Abstract
Inflammasomes are caspase-1 activating, molecular inflammatory machines that proteolytically mature pro-inflammatory cytokines and induce pyroptotic cell death during innate immune responses. Recent structural studies of proteins that constitute inflammasomes have yielded fresh insights into their assembly mechanisms. In particular, these include a crystal structure of the CARD-containing NOD-like receptor NLRC4, the crystallographic and electron microscopy (EM) studies of the dsDNA sensors AIM2 and IFI16, and of the regulatory protein p202, and the cryo-EM filament structure of the PYD domain of the inflammasome adapter ASC. These data suggest inflammasome assembly that starts with ligand recognition and release of autoinhibition followed by step-wise rounds of nucleated polymerization from the sensors to the adapters, then to caspase-1. In this elegant manner, inflammasomes form by an “all-or-none” cooperative mechanism, thereby amplifying the activation of caspase-1. The dense network of filamentous structures predicted by this model has been observed in cells as micron-sized puncta.
Introduction
The immune system protects organisms from infections and other types of insults; it consists of an innate immune component and an adaptive immune component. Innate immunity offers the first line of defense and is mediated by germ line encoded pattern recognition receptors (PRRs) that recognize pathogen-associated and damage-associated molecular patterns (PAMPs and DAMPs) [1–3]. PRRs include cell surface and endosomal Toll-like receptors (TLRs), as well as cytosolic PRRs such as RIG-I-like receptors (RLRs), AIM2-like receptors (ALRs), NOD-like receptors (NLRs) and cyclic GMP-AMP synthase (cGAS) [4]. PRR signal transduction induces a plethora of cellular reactions to counter immediate dangers, including cytokine secretion, cell death and interferon response. It also helps to initiate adaptive immunity for antigen-specific defense mechanisms.
Over the last decade, the classical model of signal transduction as a series of binding events that trigger the allosteric activation of enzymes and other downstream effector molecules has been expanded to include the formation of large supramolecular assemblies that explain the threshold kinetics and signal amplification observed in many innate, as well as adaptive, immune signaling pathways [5–8]. These assemblies, which we recently named supramolecular organizing centers (SMOCs), often manifest as large, heterogeneous micron-sized puncta in cells [6]. Recent advances in cryo-electron microscopy (cryo-EM) and single-molecule fluorescence microscopy have begun to bring the structural characterization of these puncta from the micrometer-scale to the sub-nanometer-scale [9, 10]. In this review, we will focus on one class of SMOCs, known as inflammasomes, that are caspase-1 activating machines [11, 12]. The hierarchical assembly mechanism illustrated here, involving successive steps of nucleated polymerization, may be general to other innate immune signaling pathways.
Canonical inflammasomes are formed by the assembly of three classes of molecules: sensors in the ALR and NLR family, adapters such as apoptosis-associated, speck-like protein containing a CARD (ASC) [13], and effectors such as caspase-1 (Figure 1). Caspase-1 activation leads to processing of the pro-forms of interleukin-1β (IL-1β) and IL-18 into their mature forms for secretion, and induces an inflammatory cell death known as pyroptosis [11, 12]. ALRs include absent in melanoma 2 (AIM2) and interferon-inducible protein 16 (IFI16), and are composed of an N-terminal pyrin domain (PYD) and a C-terminal, ~200 amino acid hematopoietic, interferon-inducible, nuclear localization (HIN) domain [14–17]. While the HIN domain detects phagocytosed or actively replicating viral dsDNA in the cytosol, the PYD recruits the adapter ASC through homotypic PYD/PYD interactions. Negative regulators of ALR inflammasome formation, such as p202, have only two HIN domains and lack the PYD [14]. ASC also contains a CARD domain, which is responsible for caspase-1 recruitment and activation [13]. NLRs have a more complex domain architecture with variable N-terminal domains, a central nucleotide binding and oligomerization (NACHT) domain that shares homology with the AAA+ superfamily of ATPases, and a C-terminal leucine rich repeat (LRR) domain [11, 12]. The biggest subfamily of NLRs is the NLRP family, with the “P” representing the N-terminal PYD domain. The NLRP inflammasomes require the adapter ASC to mediate caspase-1 activation. The N-terminal BIR domain-containing NLRs, such as NAIPs, detect bacterial flagellin and type III secretion proteins: NAIP5 detects flagellin, and NAIP2 specifically detects the rod protein of the type III secretion system [18–20]. Although named as an NLR, the N-terminal CARD-containing protein NLRC4 is now known to function as an adapter to the NAIP sensors [18–22]. Upon ligand binding, NAIPs can interact with NLRC4 to form a NAIP/NLRC4/caspase-1 inflammasome in the absence of ASC [18–22]. However, recent studies suggest that NLRC4 also interacts with ASC, and that the NAIP inflammasome co-localizes with NLRP3 in a single speck in THP-1 macrophages upon infection with Salmonella typhimurium derived flagellin [23].
