Abstract
Modular polyketide synthases produce a vast array of bioactive molecules that are the basis of many highly valued pharmaceuticals. The biosynthesis of these compounds is based on ordered assembly lines of multi-domain modules, each extending and modifying a specific chain-elongation intermediate before transfer to the next module for further processing. The first 3D structures of a full polyketide synthase module in different functional states were obtained recently by electron cryo-microscopy. The unexpected module architecture revealed a striking evolutionary divergence of the polyketide synthase compared to its metazoan fatty acid synthase homolog, as well as remarkable conformational rearrangements dependent on its biochemical state during the full catalytic cycle. The design and dynamics of the module are highly optimized for both catalysis and fidelity in the construction of complex, biologically active natural products.
Introduction
Nature has an impressive capacity for chemical synthesis, illustrated most dramatically in its ability to produce a vast diversity of secondary metabolite molecules or natural products. Polyketides and non-ribosomal peptides are natural products generated by complex enzyme assemblies that function as biosynthetic machines. Among the most fascinating machines are the many polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS) organized as assembly lines wherein pathway intermediates move from one synthetic module to the next in a defined order. Within each module, the pathway intermediate is extended by a specific 2-carbon ketide (PKS) or an amino acid (NRPS). Intermediates are tethered by thioester linkage to the phosphopantetheine cofactor of each carrier protein, and may undergo further modification after extension. In some pathways, this common strategy for intermediate tethering facilitates mixing of ketide and peptide extension units to create hybrid polyketide-peptide products.
Most drugs in clinical or veterinary use are natural products or their derivatives [1] and, given the urgent need for new drugs, secondary metabolic pathways represent an area of outstanding importance for exploring new leads [2]. Accordingly, modular PKS, NRPS and hybrid systems are subjects of intensive research in academic and industrial laboratories around the world, aiming to delineate the pathways for the production of promising compounds and also to provide the basis for bioengineering efforts [3]. Nevertheless the natural sources of bioactive molecules have barely been explored, as the number of pathways being studied is far outpaced by the abundance of pathways being identified in microbial genome sequences [4]. Although the natural functions of nearly all polyketide, nonribosomal peptide and hybrid natural products are unknown, these often secreted molecules likely tailor the local environment to benefit the producing organism in its ecological niche. Furthermore, secondary metabolites produced by PKS or hybrid pathways undoubtedly are natural contributors to human health, as several thousand pathways have been identified in the microbiomes of healthy people [5].
Structural biology studies have provided significant insights into the architecture and function of modular PKSs [6,7]. The focus of this review is the structure of a PKS module and new results [8••,9••] that inform our understanding of its function within a biosynthetic assembly line.
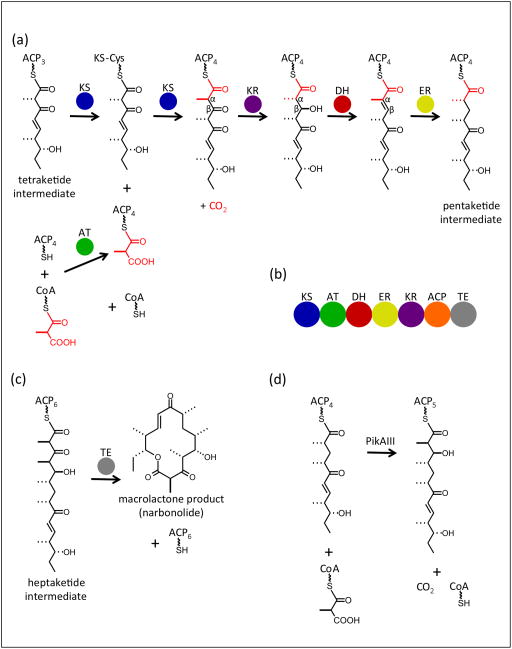
The PKS module extends a polyketide intermediate in two-carbon increments through the decarboxylative condensation of thioester-activated acyl groups. Besides the two-carbon elongation step, a module may optionally modify the extended intermediate. The obligatory extension includes acyltransferase (AT) selection of a building block from the CoA pool for loading onto an acyl carrier protein (ACP) domain, and condensation with the upstream intermediate by a ketosynthase (KS) domain to produce an elongated β-keto product (Figure 1a). In the canonical PKS module, up to three reductive reactions may then occur to generate a β-hydroxy (ketoreductase (KR) reduction of β-keto), an α,βalkene (dehydratase (DH) action on β-hydroxy), or a fully reduced intermediate (enoylreductase (ER) reduction of α,β-alkene). In the PKS module, these domains are fused in the order KS-AT-DH-ER-KR-ACP (Figure 1b). The enzymology of the individual catalytic domains is well understood, and models have been proposed to explain the catalytic fidelity of individual modules [10]. Each mega-synthase assembly line consists of several modules that typically act in a strict order. A terminal thioesterase (TE) domain in the final module of the pathway offloads the final ACP-tethered intermediate, in many cases cyclizing it to a macrolactone (Figure 1c).
Figure 1.
PKS module catalysis and organization. (a) Reactions catalyzed by a canonical PKS module with a full complement of reductive domains. The fourth extension module of the pikromycin pathway [36] uses a methylmalonyl-CoA building block to extend the tetraketide intermediate to a fully reduced pentaketide. The building block added in this extension is shown in red; the wavy bond represents phosphopantetheine. (b) Order of domains within the PKS module and the metazoan FAS-I. The offloading TE domain is present only in the terminal module (PikAIV) of PKS pathways. (c) TE offloading to generate narbonolide, the macrolactone product of the pikromycin PKS assembly line. (d) Overall conversion catalyzed by PikAIII, the fifth extension module of the pikromycin pathway.
