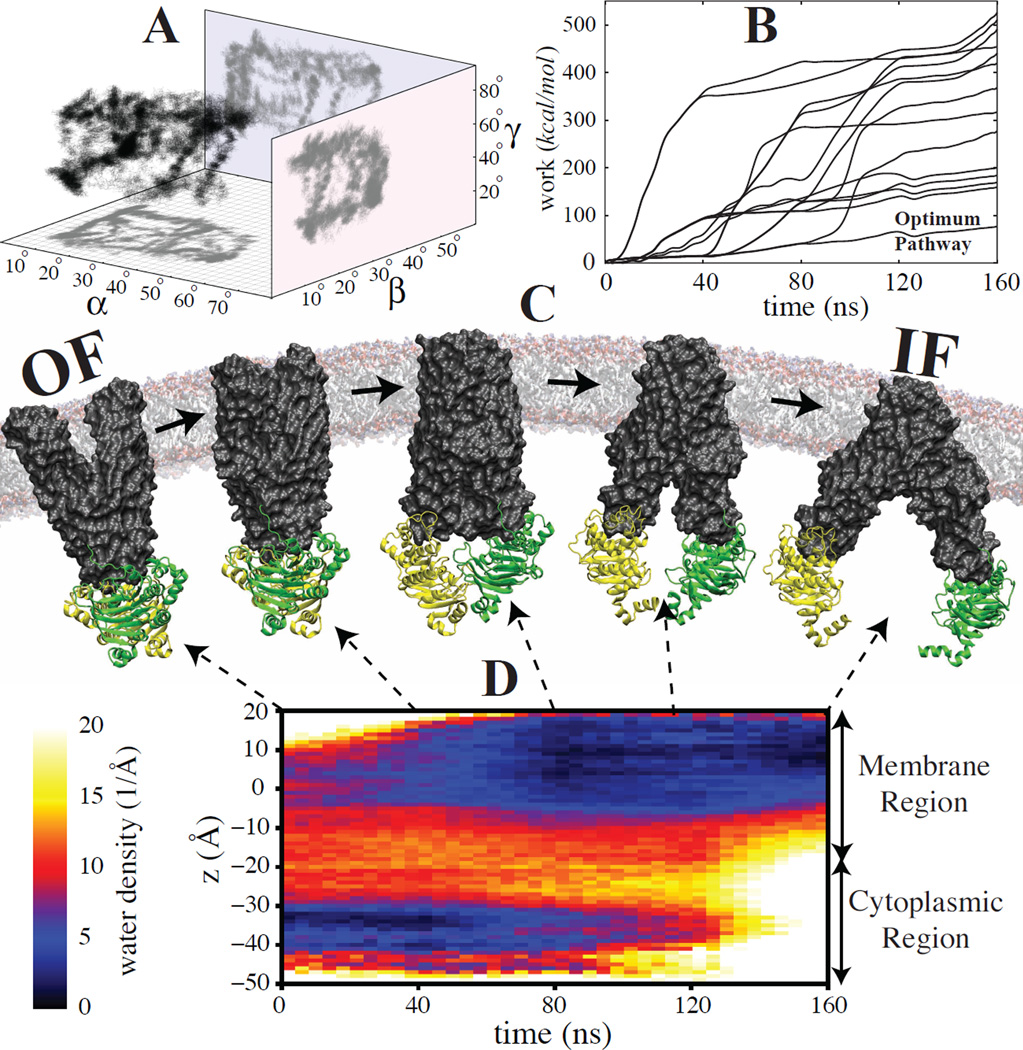
Several attempts have been made recently (11, 58, 59) to use state-of-the-art supercomputing and enhanced sampling techniques to quantitatively characterize large-scale conformational changes in fully atomistic simulations using free energy calculation methods. It is known that conventional US based on simple reaction coordinates such as RMSD may not be suitable for OF-IF transition, with a similar problem for TMD as mentioned above. One approach to relax the TMD generated pathways before performing US is using path-finding algorithms such as string method with swarms of trajectories (11). Using system-specific reaction coordinates mentioned above can also improve the US results by making the sampled regions of the conformational space more relevant to the transition of interest (58, 59). Once a reliable initial pathway is achieved one may calculate free energies using conventional US or its parallel version, i.e., bias-exchange US (BEUS) (58, 59). Unlike TMD-based pathways, a careful system-specific biasing protocol was shown to be able to generate a transition pathway for MsbA which is in line with alternating access mechanism (Fig. 4). In addition, it was shown the relative orientation of the ATPase domains in the cytoplasmic side is coupled to the conformation of the transmembrane domains on both cytoplasmic and periplasmic sides, suggesting a novel hypothesis for the conformational dynamics of ABC exporters termed the “doorknob” mechanism (58).