Figure 1.
Domain structures of Inflammasome proteins. Domain abbreviations are as follows: PYD: Pyrin domain (yellow and red symbols); HIN: Hematopoietic, Interferon-inducible, Nuclear localization domain (dark blue rounded rectangles); NACHT: Nucleotide-binding and oligomerization domain (cyan elongated ovals); LRRs: Leucine-rich repeats (repeating orange rectangles); BIR: Baculovirus IAP repeat (dark gray overlapping diamonds); and CARD: Caspase recruitment domain (light green and orange hexagons). Caspase domain: p20 and p10 as the large and small subunits, respectively (blue and purple rounded rectangles).
Inflammasome assembly and caspase activation proceed in several steps. First, in the resting state, sensor molecules, either ALRs or NLRs, appear to exist in autoinhibited conformations. Second, upon encountering PAMPs and DAMPs such as flagellin, lipoproteins, viral dsDNA, uric acid crystals and extracellular ATP, ALR or NLR sensors undergo conformational changes that overcome the autoinhibition. Third, for sensor proteins with a PYD such as AIM2 and NLRP3, clustering of PYDs ensues, which recruits ASC and promotes multivalent PYD/PYD interactions. Alternatively, for the NAIP inflammasomes, recruitment of NLRC4 follows, which leads to clustering of NLRC4 CARDs. Fourth, clustered ASC and NLRC4 CARDs recruit pro-caspase-1, whose expression is upregulated by prior NF-κB driven transcription. Activated caspase-1 cleaves pro-IL-1β and pro-IL-18 to generate the mature forms of these proinflammatory cytokines. It may also cleave a collection of additional substrates to induce the pyroptotic cell death that is associated with swelling and rupture of cellular membranes. The following sections will use existing structural information to illustrate each of these steps in inflammasome assembly and activation.
Auto-inhibition in ALR and NLR sensor and adapters through intramolecular interactions
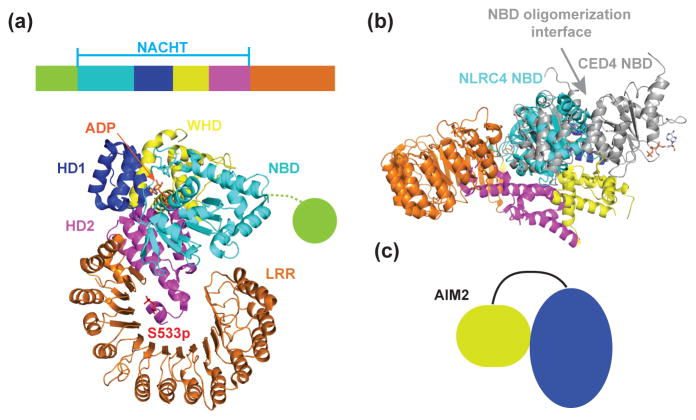
Both inflammasome sensors and adapters have been proposed to exist in an autoinhibited conformation before activation. A structure of the mouse NLRC4 adapter lacking the N-terminal CARD (ΔCARD mNLRC4) provides insights about the mechanism of autoinhibition in this NLR. NLRC4 functions as an adapter protein in NAIP/NLRC4/caspase-1 inflammasome formation [24]. The NACHT domain of NLRC4 is composed of a nucleotide-binding domain (NBD), a helical domain 1 (HD1), a winged helix domain (WHD), and a helical domain 2 (HD2) (Figure 2a). The autoinhibited mNLRC4 assumes a closed conformation with intricate intramolecular interactions that sequester mNLRC4 in a monomeric state (Figure 2a). Without oligomerization, the CARD of NLRC4 is unable to recruit caspase-1 through CARD-CARD interactions, thereby affording a safety mechanism against auto-activation in the absence of proper stimulation. In the autoinhibited mNLRC4 structure, an ADP molecule is bound at the interface between the NBD and WHD (Figure 2a). The HD2 interacts with a functionally important site in NBD and caps the N-terminal side of LRR through a phosphorylated serine residue