Much of the remarkable diversity of polyketide natural products is due to the specific sequence of modules within a pathway together with the modification domains within each module that produce unique β-keto, β-hydroxy, α,β-alkene or fully reduced intermediates. Given Nature’s predisposition for gene duplication and pathway expansion, it is not surprising that more substantial modifications of the canonical PKS module are also common, including the insertion of sequences that encode functionalities for methyl transfer [11], halogenation [12] and β-branching [13]. Additionally, some gene clusters do not encode an AT domain within each module, but instead use a pathway-associated in trans AT to supply a common building block at each extension step [14]. Most PKS gene clusters also encode tailoring enzymes that convert the released polyketide to the final biologically active natural product in reactions such as glycosylation [15], methylation [11], epoxidation and hydroxylation [16].
Operation of the assembly line - docking domains
To synthesize a specific product, the modules of a PKS pathway must operate in a defined sequence. This requires that each ACP domain deliver the product of its module to the KS domain of the next module in the assembly line. Intermediate delivery to the next module is straightforward when modules are fused within a single polypeptide. In other cases, pathway fidelity is assured by weak, but specific, non-covalent association of short “docking domains” at the ACP C-terminus of the upstream module and the KS N-terminus of the downstream module [17]. Two docking architectures have been characterized [18,17,19], both involving the docking of a C-terminal helix in the upstream module to an N-terminal coiled-coil helix dimer of the downstream module. In actinobacterial pathways, the “class 1” C-terminal docking helix is preceded by a helical dimerization element [18], whereas in cyanobacterial and myxobacterial pathways, the “class 2” C-terminal docking peptide is monomeric [19].
Relationship of PKS module to metazoan FAS
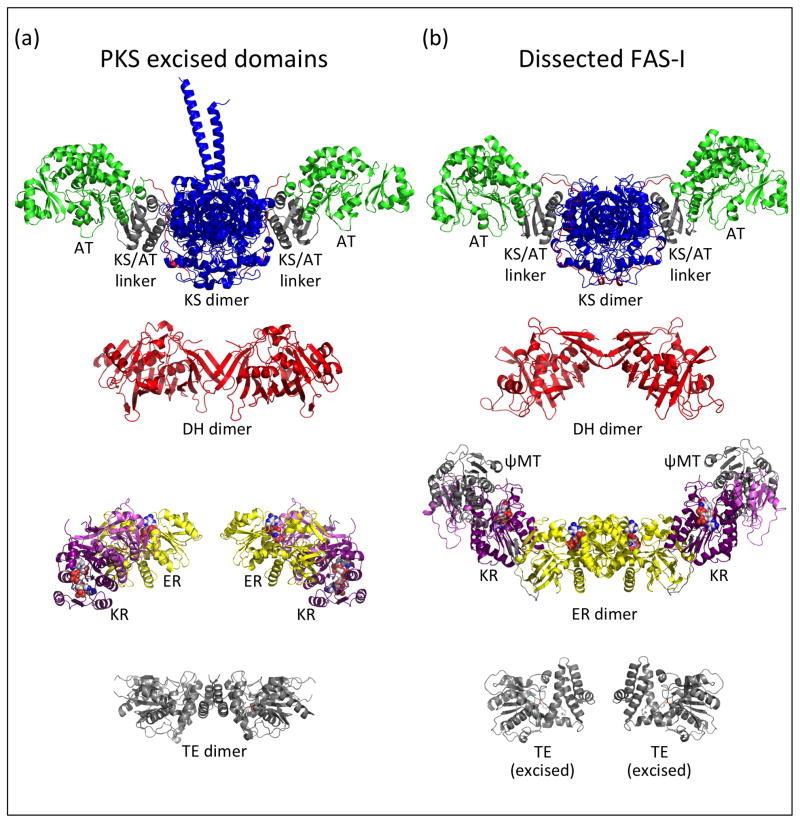
The type I PKS module and the metazoan (type I) fatty acid synthase (FAS-I) have a common evolutionary ancestor. In addition to homologous catalytic and ACP domains, these megasynthases possess a common domain order (Figure 1b) and homodimeric association [20]. However, PKS modules within a multi-component pathway act in a defined sequence in which each catalytic domain is typically used only once, with the order of modular interactions determined by cognate docking domains. By contrast, the FAS-I is a singular entity that always functions iteratively without need for docking domains. In 2006, three crystal structures (porcine FAS-I [21], PKS KS-AT di-domain [22], PKS KR domain [23]) revealed several previously unknown architectural features common to the PKS and FAS systems (Figure 2). Based on these landmark results, the dimeric metazoan FAS-I became the de facto model for the dimeric PKS module, effectively replacing two earlier models [24,25]. Arguably the strongest evidence for a common PKS-FAS architecture came from crystal structures of KS-AT di-domains excised from PKS modules [22,26,19], which are strikingly similar to the KS-AT of FAS-I. Both dimers are elongated with the KS domains (blue) at the dimer interface, the AT domains (green) at the periphery and an 80-amino-acid KS/AT linker domain (gray) between KS and AT (Figure 2). In both systems, a 30-amino-acid “post-AT linker” (red) extends from the AT, crosses the KS/AT linker domain surface, and threads between the KS/AT linker domain and the KS domain to terminate near the dimer interface. The PKS KR structure led to the important realizations that the KR of both PKS and FAS-I is the product of a gene duplication, resulting in “structural” (KRS) and “catalytic” (KRC) sub-domains, and that the ER domain is inserted between KRS and KRC [23]. A higher-resolution porcine FAS-I structure [27] led to the identification of a pseudo-methyltransferase domain (ψMT) between the DH and ER domains (Figure 2b), where bona fide MT domains are found in some PKS modules. The FAS-I-based PKS model appears to be well organized for genetic manipulation, as the obligatory extension domains (KS-AT) form one wing of the structure and the reductive domains (DH-ER-KR) form a second wing in which one, two or all three domains may be absent. The small ACP domain, which must transit all the active sites, and the terminal TE domain were not positioned in the model, as they were present but disordered in the porcine FAS-I crystal structure.