Figure 4.
Investigating the OF→IF transition in MsbA transporter using system-specific reaction coordinates α, β, and γ, describing protein global conformation (58). (A) Multiple nonequilibrium MD trajectories generated using different biasing protocols and projected onto the reduced space of (α, β, γ). (B) Nonequilibrium work required to induce the OF→IF transition associated with trajectories shown in (A). (C) Snapshots of MsbA structure along the optimum pathway shown in (B). (D) The water density along the pore measured for the optimum pathway shown in (C). These results are in line with alternating access mechanism.
Conclusions and Outlook
Molecular modeling and simulation technologies have contributed considerably to our understanding of the underlying molecular mechanism of membrane transporters. These computational techniques are highly complementary to experimental biophysical methods, owing to their advantageous spatial and temporal resolutions that enable them to provide the most detailed perspective on complex molecular processes involved in the transport cycle. A wide range of functionally relevant molecular events can be successfully described by equilibrium MD simulations, which can nowadays be performed rather routinely on the order of µs for an average-sized, atomistically represented membrane transporter within the natural environment of a fully solvated membrane. Complemented by enhanced sampling techniques, equilibrium simulations provide adequate data to achieve both a mechanistic picture and a quantitative description for localized events such as hydration/dehydration, substrate/ion binding and translocation, and gating motions at the level of side chains and small structural elements.
Taking advantage of specialized hardware, it has been possible to further extend the scope of equilibrium simulations and sampling to several hundreds of µs for membrane proteins, although such resources have been so far rather limited not allowing their widespread use. Coarse-grained simulations appear to offer another alternative to help fill the timescale gap of atomistic simulations. However, given the extremely high relevance of chemical details (e.g., gating motions of side chains, coupled translocation of substrate and ions, hydration, etc) in the mechanism of membrane transporters, the application of such reduced models to study mechanistic details has been rather limited. An effective approach might be to leverage the low-resolution results obtained from coarse-grained methods, e.g., on global structural changes of a membrane transporter, and use them to inform and guide detailed atomistic simulations.
Based on the large number of structures characterized experimentally, particularly over the recent years, the alternating access mechanism in membrane transporter for the large part hinges on large-scale (global) protein structural transitions. In order to describe these slow motions, which are refractory to equilibrium simulations, non-equilibrium methods such as targeted or steered MD offer effective alternative strategies, and have been employed to several membrane transporters, especially those for which multiple functional states have been structurally solved. However, for complex structural transitions in membrane transporters, careful design of an optimal pathway appears to be the most critical aspect for non-equilibrium simulations. Optimizing the pathway by selecting specific reaction coordinates (instead of generic coordinates such as RMSD), combining collective coordinates, and a more comprehensive search of the space during the initial phase of non-equilibrium simulations, have proven to be essential for obtaining meaningful transitions for membrane transporters.
Computational techniques have already become an indispensable component of structural biological and biophysical studies of complex molecular systems. Empowered by larger supercomputing resources, more effective computational algorithms, and more efficient enhanced sampling techniques, only a deeper integration of computational techniques into mechanistic studies of membrane transporters can be expected in years to come. Computational studies will soon be able to provide free energy descriptions of various steps and transitions involved, together with structural characterization of the system at an atomic level, for the entire transport cycle of membrane transporters. Such an unprecedented level of integrated thermodynamic and structural characterizations of the system allows for detailed description of elusive intermediate states, which is the ultimate goal of structural biological studies of these molecular machines. Availability of more intermediates would markedly expand the scope of structure-based drug design efforts in membrane transporters, which constitute one of the leading targets in pharmacological intervention of CNS and metabolic disorders.