S533p. The C-terminal LRR domain directly binds one side of the NBD to sterically prevent it from self-oligomerization. Maintenance of the autoinhibited state must be cooperative as disruption of any of the above mentioned interactions cause constitutive activation of mNLRC4 and caspase-1 [24]. In addition, the mNLRC4 NBD is in a conformation that is incompatible with oligomerization as shown by its superposition with two adjacent NBDs in the octameric CED-4 [25]. Several helices of the NBD would clash with a neighboring NBD molecule in the octameric assembly (Figure 2b). Presumably, other NLRs, whether they are sensors or adapters, may use a similar structural mechanism for their autoinhibition.
Figure 2.
Structure of monomeric, inactive Δ CARD mouse NLRC4. (a) The NACHT domain of NLRC4 is divided as follows: NBD: nucleotide-binding domain (cyan); HD1: helical domain 1 (blue); WHD: winged helix domain (yellow); and HD2: helical domain 2 (magenta). LRR: leucine-rich repeat (orange). (b) Structural superposition of the inactive NLRC4 NBD with two neighboring NBDs (gray) of active CED-4 in the C. elegans caspase activating complex apoptosome (PDB ID: 3LQQ) [25]. The inactive NLRC4 NBD is in a conformation incompatible with NBD oligomerization. (c) Schematic model shows autoinhibition of AIM2 through intramolecular interactions between the PYD and the HIN200 domain.
Although the autoinhibition mechanism of ALR sensors such as AIM2 has not been directly visualized through structural studies, it has been proposed that the N-terminal PYD and the C-terminal HIN domains form intramolecular interactions to prevent PYD oligomerization (Figure 2c). For AIM2, its PYD itself is able to form filaments and therefore promotes inflammasome assembly and activation in the absence of the HIN domain [26, 27]. The direct interaction between AIM2 PYD and its HIN domain has been measured and confirmed [28], consistent with a mechanism of autoinhibition through intramolecular interactions. A docking model shows that AIM2 PYD may bind AIM2 HIN at the same surface that the HIN domain binds dsDNA; thus DNA binding may release autoinhibited PYD from the HIN domain, thereby facilitating AIM2 PYD oligomerization and activating ASC through PYD/PYD interactions [28, 29]. There is a long, ~50-residue linker between the PYD and the HIN domain, which should allow structural rearrangement upon activation.
DNA recognition and regulation by ALR HIN domains
Ligand binding may be a general mechanism to overcome autoinhibition, leading to initiation of inflammasome assembly. While it is presumed that the C-terminal domain of an NLR is the receptor for stimulation and acts to relieve autoinhibition, the molecular basis for this function is unclear. In contrast, for ALRs, the mode of direct sensing of cytosolic dsDNA by their HIN domains has been revealed from a series of structural studies. Upon binding dsDNA, ALRs apparently undergo a conformational change to disengage from their autoinhibited states to allow the freed N-terminal PYD to cluster and initiate inflammasome activation [26, 28, 29].
