Abstract
Plus strand RNA viruses use different mechanisms to initiate the synthesis of their RNA chains. The Picornaviridae family constitutes a large group of plus strand RNA viruses that possess a small terminal protein (VPg) covalently linked to the 5’-end of their genomes. The RNA polymerases of these viruses use VPg as primer for both minus and plus strand RNA synthesis. In the first step of the initiation reaction the RNA polymerase links a UMP to the hydroxyl group of a tyrosine in VPg using as template a cis-replicating element (cre) positioned in different regions of the viral genome. In this review we will summarize what is known about the intiation reaction of protein-primed RNA synthesis by the RNA polymerases of the Picornaviridae. As an example we will use the RNA polymerase of poliovirus, the prototype of Picornaviridae. We will also discuss models of how these nucleotidylylated protein primers might be used, together with viral and cellular replication proteins and other cis-replicating RNA elements, during minus and plus strand RNA synthesis.
Keywords: Picornavirus, Terminal protein VPg, Uridylylation of VPg, Cis-replicating RNA element (cre), RNA polymerase, RNA replication
1. Introduction
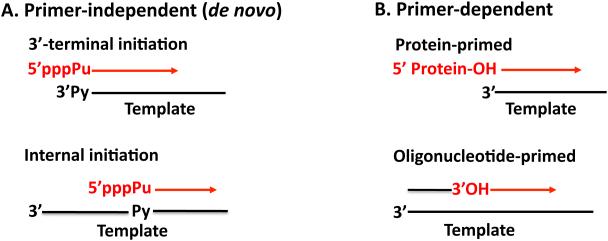
RNA templated RNA synthesis is a central step in the life cycle of plus strand RNA viruses, which replicate and transcribe their genomes in the cytoplasm of the infected host cell. RNA replication is carried out on membranous structures by the viral RNA-dependent RNA polymerase, in conjunction with other viral and cellular proteins and cis-replicating RNA elements (cres). The genomes of plus strand RNA viruses are linear molecules that have to utilize special steps to retain the integrity of the 5’-end of their genomes. The very first stage in RNA replication involves initiation of minus and plus strand RNA synthesis, processes important to preserve the integrity of the ends of the RNA genome. Although different plus strand viruses use a variety of replication strategies there are only two primary mechanisms by which RNA synthesis can be initiated (Fig. 1) (162). The first is a de novo, or primer-independent, initiation in which the 3’-hydroxyl group of the starting nucleotide, usually a purine triphosphate, serves as the acceptor of the next nucleotide. This is followed by the addition of subsequent nucleotides yielding a full-length RNA chain (17, 69). The second is the so-called primer-dependent mechanism that employs either a free oligonucleotide (99) or a protein primer that provide the hydroxyl group for the formation of a phosphodiester bond with the first nucleotide (115).
Fig. 1.
Initiation mechanisms of RNA synthesis by plus strand RNA viruses. Two basic types of mechanisms for RNA synthesis by plus strand RNA virus RNA polymerases are shown (162). (A) Primer-independent (de novo) initiation. De novo initiation involves starting an RNA chain, usually with a purine nucleoside triphosphate, templated by a pyrimidine at the 3’-end of the template strand or at an internal site. (B) Primer-dependent initiation. The primer is either an oligonucleotide or a protein. A 3’-hydroxyl group of a nucleotide or the hydroxyl group of a tyrosine or serine residue of a peptide/protein provides the hydroxyl group for the formation of a phosphodiester bond with the first nucleotide.
Most plus strand RNA viruses that use de novo initiation for both replication and transcription, initiate synthesis at the 3’- or 5’-ends of the genomes. However, some viruses employ an internal site for the process of transcription (93, 105). The very first step in protein-primed initiation, the nucleotidylylation of the terminal protein, is usually templated, as in the case of picornaviruses (82, 111, 144). The template is contained within the loop of a small RNA hairpin that is located at different positions of the RNA genome, in different genera of Picornaviridae. The nucleotidylylation reaction can, however, also occur in a template-independent manner, for example with caliciviruses (54).
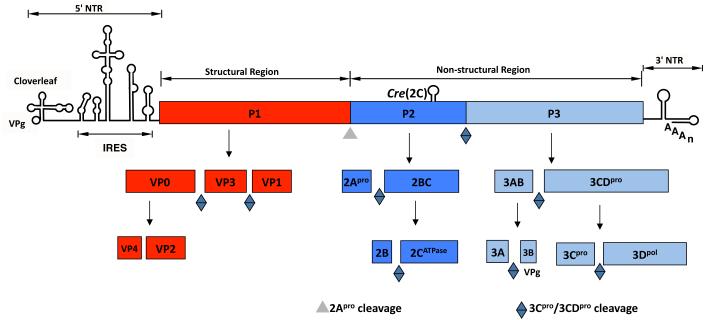
The most thoroughly studied family within the plus strand RNA viruses is the Picornaviridae. This family includes a large and diverse group of medically important human and animal pathogens such as poliovirus, rhinovirus, foot-and-mouth disease virus and hepatitis A virus. The family currently contains 26 genera of which the best known is the Enterovirus genus (PV: poliovirus) that also includes a large number of rhinoviruses, e.g. HRV2 and HRV14 (www.picornaviridae.com). Other important genera and member viruses include the Aphthovirus (FMDV: foot-and-mouth disease virus), Hepatovirus (HAV: hepatitis A virus) and Cardiovirus (EMCV: encephalomyocarditis virus). Although there are many different picornaviruses they all share several common properties. They possess a plus strand RNA genome (7.5-8 kb) with a small peptide, VPg linked to the 5’-end (Fig. 2). Their genome organization is similar with a long highly structured 5’ nontranslated region (NTR), a single large open reading frame (ORF) and a short 3’NTR, terminated with a poly(A) tail (20 nt to 150 nt long). The ORF is translated in the cytoplasm of the host cell into a polyprotein that contains one structural (P1) and two nonstructural domains (P2 and P3). The polyprotein is processed into precursor and mature proteins primarily by two viral proteinases, 2Apro and 3CDpro (Fig. 2). The “maturation cleavage” between VP4 and VP2 occurs by an autocatalytic process during maturation of the provirion (68). The role of the P2 proteins is primarily to induce the biochemical and biophysical changes that occur in the infected cell. The P3 proteins are more directly involved in the process of RNA synthesis.
Fig. 2.
Genome structure of poliovirus and processing of the polyprotein. The RNA genome of poliovirus contains a long 5’ NTR, a single large open reading frame, and a short 3’NTR, which is terminated by a poly(A) tail. At its 5’-end the RNA is covalently linked to a small peptide called VPg. The polyprotein made during translation of the RNA contains one structural (P1) and two nonstructural precursors (P2, P3). The polyprotein is processed into precursor and mature proteins by proteinases 2Apro and 3Cpro/3CDpro. The maturation cleavage of VP0 into VP2 and VP4 occurs by an autocatalytic mechanism.
It is generally accepted that picornavirus RNA replication proceeds by the following steps:
The RF consists of a double stranded structure, which has a specific infectivity 30 fold higher than ssRNA (20). It is not yet known how RF molecules initiate an infectious cycle but it is presumed that the 2 strands must be separated (74). Interestingly, the infectivity of the RF is dependent on a nuclear factor since enucleated HeLa cells cannot be transfected with PV RF (36, 74). Although the RF is normally considered to be a true intermediate during RNA replication, the possibility cannot be ruled out that it is a byproduct of rapid hybridization between plus and minus strands occurring during isolation. The RI, formed in small amounts during plus strand RNA synthesis, consists of a full-length minus-strand template strand with 6-8 nascent positive strands (9). It contains both double and single stranded regions as shown by their partial resistance to ribonuclease digestion (74, 171). The opposite polarity of RI, with a full-length positive strand RNA and incomplete nascent minus strands, has never been observed (21). It is important to note that since the picornavirus RNA genome contains two different types of termini the RNA polymerase has to be able to initiate RNA synthesis at different ends. The 5’-end consists of VPg linked to a heteropolymeric sequence that starts with two uridylates (4, 77, 130). At the 3’-end the RNA is terminated with a genetically encoded poly(A) tail (38).
In this review an attempt will be made to summarize what is known about the initiation of protein-primed RNA synthesis by the RNA polymerases of picornaviruses, which involves the uridylylation of terminal protein VPg. In addition, a model will be proposed to explain how the nucleotidylylated protein primer might be utilized for minus and plus strand RNA synthesis. It should be emphasized that these models are highly speculative and may change in the future. The review is organized such that the reader can easily find any particular topic of interest but unfortunately this has lead to some redundancy in the discussion of a few subjects, for which we apologize. It will concentrate on poliovirus, the prototype of Picornaviridae, simply because of the greater abundance of information with this virus. However, wherever important differences exist among members of the family, these will be discussed separately. Specifically, we will discuss the following topics: (i) viral and cellular factors involved in the initiation reaction; (ii) what is known about the initiation reaction and the “slide-back” model of VPgpUpU synthesis; (iii) models of protein-primed minus and plus strand RNA synthesis; (iv) unanswered questions about the initiation and elongation reactions of RNA synthesis. We regret that because of space limitations it will not be possible to discuss every publication that deals with this topic or to give credit to all of those investigators who have studied this intricate step in the life cycle of picornaviruses. However, we hope that this article will provide the reader with a summary of the accomplishments and unsolved questions of this topic.
2. Factors involved in the initiation of protein-primed RNA synthesis
2.1. RNA structures
A requirement for cis-acting RNA elements (cres) for RNA synthesis by picornavirus RNA polymerases was already suspected from early in vitro studies, which showed that purified poliovirus RNA polymerase was able to copy many different RNAs with a complementary oligonucleotide primer (46). However, in vivo the enzyme is highly specific for the copying only of its own RNA template suggesting that specific RNA structures must be present within the genome that provide specificity to this process. After years of extensive analyses it has become clear that at least 3 different cis-acting RNA elements are required for the initiation of picornavirus RNA replication. Here we will summarize what is known about these elements in enteroviruses, with an emphasis on poliovirus RNA.
2.1.1. The 5’-terminal cloverleaf
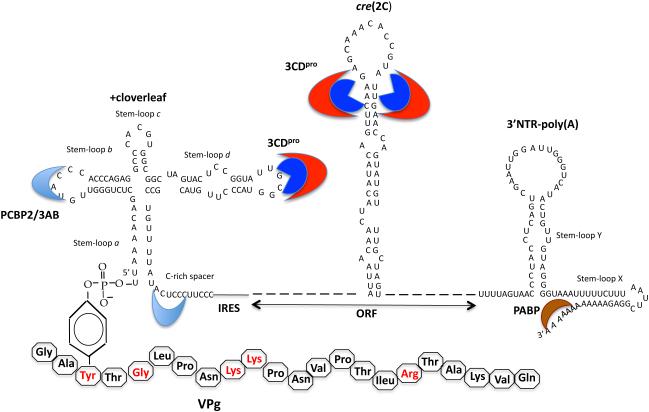
All picornaviruses contain complex RNA structures at the 5’-end of their RNA genomes. In enteroviruses and rhinoviruses, the 5’-terminal structure is similar to that of a cloverleaf (Fig. 3) (5, 6, 124) while with cardioviruses, aphthoviruses and hepatoviruses these structures are less well-defined (172). With all picornaviruses the 5’-terminal UMP of the plus strand cloverleaf is covalently linked to a hydroxyl group of a tyrosine in a small peptide called VPg (Fig. 3) (4, 77, 130).
Fig. 3.
RNA structures involved in poliovirus RNA replication. There are three RNA structures in poliovirus RNA that are required or important for RNA replication (section 2.1) (131). A cloverleaf-like structure is located at the very 5’-end of poliovirus RNA. It contains the terminal UMP that is linked to the hydroxyl group of tyrosine in VPg. The amino acid sequence of VPg is shown below and the residues important for RNA replication are shown in red. The cloverleaf binds viral protein 3CDpro to stem-loop d and cellular protein PCBP2 to stem-loop b and a C-rich spacer. Viral protein 3AB also binds to stem loop b. The cre(2C) is a hairpin located in the coding sequence of 2CATPase. It is the template for the linkage of 2 UMPs to VPg during the initiation of RNA synthesis. The third RNA structure used during PV RNA replication is the 3’NTR, which contains two stem-loops, X and Y. These stem loops are involved in a “kissing” interaction (not shown). The binding of cellular poly(A) binding protein to the poly(A) tail is believed to be important for the circularization of the RNA genome.