In the future, we should witness an even closer interaction between experiment and computation. Although such concerted efforts have already been reported in recent years, with several examples highlighted in this review, we have just begun to exploit the potential of this combined approach.
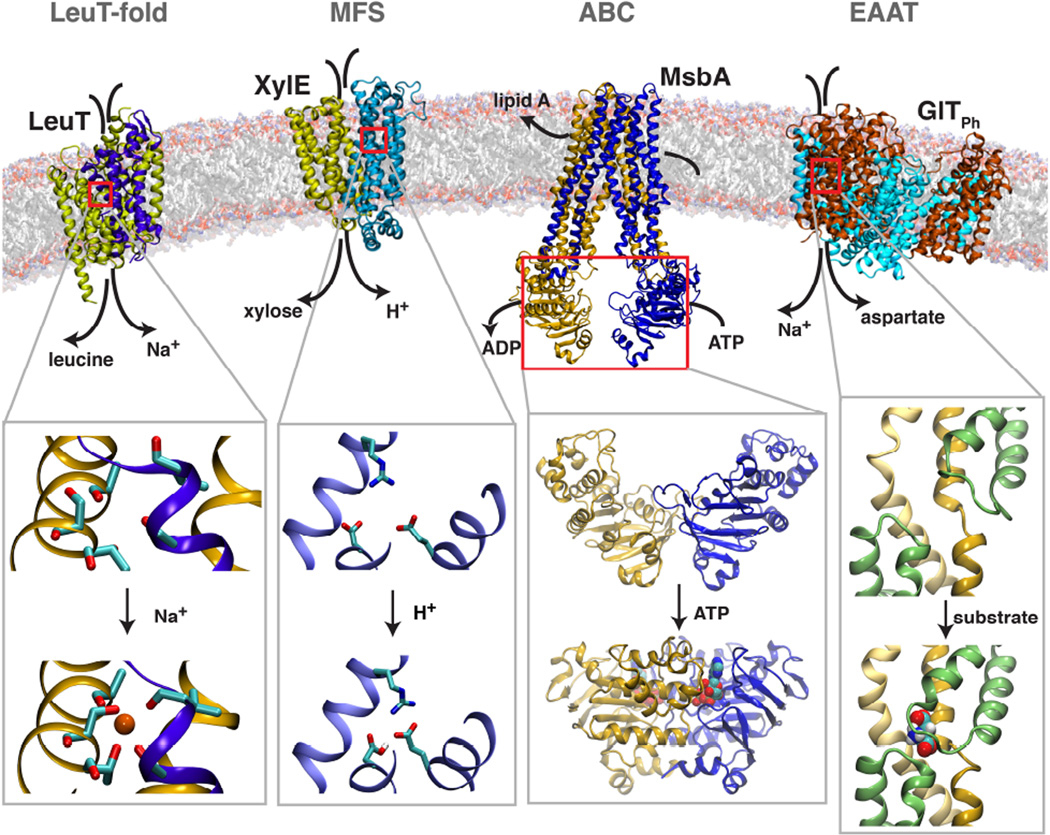
Figure 1.
Representative membrane transporters and functional events studied recently. Proteins are shown in cartoon representation, with the lipid bilayer in the background. Each transporter respectively represents a well-documented superfamily/family of membrane transporters in recent computational studies. Four close-up views of local conformations relevant with several critical functional events, i.e., Na+-binding (left), H+-binding (middle left), ATP binding (middle right), and substrate binding (right).
Highlights.