The HIN domain is comprised of tandem oligonucleotide/oligosaccharide binding (OB) folds (Figure 3a). Crystal structures of dsDNA-bound HIN domains from AIM2 and IFI16 show a similar mode of interaction that involves both OB folds and the intervening linker [29, 30]. Highly positively charged surfaces of AIM2 HIN and IFI16 HIN-B domains contact the dsDNA via mostly electrostatic interactions (Figure 3a, 3b). In contrast to sequence-specific DNA-binding, in which the proteins directly recognize the DNA bases, the HIN domains interact almost exclusively with the backbone. AIM2 and IFI16 HIN domains bind both strands of dsDNA, explaining why dsDNA, but not ssDNA, is able to initiate innate immune responses through AIM2 and IFI16.
Figure 3.
Ligand binding, filament assembly and negative regulation of ALRs. (a) Left: structure of AIM2 PYD (yellow). Middle and right: the HIN/dsDNA complex structure shown respectively in a ribbon diagram and in a surface electrostatics illustration rotated −90° along the vertical axis. HIN: dark blue. (b) Structure of the IFI16 HIN-B/dsDNA complex shown respectively in a ribbon diagram and in a surface electrostatics illustration rotated −90° along the vertical axis. (c) EM images (top, adapted from Figure 5 of [31]) and a schematic model (bottom) of filamentous structures of the IFI16/dsDNA complex. (d) Structure of the p202 HIN1/dsDNA complex shown respectively in a ribbon diagram and in a surface electrostatics illustration rotated −90° along the vertical axis. (e) Left: p202 HIN2 tetramer structure in a ribbon diagram with monomers shown in red, yellow, cyan and pink. Right: the same structure in a surface diagram, with the red monomer superimposed with the p202 HIN1/dsDNA structure (orange) and the AIM2 HIN/dsDNA structure (gray). One HIN2 tetramerization surface is analogous to the p202 HIN1 surface for dsDNA recognition, but opposite of the AIM2 HIN surface for dsDNA recognition. (f) Schematic mechanistic models for the interference with AIM2 signaling by p202 (adapted from Figure 6 of [33]).
Interactions of ALRs with long dsDNA appear to deviate from the presumed random distribution of binding implied by a beads on a string model. Instead, long dsDNA appears to cause cooperative binding of ALRs. For example, full-length IFI16 has been shown to cooperatively bind dsDNA in a length-dependent manner into distinct IFI16/dsDNA filaments even in the presence of excess dsDNA [31] (Figure 3c), suggesting the participation of protein/protein interactions. Although there are HIN/HIN contacts in the crystal structures of HIN/dsDNA complex structures [29, 30], the PYD of IFI16 is critical for the formation of IFI16 filaments [31]. These data indicate that intermolecular interactions among the PYDs drive the cooperative assembly onto dsDNA, with concomitant increase in the IFI16/dsDNA binding affinity when compared with isolated HIN domains alone [31]. This suggests that the dsDNA filaments and PYD filaments may form concurrently, leading to a central dsDNA filament decorated by short PYD filaments along its length [26, 28] (Figure 3c).
ALR inflammasome activation is tightly regulated in cells. An example comes from the mouse p202 protein, which lacks the N-terminal PYD but contains two HIN domains [14]. Expression of p202 is induced by interferon, and as negative feedback regulation p202 inhibits the inflammatory function of AIM2 [14]. Crystal structures of the p202 HIN1/dsDNA complexes show that the two OB folds are very similar to those of AIM2, but that the dsDNA binds p202 HIN1 at an almost opposite surface compared with that used in AIM2 HIN (Figure 3d, 3e) [30, 32, 33]. Surprisingly, the HIN2 domain of p202 does not interact with dsDNA [33]. It forms a dimer-of-dimers tetramer, utilizing the equivalent surface in HIN1 for dsDNA interaction as one of the dimerization interfaces (Figure 3e). The HIN2 of p202 also interacts specifically with the AIM2 HIN domain [33]. Therefore, the inhibitory function of p202 to AIM2 may be facilitated through two mechanisms. First, p202 binds dsDNA at a higher affinity than AIM2, and therefore competes with AIM2 for access to dsDNA. Second, the direct interaction between p202 HIN2 and AIM2 HIN domains may allow incorporation of p202 onto the same dsDNA that AIM2 interacts with, thereby diluting the effective concentration of AIM2 on the dsDNA [30, 32, 33] (Figure 3f). Thus, AIM2 PYD oligomerization and downstream signaling may be inhibited.