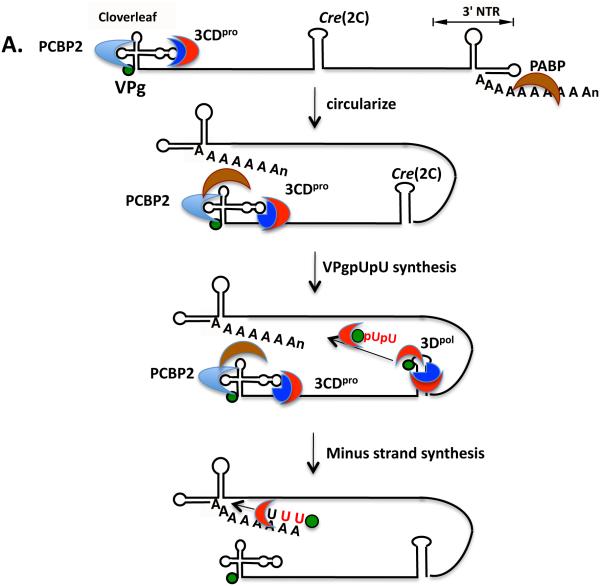
The role of the plus strand PV cloverleaf in RNA replication was discovered by Andino and coworkers (5, 6). Subsequently, the binding of 3CDpro to stem-loop d and of cellular proteins PCBP1/PCBP2 and of 3AB to stem-loop b was demonstrated and proposed to have a function in the initiation of minus strand RNA synthesis (22, 50, 51, 63). Poly(A) binding protein (PABP), which binds to the poly(A) tail, also interacts with 3CDpro and is believed to facilitate the circularization of the RNA genome prior to minus strand RNA synthesis (15, 64). The observation that minus strand RNA synthesis requires the cloverleaf suggests that circularization of the genome is a necessary part of the replication process.
The sequence complementary to the plus strand cloverleaf forms a similar structure, the minus strand cloverleaf, which serves as the template for the initiation of plus strand RNA synthesis. Viral proteins 2CATPase and 2BCATPase were shown to bind specifically to the minus strand cloverleaf but the biological significance of this interaction has not yet been determined (10, 11). Similarly, specific binding of cellular protein hnRNPC1/C2 to the 3’-end of minus strand cloverleaf RNA was demonstrated (25). These proteins interact with 3CDpro and the P2 and P3 precursor polypeptides and this interaction has been found to stimulate plus strand RNA synthesis (24, 25). The importance of the two As at the 3’end of the minus strand cloverleaf for efficient VPgpUpU-primed plus strand RNA synthesis in preinitiation replication complexes (section 3.5) was suggested by Sharma and coworkers (138).
It has been known for some time that the cloverleaf structure is a critical RNA element for both minus and plus strand RNA synthesis but determining the exact role of each cloverleaf separately has been difficult. To dissect the function of the cloverleaf, Vogt and Andino has used a novel construct of PV with dual cloverleaves in the 5’NTR, one of which regulated minus strand synthesis while the other mediated plus strand RNA synthesis (167). Using this construct they demonstrated that the entire 5’-plus strand cloverleaf, including intact stem-loops b and d for PCBP and 3CDpro binding, were required for the initiation of plus strand RNA synthesis. Moreover, it was shown that specific sequences in stem loop a were required for this process but not for minus strand RNA synthesis.
2.1.2. The 3’NTR-poly(A)
The 3’NTRs of picornaviruses are highly diverse. Those of enteroviruses form a highly ordered structure with 2 stem-loops (X, Y) (Fig. 3), which are involved in a “kissing” interaction (91, 121). Other picornaviruses such a rhinoviruses contain only a single stem-loop in their 3’NTRs while the 3’NTRs of coxsackieviruses and echoviruses consist of three stem-loops (X, Y, Z) (120). The importance of the 3’NTR in RNA replication was first suggested by mutational analyses (91, 120, 129). However, subsequent studies have shown that this domain is interchangeable between PV, CVB4 and HRV14 (129). In addition, it was shown that the 3’NTRs of poliovirus, HRV14 and CVB3 are not required for infectivity (23, 156, 163). In contrast those of FMDV and EMCV are essential for viral viability (40, 132). Deletion of the Z domain of CVB3 had no significant effect on growth in tissue culture but in mice this mutant exhibited reduced viral pathogenesis (92).
Numerous studies have attempted to find specific binding of viral or cellular proteins to the 3’NTR of picornavirus RNAs. Both PV 3AB and 3CDpro were shown to specifically interact with the PV 3’NTR (63). The RNA polymerase of EMCV binds to its cognate 3’NTR while 3AB and 3ABC of HAV interact with the 3’NTR (33, 76).
The poly(A) tail of picornaviruses, first identified by Yogo and Wimmer, is variable in length (180). That of poliovirus is reported to be on average between 50 and 90 nucleotides long (70, 180). It is genetically encoded, that is, it is transcribed from poly(U) in the minus strand (38) and that seems to be true for all picornaviruses. It was recently reported for poliovirus that 3Dpol polyadenylylates viral RNA during RNA replication using a reiterative transcription mechanism (70, 147). Previous evidence indicated that shortening the poly(A) tail reduces the infectivity of genomic RNA (143). The poly(A) tail was also proposed to play a role in the formation of a circularized RNP complex, which is stabilized by protein/protein interactions (64). In the in vitro translation/RNA replication system a poly(A)13-20 RNA was required to achieve wt-like minus strand RNA synthesis (141). This is also about the same length necessary for human PABP to interact with poly(A) in vitro (64).
2.1.3. The cre elements of picornaviruses
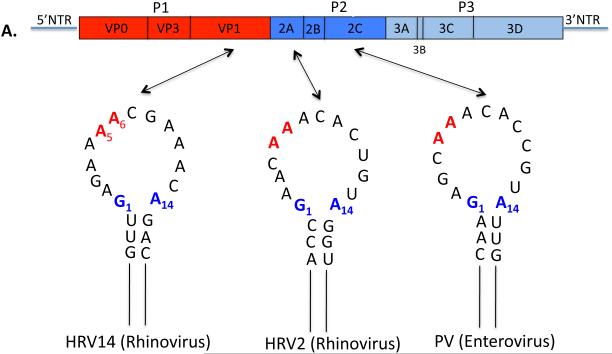
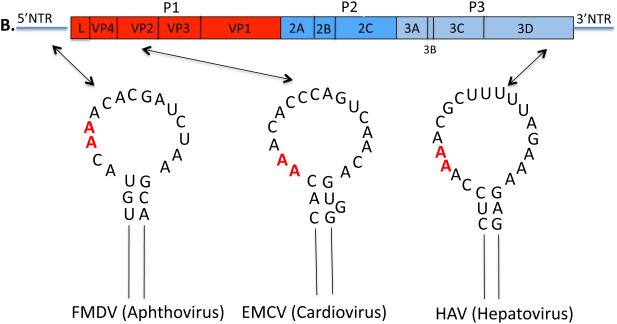
For many years only the 5’ and 3’NTRs of picornaviral genomes were considered as ample sources of cis-replicating RNA elements for RNA replication. The first indication of a cis-replicating RNA element in the coding sequences of a picornavirus came from the finding that deletion of the P1 coding region of HRV14 resulted in the replicons’ inability to replicate (90). A hairpin structure in VP1 was identified that was absolutely required for genome replication. This genetic element was named as a cis-acting replication element, cre. Subsequently, Goodfellow and coworkers reported the presence of an essential RNA hairpin, cre(2C), in the coding sequence of poliovirus 2CATPase ( Fig. 3) (55). The biological function of cre(2C) was demonstrated both by biochemical and genetic experiments to be the template for the uridylylation of VPg, generating the primer (VPgpUpU) for RNA synthesis (113, 114, 123). Since then cre elements with the same function have been discovered or predicted to exist in the coding sequences of various picornaviruses. These structures are similar in the different genera of Picornaviridae although the location of these elements varies and overall their nucleotide sequences are highly divergent (Figs. 4A, 4B): cres of PV and CVB3 (Enterovirus) in 2CATPase (55, 164); cres of HRV14, HRV3 and HRV76 (Enterovirus) in VP1 (89, 90); cres of HRV2, HRV1a, HRV16 (Enterovirus) (52, 89); cre of species C Rhinovirus (Enterovirus) in VP2 (30); cre of TMEV and Mengovirus (Cardiovirus) in VP2 (83); cre of HAV (Hepatovirus) in 3Dpol (178); cre of HPeV (Parechovirus) in VP0 (2); cre of Porcine Sapelovirus (Sapelovirus) in 2CATPase (142). The only known exception is FMDV (Aphthovirus) whose cre is located in the 5’NTR rather than the coding sequences (88).
Fig. 4.
Secondary structures of the picornavirus cre elements and their locations in the genome. (A) A simplified version of the enterovirus and rhinovirus RNA genome is shown on top. Open loops of the PV1, HRV2 and HRV14 cre elements (section 2.1.3) are shown in detail with G1 and A14 shown in blue and A5 and A6 that template VPgpUpU synthesis are shown in red. Only the upper part of the stem is shown. (B) A simplified version of the cardiovirus and aphthovirus RNA genomes is shown on top. The hepatovirus genome is the same except it lacks the L protein. The loops of the EMCV, FMDV, and HAV cre elements (section 2.1.3) are shown in detail and A5 and A6 that template VPgpUpU synthesis are shown in red.
The diversity of the location of cre elements in different picornavirus genomes is astounding since, as we will show below, their basic function is identical. Since cre elements function strictly by their RNA structure that, as sequence, has also to accommodate the coding activity of the viral mRNA, the location may be determined by the best “fit”into messenger function of the viral open reading frame. Equally surprising, the location of the functioning cre elements by genetic engineering can be changed within a specific genome. It has been shown for several viruses (PV, FMDV and HRV14) that the location of the cre elements can be changed within the genomes (55, 88, 178, 179). In this case, the endogenous cre function can be completely inactivated by mutation without loss of viral proliferation. In poliovirus, a synthetic PV-specific cre element can even be moved to the 5’NTR (into spacer I between the cloverleaf and IRES) (179). In this construct the inactivation of the cre(2C) by mutation has very little influence on the replication in HeLa cells although, remarkably, its neurovirulence in CD155 tg mice was lowered five orders of magnitude (34, 160). It should be stressed, however, that the location of the cre is conserved within each particular picornavirus species (30).
The cre elements of human enteroviruses and rhinoviruses are characterized by a loop that is 14 nt long and contains a conserved motif: R1NNNA5A6RNNNNNNR14 (Fig. 4) (176, 179). The first base, the 7th base and the 14th base are all purines (R) and the As at positions 5 and 6 in the loop are involved in templating the two Us during the synthesis of VPgpU/VPgpUpU (52, 100, 123, 164, 176, 178). The other genera of Picornaviridae where cre elements have been identified possess cre structures in which the length of the loop is more variable (15-23 nt) except for the core sequence of AAA/GC. The double stranded stem of the cres with internal bulges and loops are variable in length and in sequence. The minimal functional HRV14 cre(VP1) consists of the 14 nt loop and a 9 bp stem (154, 176). With PV cre(2C) the minimal structure that retained activity in vitro was about 26-29 nt long, including the 14 nt loop (108, 179).
2.2. Viral and cellular proteins
In the simplest in vitro reaction on a poly(A) template only two purified poliovirus proteins (3Dpoland VPg) are required for the synthesis of the nucleotidylylated protein primer, VPgpU/VPgpUpU, while on the cre(2C) template the optimal reaction also requires 3CDpro (section 3.2). However, in preinitiation replication complexes (PIRCs) (section 3.5), the initiation reaction appears to be much more complicated because VPg-uridylylation requires the cloverleaf (85), the ATPase activity of protein 2CATPase (13, 14) and membranes (47). This section of the review will discuss briefly what is known about the role of the viral and cellular proteins that are involved in the initiation reaction.