Membrane transporters rely on diverse conformational transitions for their function
Advanced computational techniques successfully describe membrane transporter dynamics
Major molecular events and processes described by simulation studies are discussed
Energetics of various steps involved in transport can be described computationally
Acknowledgements
The authors would like to acknowledge support received from the National Institutes of Health (grants R01–GM086749, U54–GM087519, and P41–RR05969), and supercomputing allocation through Xsede (grant MCA06N060), PSC Anton, and the Blue Waters. The authors also thank Dr. Wei Han, Josh Vermaas and Tao Jiang for their assistance with the figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors have no conflicts of interest to declare.
References
- 1.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180:134–136. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- 2.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 3. Marinelli F, Almagor L, Hiller R, Giladi M, Khananshvili D, Faraldo-Gómez JD. Sodium recognition by the Na+/Ca2+ exchanger in the outward-facing conformation. Proc Natl Acad Sci USA. 2014;111:E5354–E5362. doi: 10.1073/pnas.1415751111. • Combining structural analysis, MD and FEP, this study reports a detailed thermodynamic analysis of ion binding and specificity in a complex exchanger (Na+/Ca2+ exchanger)
- 4. Khafizov K, Perez C, Koshy C, Quick M, Fendler K, Ziegler C, Forrest LR. Investigation of the sodium-binding sites in the sodium-coupled betaine transporter BetP. Proc Natl Acad Sci USA. 2012;109:E3035–E3044. doi: 10.1073/pnas.1209039109. • Structural analysis and MD simulations were used to characterize elusive ion binding sites in a secondary transporter, with the computational results validated experimentally.
- 5.Perez C, Faust B, Mehdipour AR, Francesconi KA, Forrest LR, Ziegler C. Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na+ coupling. Nat Commun. 2014;5:4231. doi: 10.1038/ncomms5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazmier K, Sharma S, Islam SM, Roux B, Mchaourab HS. Conformational cycle and ion-coupling mechanism of the Na+/hydantoin transporter Mhp1. Proc Natl Acad Sci USA. 2014;111:14752–14757. doi: 10.1073/pnas.1410431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kazmier K, Sharma S, Quick M, Islam SM, Roux B, Weinstein H, Javitch JA, Mchaourab HS. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat Struct Mol Biol. 2014;21:472–479. doi: 10.1038/nsmb.2816. •• A closely concerted experimental and computational study investigating ion/substrate-coupled dynamics of LeuT.
- 8.Zomot E, Gur M, Bahar I. Microseconds simulations reveal a new sodium-binding site and the mechanism of sodium-coupled substrate uptake by LeuT. J Biol Chem. 2015;290:544–555. doi: 10.1074/jbc.M114.617555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao C, Stolzenberg S, Gracia L, Weinstein H, Noskov S, Shi L. Ion-controlled conformational dynamics in the outward-open transition from an occluded state of LeuT. Biophys J. 2012;103:878–888. doi: 10.1016/j.bpj.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisha I, Laio A, Magistrato A, Giorgetti A, Sgrignani J. A candidate ion-retaining state in the inward-facing conformation of sodium/galactose symporter: Clues from atomistic simulations. J Chem Theor Comp. 2013;9:1240–1246. doi: 10.1021/ct3008233. [DOI] [PubMed] [Google Scholar]
- 11.Zhao C, Noskov SY. The molecular mechanism of ion-dependent gating in secondary transporters. PLoS Comput Biol. 2013;9:e1003296. doi: 10.1371/journal.pcbi.1003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grazioso G, Limongelli V, Branduardi D, Novellino E, Micheli CD, Cavalli A, Parrinello M. Investigating the mechanism of substrate uptake and release in the glutamate transporter homologue GltPh through metadynamics simulations. J Am Chem Soc. 2012;134:453–463. doi: 10.1021/ja208485w. • Free energy profiles for substrate/ion uptake and release in glutamate transporter GltPh are computed using metadynamics simulations, proposing a sequence for binding events and their coupling.