The nucleated polymerization mechanism of inflammasome assembly
The PYDs and CARDs involved in inflammasome assembly both belong to the death domain superfamily that mediates the formation of oligomeric signaling complexes important in cell death and innate immune signaling [34–37]. The subunit structures of these domains exhibit a globular, six-helix bundle fold, as illustrated here for ASC PYD [27] (Figure 4a). For ASC-dependent inflammasomes, the central organizing scaffold is formed by PYD/PYD and CARD/CARD interactions through a common helical assembly mechanism that was first elucidated for death domain complexes such as the oligomeric MyD88/IRAK4/IRAK2 complex and the PIDD/RAIDD complex [34, 37].
Figure 4.
ASC PYD filament structure and mechanism of nucleation dependent polymerization in Inflammasome formation. (a) Near atomic resolution cryo-EM structure of a protomer from the ASC PYD filament (PDB ID: 3J63). (b) ASC PYD helical filament structure is depicted as a surface representation in two orthogonal views, and is composed of three helical strands denoted by the colors green, blue, and red. (c) The AIM2 PYD structure (PDB ID: 3VD8) is modeled as a cluster with C3 symmetry (depicted as protomers in shades of yellow to denote subunit boundaries) at the end of the ASC PYD filament. An electron micrograph with streptavidin-gold labeling of biotin-AIM2 PYD/ASC PYD filaments shows that AIM2 PYD localizes to one end of the ASC PYD filament (adapted from Figure 1D of [27]). (d) The full-length ASC solution structure (PDB ID: 2KN6) is modeled as a cartoon-ribbon representation onto the ASC PYD filament structure (transparent surface representation) showing the outward projecting ASC-CARD domains. The AIM2 PYD cluster is also modeled in shades of yellow. Two orthogonal views are shown. (e). Electron micrographs of the His-GFP-caspase-1 CARD/ASC/AIM2 PYD ternary complex with anti-ASC gold and Ni-NTA gold labeling respectively, adapted from Figure 6 of [27]. (f) Electron micrographs of the flagellin/NAIP5/NLRC4 complex purified from HEK293E cells adapted from Figure 8 of [50].
Because of the importance of PYDs in inflammasome assembly, many structural studies have been performed on these domains. However, like many death domain superfamily proteins, PYDs are multivalent with a high propensity to aggregate, which makes structural studies difficult. To reduce aggregation for NMR and X-ray crystallography, many groups have prepared PYD proteins at low pH (~pH 4), which likely modifies the surface potential and prevents charge complementarity-induced aggregation. To date twelve monomeric PYD domain structures have been solved including ASC [38, 39], AIM2 [26, 28], NLRP1 [40], NLRP3 [41], NLRP4 [42], NLRP7 [43], NLRP10 (human) [44], NLRP10 (mouse, PDB ID 2DO9), NLRP12 [45], ASC2 (a regulatory PYD only protein) [46, 47], myeloid nuclear differentiation antigen (MNDA, PDB ID 2DBG), and Pyrin (PDB ID 2MPC, mutations of which cause familial Mediterranean fever).
Recent studies show that ASC PYD forms filamentous structures [27, 48], and that AIM2 PYD and NLRP3 can both form a complex with ASC PYD [27]. Electron microscopy imaging shows that the AIM2-PYD/ASC-PYD and NLRP3/ASC-PYD complexes are also filamentous [27]. Gold-particle labeling of AIM2 PYD and NLRP3 in these complexes reveal that they only reside at one end of the filamentous structures that are composed mostly of ASC PYD (Figure 4b, 4c), suggesting that the inflammasome sensors nucleate filament formation of the ASC adapter [27]. Indeed, fluorescence polarization assays reveal that sub-stoichiometric amounts of the AIM2/dsDNA complex or oligomerized NLRP3 can robustly nucleate the formation of the ASC-PYD filament [27].