2.2.1. RNA polymerase 3Dpol
The central player in RNA synthesis is 3Dpol, the RNA-dependent RNA polymerase, which catalyzes both the VPg-uridylylation and RNA chain elongation reactions (46, 115). Three dimensional structures have been reported for several members of the virus family such as PV, HRV14, CVB3, FMDV, EV-71 (7, 27, 43, 44, 59, 61). The 3Dpol of Picornaviridae are 460-470 amino acids long and possess high primary sequence similarity and structural homology (26). They adopt a fold resembling a cupped right hand with fingers, thumb and palm domains and a strong interaction between the fingers and thumb domains accounts for the closed protein structure. The palm domain is the most conserved where two aspartic acid coordinate two metal ions essential for catalysis. The roles of the fingers and thumb domains are in primer, NTP and template binding. In vitro the protein oligomerizes through an interaction between the palm and thumb domains, which enhances the RNA binding and elongation acitivities of the polypeptide (65, 107). Although in infected cells most of the protein is in a soluble form, a fraction of it is anchored to the membranous replication complexes, possibly via an interaction with the 3B domain of membrane protein 3AB (66, 173). Recently, it was proposed that PI4P (phosphatidylinositol 4-phosphate) lipids, produced by cellular protein PI4KIIIβ (phosphatidylinositol-4 kinase III beta), recruit 3Dpol to the membranes for coxsackie virus B3 (CVB3) RNA replication (67). However, CVB3 mutants that, interestingly, can bypass the requirement for PI4KIIIβ have been described (161), which suggests that the current model for enterovirus genome replication in infected tissue culture cells may have to be modified (67).
2.2.2. Terminal protein VPg and its precursors
A 5’-terminal viral protein VPg that is covalently bound to the genome of poliovirus was first identified by Wimmer and his colleagues (77). They are small, basic peptides 19-26 amino acids in length (Table 1) (148). They contain a fully conserved tyrosine at position 3 of the VPg amino acid sequence, the attachment site to the RNA. With the exception of EMCV, picornavirus VPgs also contain a conserved glycine at position 5 (Table 1). It should be noted that the conserved spacing of Y3 and G5, downstream of the 3A/3B cleavage site, can be potentially used in the identification of VPg sequences in newly identified picornaviral polyproteins. Other residues required in PV VPg for viral growth are the glycine at position 5, two lysines at positions 9 and 10 and an arginine at position 17 (Fig. 3) (53, 75, 113, 122). All of these residues are important for an interaction of VPg with 3Dpol of PV when assayed in a yeast two-hybrid system (113). Interestingly, enterovirus VPgs contain 3 prolines at positions, 7, 11 and 14. The structure in solution of PV VPg in the presence of an organic solvent, TMAO, was determined by NMR (136). The structure consists of a long loop (residues 1-14) and a short C-terminal helix. An NMR structure of chemically synthesized VPgpU was more stable than that of free VPg and exhibited a defined globular structure (135). FMDV is unique in that it encodes 3 distinct VPg sequences all of which function as substrates in vitro for uridylylation and all of them can be observed attached to viral RNAs (71, 100). Interestingly, the VPg of PV could be replaced by that of HRV14 both in the in vitro uridylylation assay and in vivo, yielding a viable virus (113) that acquired a mutation (L11P) in HRV14 VPg after passages. Similarly, the VPg of HRV16 was also compatible with the growth of PV but HRV16 replication did not tolerate the VPg of PV (28).
Table 1.
VPg sequences of Picornaviridae
| Genus | Virus | VPg amino acid sequence* |
|---|---|---|
|
| ||
| Enterovirus | ||
| PV1(M) | GAYTGLPNKKPNVPTIRTAKVQ | |
| PV2s | GAYTGLPNKRPNVPTIRTAKVQ | |
| PV3s | GAYTGLPNKRPNVPTIRAAKVQ | |
| CAV21 | GAYTGLPNKKPNVPTIRIAKVQ | |
| CVB3 | GAYTGVPNQKPRVPTLRQAKVQ | |
| EV71 | GAYSGAPNQVLKKPVLRTATVQ | |
| Echo6 | GAYTGMPNQKPKVPTLRQAKVQ | |
| Rhinovirus | ||
| HRV14 | GPYSGNPPHNKLKAPTLRPVVVQ | |
| HRV2 | GPYSGEPKPKTKIPERRVVTQ | |
| Aphthovirus | ||
| FMDV1 | GPYAGPLERQKPLKVRAKLPQQE | |
| FMDV2 | GPYAGPMERQKPLKVKAKAPVVKE | |
| FMDV3 | GPYEGPVKKPVALKVKAKNLIVTE | |
| Cardiovirus | ||
| EMCV | GPYNETARVKPKTLQLLDIQ | |
| Hepatovirus | ||
| HAV | GVYHGVTKPKQVIKLDADPVESQ | |
| Sapelovirus | ||
| SPV | GAYTGAPKPETRKPVLRKAVVQ | |
The conserved Tyrosine at position 3 is shown in red and bold.
There are 5 precursors of VPg (3AB, 3ABC, 3BC, 3BCD and P3) and it is not yet known whether in vivo VPg itself is uridylylated by 3Dpol or one of its precursors. 3AB, a small basic protein (12 kDa) has multiple functions in vitro. It is a nonspecific RNA binding protein but when complexed with 3CDpro it specifically interacts with the 5’ cloverleaf of PV RNA and with the 3’NTR (63, 174, 175). In addition it possesses in vitro chaperone and helix destabilizing activities (35). Experiments to uridylylate 3AB in vitro, however, have failed (49). Interestingly, mutations in 3AB that cause a severe RNA replication defect can be complemented in trans only by P3 but not by the mature 3AB polypeptide (157).
Polypeptide 3BC is normally not observed in PV-infected cells but it functions efficiently as substrate for 3Dpol in in vitro VPg-uridylylation reactions on a cre(2C) template and it can also replace the function of 3CDpro in the reaction (110). The larger 3B-containing precursors, 3BCD and P3, can be observed in small amounts as uncleaved polypeptides in PV-infected cells. 3BCD can also be uridylylated by 3Dpol in vitro though less efficiently than either VPg or 3BC (110). A 3BC cleavage site mutant replicated in vivo and produced 3BC-linked RNA (104). Infectious virus, however, was not produced. A pseudorevertant of this virus, which partially restored 3BC cleavage, also restored virus infectivity.
2.2.3. Proteases 3Cpro and 3CDpro
Protein 3Cpro is a Cys-reactive proteinase with a chymotrypsin like structure (97), which is also an important RNA-binding protein during RNA replication (108, 177, 179). 3Cpro and its precursor 3CDpro are responsible for the processing of most of the polyprotein at Q/G cleavage sites and they also affect several cellular functions (39, 87). The crystal structures of the 3Cpro polypeptide have been determined for HRV14, PV, HRV2, HAV and FMDV (87). 3CDpro is the precursor of both proteinase 3Cpro and of RNA polymerase 3Dpol. Although, it possesses proteinase activity it totally lacks RNA polymerase activity. As noted above (section 2.1.1), as an RNA-binding protein 3CDpro interacts with the 5’ cloverleaf in a complex with PCBP2 or 3AB, with the cre(2C) RNA element in a complex with VPg and 3Dpol, and with the 3’NTR in a complex with 3AB (50, 63, 108, 123). The RNA-binding activity of 3CDpro resides in the 3Cpro domain but the activity is modulated by the 3Dpol domain of the polypeptide (31). The crystal structure of 3CDpro revealed that the 3C/3D cleavage site in the polypeptide is on the opposite side from the catalytic site of the 3Cpro protein (86). The RNA binding activity of 3CDpro appears to be also important for virus maturation (48).
2.2.4. 2CATPase
The 329 amino acid long 2CATPase is a complex multifunctional polypeptide, which contains an important ATP binding domain and possesses ATPase activity in vitro that is inhibited by guanidine hydrochloride (94, 119, 126). It was classified as a member of superfamily III helicases with 3 conserved motifs two of which are related to ATP binding (57). However, so far no helicase activity has been detected in in vitro assays with the purified protein (119). The N-terminal domain of 2CATPase, which anchors the protein to membranes, contains an amphipathic helix, an oligomerization domain and an RNA-binding domain (1, 41, 112, 125). Near the C-terminus there is a Zn++ binding domain and another amphipathic helix that is likely involved in membrane binding (118, 152). Genetic studies have implicated 2CATPase in numerous functions in the viral life cycle, such as uncoating (78), host cell membrane rearrangements (3, 29, 152), RNA binding (125), RNA replication (14, 79, 112, 153), and encapsidation (81, 165, 169).
2.2.5. Poly(rC) binding protein 2: PCBP2
PCBP2 is a cellular RNA binding protein belonging to a family of proteins that contain K-homologous domains. It recognizes and binds poly(rC) residues and interacts specifically (i) with the IRES of PV to enhance translation and (ii) with the cloverleaf and a C-rich sequence (see below) to facilitate minus strand RNA synthesis (16, 22, 50, 51, 106, 168). During PV infection PCBP2 is cleaved by proteinase 3Cpro/3CDpro (117). Mammalian cell extracts depleted of PCBP2 can rescue PV translation and RNA replication. PCBP1, another member of the PCBP group, also binds to the 5’NTR of PV but with less affinity than PCBP2 (131). PCBP1 can rescue only RNA replication in extracts depleted of this protein.
Recent studies have shed new light to the binding of PCBP2 to the PV cloverleaf (158, 181). It was shown that PCBP2 needs two cytosine-rich clusters just downstream of the cloverleaf (mapping to spacer between the cloverleaf and the IRES) in order to form a PCBP2 /cloverleaf complex for function in RNA replication (158).
2.2.6. Heterogeneous nuclear ribonucleoprotein C: hnRNP C
Cellular protein hnRNPC is a primarily nuclear polypeptide that exists in human cells in two spliced isoforms C1 and C2 (25). It forms heterotetramers consisting of 3 C1 molecules and one C2 molecule, which bind RNA (58). Previous studies have identified RNP complexes between hnRNP C and the 3’-end of PV minus strand RNA (127). The protein also interacts with 3CDpro and with the P2 and P3 precursor polypeptides (24). More recently Ertel and coworkers have demonstrated that hnRNP C also binds to the 5’-end of PV minus strand RNA (42). In addition, it was shown that depletion of hnRNP C in cell culture results in reduced plus strand RNA synthesis and virus yields. These authors proposed that the role of hnRNP C in PV replication is to bind to the 5’- and 3’-ends of the genome and to stabilize the interactions between them. This would facilitate the initiation of plus strand RNA synthesis at the 3’-end of the minus strand.
2.2.7. Poly(A) binding protein: PABP
A role for PABP1 in poliovirus RNA replication was first proposed by Herold and Andino (64). These authors have observed that PABP interacts in vitro with both the poly(A) tail of PV RNA and also with proteins PCBP and 3CDpro. It was suggested that these interactions are required for the circularization of the genome and initiation of poliovirus minus strand RNA synthesis. Using preinitiation replication complexes Silvestri and coworkers have determined the minimum length of the poly(A) tail on PV RNA as 12 nt in length for optimal binding of PABP, for VPg uridylylation, and for negative strand RNA synthesis (141).
3. Initiation of protein-primed picornavirus RNA synthesis
Prior to our studies on the intitation of poliovirus RNA synthesis, DNA synthesis by a protein-primed mechanism was already demonstrated for a number of viruses. These viruses all possess linear double-stranded DNA genomes such as adenovirus, phages Φ29, PRD1 and GA-1 (133, 134). The DNA polymerases of these viruses use as template the 3’-terminus of their DNA strands for the nucleotidylylation of their terminal or preterminal proteins. In addition, it was known that the reverse transcriptase of hepatitis B virus uses a nucleotidylylated protein-primer for cDNA synthesis (137). With RNA viruses protein-primed RNA synthesis was first demonstrated in vitro with a double stranded RNA virus (infectious necrosis virus) whose RNA polymerase itself becomes guanylylated and serves as primer for RNA synthesis (37).
3-1. Discovery of the in vitro VPg uridylylation reaction
The role of VPg as a primer for poliovirus RNA synthesis was already proposed more than thirty years ago (170) shortly after the discovery of VPg covalently linked to both minus and plus strands of poliovirus RNA (4, 77, 130). This model was consistent with the absolute dependence of RNA synthesis by the RNA polymerase on a primer (46). Subsequently, the presence of free VPg and of VPgpUpU in poliovirus-infected cells was demonstrated (32). In addition, the synthesis of VPgpU/VPgpUpU in crude replication complexes was observed and the elongation of these precursors into plus strand RNAs could also be demonstrated (149, 150, 159). Although these results strongly supported the possibility that VPg is the primer for RNA synthesis direct evidence for this model remained elusive. Moreover, alternative models of primed RNA synthesis in poliovirus RNA replication were proposed: amongst them initiation by elongation of a snap-back oligo(U) tail that the 3Dpol polymerized onto the 3’ poly(A) of plus strands (155).