- 13.Heinzelmann G, Bastug T, Kuyucak S. Mechanism and energetics of ligand release in the aspartate transporter GltPh. J Phys Chem B. 2013;117:5486–5496. doi: 10.1021/jp4010423. [DOI] [PubMed] [Google Scholar]
- 14.Vanni S, Campomanes P, Marcia M, Rothlisberger U. Ion binding and internal hydration in the multidrug resistance secondary active transporter NorM investigated by molecular dynamics simulations. Biochemistry. 2012;51:1281–1287. doi: 10.1021/bi2015184. [DOI] [PubMed] [Google Scholar]
- 15.Lee C, Kang HJ, von Ballmoos C, Newstead S, Uzdavinys P, Dotson DL, Iwata S, Beckstein O, Cameron AD, Drew D. A two-domain elevator mechanism for sodium/proton antiport. Nature. 2013;501:573–577. doi: 10.1038/nature12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C, Yashiro S, Dotson DL, Uzdavinys P, Iwata S, Sansom MSP, von Ballmoos C, Beckstein O, Drew D, Cameron AD. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J Gen Physiol. 2014;144:529–544. doi: 10.1085/jgp.201411219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson M, Bondar AN, Freites JA, Tobias DJ, Kaback HR, White SH. Proton-coupled dynamics in lactose permease. Structure. 2012;20:1893–1904. doi: 10.1016/j.str.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doki S, Kato HE, Solcan N, Iwaki M, Koyama M, Hattori M, Iwase N, Tsukazaki T, Sugita Y, Kandori H, Newstead S, Ishitani R, Nureki O. Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc Natl Acad Sci USA. 2013;110:11343–11348. doi: 10.1073/pnas.1301079110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wisedchaisri G, Park MS, Iadanza MG, Zheng H, Gonen T. Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat Commun. 2014;5:4521. doi: 10.1038/ncomms5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kantcheva AK, Quick M, Shi L, Winther AML, Stolzenberg S, Weinstein H, Javitch JA, Nissen P. Chloride binding site of neurotransmitter sodium symporters. Proc Natl Acad Sci USA. 2013;110:8489–8494. doi: 10.1073/pnas.1221279110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzelmann G, Kuyucak S. Molecular dynamics simulations elucidate the mechanism of proton transport in the glutamate transporter EAAT3. Biophys J. 2014;106:2675–2683. doi: 10.1016/j.bpj.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen PC, Verhalen B, Wilkens S, Mchaourab HS, Tajkhorshid E. On the origin of large flexibility of P-glycoprotein in the inward-facing state. J Biol Chem. 2013;288:19211–19220. doi: 10.1074/jbc.M113.450114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi T, Chiba S, Kaneta Y, Furuta T, Sakurai M. ATP-induced conformational changes of nucleotide-binding domains in an ABC transporter. importance of the water-mediated entropic force. J Phys Chem B. 2014;118:12612–12620. doi: 10.1021/jp507930e. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Ojeda-May P, Pu J. H-loop histidine catalyzes ATP hydrolysis in the E. coli ABC-transporter HlyB. Phys Chem Chem Phys. 2014;15:15811–15815. doi: 10.1039/c3cp50965f. [DOI] [PubMed] [Google Scholar]
- 25.George AM, Jones PM. An asymmetric post-hydrolysis state of the ABC transporter ATPase dimer. PLoS One. 2013;8:e59854. doi: 10.1371/journal.pone.0059854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons KJ, Jackson SM, Brueckner F, Patching SG, Beckstein O, Ivanova E, Geng T, Weyand S, Drew D, Lanigan J, Sharples DJ, Sansom MS, Iwata S, Fishwick CW, Johnson AP, Cameron AD, Henderson PJ. Molecular mechanism of ligand recognition by membrane transport protein, Mhp1. EMBO J. 2014;33:1831–1844. doi: 10.15252/embj.201387557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Tajkhorshid E. A gate-free pathway for substrate release from the inwardfacing state of the Na+-galactose transporter. Biochim Biophys Acta Biomembr. 2012;1818:263–271. doi: 10.1016/j.bbamem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bisha I, Rodriguez A, Laio A, Magistrato A. Metadynamics simulations reveal a Na+ independent exiting path of galactose for the inward-facing conformation of vSGLT. PLoS Comput Biol. 2014;10:e1004017. doi: 10.1371/journal.pcbi.1004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zomot E, Bahar I. A conformational switch in a partially unwound helix selectively determines the pathway for substrate release from the carnitine/-γ butyrobetaine antiporter CaiT. J Biol Chem. 2012;287:31823–31832. doi: 10.1074/jbc.M112.397364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng MH, Bahar I. Coupled global and local changes direct substrate translocation by neurotransmitter-sodium symporter ortholog LeuT. Biophys J. 2013;105:630–639. doi: 10.1016/j.bpj.2013.