The cryo-EM structure of the ASC-PYD filament provides a first view of the inflammasome assembly mechanism at near atomic resolution [27]. The filament resembles a cylinder with outer and inner diameters of ~90Å and ~20Å, respectively, and is built from three helical strands related by a three-fold symmetry along the helical axis (Figure 4b). Three types of repeating asymmetric interfaces that are characteristic of death domain superfamily proteins (Type I, II, and III) are used to form the filament from individual ASC-PYD protomers [34]. Upon incorporation into the filament, significant conformational changes occur in ASC-PYD, especially at the α2-α3 loop and the α3 helix, suggesting a directional elongation in ASC-PYD filament formation [27].
Because AIM2-PYD also forms filaments when expressed alone [26], it is anticipated that the filament will match the symmetry of ASC-PYD and therefore is able to nucleate ASC-PYD filament formation upon activation by cytosolic dsDNA. A model of the AIM2-PYD filament can be obtained by superimposing the AIM2-PYD protomer structure onto the ASC-PYD filament (Figure 3c). Connecting the short AIM2-PYD filament that is likely to form when wrapped around dsDNA (Figure 3c) and the long ASC-PYD filament would recapitulate the end-labeled AIM2/ASC complex observed by electron microscopy (Figure 4c), thereby providing the molecular basis for the nucleation and polymerization relationship between AIM2 and ASC.
In addition to the N-terminal PYD, ASC is a bifunctional molecule with a C-terminal CARD. The NMR structure of full-length ASC determined at a non-aggregating condition shows that the two domains are flexibly linked but with some preference in the relative orientation [38]. Superimposing the full-length ASC structure onto the ASC-PYD filament structure generates a model of the full-length ASC filament in which the PYD localizes at the core and the CARD forms the outer layer of the filament (Figure 4d). Radially positioned CARDs can then further cluster, recruit and nucleate filament formation of caspase-1, which is upregulated from previous NF-κB driven transcription, through CARD/CARD interactions of currently unknown mechanisms. The reconstituted ternary AIM2-PYD/ASC/caspase-1-CARD complex shows star-shaped structures under electron microscopy in which AIM2 and ASC locate near the center while caspase-1 forms the main bodies of the filamentous extensions, as shown by specific gold-labeling [27] (Figure 4e). The ASC-dependent NLRP3 inflammasome assembles through a similar mechanism. The initial star-shaped structures likely further coalesce to become a dense single perinuclear punctum per cell composed of networks of ASC and casapse-1 filaments [15, 27, 49].
The mechanism of ASC-independent NAIP/NLRC4/caspase-1 inflammasome assembly is still unclear as there are no atomic resolution structures currently available. A preliminary electron microscopy study shows that the Flagellin/NAIP5/NLRC4 complex forms a striking double disk structure with 11- or 12- fold symmetry [50] (Figure 4f). Presumably, the CARDs of NLRC4 in the double disk structure are clustered to allow the recruitment of caspase-1 and nucleation of caspase-1 filament formation, leading to proximity-induced, auto-proteolytic cleavage of caspase-1 to yield its active form. Because ASC appears to enhance NAIP inflammasome formation, it is likely that the NLRC4 CARD can interact with the ASC CARD, which in turn recruits more caspase-1 through multiple CARD/CARD interactions to amplify the activation signal.
Conclusions and perspectives
Recent structural studies have now provided a framework for understanding inflammasomes through steps of hierarchical assembly involving ligand recognition, release of autoinhibition, and nucleated polymerization (Figure 5). The process is likely highly cooperative, generating an all-or-none mechanism of inflammasome activation, which is a common emerging theme among SMOCs in innate immunity [5, 6]. The increase in stoichiometry from the ALR and NLR sensors, to the adapter ASC, and then to caspase-1 in the ternary inflammasomes, gives an effective amplification mechanism for the signal transduction. For the NLRP3 inflammasome, the efficient assembly and amplification is reflected in their low activation threshold in dendritic cells [51]. Although we have progressed a great deal, many more structural studies are required to elucidate the detailed molecular mechanism for the assembly and activation of each member of the large inflammasome family, undoubtedly with surprises awaiting.