Our studies with the in vitro synthesis of VPgpU/VPgpUpU started when the late Jacques van Boom of Leiden University sent us a sample of chemically synthetized VPgpU. We first tried to elongate it to VPgpUpU on a poly(A) template with purified PV 3Dpol and 32P-UTP, under conditions normally used for the elongation of oligonucleotides on RNA templates. These experiments, however, were unsuccessful. After testing various reaction conditions to achieve an elongation reaction we accidentally discovered that the only change needed was an increase in 3Dpol concentration from nanomolar to micromolar concentrations. As a control, we also tested synthetic VPg under the same reaction conditions and observed 32P-labeling of VPg to yield products migrating at the expected positions of VPgpU, VPgpUpU and VPg-poly(U) on polyacrylamide gels (115). This critical experiment proved that the viral RNA polymerase was able to uridylylate VPg and then elongate it by transcription of the poly(A). In fact, uridylylation itself was critically dependent on a poly(A) template (115).
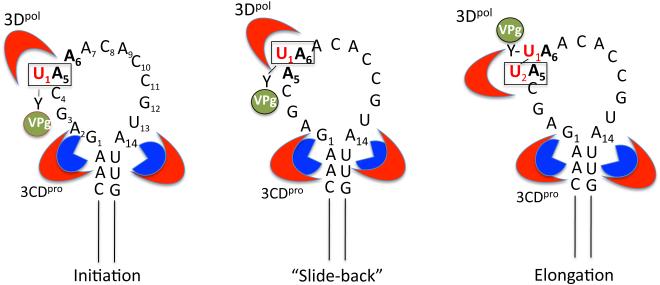
Although the in vitro reaction on a poly(A) template could in principle explain the initiation of minus strand RNA synthesis at the 3’-end of genomic RNA the yield was poor even with full length PV transcript RNAs terminated by poly(A). More importantly, considering that in the infected cell mRNA molecules carrying poly(A) tails are abundant, the uridylylation of VPg did not account for the observed specificity in vivo. Surprisingly, transcripts from which the poly(A) tail was removed worked equally well suggesting that the real template for the uridylylation reaction was somewhere else in the genome. The most likely location of the template sequence was in the 5’ or 3’NTR but there was no detectable VPg-linked product made in vitro with transcripts of these RNA sequences/structures. At about the same time there was a report of an RNA hairpin in the 2CATPase coding sequence, called cre(2C), that is required for RNA replication (55). To our surprise transcripts of this hairpin yielded the expected VPgpU/VPgpUpU products in the in vitro reaction suggesting the possibility that the unknown template sequence resided in this RNA structure (55, 114). Since the yield of the product was still relatively low we tried to stimulate the synthesis of the VPg-linked products with various purified viral and cellular proteins that were available in our lab. These experiments led to the discovery that 3CDpro stimulates the in vitro reaction about 100 fold (114). Subsequent mutational analyses of the cre(2C) led us to suggest that 2 consecutive A residues, A5 and A6 in a conserved motif (A5A6A7C8A9) located in the loop of cre(2C), are of critical importance to cre function (Figs. 4A and 5) (55, 113, 114, 123).
Fig. 5.
“Slide-back” model for VPgpUpU synthesis during initiation of RNA synthesis. The upper stem of cre(2C) interacts with 2 molecules of 3CDpro (or 3Cpro) (section 3.7). 3CDpro binds VPg with the back side of its 3Dpol domain, where another molecule of 3Dpol links UMP to the hydroxyl group of tyrosine in VPg. A5 in the loop of cre(2C) is the template for the linkage of the first UMP (U1) to VPg yielding VPgpU1 . VPgpU1 “slides-back” to hydrogen bond with A6 and the second UMP (U2) is templated again by the A5 nucleotide during the elongation step yielding VPgpU1U2. The nucleotides involved in the “slide back” are boxed. Nucleotides A5 and A6 of the conserved motif are shown in bold.
3.2. Characterization of the in vitro VPg uridylylation reaction catalyzed by PV 3Dpol
When purified PV RNA polymerase 3Dpol is incubated with a PV RNA template or with other 3’-adenylylated RNA templates, NTPs, Mg++ and an oligo(U) primer, full length minus strand RNA copies can be synthesized (12). Poly(A) also serves as a template for the uridylylation of VPg in vitro by 3Dpol to yield VPgpU and VPgpUpU (115). The final product is VPg-poly(U), the 5’-end of minus strand RNA:
The poly(A) templated in vitro reaction can be enhanced 100 fold by replacing Mg++ by Mn++, which, however, is known to decrease the specificity of RNA polymerases (8, 113).
When full length PV RNA is used in the in vitro reaction, instead of poly(A)-RNA, the template for the synthesis of VPgpU and VPgpUpU is the cre(2C) hairpin (114), located in the coding sequence of protein 2CATPase (55). Protein 3CDpro strongly stimulates the cre(2C) templated reaction and this reaction is equally active with Mg++ and Mn++:
Protein 3CDpro can be replaced by 3Cpro, 3BC or by 3BCD (108, 110, 179). It was proposed that the 3Cpro domain of 3CDpro determines the specificity of the reaction while the 3Dpol domain enhances affinity (108). The cre RNAs of HRV14 and to a small extent that of HRV2 can also serve as template in the uridylylation reaction of PV VPg by PV 3Dpol and 3CDpro (52, 114). The template nucleotide in vitro is A5 while A6 provides specificity to the reaction, and A7 only serves to enhance the uridylylation reaction (113, 116, 123). The elongation of VPgpU on cre(2C) RNA is aborted after the addition of the second U most likely because of structural restrictions imposed by the uridylylation site of the enzyme. Interestingly, an exact spacing of A5 and A6 within the loop of cre(2C) is required for optimal uridylylation of VPg in vitro (Fig. 4A). The optimal size of the cre(2C) loop is 14 nt (Fig. 4A) in accordance to previous findings with HRV14 cre(VP1) (176, 179). Mutations that lead to base pairing with A5 and A6 within the loop abolished cre function in the reaction (179). In addition to A5 and A6 in the loop A1 and G14 at the bottom of the loop are also required for efficient production of the VPg-linked precursors (179). Transversions such as G1C or G1U or A14C are highly detrimental to uridylylation activity while transitions (G1A or A14G) are better tolerated at these nucleotide positions. Studies analyzing the optimal length of the cre(2C) element revealed that the upper stem and the loop with a total length of about 26 nt carry the minimal structural requirements for a uridylylation complex formation (108, 179). The upper stem provides the specificity for protein binding and the lower stem plays a structural role (110).
A mutational analysis of PV VPg identified several amino acids that are critical or important for VPg uridylylation by PV 3Dpol in vitro either on a poly(A) or a cre(2C) template (Fig. 3) (113). The most important residues in VPg are Y3, the attachment site of the peptide to genomic RNA, G5, K9K10 and R17. The same residues were found to be important for uridylylation of HRV2 VPg by its cognate 3Dpol (53). PV 3Dpol is able to use as substrate, in addition to its own VPg, the VPg of HRV14 but the yield of product is reduced. In contrast the VPgs of HRV2 and HRV89 had no detectable substrate activities in the in vitro assay. The 3Dpol of HRV2 on the other hand was able to use all four viral VPgs (PV, HRV14, HRV2 and HRV89) as substrates with nearly equal efficiency (53).
To demonstrate that the VPg-uridylylation is a common mechanism used by picornaviral RNA polymerases during the initiation of RNA synthesis we also examined the reactions catalyzed by the RNA polymerase of HRV2. We have demonstrated that the purified 3Dpol of HRV2 catalyzes the same type of synthetic reactions with either poly(A) or a cre RNA template, located in the coding sequence of 2Apro template [cre(2A)] (52, 53). Subsequent in vitro studies with purified RNA polymerases of other picornaviruses have confirned these results. The RNA polymerase of HRV14 uridylylates VPg on a cre located in the VP1 capsid protein coding sequence (176), which was first identified by genetic experiments by McKnight and Lemon (90). In contrast the FMDV RNA polymerase uses a cre element derived from the 5’NTR for the in vitro uridylylation of all of its three distinct VPgs, 3B1, 3B2, 3B3 (88, 100). The reaction is enhanced not only by FMDV 3CDpro but also to a lesser extent by 3Cpro and precursors 3B33C and 3B1233C. The residues within 3Cpro involved in the interaction with the cre have been identified and are located on the face of the polypeptide opposite of the catalytic site (101). In contrast to the polymerases of PV and HRV2 the FMDV enzyme does not use poly(A) as template in vitro either in the presence of Mg++ or Mn++ (100).
3.3. Correlation of VPg uridylylation in vitro with viral growth phenotypes in vivo
Complete mutational analyses of the A5 and A6 positions of the PV and CVB3 cre(2C) loops, and the HRV2 cre(2A) loop, yielding either lethal or quasi-infectious progeny, fully correlated with the inability of such mutants to uridylylate VPg in vitro (53, 123, 164). Substitution of A7 with pyrimidines resulted in reduced synthesis of VPg-linked products in vitro and to a moderately defective growth in vivo. We have confirmed these results with a dual cre virus in which the endogenous cre was fully inactivated by an A5C mutation and an extra wt copy of the cre was inserted between P1 and P2 (179). The results with this virus indicated that the positively charged lysine encoded by the A5A6A7 sequence in the 2CATPase protein could be functionally fully replaced by either Gln or Glu and to a smaller extent by Asn (116). Therefore the lethal growth phenotype of an A5C mutant is not related to 2CATPase protein function but rather to cre function.
A subsequent mutational analysis of the HRV14 cre(VP1) was also in full agreement with our results (176). In addition, it pointed out the functional importance of purines at positions 1 and 14 of the enteroviral and rhinoviral cre elements, forming a non-Watson-Crick closing pair at the base of the loop, which is possibly contributing to the structure of the cre loop.
FMDV is so far the only exception among picornaviruses in that its cre is not located in the ORF but rather in the 5’NTR, contiguous with domain I of the IRES (88). Mutations introduced into this hairpin resulted in reduced RNA replication but did not influence viral mRNA translation. The position of the cre in the 5’NTR was not important since functional replicons and viruses could be obtained if a wt cre was added to the genome following the 3Dpol coding sequences. Genetic experiments with the FMDV cre have shown that the cre can function in “trans” and for this reason it has been suggested that the name cre be replaced by “bus” (3B-uridylylation site) (100).
Mutations in PV VPg or the HRV2 VPg, which reduced uridylylation in vitro were found to abolish or reduce viral growth (53, 113). A chimeric PV with the VPg of HRV14 was viable but substitutions with the VPgs of HRV2 or HRV89 were lethal (113).
3.5. VPg-uridylylation in vivo and in preinitiation replication complexes
As noted above (section 3.2), the first indication that VPg might serve as a protein primer for the RNA polymerase was that VPgpUpU was identified in PV-infected HeLa cells (32). In addition, it was observed that in crude replication complexes isolated from PV-infected cells, VPg was uridylylated in the presence of UTP to yield VPgpU and VPgpUpU (149, 150, 159). These precursors could be chased into VPg-linked UUAAAACAG, the first nine nucleotides of plus strand RNA and into full length plus strand RNAs. The treatment of such crude replication complexes with detergent abolished the synthesis of VPgpUpU suggesting that the initiation reaction is dependent on the integrity of membranes (47, 150, 159). This is in contrast to the in vitro reaction where 3Dpol is able to uridylylate VPg in the absence of membranes (53, 114, 115). It has been suggested that in vivo membranes might enhance the concentration of ingredients that is required for the uridylylation reaction (107, 111).
In addition to isolating replication complexes from infected cells they can be also made in vitro in a translation and RNA replication system in a HeLa cell free extract, which produces viable poliovirus (95). Preinitiation replication complexes (PIRCs), formed in the presence of GnHCl, an inhibitor of RNA replication, are isolated and used for studies of poliovirus RNA replication (13, 14, 85). Upon removal of GnHCl the PIRCs immediately initiate both cre-dependent VPg uridylylation and minus strand RNA synthesis. Both VPg uridylylation and minus strand synthesis were found to require the GnHCl-inhibited activity of protein 2CATPase (13, 14). It was suggested that PV RNA functions coordinately as a template for both RNA replication and cre-dependent VPgpUpU synthesis (85, 145). Whether or not 2CATPase has a function directly in the initiation reaction itself or indirectly in the production of membranous replication complexes required for the reaction is not yet known. In PIRCs negative strand RNA is made before positive strand RNA and RNA replication is asymmetric, with an excess of plus strand RNA made for each negative strand RNA template. During the initiation reaction hundreds of excess molecules of VPgpUpU per minus strand are made (85) and each minus strand is copied into 40-70 copies of plus strand progeny RNA (102). Barton and coworkers have demonstrated that PIRCs produced RF RNA and VPg-linked genome length plus strand RNA in a large excess over minus strands (13). Unlike in the simple in vitro reaction that uses purified components, the synthesis of VPgpUpU in PIRCs is sensitive to detergents suggesting a requirement for intact membranes in this process (47).