Figure 2.
Comparison of domain structures from PKS and metazoan FAS mega-enzymes. (a) Domains excised from PKS modules, including the CurL KS-AT (PDB 4MZ0, [19]) and CurH DH (PDB 3KG7, [29]) from the curacin pathway, ER2-KR2 (PDB 3SLK, [28]) from the spinosyn pathway, and the pikromycin TE (PDB 2HFJ, [29]). (b) Domains of the metazoan FAS, including the KS-AT, DH and ψMT-ER-KR from the porcine FAS (PDB 2VZ9, [27]) and the TE (PDB 3TJM, [31]) excised from the human FAS. The KS domains are in blue, AT green, DH red, ER yellow, KR dark (catalytic) and light (structural) purple; the KS/AT linker, ψMT and TE domains are in gray. Cofactors and active site residues are rendered as spheres in atomic coloring. The post-AT linker peptides are shown in red in the KS-AT images. In all images, the PKS module or FAS-I twofold axis is vertical. To facilitate comparison, the PKS and FAS-I KS domains are oriented identically, as are the ER monomers and the core domains of TE monomers.
Other aspects of the PKS module and FAS-I differ, particularly in the reductive domains (Figure 2). The PKS inter-domain linkers are shorter than the corresponding FAS-I linkers and dissimilar in sequence. The linker peptides connecting the ER and KR domains form a two-stranded β-sheet in the FAS-I that does not exist in an excised PKS ER-KR di-domain [28], and the orientation of ER relative to KR is radically different in the PKS and FAS-I. The quaternary structures of some catalytic domains also differ in the two systems. The FAS-I ER is dimeric whereas the PKS ER domain is monomeric with a helix that blocks dimer formation [28]. Despite their common fold, the dimeric DHs have strikingly different monomer orientations in PKS [29] and FAS-I, and the PKS DH domain remains dimeric when excised whereas the excised FAS-I DH behaves as a monomer. Moreover, the terminal PKS TE domain is a dimer [30] while the FAS-I TE is monomeric [31].
Segue from crystallography to single-particle electron cryo-microscopy
Structures of excised domains from PKS modules have accumulated in the past decade, but structures of full modules have remained elusive. This is despite heroic attempts at crystallization in many labs, including our own. Models have been proposed to explain multi-domain catalysis [10] or to fit small-angle x-ray scattering data for PKS modules in solution [32,33]. Likewise, knowledge of the metazoan FAS-I structure remains limited to a single snapshot of the porcine FAS-I in one crystal form [27] as well as maps from low-resolution negative-stain electron microscopy (EM) showing flexibility in the region connecting the upper KS-AT wing and the lower reductive wing [34,35]. The working assumption of the field has been that inherent flexibility poses great challenges for crystallization of both PKS modules and FAS-I. More recently, we explored the feasibility of electron cryo-microscopy (cryo-EM) to study PKS module structure and quickly discovered that a few “canonical” PKS modules had remarkably homogeneous conformations. For detailed studies, we chose the native monomodular polypeptide, PikAIII (module 5), from the well characterized pikromycin PKS (Pik) of Streptomyces venezuelae [36], a canonical pathway closely related to the prototypical erythromycin PKS (DEBS) of Saccharopolyspora erythraea [37]. PikAIII catalyzes the extension of a pentaketide intermediate and transfers its hexaketide product to the PikAIV termination module 6 (Figure 1d). Effective biochemical assays have been developed for PikAIII and for coupled PikAIII-PikAIV catalysis [38,39], which enabled us to evaluate our findings and interpretations through a series of biochemical experiments involving site-directed mutagenesis.
Single-particle cryo-EM capabilities and advances
The resolution achievable by cryo-EM has steadily improved over the past 10–20 years thanks to advances in instrumentation and image processing methodologies. Primarily propelled by the recent development of direct-electron detectors that facilitate the recording of images with minimal noise and blurring [40,41], cryo-EM has now entered the “club” of high-resolution structure determination methods [42–44]. Importantly, near atomic-resolution cryo-EM structures are not limited to large macromolecular complexes such as ribosomes or highly symmetric assemblies such as viruses, but can also be achieved for complexes well below 400 kDa [45•,46•]. As the community has gained experience in using single-particle EM for macromolecular complexes, the requirements for sample quality have become clear and techniques for sample preparation have been further refined. The ability to obtain such structures in solution at high resolution is a game changer in many areas of molecular biology and alleviates many of the bottlenecks associated with X-ray crystallography or NMR. Nevertheless, the achievable resolution by single-particle cryo-EM is fundamentally limited by the inherent dynamics of the system and the extent to which compositional and conformational heterogeneity can be effectively addressed computationally through particle classification procedures. It is thus expected that 3D reconstructions of relatively small macromolecular complexes with a near continuum of conformations will be limited in resolution, and may in fact be inaccurate at even low resolution.
Given these considerations, it was surprising that PikAIII, a canonical PKS module composed of four catalytic domains, KS-AT-KR-ACP, was highly homogeneous in solution and yielded 3D reconstructions with resolutions as high as 7 Å from data recorded on a conventional CCD detector [8••,9•• ]. At 300-kDa, the PikAIII dimer is considered a relatively small cryo-EM target and, if compounded by significant flexibility, would yield 3D reconstructions of limited accuracy and resolution. However, in this case the resolution of the final EM maps was limited more by the instrumentation than by the sample. The EM maps were of sufficient detail for construction of pseudo-atomic PikAIII models, which were created by rigid-body positioning the high-resolution crystal structures of individual domains (KS dimer, AT, KR, ACP) into the well resolved EM density.