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng MH, Bahar I. Complete mapping of substrate translocation highlights the role of LeuT N-terminal segment in regulating transport cycle. PLoS Comput Biol. 2014;10:e1003879. doi: 10.1371/journal.pcbi.1003879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeVine MV, Weinstein H. NbIT - a new information theory-based analysis of allosteric mechanisms reveals residues that underlie function in the leucine transporter LeuT. PLoS Comput Biol. 2014;10:e1003603. doi: 10.1371/journal.pcbi.1003603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zomot E, Bahar I. Intracellular gating in an inward-facing state of aspartate transporter GltPh is regulated by the movements of the helical hairpin HP2. J Biol Chem. 2013;288:8231–8237. doi: 10.1074/jbc.M112.438432. • A very large data set composed of several multi-μs simulations used to characterize gating elements in glutamate transporter.
- 34.Wise JG. Catalytic transitions in the human MDR1 P-glycoprotein drug binding sites. Biochemistry. 2012;51:5125–5141. doi: 10.1021/bi300299z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prajapati R, Sangamwar AT. Translocation mechanism of P-glycoprotein and conformational changes occurring at drug-binding site: Insights from multi-targeted molecular dynamics. Biochim Biophys Acta Biomembr. 2014;1838:2882–2898. doi: 10.1016/j.bbamem.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Corradi V, Singh G, Tieleman DP. The human transporter associated with antigen processing: Molecular models to describe peptide binding competent states. J Biol Chem. 2012;287:28099–28111. doi: 10.1074/jbc.M112.381251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han W, Cheng RC, Maduke MC, Tajkhorshid E. Water access points and hydration pathways in CLC H+/Cl− transporters. Proc Natl Acad Sci USA. 2014;111:1819–1824. doi: 10.1073/pnas.1317890111. • Extended equilibrium simulations capturing spontaneous water wire formation and its Cl− dependence in a CLC transporter, offering a novel coupling mechanism.
- 38.Cheng MH, Coalson RD. Molecular dynamics investigation of Cl− and water transport through a eukaryotic ClC transporter. Biophys J. 2012;102:1363–1371. doi: 10.1016/j.bpj.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li J, Shaikh SA, Enkavi G, Wen PC, Huang Z, Tajkhorshid E. Transient formation of water-conducting states in membrane transporters. Proc Natl Acad Sci USA. 2013;110:7696–7701. doi: 10.1073/pnas.1218986110. #x02022;• Simulation of several membrane transporters in different functional states proposing universality of water leak in the transport mechanism, offering a revised structural framework for the alternating access model.
- 40.Adelman JL, Sheng Y, Choe S, Abramson J, Wright EM, Rosenberg JM, Grabe M. Structural determinants of water permeation through the sodium-galactose transporter vSGLT. Biophys J. 2014;106:1280–1289. doi: 10.1016/j.bpj.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondal S, Khelashvili G, Shi L, Weinstein H. The cost of living in the membrane: a case study of hydrophobic mismatch for the multi-segment protein LeuT. Chem Phys of Lipids. 2013;169:27–38. doi: 10.1016/j.chemphyslip.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoonens M, Comer J, Masscheleyn S, Pebay-Peyroula E, Chipot C, Miroux B, Dehez F. Dangerous liaisons between detergents and membrane proteins. the case of mitochondrial uncoupling protein 2. J Am Chem Soc. 2013;135:15174–15182. doi: 10.1021/ja407424v. [DOI] [PubMed] [Google Scholar]
- 43. Khelashvili G, LeVine MV, Shi L, Quick M, Javitch JA, Weinstein H. The membrane protein LeuT in micellar systems: aggregation dynamics and detergent binding to the S2 site. J Am Chem Soc. 2013;135:14266–14275. doi: 10.1021/ja405984v. • Capturing detergent binding to a specific site in LeuT, the study provides a clear case for the effect of detergents on the function of membrane transporters.