Figure 5.
A schematic diagram for the nucleation dependent polymerization mechanism of AIM2 and NLRP3 inflammasome formation, with proximity-induced autoproteolytic maturation of caspase-1. Proteins and domains are labeled.
Highlights.
The crystal structure of an inflammasome adapter, NLRC4, reveals an autoinhibited conformation.
Structural studies on DNA binding and inhibition of AIM2-like receptors suggest cooperative recognition and regulatory mechanisms.
The cryo-EM structure of the ASC-PYD filament shows the inflammasome formation by nucleated polymerization.
Acknowledgments
We apologize for incomplete coverage due to the space limitations and the vast data in 0the highly active field of inflammasome biology. The work was supported by grants from NIH to HW.
Footnotes
Conflict of interest statement
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, et al. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 4*.Yin Q, et al. Structural Biology of Innate Immunity. Ann Rev Immunology. 2015 doi: 10.1146/annurev-immunol-032414-112258. in press This review gives a structural overview of proteins that constitute various supramolecular signaling complexes in the innate immune response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. This paper describes the growing viewpoint that innate immune signaling is dominated by the cooperative assembly of large, multimeric protein complexes, which drive proximity-driven enzyme activation instead of the classical model of signal transduction as a cascade of protein-protein binding and enzyme activation events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Kagan JC, et al. Supramolecular organizing centres: location-specific higher-order signalling complexes that control innate immunity. Nat Rev Immunol. 2014;14:821–6. doi: 10.1038/nri3757. This review coins the term, supramolecular organizing centers (SMOCs), as a way to describe spatially resolved higher-order immune signaling complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman E, et al. Super-resolution characterization of TCR-dependent signaling clusters. Immunol Rev. 2013;251:21–35. doi: 10.1111/imr.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul S, Schaefer BC. A new look at T cell receptor signaling to nuclear factor-kappaB. Trends Immunol. 2013;34:269–281. doi: 10.1016/j.it.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhlbrandt W. Cryo-EM enters a new era. eLife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godin AG, et al. Super-resolution Microscopy Approaches for Live Cell Imaging. Biophys J. 2014;107:1777–1784. doi: 10.1016/j.bpj.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. This review describes canonical and non-canonical inflammasome biology with overviews of NLR and ALR inflammasome assembly and activation, and of implications on therapeutic applications. [DOI] [PubMed] [Google Scholar]
- 12.Rathinam VA, et al. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masumoto J, et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 14.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 15.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri T, et al. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 18.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, et al. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A. 2013;110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poyet JL, et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 22.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 23.Man SM, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. This paper reports the first crystal structure of a CARD-containing NOD-like receptor. The observed closed conformation mediated by extensive intramolecular interactions suggest an autoinhibition mechanism that prevents oligomerization. [DOI] [PubMed] [Google Scholar]
- 25.Qi S, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 26*.Lu A, et al. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J Mol Biol. 2014;426:1420–1427. doi: 10.1016/j.jmb.2013.12.029. This study describes the filament forming ability of AIM2-PYD and the crystal structure of the monomeric F27G AIM2-PYD mutant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. This paper reports the first near atomic resolution cryo-EM structure of the ASC-PYD filament, and in vitro reconstitution of the PYD/PYD and CARD/CARD interactions in the AIM2 and NLRP3 inflammasome. These studies reveal a stepwise nucleated polymerization mechanism of inflammasome assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Jin T, et al. Structure of the AIM2 pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013 doi: 10.1074/jbc.M113.468033. This article presents the MBP-fused AIM2-PYD crystal structure and provides evidence for an intramolecular interaction between the PYD and the HIN domain of AIM2 in autoinhibition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. The crystal structures of AIM2-HIN200 and IFI16-HIN-B each bound to dsDNA are presented. HIN binding to dsDNA is shown to be non-specific and dependent on charge/charge interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Ru H, et al. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 2013;23:855–8. doi: 10.1038/cr.2013.52. This paper presents crystal structures of AIM2-HIN200/dsDNA and p202-HIN1/dsDNA complexes. It further describes the molecular basis for the regulatory function of p202 by comparing the DNA binding modes of AIM2-HIN200 vs. p202-HIN1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Morrone SR, et al. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A. 2014;111:E62–71. doi: 10.1073/pnas.1313577111. This report presents EM and quantitative binding data, which suggest that IFI16 binds dsDNA in a cooperative manner and that cooperative binding of the HIN domains to dsDNA is driven by the PYD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling. Acta crystallographica Section F, Structural biology communications. 2014;70:21–29. doi: 10.1107/S2053230X1303135X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Yin Q, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. Crystallographic and EM structures of tetrameric p202 and the p202-HIN1 domain bound to dsDNA are presented. These structures, along with quantitative binding data showing a direct interaction between p202-HIN2 and AIM2-HIN200 domains, suggest an inhibitory mechanism in which p202 tetramers bind both dsDNA and AIM2, preventing cooperative clustering of AIM2-PYDs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. This review describes the conserved three types of interactions that mediate the helical assembly of death domain containing proteins including the 5 PIDD/7 RAIDD, 6 MyD88/4 IRAK4/4 IRAK2, and 5 Fas/5 FADD signaling complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat Struct Mol Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Lin SC, et al. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. The crystal structure of the 14-subunit MyD88/IRAK4/IRAK2 death domain complex is reported, revealing a helical assembly mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J Biol Chem. 2009;284:32932–32941. doi: 10.1074/jbc.M109.024273. This article presents the solution structure of full-length ASC. Despite a long flexible linker connecting the PYD and CARD domains, a preferred back-to-back domain orientation is observed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liepinsh E, et al. The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol. 2003;332:1155–1163. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Hiller S, et al. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure (Camb) 2003;11:1199–1205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Bae JY, Park HH. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J Biol Chem. 2011;286:39528–39536. doi: 10.1074/jbc.M111.278812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eibl C, et al. Structural and Functional Analysis of the NLRP4 Pyrin Domain. Biochemistry. 2012;51:7330–7341. doi: 10.1021/bi3007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinheiro AS, et al. Three-dimensional structure of the NLRP7 pyrin domain: insight into pyrin-pyrin-mediated effector domain signaling in innate immunity. J Biol Chem. 2010;285:27402–27410. doi: 10.1074/jbc.M110.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su MY, et al. Three-dimensional structure of human NLRP10/PYNOD pyrin domain reveals a homotypic interaction site distinct from its mouse homologue. PLoS One. 2013;8:e67843. doi: 10.1371/journal.pone.0067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinheiro AS, et al. The NLRP12 pyrin domain: structure, dynamics, and functional insights. J Mol Biol. 2011;413:790–803. doi: 10.1016/j.jmb.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natarajan A, et al. Structure and dynamics of ASC2, a pyrin domain-only protein that regulates inflammatory signaling. J Biol Chem. 2006;281:31863–31875. doi: 10.1074/jbc.M605458200. [DOI] [PubMed] [Google Scholar]
- 47.Espejo F, Patarroyo ME. Determining the 3D structure of human ASC2 protein involved in apoptosis and inflammation. Biochem Biophys Res Commun. 2006;340:860–864. doi: 10.1016/j.bbrc.2005.12.087. [DOI] [PubMed] [Google Scholar]
- 48*.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. Presents data showing PYD and CARD containing proteins such as ASC and MAVS have inducible prion-like behavior in yeast and mammalian cell reconstitution assays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J, et al. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J Clin Immunol. 2010;30:693–702. doi: 10.1007/s10875-010-9425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halff EF, et al. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J Biol Chem. 2012 doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Latz E, et al. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]