The PikAIII structures were full of surprises. The two most significant are a substantially different overall architecture from the prevailing FAS-based model, and the substrate-dependent localization of the ACP domain. In retrospect, both of these features are highly evolved and facilitate pathway throughput of the polyketide chain elongation intermediate as it is extended and reductively processed during a single catalytic cycle.
Overall architecture
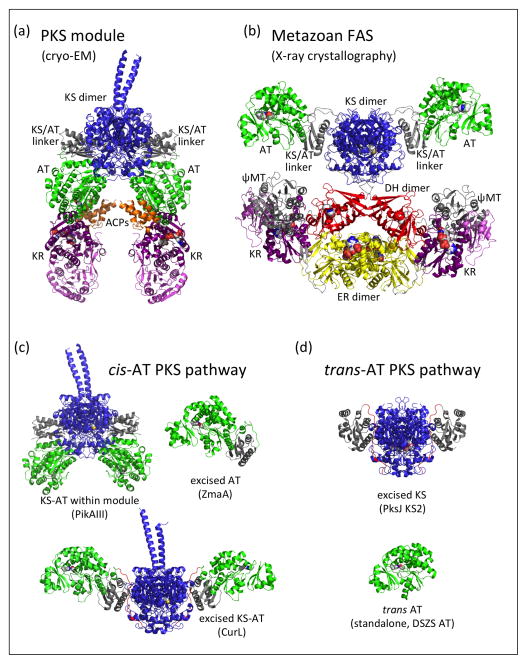
The PKS module has an overall arch shape (Figure 3a), a major departure from the more open architecture visualized in the porcine FAS-I crystal structure [27] (Figure 3b). The arch shape of the PKS module creates a single reaction chamber for intermediates tethered to the ACP, as the active sites of catalytic domains face into the chamber. The PKS and FAS structures have identical KS dimers, but differ most dramatically in the positions of the AT domains relative to the KS dimers. The arch shape and single reaction chamber of the PKS module are created by an “AT-down” conformation whereas the FAS-I open architecture is due to an “AT-out” conformation. The position of the post-AT linker peptide, which connects the AT to the KR domain of PikAIII, could not be determined in the current EM maps.
Figure 3.
Comparison of PKS and FAS overall structures. (a) PikAIII with β-ketohexaketide-ACP (EMD-5664, [9••]). (b) Porcine FAS (PDB 2VZ9, [27]). (c) Linker-AT unit within cis-AT PKS pathways, including within the PikAIII module [8••], excised from a zwittermicin module (PDB 4QBU, [48]) and excised as a KS-AT (PDB 4MZ0, [19]). (d) KS/AT linker domain association with KS in trans-AT PKS pathways, including KS2 excised from the bacillaene PKS (PDB 4NA2, [51]) and the trans AT from the disorazole pathway (PDB 3RGI, [49]). Structures are rendered with KS dimers in a common orientation; the excised ZmaA AT in (c) and the trans AT in (d) are shown in the same orientation as the left-most AT of CurL KS-AT. Domains are colored as in Figure 2.
The dramatic difference in architecture of the PKS module and FAS-I is due to a radically different interface of the KS domain with the 80-amino-acid KS/AT linker domain. The KS/AT linker domain (gray) and the AT domain (green) are positioned identically to each other in the PikAIII module (Figure 3a) and in the excised KS-AT di-domains (Figure 2a), as an excellent fit to the EM density was obtained without internal adjustment of the KS/AT linker domain to the AT domain. Thus we refer to these two domains together as the “linker-AT unit”. Relative to the KS, the linker-AT unit of the PKS module differs by an ~120º rotation compared to its position the porcine FAS-I or in structures of excised PKS KS-AT di-domains [22,26,19], representing an ~40 Å difference in the AT active site position. The linker-AT unit appears to be a fixed unit in cis-AT PKS pathways. For example, guided by the domain architecture from the original KS-AT di-domain structures, two groups excised linker-AT units and obtained crystal structures [47,48] in which the interfaces of KS/AT linker domain and AT domain are identical to those of the PikAIII full module (Figure 3c). In contrast to the KS/AT linker domain, a small ferredoxin-like sub-domain of the AT occupies more varied positions relative to the AT core. Moreover, the structures of KS-ATs excised from two DEBS modules [22,26] and a curacin module [19] reveal evidence for flexibility of the linker-AT unit relative to the KS, although all are in an “AT-out” position (Figure 3c). The orientation of linker-AT unit relative to KS also differed among the biochemical states of PikAIII, although all are in an “AT-down” position. Intriguingly, the KS/AT linker domain is clearly a feature of PKS modules. In trans-AT pathways, the KS/AT linker domain is not part of the standalone trans AT enzyme [49], but is instead associated with the KS domain, as seen in two recent KS crystal structures [50,51] (Figure 3d).
The PKS module is inherently dimeric. Nature employs at least two types of dimerization elements in addition to the dimeric KS, DH and TE catalytic domains. Surprisingly, in many cases the extensive dimer interface of the KS domain is insufficient for module dimerization. We found that PikAIII dimerization requires the small post-ACP dimerization element of the class 1 docking domain [8••]. The proximity of the dimerization element to the ACP may explain why the ACP domains were faithful to the module dimer symmetry in all EM maps. Consistent with this observation, retention of the post-ACP dimerization element was essential for efficient di-module catalysis (PikAIII::PikAIV) when the native PikAIII class 1 docking domain was replaced with a monomeric class 2 docking domain [19]. Another type of dimerization element was discovered recently in the spinosyn pathway, consisting of a 55-amino-acid insert between the post-AT linker and the KR domain [52]. Similar dimerization elements have been detected in pathways with monomeric class 2 post-ACP docking domains [19].