3.6. Protein/protein and protein/RNA interactions involved in the VPg-uridylylation reaction
In vitro three proteins are required for the uridylylation of PV VPg on cre(2C) RNA: RNA polymerase 3Dpol, the substrate VPg, and an RNA binding protein 3Cpro/3CDpro (section 3.2) (114). While 3Cpro alone can stimulate the reaction it is not as effective as 3CDpro suggesting that the 3Dpol domain also contributes to its function (101, 109, 140, 177, 179). The enhancing effect can be abolished by mutations within the RNA–binding domain of PV 3Cpro (R84S, I86A) (114). Direct binding between 3Cpro and cre(2C) transcript RNA was demonstrated in vitro by gel-shift and filter binding assays (179). Both structural and biochemical studies have shown that PV 3Cpro can form dimers in vitro and furthermore genetic studies indicate that PV 3Cpro dimer functions in VPg-uridylylation (108). Recent studies have suggested that 3Cpro binding to cre(2C) occurs in two steps. First a dimer of 3Cpro binds to the upper stem of cre(2C) and unwinds it such that each 3C-domain interacts with a strand of the stem in a sequence specific way (108). Subsequently, 3Dpol is recruited to this complex by an interaction between the back of the thumb subdomain of 3Dpol and the top of the 3Cpro dimer (26, 110).
Since in vitro VPg functions well as substrate for uridylylation by 3Dpol, one would expect that it binds into the active site of the enzyme. However, previous genetic studies have indicated the presence of a second VPg-binding site (in the context of 3AB) on PV 3Dpol on the palm subdomain near motif E, far from the active site (84). This finding was supported by structural studies of CVB3 in which 3Dpol was cocrystalized with VPg and the peptide was found to be bound to the back of the thumb, again distant from the active site (59). One possible explanation of how VPg could be uridylylated at this site is that the nucleotidylylation reaction is carried out by a second molecule of 3Dpol (59, 103, 151). Such a “trans” uridylylation mechanism was supported also from structural studies of the EV71 polymerase complexed with VPg, in which VPg bound at the bottom of the palm domain could not access the catalytic site (27). It should be noted, however, that a front-loading model of VPg-binding and a “cis” uridylylation mechanism was proposed for FMDV 3Dpol (43) and for the RNA polymerase of rhinovirus (7). These conflicting results suggest that there exists some variation in the uridylylation reaction among members of the Picornaviridae. A recent study with PIRCs have provided strong evidence that PV VPg binds at the back site in the 3Dpol domain of PV 3CDpro, and that a second molecule of 3Dpol carries out the linkage of UMP to the tyrosine of VPg (45).
3.7. A “slide-back” mechanism of VPg uridylylation
In our initial studies on VPg-uridylylation we demonstrated that A5 and A6 in the conserved A5A6ACA sequence in the cre(2C) loop are essential not only for the in vitro synthesis of the VPgpU/VPgpUpU precursors but also for RNA replication in vivo and for viral growth (113, 114, 123). Since RNA polymerases synthesize new chains in the 5’----✧ 3’ direction we expected that A6 would serve as template for the addition of the first UMP to VPg followed by the addition of the second UMP on the A5 template nucleotide. In addition, we assumed that the RNA polymerase possesses a strict specificity toward UTP in the nucleotidylylation reaction since in both minus and plus strand RNAs VPg is linked to UMP, the first nucleotide of the RNA chains. Surprisingly, neither of these predictions turned out to be correct. We observed that beside U, 3Dpol could also link C, G, or A to VPg on mutant templates that contained G, C, or U at the A5 position of the loop, respectively (Fig. 5) (116). The nucleotidylylated peptides, VPgpN (where N is C, G or A) could not be efficiently elongated into a dinucleotide such as VPgpNpN or VPgpNpU. In contrast, nucleotide changes at the A6 position of the conserved motif did not lead to any linking of C, G, or A to VPg but resulted in a much reduced yield of products, particularly of VPg-linked dinucleotides.
These results are consistent with a “slide-back” mechanism for the synthesis of VPgpUpU, similar to what was proposed for the initiation of protein-primed DNA synthesis by the DNA polymerases of adenovirus or phages Φ29, PRD1 or G-1 (Fig. 5) (111, 133, 134). According to this model the first UMP (U1) is linked to the hydroxyl group of Y3 in VPg on the A5 template nucleotide of the conserved cre(2C) sequence in the loop, yielding VPgpU (116). In the next step VPgpU1 “slides-back” so that U1 can hydrogen bond to A6. This is followed by the addition of a second UMP (U2), again templated by A5, producing the final product VPgpU1pU2. In this model A5 is the template for the addition of both U1 and U2 to VPg and A6 provides the specificity to the reaction. Although A6 itself is not used as a template the role of this nt is very important because it ensures that the only dinucleotide produced will be VPgpUpU. The polymerase 3Dpol has a relaxed nucleotide specificity in the nucleotidylylation reaction but it does not possess a proof-reading activity. Therefore the enzyme would be unable to remove any incorrect nucleotide from VPgpN (N is C, G or A), if it were produced by mistake. However, during the “slide-back” step VPgpN cannot hydrogen bond with A6 therefore it will not be converted into a dinucleotide and only VPgpU will be elongated into VPgpUpU by the enzyme.
The elongation of VPgpU is aborted after the addition of the second U on the cre(2C) template, to produce VPgpUpU (116). For example, the polymerase is not able to synthesize VPgpUpUpU on a mutated template in which C4 is substituted with an A (116). In addition, further copying of the loop sequence in cre(2C) would result in VPgpUpU being linked to a G and then a C, products that would not be complementary to the 3’ sequence of the minus strands during plus strand RNA synthesis. Therefore the termination of VPgpUpU elongation is likely due to some structural characteristics of 3Dpol associated with the nucleotidylylation reaction. Even in the poly(A)-templated reaction the primary products are VPgpU/VPgpUpU and only a small fraction of the precursors are elongated into VPg-poly(U) (115). Such abortive synthesis of VPgpUpU is also evident in crude replication complexes isolated from PV-infected cells (149, 150, 159) and in PV-infected HeLa cells in vivo (32).
Both biochemical and genetic experiments suggest that VPg is delivered to the replication complex in the form of one of its precursors (80, 110, 157). However, the identity of the precursor that is used as substrate for uridylylation remains to be elucidated. Evidence for 3AB (or a 3AB precursor) as the VPg-precursor in vivo was provided by Liu and coworkers who showed that a replication defect caused by an Y3F mutation in VPg could only be rescued by wt P3 (80). Other in vivo studies showed that a mutation in the 3A domain of 3AB that caused a replication defect that could only be rescued by the P3 polypeptide and not by 3AB (157). Surprisingly, 3AB can not be uridylylated by 3Dpol in vitro on the cre(2C) template with Mg++ as the cofactor while 3BC is an excellent substrate for this reaction under the same conditions (110). Although 3BC is not normally observed in infected cells, 3BC-linked RNA was found with a PV replicon that contains a mutation in the 3B/3C cleavage site (104). This suggests the possibility that the VPg precursor in vivo is 3BC or 3BCD or P3.
3.8. Is VPgpUpU synthesized on the cre required for both plus and minus strand RNA synthesis?
The fact that the picornaviral cre templates the synthesis of VPgpU and VPgpUpU for priming RNA synthesis has been well established for several viruses by both biochemical and genetic experiments (52, 100, 114, 123, 176, 178). However, it was not clear whether the VPgpUpU made on the cre was used in vivo for both minus and plus strand RNA synthesis. As far as minus strand RNA synthesis is concerned, beside cre(2C), the 5’-end of the poly(A) tail could also be considered a potential template for the uridylylation of VPg (98, 115). Similarly, instead of cre(2C), the two As at the 3’end of minus strands might serve as templates for VPgpUpU synthesis during plus strand RNA synthesis (138). However, early studies in vivo with full-length transcript RNAs of PV or CVB3, lacking the first two Us of the genome, indicated that they were infectious but the two Us were regained during replication in transfected cells (62, 73). These observations suggested that the first two As at the 3’-end of minus strand RNA are not essential as templates for VPgpUpU synthesis in vivo during plus strand RNA synthesis. Subsequently, PIRCs have provided useful, though sometimes conflicting information, on this subject (14, 56, 164). Initial experiments with this system suggested that the VPgpUpU made on the cre(2C) was used only for plus strand RNA synthesis (56, 96, 98). Subsequently, Steil and Barton have shown that cre-dependent VPgpUpU is made both before and during minus strand RNA synthesis, with 100-400 molecules of VPgpUpU made per negative-strand (145). This observation is consistent with the possibility that VPgpUpU also primes negative strand RNA synthesis. The hypothesis was further supported by several lines of evidence: (i) guanidine, a potent inhibitor of RNA replication and 2CATPase activity, reversibly inhibits both cre-dependent VPg uridylylation and minus strand RNA synthesis (98, 119); (ii) the A5C mutation in the CVB3 cre blocked both VPg-uridylylation in vitro and minus strand RNA synthesis in PIRCs (164); (iii) under conditions of low UTP concentration VPgpUpU made on the cre is the preferred primer for negative strand RNA synthesis in PIRCs (146). The consensus opinion emerging on this topic is that cre(2C) templates VPgpUpU synthesis for the production of both minus and plus strands of PV RNA.
4. Models of poliovirus minus and plus strand RNA synthesis
The following models aim to integrate all the data currently available from both genetic and biochemical experiments, including preinitiation replication complexes (72, 111, 131, 144). It is clear that picornavirus RNA replication is an extremely complex process, which takes place on membranous structures (19) where several cis-acting RNA elements and viral and cellular proteins have to function in a coordinated fashion. The membranous structures involved in this process are not yet well understood and will not be discussed in the models presented below. Therefore the models, as presented, are highly simplified versions of what might happen in vivo.
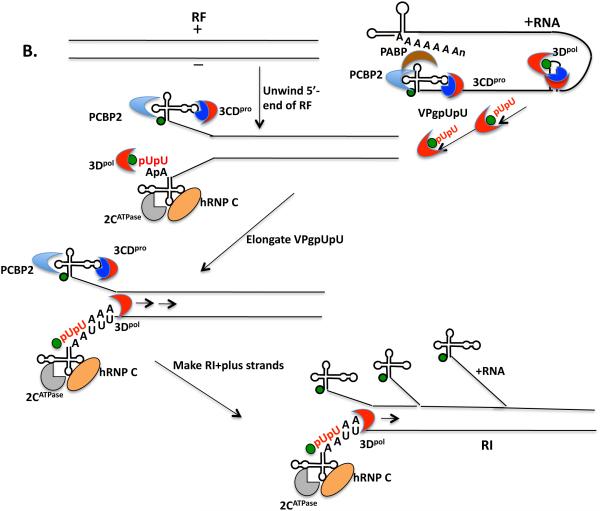
Minus strand RNA synthesis begins with the circularization of the PV plus strand RNA via interaction between 3CDpro and PCBP2 (and/or 3AB) bound to the 5’cloverleaf and PABP, which interacts with the poly(A) tail (Fig. 6A). Whether the interaction between the RNP complexes at the 5’- and 3’-ends of the RNA occurs only in cis or can also take place in trans is not yet known. VPg or one of its precursors is uridylylated on the cre(2C) template to yield VPgpUpU. This is then transferred to the 3’-end of the poly(A) tail but the mechanism of transfer and what provides specificity to this process are not yet known. Since the two uridylates attached to VPg are insufficient to provide sequence specificity to the transfer reaction it is likely that this is accomplished through binding of VPgpUpU to 3Dpol alone or complexed with 3CDpro, which could recognize a sequence or structure near the 5’-end of the genome. In the next step 3Dpol elongates the VPgpUpU primer on the plus strand RNA template into a VPg-linked minus strand yielding a double stranded intermediate RNA (RF). In order to begin plus strand RNA synthesis the end of the RF structure has to be unwound or at least destabilized so that the plus strand cloverleaf can refold and interact with a 3CDpro/PABP/PCBP2 complex (Fig. 6B). This might be facilitated by the binding of 2C/2BCATPase to the 3’-end of the minus strand cloverleaf. A cellular protein hnRNP C then might bind to both the 5’- and 3’-ends of the minus strand thereby stabilizing the interactions between the termini of the RF. Alternatively, 3AB, which possesses RNA chaperone and helix destabilizing activity, and binds in a complex with 3CDpro to the plus strand cloverleaf, assists in the unwinding process. VPgpUpU, made on cre(2C) prior to the formation of RF, is then transferred to the 3’-end of the minus strand cloverleaf. As noted above, it is likely that VPgpUpU is in a complex with 3Dpol and/or 3CDpro during the translocation step. The two terminal adenylate residues align with the two Us of VPgpUpU and 3Dpol elongates the plus strand RNAs on the minus strand template. Multiple plus strands are made off the same minus strand template yielding a replicative intermediate (RI) and finally the full length plus strands are completed.