- 44.O’Mara ML, Mark AE. The effect of environment on the structure of a membrane protein. P-glycoprotein under physiological conditions. J Chem Theor Comp. 2012;8:3964–3976. doi: 10.1021/ct300254y. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465:188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Terry DS, Shi L, Quick M, Weinstein H, Blanchard SC, Javitch JA. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474:109–113. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen MØ, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 48.Kohlhoff KJ, Shukla D, Lawrenz M, Bowman GR, Konerding DE, Belov D, Altman RB, Pande VS. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat Chemistry. 2014;6:15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pendse PY, Brooks BR, Klauda JB. Probing the periplasmic-open state of lactose permease in response to sugar binding and proton translocation. J Mol Biol. 2010;404:506–521. doi: 10.1016/j.jmb.2010.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stolzenberg S, Khelashvili G, Weinstein H. Structural intermediates in a model of the substrate translocation path of the bacterial glutamate transporter homologue GltPh. J Phys Chem B. 2012;116:5372–5383. doi: 10.1021/jp301726s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das A, Gur M, Cheng MH, Jo S, Bahar I, Roux B. Exploring the conformational transitions of biomolecular systems using a simple two-state anisotropic network model. PLoS Comput Biol. 2014;10:e1003521. doi: 10.1371/journal.pcbi.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adelman JL, Dale AL, Zwier MC, Bhatt D, Chong LT, Zuckerman DM, Grabe M. Simulations of the alternating access mechanism of the sodium symporter Mhp1. Biophys J. 2011;101:2399–2407. doi: 10.1016/j.bpj.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stelzl LS, Sansom MSP, Beckstein O. Flexible gates generate occluded intermediates in the transport cycle of LacY. J Mol Biol. 2014;426:735–751. doi: 10.1016/j.jmb.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimamura T, Weyand S, Beckstein O, Rutherford NG, Hadden JM, Sharples D, Sansom MSP, Iwata S, Henderson PJF, Cameron AD. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328:470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehmood S, Domene C, Forest E, Jault JM. Dynamics of a bacterial multidrug ABC transporter in the inward- and outward-facing conformations. Proc Natl Acad Sci USA. 2012;109:10832–10836. doi: 10.1073/pnas.1204067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Wonderen JH, McMahon RM, O’Mara ML, McDevitt CA, Thomson AJ, Kerr ID, MacMillan F, Callaghan R. The central cavity of ABCB1 undergoes alternating access during ATP hydrolysis. FEBS J. 2014;281:2190–2201. doi: 10.1111/febs.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaikh SA, Tajkhorshid E. Modeling and dynamics of the inward-facing state of a Na+/Cl− dependent neurotransmitter transporter homologue. PLoS Comput Biol. 2010;6:e1000905. doi: 10.1371/journal.pcbi.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Moradi M, Tajkhorshid E. Mechanistic picture for conformational transition of a membrane transporter at atomic resolution. Proc Natl Acad Sci USA. 2013;110:18916–18921. doi: 10.1073/pnas.1313202110. •• This study introduces an efficient non-equilibrium approach to describe global structural transitions in membrane transporters at an atomic level.
- 59.Moradi M, Tajkhorshid E. Computational recipe for efficient description of large-scale conformational changes in biomolecular systems. J Chem Theor Comp. 2014;10:2866–2880. doi: 10.1021/ct5002285. [DOI] [PMC free article] [PubMed] [Google Scholar]