The single reaction chamber of the PKS module effectively sequesters the AT and KR active sites away from the ACPs of other modules, a potentially significant factor in pathway fidelity. The reaction chamber also protects the module ACP from other reactants. For example, to generate the pentaketide-KS state of PikAIII (Figure 4b), the pentaketide intermediate was delivered as the thiophenol-activated mimic [53] of pentaketide-ACP4. The small-molecule mimic reacted with the PikAIII KS catalytic cysteine, but no adduct at the PikAIII ACP5 phosphopantetheine thiol group was detected by mass spectrometry.
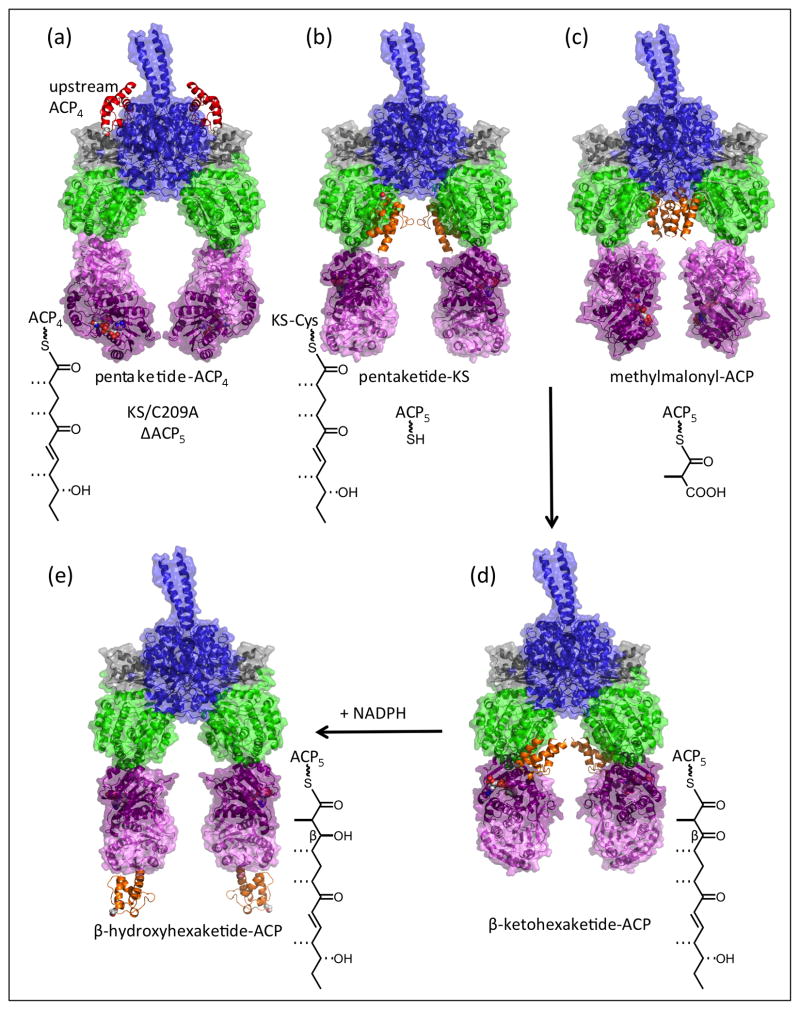
Figure 4.
Travels of ACP in the catalytic cycle of PikAIII PKS module [8••,9••]. (a) Delivery of the pentaketide intermediate by ACP4 (red) of the upstream module to the exterior entrance to the KS active site (EMD-5651). (b) Pre-extension pentaketide-loaded KS with holo-ACP docked at the AT active site entrance (EMD-5663). (c) Methylmalonyl-ACP docked at the KS active site entrance within the reaction chamber (EMD-5653). (d) Post-extension β-ketohexaketide-ACP docked at the KR active site (EMD-5664). (e) Post-reduction β-hydroxyhexaketide-ACP outside the reaction chamber where it is accessible to the next module in the pathway (EMD-5666). Domains are colored as in Figure 2 with transparent surface rendering for all but the ACP domains. The biochemical state of the module is shown adjacent to the molecular drawing.
Travels of ACP
Another major surprise of the PikAIII structures was the substrate-dependent localization of the ACP domain. Even without an acyl group (holo-ACP), the ACP was well localized within the reaction chamber at two distinct positions distant from the active sites. The ACP traveled to active site entrances dependent on its acylation state and on the biochemical state of the module, generally residing at the active site entrance appropriate to the next reaction of its cargo. The series of structures provides significant new information about the dynamics of ACP and its interaction with the catalytic domains (Figure 4), as well as a new baseline for future analyses relating to substrate selectivity and catalytic efficiency.
ACP localization led to the important discovery of a second, previously unknown entrance to the KS active site. When loaded with a methylmalonyl building block, the PikAIII ACP localizes to a KS active site entrance within the reaction chamber where it awaits the intermediate from the upstream module (Figure 4c). The ACP of the upstream module (PikAII, module 4) delivers its pentaketide intermediate through another entrance, which is outside the reaction chamber (Figure 4a). After pentaketide delivery, the outside entrance is blocked by a slight shift in the linker-AT unit. This dual-entry scheme cleverly avoids mix-ups within KS domains that must interact with two different ACPs. The outside entrance was well established by structures of a number of enzymes from the thiolase superfamily, but the entrance within the reaction chamber was unanticipated. Re-examination of the crystal structures of excised PKS KS-AT didomains [22,26,19] revealed an open or flexibly covered KS active site entrance at the position of the intra-module ACP entrance in PikAIII, and its functional role was established through mutagenesis. In contrast, KS domains or discrete forms of the enzyme (e.g. from FAS, type II PKS) that interact with only one ACP or with acyl-CoA substrates have only the previously identified “side” entrance.