Fig. 6.
Models of minus and plus strand RNA synthesis. (A) The poliovirus RNA circularizes through the interaction of the cloverleaf/PCBP2/3CDpro complex with PABP that is bound to the poly(A) at the 3’-end of the genome (section 4). VPgpUpU is synthesized on the cre(2C) and is probably transferred in a complex by 3Dpol and/or 3CDpro to the 3’-end of the poly(A) tail. There it serves as the primer for minus strand RNA synthesis yielding a double-stranded replicative form (RF). (B) The 5’-end of the RF is destabilized by the binding of viral and cellular proteins. 3CDpro in a complex with PCBP2 or 3AB binds to the plus strand cloverleaf after destabilizing the 5’-end of the RF. 2CATPase and hRNP C bind to and stabilize the minus strand cloverleaf (section 4). VPgpUpU that was made prior to or during minus strand synthesis is transferred in a complex with 3Dpol and or 3CDpro to the 3’-terminal two As of the minus strand. There VPgpUpU is elongated by 3Dpol yielding a replicative intermediate (RI) that contains multiple plus strands at various stages of elongation. The final products are the full length VPg-linked plus strands.
5. Unanswered questions about the initiation of picornavirus RNA replication
Although great progress has been made in identifying the viral and cellular factors involved in the initiation of picornavirus minus and plus strand RNA synthesis, many important questions remain unanswered:
what is the true protein substrate in vivo for uridylylation?
why are the cre elements of picornaviruses located in different regions of the RNA genomes?
how is VPgpUpU translocated from the cre to the 3’-end of plus strand RNA and 3’-end of minus strand RNA?
what provides specificity to the transfer of VPgpUpU from the cre to the ends of plus and minus strand RNAs?
what factors (viral and/or cellular) are required for the transfer reaction?
what is the role of membranes in VPg uridylylation in vivo?
why is there a large excess of VPgpUpU molecules synthesized relative to the number of progeny RNAs made?
how is the end of the RF unwound prior to plus strand RNA synthesis?
what is the role of 2CATPase in VPgpUpU synthesis?
6. Conclusions
As we discussed above (section 3), it is well established that picornavirus RNA polymerases utilize a protein-primed reaction to synthesize both their minus and plus RNA strands. Although initially the poly(A) tail was proposed to template VPgpUpU synthesis for minus strand RNA synthesis, it is now generally accepted from both in vivo studies and those with PIRCs that VPgpUpU made on the cre is used to prime both minus and plus strand RNA synthesis. Assuming that the mechanism of protein-primed RNA synthesis is similar with all viruses that possess terminal proteins this model is consistent with the existence of some plant viruses (Sobemovirus, Luteovirus, Enamovirus and Barnavirus) with VPg-linked genomes that do not possess poly(A) tails (139). It is also clear that the template for the linkage of two UMPs to VPg (or VPg precursor) is a conserved sequence in the loop of a cre element, located usually in the ORF, and the reaction is strongly enhanced by the presence of viral protein 3CDpro. The PV VPg-uridylylation reaction is strictly dependent on the presence of the cloverleaf, of 2CATPase function, and of membranes in vivo and in PIRCs but not in vitro. The “slide back” mechanism of VPg uridylylation ensures the integrity of the ends of the viral RNA genomes.
Studies with other plus strand viruses are limited but so far they suggest that protein-primed RNA synthesis is a general mechanism used by viruses that contain protein-linked genomes. Recently, the RNA polymerase and/or RNApolymerase precursor of norovirus was shown to nucleotidylylate its VPg protein (18, 60, 128). There are a large number of RNA virus families, in addition to Picornaviridae, that possess genome-linked proteins (82): Caliciviridae, Astroviridae, Potyviridae, Comoviridae, Dicistroviridae and Sequiviridae (133). The terminal proteins of these viruses vary in length from 2 kDa to ~24 kDa and also vary in amino acid sequence.
Cre elements have been either discovered or their existence has been predicted for several but not all genera of Picornaviridae (144). It should be noted that the conserved AAA/GC sequence within the loop of the typical picornavirus cre hairpin, together with conserved RNA structure predictions, might be useful in the future in predicting the location of unknown cre elements in newly identified genera of Picornaviridae. Outside of picornaviruses there is very little known about cre elements in members of other plus strand RNA virus families that might be templates for nucleotidylylation of their VPgs. However, stimulation of the VPg uridylylation reaction by RNA sequences in the genome of norovirus, a member of Caliciviridae, was reported (18, 60). In addition, the presence of a cre-like sequence was observed in the genome of norovirus (166).
The insights we have gained from studies of protein/protein and protein/RNA interactions involved in the process of the initiation step of picornavirus RNA replication will hopefully facilitate studies of RNA replication of other plus strand RNA viruses with genome-linked proteins. Since protein-primed RNA replication normally does not occur in eukaryotic cells this unique reaction is particularly well suited for studies of drug development to fight picornaviral infections.
Highlights.
Picornaviruses use a protein-primed mechanism of RNA synthesis
The viral RNA polymerase links two UMPs to a tyrosine in VPg, the terminal protein
Sequences in an RNA hairpin are the templates for the linkage of the UMPs to VPg
VPgpUpU is the primer for the elongation of RNA chains by the RNA polymerase
7. Acknowledgements
This work was supported in part by NIH grant RO1AI015122-35.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adams P, Kandiah E, Effantin G, Steven AC, Ehrenfeld E. Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J Biol Chem. 2009;284:22012–21. doi: 10.1074/jbc.M109.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Sunaidi M, Williams CH, Hughes PJ, Schnurr DP, Stanway G. Analysis of a new human parechovirus allows the definition of parechovirus types and the identification of RNA structural domains. J Virol. 2007;81:1013–21. doi: 10.1128/JVI.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldabe R, Carrasco L. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem Biophys Res Commun. 1995;206:64–76. doi: 10.1006/bbrc.1995.1010. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V, Baltimore D. Protein is linked to the 5' end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem. 1978;253:5263–6. [PubMed] [Google Scholar]
- 5.Andino R, Rieckhof GE, Achacoso PL, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5'-end of viral RNA. EMBO J. 1993;12:3587–98. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990;63:369–80. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 7.Appleby TC, Luecke H, Shim JH, Wu JZ, Cheney IW, Zhong W, Vogeley L, Hong Z, Yao N. Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer. J Virol. 2005;79:277–88. doi: 10.1128/JVI.79.1.277-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold JJ, Ghosh SK, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem. 1999;274:37060–9. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 9.Baltimore D. The biochemistry of viruses. Marcel Dekker; New York: 1969. [Google Scholar]
- 10.Banerjee R, Echeverri A, Dasgupta A. Poliovirus-encoded 2C polypeptide specifically binds to the 3'-terminal sequences of viral negative-strand RNA. J Virol. 1997;71:9570–8. doi: 10.1128/jvi.71.12.9570-9578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee R, Tsai W, Kim W, Dasgupta A. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3' terminus negative-strand cloverleaf requires an intact stem-loop b. Virology. 2001;280:41–51. doi: 10.1006/viro.2000.0770. [DOI] [PubMed] [Google Scholar]
- 12.Baron MH, Baltimore D. In vitro copying of viral positive strand RNA by poliovirus replicase. Characterization of the reaction and its products. J Biol Chem. 1982;257:12359–66. [PubMed] [Google Scholar]
- 13.Barton DJ, Black EP, Flanegan JB. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–27. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barton DJ, Flanegan JB. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–9. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barton DJ, O'Donnell BJ, Flanegan JB. 5' cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–48. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedard KM, Walter BL, Semler BL. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA. 2004;10:1266–76. doi: 10.1261/rna.7070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beerens N, Selisko B, Ricagno S, Imbert I, van der Zanden L, Snijder EJ, Canard B. De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J Virol. 2007;81:8384–95. doi: 10.1128/JVI.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belliot G, Sosnovtsev SV, Chang KO, McPhie P, Green KY. Nucleotidylylation of the VPg protein of a human norovirus by its proteinase-polymerase precursor protein. Virology. 2008;374:33–49. doi: 10.1016/j.virol.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bienz K, Egger D, Pfister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–7. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop JM, Koch G. Purification and characterization of poliovirus-induced infectious double-stranded ribonucleic acid. J Biol Chem. 1967;242:1736–43. [PubMed] [Google Scholar]
- 21.Bishop JM, Koch G, Evans B, Merriman M. Poliovirus replicative intermediate: structural basis of infectivity. J Mol Biol. 1969;46:235–49. doi: 10.1016/0022-2836(69)90419-7. [DOI] [PubMed] [Google Scholar]
- 22.Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5' noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci U S A. 1996;93:11115–20. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown DM, Cornell CT, Tran GP, Nguyen JH, Semler BL. An authentic 3' noncoding region is necessary for efficient poliovirus replication. J Virol. 2005;79:11962–73. doi: 10.1128/JVI.79.18.11962-11973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunner JE, Ertel KJ, Rozovics JM, Semler BL. Delayed kinetics of poliovirus RNA synthesis in a human cell line with reduced levels of hnRNP C proteins. Virology. 2010;400:240–7. doi: 10.1016/j.virol.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunner JE, Nguyen JH, Roehl HH, Ho TV, Swiderek KM, Semler BL. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J Virol. 2005;79:3254–66. doi: 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron CE, Oh HS, Moustafa IM. Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future Microbiol. 2010;5:867–81. doi: 10.2217/fmb.10.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Wang Y, Shan C, Sun Y, Xu P, Zhou H, Yang C, Shi PY, Rao Z, Zhang B, Lou Z. Crystal structure of enterovirus 71 RNA-dependent RNA polymerase complexed with its protein primer VPg: implication for a trans mechanism of VPg uridylylation. J Virol. 2013;87:5755–68. doi: 10.1128/JVI.02733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheney IW, Naim S, Shim JH, Reinhardt M, Pai B, Wu JZ, Hong Z, Zhong W. Viability of poliovirus/rhinovirus VPg chimeric viruses and identification of an amino acid residue in the VPg gene critical for viral RNA replication. J Virol. 2003;77:7434–43. doi: 10.1128/JVI.77.13.7434-7443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–45. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 30.Cordey S, Gerlach D, Junier T, Zdobnov EM, Kaiser L, Tapparel C. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA. 2008;14:1568–78. doi: 10.1261/rna.1031408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornell CT, Semler BL. Subdomain specific functions of the RNA polymerase region of poliovirus 3CD polypeptide. Virology. 2002;298:200–13. doi: 10.1006/viro.2002.1481. [DOI] [PubMed] [Google Scholar]
- 32.Crawford NM, Baltimore D. Genome-linked protein VPg of poliovirus is present as free VPg and VPg-pUpU in poliovirus-infected cells. Proc Natl Acad Sci U S A. 1983;80:7452–5. doi: 10.1073/pnas.80.24.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui T, Porter AG. Localization of binding site for encephalomyocarditis virus RNA polymerase in the 3'-noncoding region of the viral RNA. Nucleic Acids Res. 1995;23:377–82. doi: 10.1093/nar/23.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Jesus N, Franco D, Paul A, Wimmer E, Cello J. Mutation of a single conserved nucleotide between the cloverleaf and internal ribosome entry site attenuates poliovirus neurovirulence. J Virol. 2005;79:14235–43. doi: 10.1128/JVI.79.22.14235-14243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeStefano JJ, Titilope O. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J Virol. 2006;80:1662–71. doi: 10.1128/JVI.80.4.1662-1671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Detjen BM, Lucas J, Wimmer E. Poliovirus single-stranded RNA and double-stranded RNA: differential infectivity in enucleate cells. J Virol. 1978;27:582–6. doi: 10.1128/jvi.27.3.582-586.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobos P. Protein-primed RNA synthesis in vitro by the virion-associated RNA polymerase of infectious pancreatic necrosis virus. Virology. 1995;208:19–25. doi: 10.1006/viro.1995.1125. [DOI] [PubMed] [Google Scholar]
- 38.Dorsch-Hasler K, Yogo Y, Wimmer E. Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded. J Virol. 1975;16:1512–7. doi: 10.1128/jvi.16.6.1512-1517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dougherty JD, Park N, Gustin KE, Lloyd RE. Interference with cellular gene expression. ASM Press; Washington, D.C.: 2010. [Google Scholar]