When the PikAIII KS catalytic cysteine is loaded with the upstream intermediate, the outside entrance is blocked by a slight shift in the linker-AT unit compared to its position in other biochemical states of the module. Two recent KS crystal structures provide the first views of bona fide substrates as covalent intermediates in the KS active site. One structure is for a PKS that acts on acyl-CoA substrates [54], and the other is excised from a trans-AT pathway [51]. The spacious KS active sites appear to provide binding pockets adapted to their substrates, and based on these high-resolution substrate complexes, it is challenging to predict the position of a new building block delivered through the entrance for the module ACP. Interestingly, recent structures of excised KS domains from two trans-AT pathways [50,51] do not indicate the presence of a second active site entrance. As the AT domain forms one wall of the PikAIII reaction chamber, we anticipate unique features in the module architecture for the trans-AT PKSs.
After the PikAIII KS extends the pentaketide intermediate, the β-ketohexaketide-ACP moves to the KR active site where it stalls in absence of the NADPH cofactor (Figure 4d). The ACP contacts the KR “cofactor lid”, which directly overlooks the NADPH nicotinamide site and is consistent with a recent hexanoyl-CoA-KR complex structure [55]. The direction from which a β-keto substrate enters the KR active site is thought to establish the chirality of the β-hydroxy KR product [56], and the ACP is in a perfect position to introduce its substrate under the lid into the active site from either direction. Two sequence motifs in PKS KR domains are strongly correlated with specific stereochemical outcomes [57] through a mechanism that is not apparent in structures of excised KR domains but may involve blocking access from one direction or the other after cofactor binding [58].
Finally, the ACP is expelled from the reaction chamber after KR-catalyzed reduction of the β-ketohexaketide to the β-hydroxyhexaketide (Figure 4e). The module product is thus positioned to interact with the outside entrance to the KS active site of the next module (PikAIV) in the pathway and completion of the biosynthetic process that results in formation of narbonolide (Figure 1c).
Summary and remaining questions
The unanticipated structures and dynamics of the PikAIII module provide long-awaited explanations for the astounding pathway throughput and fidelity in type I PKS systems. The stable localization of ACP according to the ketide intermediate it carries can contribute substantially to pathway throughput by placing it in an optimal position for the next reaction of the biochemical cycle. Thus, the ACP does not diffuse freely in the vicinity of the module active sites, as was generally thought. How much ketide variability can the module tolerate without loss of ACP localization, and does this differ among catalytic domains? Current efforts indicate that pathway throughput is substantially slowed by modest changes to the natural pentaketide substrate (Hansen, D. A. and Sherman, D. H. unpublished), but is this due to bottlenecks at specific domains or general lack of ACP localization? These critical questions must be answered if we are ever to achieve the long-standing goals of scalable chemoenzymatic synthesis using modular PKSs, or efficient structural diversification using combinatorial biosynthesis with engineered PKS pathways.
ACP localization also contributes to pathway fidelity. The ACP is sequestered within the reaction chamber until the final synthetic step is complete, then ejected from the chamber and made accessible to the next module (Figure 4e). This mechanism prevents the accumulation of aberrant pathway intermediates by protecting the ACP from erroneous reactions in the catalytic domains of other modules, of which the KS of the downstream module may be the most critical. Specificity in the ACP::KS protein-protein interaction of discrete cognate modules (in addition to the docking domain interaction) has been reported and predicted to contribute to pathway fidelity [59]. In such cases, it may be especially important to sequester the upstream ACP away from the downstream KS until catalysis is complete in the upstream module. It is noteworthy that the upstream ACP4 for the PikAIII module did not localize in any position with respect to the downstream KS5 in the absence of bound pentaketide intermediate [8 ], indicating that any ACP4::KS5 specificity is weak in this case.
Many details remain to be established for the remarkable progression of conformational changes and ACP travels throughout the PikAIII biosynthetic cycle. The domain connectivity within the dimeric module was not established with certainty, as the inter-domain peptides were not discernible in the EM maps. The 8-20 amino-acid connector from the KS to the linker-AT unit is also disordered in all crystal structures of excised KS-AT didomains. In the excised PKS KS-AT didomains and the FAS-I, the post-AT linker occupies remarkably similar positions along the KS surface, but the radically different interface of KS to linker-AT unit requires that the post-AT linker be elsewhere in the PKS module. The EM maps revealed several structural changes associated with the PikAIII module biochemical state, including shifts in the linker-AT unit, changes at the intra-module KS entrance, and an end-to-end (i.e. 180º) flip of the KR domain. Understanding these dynamic features and their functional consequences will require structural snapshots at higher resolution than in the initial cryo-EM study.
Finally, the PKS module structures raise several global questions. Are all PKS modules from cis-AT pathways like PikAIII, or do other PKS modules have different conformations? How are DH, ER and the growing number of non-canonical domains (e.g. halogenase [60], sulfotransferase [61]) accommodated in the module architecture? Does the post-ACP dimerization element at the module C-terminus contribute to the overall arch shape? How flexible is the interface between the KS and the linker-AT unit? Does the metazoan FAS exist in any PKS-like conformation? The new structures are an excellent starting point to answer these and other questions about PKS module architecture and function.
Highlights.
The first structures of a polyketide synthase module were solved by cryo-EM.
The module structure is strikingly different than its fatty acid synthase homolog.
Structures illustrate the dynamics of the module during its catalytic cycle.
The carrier protein domain localizes optimally for the next reaction of its cargo.
Acknowledgments
The research in our laboratories on the topic of this review is supported by the US National Institutes of Health (DK042303 to JLS, GM076477 to DHS and JLS, DK090165 to GS) and by the Martha L. Ludwig Professorship to JLS, the Pew Scholars Program to GS, and the Hans W. Vahlteich Professorship to DHS.