- 40.Duque H, Palmenberg AC. Phenotypic characterization of three phylogenetically conserved stem-loop motifs in the mengovirus 3' untranslated region. J Virol. 2001;75:3111–20. doi: 10.1128/JVI.75.7.3111-3120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Echeverri A, Banerjee R, Dasgupta A. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 1998;54:217–23. doi: 10.1016/s0168-1702(98)00016-1. [DOI] [PubMed] [Google Scholar]
- 42.Ertel KJ, Brunner JE, Semler BL. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J Virol. 2010;84:4229–42. doi: 10.1128/JVI.02198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrer-Orta C, Arias A, Agudo R, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 2006;25:880–8. doi: 10.1038/sj.emboj.7600971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Flanegan J, Morasco J, Ogram S, O'Donnell B, Boone C, McKenna R. Intermolecular uridylylation of VPg by 3Dpol during picornavirus RNA replication. EUROPIC Abstracts. 2014:39. [Google Scholar]
- 46.Flanegan JB, Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A) Proc Natl Acad Sci U S A. 1977;74:3677–80. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fogg MH, Teterina NL, Ehrenfeld E. Membrane requirements for uridylylation of the poliovirus VPg protein and viral RNA synthesis in vitro. J Virol. 2003;77:11408–16. doi: 10.1128/JVI.77.21.11408-11416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franco D, Pathak HB, Cameron CE, Rombaut B, Wimmer E, Paul AV. Stimulation of poliovirus RNA synthesis and virus maturation in a HeLa cell-free in vitro translation-RNA replication system by viral protein 3CDpro. Virol J. 2005;2:86. doi: 10.1186/1743-422X-2-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujita K, Krishnakumar SS, Franco D, Paul AV, London E, Wimmer E. Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication. Biochemistry. 2007;46:5185–99. doi: 10.1021/bi6024758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamarnik AV, Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5' untranslated region of the poliovirus genome. J Virol. 2000;74:2219–26. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerber K, Wimmer E, Paul AV. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2A(pro) J Virol. 2001;75:10979–90. doi: 10.1128/JVI.75.22.10979-10990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerber K, Wimmer E, Paul AV. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: purification and enzymatic analysis of the RNA-dependent RNA polymerase 3D(pol) J Virol. 2001;75:10969–78. doi: 10.1128/JVI.75.22.10969-10978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodfellow I. The genome-linked protein VPg of vertebrate viruses - a multifaceted protein. Curr Opin Virol. 2011;1:355–62. doi: 10.1016/j.coviro.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond JW, Barclay W, Evans DJ. Identification of a cis-acting replication element within the poliovirus coding region. J Virol. 2000;74:4590–600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodfellow IG, Polacek C, Andino R, Evans DJ. The poliovirus 2C cis-acting replication element-mediated uridylylation of VPg is not required for synthesis of negative-sense genomes. J Gen Virol. 2003;84:2359–63. doi: 10.1099/vir.0.19132-0. [DOI] [PubMed] [Google Scholar]
- 57.Gorbalenya A. Helicases:amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. K. E. [Google Scholar]
- 58.Gorlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–95. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gruez A, Selisko B, Roberts M, Bricogne G, Bussetta C, Jabafi I, Coutard B, De Palma AM, Neyts J, Canard B. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J Virol. 2008;82:9577–90. doi: 10.1128/JVI.00631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han KR, Choi Y, Min BS, Jeong H, Cheon D, Kim J, Jee Y, Shin S, Yang JM. Murine norovirus-1 3Dpol exhibits RNA-dependent RNA polymerase activity and nucleotidylylates on Tyr of the VPg. J Gen Virol. 2010;91:1713–22. doi: 10.1099/vir.0.020461-0. [DOI] [PubMed] [Google Scholar]
- 61.Hansen JL, Long AM, Schultz SC. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–22. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 62.Harmon SA, Richards OC, Summers DF, Ehrenfeld E. The 5'-terminal nucleotides of hepatitis A virus RNA, but not poliovirus RNA, are required for infectivity. J Virol. 1991;65:2757–60. doi: 10.1128/jvi.65.5.2757-2760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–14. [PubMed] [Google Scholar]
- 64.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7:581–91. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001;20:1153–63. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hope DA, Diamond SE, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol. 1997;71:9490–8. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. Picornavirus morphogenesis. Microbiol Mol Biol Rev. 2014;78:418–37. doi: 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kao CC, Singh P, Ecker DJ. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–60. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 70.Kempf BJ, Kelly MM, Springer CL, Peersen OB, Barton DJ. Structural features of a picornavirus polymerase involved in the polyadenylation of viral RNA. J Virol. 2013;87:5629–44. doi: 10.1128/JVI.02590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.King AM, Sangar DV, Harris TJ, Brown F. Heterogeneity of the genome-linked protein of foot-and-mouth disease virus. J Virol. 1980;34:627–34. doi: 10.1128/jvi.34.3.627-634.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirkegaard K, Semler B. Genome replication II: The process. ASM Press; Washington, DC: 2010. [Google Scholar]
- 73.Klump WM, Bergmann I, Muller BC, Ameis D, Kandolf R. Complete nucleotide sequence of infectious Coxsackievirus B3 cDNA: two initial 5' uridine residues are regained during plus-strand RNA synthesis. J Virol. 1990;64:1573–83. doi: 10.1128/jvi.64.4.1573-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuhn R, Wimmer E. The replication of picornaviruses. Academic Press; London: 1987. [Google Scholar]
- 75.Kuhn RJ, Tada H, Ypma-Wong MF, Semler BL, Wimmer E. Mutational analysis of the genome-linked protein VPg of poliovirus. J Virol. 1988;62:4207–15. doi: 10.1128/jvi.62.11.4207-4215.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kusov YY, Morace G, Probst C, Gauss-Muller V. Interaction of hepatitis A virus (HAV) precursor proteins 3AB and 3ABC with the 5' and 3' termini of the HAV RNA. Virus Res. 1997;51:151–7. doi: 10.1016/s0168-1702(97)00089-0. [DOI] [PubMed] [Google Scholar]
- 77.Lee YF, Nomoto A, Detjen BM, Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci U S A. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li JP, Baltimore D. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J Virol. 1990;64:1102–7. doi: 10.1128/jvi.64.3.1102-1107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li JP, Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988;62:4016–21. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y, Franco D, Paul AV, Wimmer E. Tyrosine 3 of poliovirus terminal peptide VPg(3B) has an essential function in RNA replication in the context of its precursor protein, 3AB. J Virol. 2007;81:5669–84. doi: 10.1128/JVI.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, Wang C, Mueller S, Paul AV, Wimmer E, Jiang P. Direct interaction between two viral proteins, the nonstructural protein 2C and the capsid protein VP3, is required for enterovirus morphogenesis. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lobert PE, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc Natl Acad Sci U S A. 1999;96:11560–5. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lyle JM, Clewell A, Richmond K, Richards OC, Hope DA, Schultz SC, Kirkegaard K. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J Biol Chem. 2002;277:16324–31. doi: 10.1074/jbc.M112429200. [DOI] [PubMed] [Google Scholar]
- 85.Lyons T, Murray KE, Roberts AW, Barton DJ. Poliovirus 5'-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J Virol. 2001;75:10696–708. doi: 10.1128/JVI.75.22.10696-10708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81:3583–96. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martinez-Salas E. a. R. Translation and protein processing. ASM Press; Washington, DC: 2010. M. [Google Scholar]
- 88.Mason PW, Bezborodova SV, Henry TM. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J Virol. 2002;76:9686–94. doi: 10.1128/JVI.76.19.9686-9694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKnight KL. The human rhinovirus internal cis-acting replication element (cre) exhibits disparate properties among serotypes. Arch Virol. 2003;148:2397–418. doi: 10.1007/s00705-003-0177-7. [DOI] [PubMed] [Google Scholar]
- 90.McKnight KL, Lemon SM. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–84. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Melchers WJ, Hoenderop JG, Bruins Slot HJ, Pleij CW, Pilipenko EV, Agol VI, Galama JM. Kissing of the two predominant hairpin loops in the coxsackie B virus 3' untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–96. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Merkle I, van Ooij MJ, van Kuppeveld FJ, Glaudemans DH, Galama JM, Henke A, Zell R, Melchers WJ. Biological significance of a human enterovirus B-specific RNA element in the 3' nontranslated region. J Virol. 2002;76:9900–9. doi: 10.1128/JVI.76.19.9900-9909.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller WA, Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- 94.Mirzayan C, Wimmer E. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology. 1994;199:176–87. doi: 10.1006/viro.1994.1110. [DOI] [PubMed] [Google Scholar]
- 95.Molla A, Paul AV, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–51. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 96.Morasco BJ, Sharma N, Parilla J, Flanegan JB. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive-but not negative-strand RNA synthesis. J Virol. 2003;77:5136–44. doi: 10.1128/JVI.77.9.5136-5144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mosimann SC, Cherney MM, Sia S, Plotch S, James MN. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol. 1997;273:1032–47. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 98.Murray KE, Barton DJ. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J Virol. 2003;77:4739–50. doi: 10.1128/JVI.77.8.4739-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nagy PD, Carpenter CD, Simon AE. A novel 3'-end repair mechanism in an RNA virus. Proc Natl Acad Sci U S A. 1997;94:1113–8. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nayak A, Goodfellow IG, Belsham GJ. Factors required for the Uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro. J Virol. 2005;79:7698–706. doi: 10.1128/JVI.79.12.7698-7706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nayak A, Goodfellow IG, Woolaway KE, Birtley J, Curry S, Belsham GJ. Role of RNA structure and RNA binding activity of foot- and-mouth disease virus 3C protein in VPg uridylylation and virus replication. J Virol. 2006;80:9865–75. doi: 10.1128/JVI.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Novak JE, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–7. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ogram SA, Boone CD, McKenna R, Flanegan JB. Amiloride inhibits the initiation of Coxsackievirus and poliovirus RNA replication by inhibiting VPg uridylylation. Virology 464. 2014;465:87–97. doi: 10.1016/j.virol.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oh HS, Pathak HB, Goodfellow IG, Arnold JJ, Cameron CE. Insight into poliovirus genome replication and encapsidation obtained from studies of 3B-3C cleavage site mutants. J Virol. 2009;83:9370–87. doi: 10.1128/JVI.02076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Panavas T, Pogany J, Nagy PD. Internal initiation by the cucumber necrosis virus RNA-dependent RNA polymerase is facilitated by promoter-like sequences. Virology. 2002;296:275–87. doi: 10.1006/viro.2002.1422. [DOI] [PubMed] [Google Scholar]
- 106.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. Poly (rC) binding protein 2 forms a ternary complex with the 5'-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–34. [PMC free article] [PubMed] [Google Scholar]
- 107.Pata JD, Schultz SC, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–77. [PMC free article] [PubMed] [Google Scholar]
- 108.Pathak HB, Arnold JJ, Wiegand PN, Hargittai MR, Cameron CE. Picornavirus genome replication: assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J Biol Chem. 2007;282:16202–13. doi: 10.1074/jbc.M610608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pathak HB, Ghosh SK, Roberts AW, Sharma SD, Yoder JD, Arnold JJ, Gohara DW, Barton DJ, Paul AV, Cameron CE. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J Biol Chem. 2002;277:31551–62. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- 110.Pathak HB, Oh HS, Goodfellow IG, Arnold JJ, Cameron CE. Picornavirus genome replication: roles of precursor proteins and rate-limiting steps in oriI-dependent VPg uridylylation. J Biol Chem. 2008;283:30677–88. doi: 10.1074/jbc.M806101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Paul A. Possible unifying mechanism of Picornavirus genome replication. ASM Press; Washington, DC: 2002. [Google Scholar]
- 112.Paul AV, Molla A, Wimmer E. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology. 1994;199:188–99. doi: 10.1006/viro.1994.1111. [DOI] [PubMed] [Google Scholar]
- 113.Paul AV, Peters J, Mugavero J, Yin J, van Boom JH, Wimmer E. Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J Virol. 2003;77:891–904. doi: 10.1128/JVI.77.2.891-904.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–70. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–4. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 116.Paul AV, Yin J, Mugavero J, Rieder E, Liu Y, Wimmer E. A "slide-back" mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J Biol Chem. 2003;278:43951–60. doi: 10.1074/jbc.M307441200. [DOI] [PubMed] [Google Scholar]
- 117.Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J Virol. 2007;81:8919–32. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pfister T, Jones KW, Wimmer E. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J Virol. 2000;74:334–43. doi: 10.1128/jvi.74.1.334-343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 120.Pilipenko EV, Maslova SV, Sinyakov AN, Agol VI. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992;20:1739–45. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pilipenko EV, Poperechny KV, Maslova SV, Melchers WJ, Slot HJ, Agol VI. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary ('kissing') interactions. EMBO J. 1996;15:5428–36. [PMC free article] [PubMed] [Google Scholar]
- 122.Reuer Q, Kuhn RJ, Wimmer E. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J Virol. 1990;64:2967–75. doi: 10.1128/jvi.64.6.2967-2975.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rieder E, Paul AV, Kim DW, van Boom JH, Wimmer E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J Virol. 2000;74:10371–80. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rivera VM, Welsh JD, Maizel JV., Jr Comparative sequence analysis of the 5' noncoding region of the enteroviruses and rhinoviruses. Virology. 1988;165:42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- 125.Rodriguez PL, Carrasco L. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem. 1995;270:10105–12. doi: 10.1074/jbc.270.17.10105. [DOI] [PubMed] [Google Scholar]
- 126.Rodriguez PL, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–10. [PubMed] [Google Scholar]
- 127.Roehl HH, Semler BL. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3' end of viral negative-strand RNA. J Virol. 1995;69:2954–61. doi: 10.1128/jvi.69.5.2954-2961.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rohayem J, Jager K, Robel I, Scheffler U, Temme A, Rudolph W. Characterization of norovirus 3Dpol RNA-dependent RNA polymerase activity and initiation of RNA synthesis. J Gen Virol. 2006;87:2621–30. doi: 10.1099/vir.0.81802-0. [DOI] [PubMed] [Google Scholar]
- 129.Rohll JB, Moon DH, Evans DJ, Almond JW. The 3' untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–44. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rothberg PG, Harris TJ, Nomoto A, Wimmer E. O4-(5'-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci U S A. 1978;75:4868–72. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rozovich J, Semler B. Genome replication: the players. ASM Press; Washington, DC: 2010. [Google Scholar]
- 132.Saiz M, Gomez S, Martinez-Salas E, Sobrino F. Deletion or substitution of the aphthovirus 3' NCR abrogates infectivity and virus replication. J Gen Virol. 2001;82:93–101. doi: 10.1099/0022-1317-82-1-93. [DOI] [PubMed] [Google Scholar]
- 133.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 134.Salas M, Miller J, Leis J, DePamphilis M. Mechanism for priming DNA synthesis. Cold Spring Harbor Press; Cold Spring Harbor, N.Y.: 1996. [Google Scholar]
- 135.Schein CH, Oezguen N, van der Heden van Noort GJ, Filippov DV, Paul A, Kumar E, Braun W. NMR solution structure of poliovirus uridylyated peptide linked to the genome (VPgpU) Peptides. 2010;31:1441–8. doi: 10.1016/j.peptides.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schein CH, Oezguen N, Volk DE, Garimella R, Paul A, Braun W. NMR structure of the viral peptide linked to the genome (VPg) of poliovirus. Peptides. 2006;27:1676–84. doi: 10.1016/j.peptides.2006.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Seeger C, Mason WS. Replication of the hepatitis virus genome. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1996. [Google Scholar]
- 138.Sharma N, O'Donnell BJ, Flanegan JB. 3'-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J Virol. 2005;79:3565–77. doi: 10.1128/JVI.79.6.3565-3577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shaw J. Plant viruses. Lippincotte-Raven; Philadelphia: 1996. [Google Scholar]
- 140.Shen M, Wang Q, Yang Y, Pathak HB, Arnold JJ, Castro C, Lemon SM, Cameron CE. Human rhinovirus type 14 gain-of-function mutants for oriI utilization define residues of 3C(D) and 3Dpol that contribute to assembly and stability of the picornavirus VPg uridylylation complex. J Virol. 2007;81:12485–95. doi: 10.1128/JVI.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Silvestri LS, Parilla JM, Morasco BJ, Ogram SA, Flanegan JB. Relationship between poliovirus negative-strand RNA synthesis and the length of the 3' poly(A) tail. Virology. 2006;345:509–19. doi: 10.1016/j.virol.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 142.Son KY, Kim DS, Kwon J, Choi JS, Kang MI, Belsham GJ, Cho KO. Full-length genomic analysis of Korean porcine Sapelovirus strains. PLoS One. 2014;9:e107860. doi: 10.1371/journal.pone.0107860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Spector DH, Baltimore D. Requirement of 3'-terminal poly(adenylic acid) for the infectivity of poliovirus RNA. Proc Natl Acad Sci U S A. 1974;71:2983–7. doi: 10.1073/pnas.71.8.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Steil BP, Barton DJ. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res. 2009;139:240–52. doi: 10.1016/j.virusres.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Steil BP, Barton DJ. Conversion of VPg into VPgpUpUOH before and during poliovirus negative-strand RNA synthesis. J Virol. 2009;83:12660–70. doi: 10.1128/JVI.01676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Steil BP, Barton DJ. Poliovirus cis-acting replication element-dependent VPg Uridylylation lowers the Km of the initiating nucleoside triphosphate for viral RNA replication. J Virol. 2008;82:9400–8. doi: 10.1128/JVI.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Steil BP, Kempf BJ, Barton DJ. Poly(A) at the 3' end of positive-strand RNA and VPg-linked poly(U) at the 5' end of negative-strand RNA are reciprocal templates during replication of poliovirus RNA. J Virol. 2010;84:2843–58. doi: 10.1128/JVI.02620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sun Y, Guo Y, Lou Z. Formation and working mechanism of the picornavirus VPg uridylylation complex. Curr Opin Virol. 2014;9C:24–30. doi: 10.1016/j.coviro.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 149.Takeda N, Kuhn RJ, Yang CF, Takegami T, Wimmer E. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J Virol. 1986;60:43–53. doi: 10.1128/jvi.60.1.43-53.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Takegami T, Kuhn RJ, Anderson CW, Wimmer E. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc Natl Acad Sci U S A. 1983;80:7447–51. doi: 10.1073/pnas.80.24.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tellez AB, Crowder S, Spagnolo JF, Thompson AA, Peersen OB, Brutlag DL, Kirkegaard K. Nucleotide channel of RNA-dependent RNA polymerase used for intermolecular uridylylation of protein primer. J Mol Biol. 2006;357:665–75. doi: 10.1016/j.jmb.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 152.Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–72. doi: 10.1128/jvi.71.12.8962-8972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Teterina NL, Kean KM, Gorbalenya AE, Agol VI, Girard M. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J Gen Virol. 1992;73:1977–86. doi: 10.1099/0022-1317-73-8-1977. Pt 8. [DOI] [PubMed] [Google Scholar]
- 154.Thiviyanathan V, Yang Y, Kaluarachchi K, Rijnbrand R, Gorenstein DG, Lemon SM. High-resolution structure of a picornaviral internal cis-acting RNA replication element (cre) Proc Natl Acad Sci U S A. 2004;101:12688–93. doi: 10.1073/pnas.0403079101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tobin GJ, Young DC, Flanegan JB. Self-catalyzed linkage of poliovirus terminal protein VPg to poliovirus RNA. Cell. 1989;59:511–9. doi: 10.1016/0092-8674(89)90034-2. [DOI] [PubMed] [Google Scholar]
- 156.Todd S, Towner JS, Brown DM, Semler BL. Replication-competent picornaviruses with complete genomic RNA 3' noncoding region deletions. J Virol. 1997;71:8868–74. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Towner JS, Mazanet MM, Semler BL. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J Virol. 1998;72:7191–200. doi: 10.1128/jvi.72.9.7191-7200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Toyoda H, Franco D, Fujita K, Paul AV, Wimmer E. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J Virol. 2007;81:10017–28. doi: 10.1128/JVI.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Toyoda H, Yang CF, Takeda N, Nomoto A, Wimmer E. Analysis of RNA synthesis of type 1 poliovirus by using an in vitro molecular genetic approach. J Virol. 1987;61:2816–22. doi: 10.1128/jvi.61.9.2816-2822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Toyoda H, Yin J, Mueller S, Wimmer E, Cello J. Oncolytic treatment and cure of neuroblastoma by a novel attenuated poliovirus in a novel poliovirus-susceptible animal model. Cancer Res. 2007;67:2857–64. doi: 10.1158/0008-5472.CAN-06-3713. [DOI] [PubMed] [Google Scholar]
- 161.van der Schaar HM, van der Linden L, Lanke KH, Strating JR, Purstinger G, de Vries E, de Haan CA, Neyts J, van Kuppeveld FJ. Coxsackievirus mutants that can bypass host factor PI4KIIIbeta and the need for high levels of PI4P lipids for replication. Cell Res. 2012;22:1576–92. doi: 10.1038/cr.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol. 2004;85:1077–93. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 163.van Ooij MJ, Glaudemans DH, Heus HA, van Kuppeveld FJ, Melchers WJ. Structural and functional integrity of the coxsackievirus B3 oriR: spacing between coaxial RNA helices. J Gen Virol. 2006;87:689–95. doi: 10.1099/vir.0.81558-0. [DOI] [PubMed] [Google Scholar]
- 164.van Ooij MJ, Vogt DA, Paul A, Castro C, Kuijpers J, van Kuppeveld FJ, Cameron CE, Wimmer E, Andino R, Melchers WJ. Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative- and positive-strand RNA synthesis. J Gen Virol. 2006;87:103–13. doi: 10.1099/vir.0.81297-0. [DOI] [PubMed] [Google Scholar]
- 165.Vance LM, Moscufo N, Chow M, Heinz BA. Poliovirus 2C region functions during encapsidation of viral RNA. J Virol. 1997;71:8759–65. doi: 10.1128/jvi.71.11.8759-8765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Victoria M, Colina R, Miagostovich MP, Leite JP, Cristina J. Phylogenetic prediction of cis-acting elements: a cre-like sequence in Norovirus genome? BMC Res Notes. 2009;2:176. doi: 10.1186/1756-0500-2-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vogt DA, Andino R. An RNA element at the 5'-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 2010;6:e1000936. doi: 10.1371/journal.ppat.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Walter BL, Parsley TB, Ehrenfeld E, Semler BL. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol. 2002;76:12008–22. doi: 10.1128/JVI.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang C, Jiang P, Sand C, Paul AV, Wimmer E. Alanine scanning of poliovirus 2CATPase reveals new genetic evidence that capsid protein/2CATPase interactions are essential for morphogenesis. J Virol. 2012;86:9964–75. doi: 10.1128/JVI.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wimmer E. Genome-linked proteins of viruses. Cell. 1982;28:199–201. doi: 10.1016/0092-8674(82)90335-x. [DOI] [PubMed] [Google Scholar]
- 171.Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 172.Witwer C, Rauscher S, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 2001;29:5079–89. doi: 10.1093/nar/29.24.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J Virol. 1998;72:6732–41. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xiang W, Cuconati A, Paul AV, Cao X, Wimmer E. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA. 1995;1:892–904. [PMC free article] [PubMed] [Google Scholar]
- 175.Xiang W, Harris KS, Alexander L, Wimmer E. Interaction between the 5'-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–67. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yang Y, Rijnbrand R, McKnight KL, Wimmer E, Paul A, Martin A, Lemon SM. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J Virol. 2002;76:7485–94. doi: 10.1128/JVI.76.15.7485-7494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yang Y, Rijnbrand R, Watowich S, Lemon SM. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J Biol Chem. 2004;279:12659–67. doi: 10.1074/jbc.M312992200. [DOI] [PubMed] [Google Scholar]
- 178.Yang Y, Yi M, Evans DJ, Simmonds P, Lemon SM. Identification of a conserved RNA replication element (cre) within the 3Dpol-coding sequence of hepatoviruses. J Virol. 2008;82:10118–28. doi: 10.1128/JVI.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yin J, Paul AV, Wimmer E, Rieder E. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J Virol. 2003;77:5152–66. doi: 10.1128/JVI.77.9.5152-5166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yogo Y, Wimmer E. Polyadenylic acid at the 3'-terminus of poliovirus RNA. Proc Natl Acad Sci U S A. 1972;69:1877–82. doi: 10.1073/pnas.69.7.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zell R, Ihle Y, Seitz S, Gundel U, Wutzler P, Gorlach M. Poly(rC)-binding protein 2 interacts with the oligo(rC) tract of coxsackievirus B3. Biochem Biophys Res Commun. 2008;366:917–21. doi: 10.1016/j.bbrc.2007.12.038. [DOI] [PubMed] [Google Scholar]