Footnotes
Conflicts of interest
Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cragg GM, Grothaus PG, Newman DJ. New horizons for old drugs and drug leads. J Nat Prod. 2014;77:703–723. doi: 10.1021/np5000796. [DOI] [PubMed] [Google Scholar]
- 3.Poust S, Hagen A, Katz L, Keasling JD. Narrowing the gap between the promise and reality of polyketide synthases as a synthetic biology platform. Curr Opin Biotechnol. 2014;30C:32–39. doi: 10.1016/j.copbio.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akey DL, Gehret JJ, Khare D, Smith JL. Insights from the sea: structural biology of marine polyketide synthases. Nat Prod Rep. 2012;29:1038–1049. doi: 10.1039/c2np20016c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keatinge-Clay AT. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 8••.Dutta S, Whicher JR, Hansen DA, Hale WA, Chemler JA, Congdon GR, Narayan AR, Håkansson K, Sherman DH, Smith JL, Skiniotis G. Structure of a modular polyketide synthase. Nature. 2014;510:512–517. doi: 10.1038/nature13423. Based on cryo-EM reconstructions of three biochemical states, the PikAIII module is found to have a overall arch shape with a single reaction chamber for the ACP domains. The structures unexpectedly reveal separate KS active site entrances for the PikAIII ACP and the ACP of the upstream module. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Whicher JR, Dutta S, Hansen DA, Hale WA, Chemler JA, Dosey AM, Narayan AR, Håkansson K, Sherman DH, Smith JL, Skiniotis G. Structural rearrangements of a polyketide synthase module during its catalytic cycle. Nature. 2014;510:560–564. doi: 10.1038/nature13409. Cryo-EM reconstructions of three additional biochemical states of PikAIII demonstrate a substrate-dependent localization of the ACP domain at active-site entrances and outside the reaction chamber following all catalytic steps. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khosla C, Herschlag D, Cane DE, Walsh CT. Assembly line polyketide synthases: mechanistic insights and unsolved problems. Biochemistry. 2014;53:2875–2883. doi: 10.1021/bi500290t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 12.Blasiak LC, Drennan CL. Structural perspective on enzymatic halogenation. Acc Chem Res. 2009;42:147–155. doi: 10.1021/ar800088r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderone CT. Isoprenoid-like alkylations in polyketide biosynthesis. Nat Prod Rep. 2008;25:845–853. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nature biotechnology. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 15.Singh S, Phillips GN, Jr, Thorson JS. The structural biology of enzymes involved in natural product glycosylation. Nat Prod Rep. 2012;29:1201–1237. doi: 10.1039/c2np20039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Podust LM, Sherman DH. Diversity of P450 enzymes in the biosynthesis of natural products. Nat Prod Rep. 2012;29:1251–1266. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchholz TJ, Geders TW, Bartley FE, 3rd, Reynolds KA, Smith JL, Sherman DH. Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains. ACS Chem Biol. 2009;4:41–52. doi: 10.1021/cb8002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem Biol. 2003;10:723–731. doi: 10.1016/s1074-5521(03)00156-x. [DOI] [PubMed] [Google Scholar]
- 19.Whicher JR, Smaga SS, Hansen DA, Brown WC, Gerwick WH, Sherman DH, Smith JL. Cyanobacterial polyketide synthase docking domains: a tool for engineering natural product biosynthesis. Chem Biol. 2013;20:1340–1351. doi: 10.1016/j.chembiol.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4. 5 Å resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Å crystal structure of a 194–kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U S A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the β-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Kao CM, Pieper R, Cane DE, Khosla C. Evidence for two catalytically independent clusters of active sites in a functional modular polyketide synthase. Biochemistry. 1996;35:12363–12368. doi: 10.1021/bi9616312. [DOI] [PubMed] [Google Scholar]
- 25.Staunton J, Caffrey P, Aparicio JF, Roberts GA, Bethell SS, Leadlay PF. Evidence for a double-helical structure for modular polyketide synthases. Nat Struct Biol. 1996;3:188–192. doi: 10.1038/nsb0296-188. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 28.Zheng J, Gay DC, Demeler B, White MA, Keatinge-Clay AT. Divergence of multimodular polyketide synthases revealed by a didomain structure. Nat Chem Biol. 2012;8:615–621. doi: 10.1038/nchembio.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akey DL, Razelun JR, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Crystal structures of dehydratase domains from the curacin polyketide biosynthetic pathway. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai SC, Miercke LJ, Krucinski J, Gokhale R, Chen JC, Foster PG, Cane DE, Khosla C, Stroud RM. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc Natl Acad Sci U S A. 2001;98:14808–14813. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Chakravarty B, Zheng F, Gu Z, Wu H, Mao J, Wakil SJ, Quiocho FA. Crystal structure of FAS thioesterase domain with polyunsaturated fatty acyl adduct and inhibition by dihomo-γ-linolenic acid. Proc Natl Acad Sci U S A. 2011;108:15757–15762. doi: 10.1073/pnas.1112334108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davison J, Dorival J, Rabeharindranto H, Mazon H, Chagot B, Gruez A, Weissman KJ. Insights into the function of trans-acyl transferase polyketide synthases from the SAXS structure of a complete module. Chem Sci. 2014;5:3081–3095. [Google Scholar]
- 33.Edwards AL, Matsui T, Weiss TM, Khosla C. Architectures of whole-module and bimodular proteins from the 6-deoxyerythronolide B synthase. J Mol Biol. 2014;426:2229–2245. doi: 10.1016/j.jmb.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asturias FJ, Chadick JZ, Cheung IK, Stark H, Witkowski A, Joshi AK, Smith S. Structure and molecular organization of mammalian fatty acid synthase. Nature Struc Mol Biol. 2005;12:225–232. doi: 10.1038/nsmb899. [DOI] [PubMed] [Google Scholar]
- 35.Brignole EJ, Smith S, Asturias FJ. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nature Struc Mol Biol. 2009;16:190–197. doi: 10.1038/nsmb.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue Y, Wilson D, Sherman DH. Genetic architecture of the polyketide synthases for methymycin and pikromycin series macrolides. Gene. 2000;245:203–211. doi: 10.1016/s0378-1119(00)00003-2. [DOI] [PubMed] [Google Scholar]
- 37.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 38.Aldrich CC, Beck BJ, Fecik RA, Sherman DH. Biochemical investigation of pikromycin biosynthesis employing native penta- and hexaketide chain elongation intermediates. J Am Chem Soc. 2005;127:8441–8452. doi: 10.1021/ja042592h. [DOI] [PubMed] [Google Scholar]
- 39.Aldrich CC, Venkatraman L, Sherman DH, Fecik RA. Chemoenzymatic synthesis of the polyketide macrolactone 10-deoxymethynolide. J Am Chem Soc. 2005;127:8910–8911. doi: 10.1021/ja0504340. [DOI] [PubMed] [Google Scholar]
- 40.Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, Pan J, Harrison SC, Potter CS, Carragher B, Grigorieff N. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20:1823–1828. doi: 10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Meth. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henderson R. Structural biology: Ion channel seen by electron microscopy. Nature. 2013;504:93–94. doi: 10.1038/504093a. [DOI] [PubMed] [Google Scholar]
- 43.Kühlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 44.Kühlbrandt W. Cryo-EM enters a new era. Elife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. The first published high-resolution (3.4 Å) reconstruction of the TRPV1 ion channel, a relatively small (280-kDa ) macromolecule with four-fold symmetry, demonstrates the power of modern cryo-EM instruments and methods to generate images at unprecedented resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, Shi Y. Three-dimensional structure of human γ-secretase. Nature. 2014;512:166–170. doi: 10.1038/nature13567. In another landmark result, a cryo-EM reconstruction was obtained for the human γ-secretase at 4.5-Å resolution. This structure is noteworthy because the 170-kDa membrane protein lacks any internal symmetry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergeret F, Gavalda S, Chalut C, Malaga W, Quemard A, Pedelacq JD, Daffe M, Guilhot C, Mourey L, Bon C. Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase Pks13. J Biol Chem. 2012;287:33675–33690. doi: 10.1074/jbc.M111.325639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park H, Kevany BM, Dyer DH, Thomas MG, Forest KT. A polyketide synthase acyltransferase domain structure suggests a recognition mechanism for its hydroxymalonyl-acyl carrier protein substrate. PLoS One. 2014;9:e110965. doi: 10.1371/journal.pone.0110965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong FT, Jin X, Mathews II, Cane DE, Khosla C. Structure and mechanism of the trans-acting acyltransferase from the disorazole synthase. Biochemistry. 2011;50:6539–6548. doi: 10.1021/bi200632j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bretschneider T, Heim JB, Heine D, Winkler R, Busch B, Kusebauch B, Stehle T, Zocher G, Hertweck C. Vinylogous chain branching catalysed by a dedicated polyketide synthase module. Nature. 2013;502:124–128. doi: 10.1038/nature12588. [DOI] [PubMed] [Google Scholar]
- 51.Gay DC, Gay G, Axelrod AJ, Jenner M, Kohlhaas C, Kampa A, Oldham NJ, Piel J, Keatinge–Clay AT. A close look at a ketosynthase from a trans-acyltransferase modular polyketide synthase. Structure. 2014;22:444–451. doi: 10.1016/j.str.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng J, Fage CD, Demeler B, Hoffman DW, Keatinge-Clay AT. The missing linker: a dimerization motif located within polyketide synthase modules. ACS Chem Biol. 2013;8:1263–1270. doi: 10.1021/cb400047s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen DA, Rath CM, Eisman EB, Narayan AR, Kittendorf JD, Mortison JD, Yoon YJ, Sherman DH. Biocatalytic synthesis of pikromycin, methymycin, neomethymycin, novamethymycin, and ketomethymycin. J Am Chem Soc. 2013;135:11232–11238. doi: 10.1021/ja404134f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gokulan K, O’Leary SE, Russell WK, Russell DH, Lalgondar M, Begley TP, Ioerger TR, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis polyketide synthase 11 (PKS11) reveals intermediates in the synthesis of methyl-branched alkylpyrones. J Biol Chem. 2013;288:16484–16494. doi: 10.1074/jbc.M113.468892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dutta D, Bhattacharyya S, Roychowdhury A, Biswas R, Das AK. Crystal structure of hexanoyl-CoA bound to β-ketoacyl reductase FabG4 of Mycobacterium tuberculosis. Biochem J. 2013;450:127–139. doi: 10.1042/BJ20121107. [DOI] [PubMed] [Google Scholar]
- 56.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 57.Caffrey P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 58.Bonnett SA, Whicher JR, Papireddy K, Florova G, Smith JL, Reynolds KA. Structural and stereochemical analysis of a modular polyketide synthase ketoreductase domain required for the generation of a cis-alkene. Chem Biol. 2013;20:772–783. doi: 10.1016/j.chembiol.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. Journal of the American Chemical Society. 2006;128:3067–3074. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu L, Wang B, Kulkarni A, Gehret JJ, Lloyd KR, Gerwick L, Gerwick WH, Wipf P, Hakansson K, Smith JL, Sherman DH. Polyketide decarboxylative chain termination preceded by o-sulfonation in curacin a biosynthesis. Journal of the American Chemical Society. 2009;131:16033–16035. doi: 10.1021/ja9071578. [DOI] [PMC free article] [PubMed] [Google Scholar]