Abstract
Signal transduction pathways and their coordination are critically important for proper functioning of animal immune systems. Our knowledge of the constituents of the intracellular signaling network in insects mainly comes from genetic analyses in Drosophila melanogaster. To facilitate future studies of similar systems in the tobacco hornworm and other lepidopteran insects, we have identified and examined the homologous genes in the genome of Manduca sexta. Based on 1:1 orthologous relationships in most cases, we hypothesize that the Toll, Imd, MAPK-JNK-p38 and JAK-STAT pathways are intact and operative in this species, as are most of the regulatory mechanisms. Similarly, cellular processes such as autophagy, apoptosis and RNA interference probably function in similar ways, because their mediators and modulators are mostly conserved in this lepidopteran species. We have annotated a total of 186 genes encoding 199 proteins, studied their domain structures and evolution, and examined their mRNA levels in tissues at different life stages. Such information provides a genomic perspective of the intricate signaling system in a non-drosophiline insect.
Keywords: insect immunity, RNA-Seq, transcriptome, gene annotation, expression profiling
1. Introduction
Insects fight against invading pathogens and parasites via their innate immune system (Gillespie et al., 1997; Lemaitre and Hoffmann, 2007). Like other physiological processes, insect immune responses involve sensors, effectors, and signal transducers, linking pathogen recognition with cellular and humoral responses. Some of the responses occur in minutes while others involve transcriptional activation of genes that are not highly expressed under normal conditions, and thus may provide responses in hours to days. In the latter case, a relay system must exist to transduce the extracellular signals of wounding or invasion into the nuclei of cells, where transcriptional regulation occurs. If pathogens are sensed by receptors (e.g. PGRP-LC) on the cell surface, responses are more direct than if recognition occurs in hemolymph. In the latter scenario, receptors (e.g. PGRP-SA) in hemolymph bind to the pathogens and initiate extracellular signal transduction to generate active cytokines. The cytokines then interact with their receptors on the cell surface to induce cellular responses including phagocytosis, encapsulation, apoptosis, autophagy, and synthesis of immune effectors (Strand, 2008; Jiang et al., 2010). Consequently, the intracellular signal transduction network is essential for mediating immune responses in insects.
Receptor-mediated Toll, Imd, MAPK-JNK-p38, JAK-STAT and other pathways are widely conserved in metazoans, functioning as regulators and mediators of humoral and cellular immune responses (Buchon et al., 2014). Extensive studies in Drosophila melanogaster have revealed many details of the signal transduction network. The Toll pathway was discovered in the screens that identified mutations in genes affecting the establishment of dorsoventral axis and later found to regulate the expression of immunity-related genes through Dorsal and Dif, transcription activators of the Rel family (Valanne et al., 2011). Gram-positive bacteria and fungi trigger this pathway via an extracellular serine protease cascade that activates the cytokine Spätzle through limited proteolysis. This activated cytokine binds to the Toll receptor, leading to antimicrobial peptide synthesis and differentiation of certain hemocytes into lamellocytes. These lamellocytes are capable of encapsulating and killing parasites such as parasitoid wasps (Sorrentino et al., 2004). In the case of Gram-negative bacteria, DAP-type peptidoglycans (PGs) elicit the Imd pathway via transmembrane PGRP-LC and intracellular signal mediators (Kaneko et al., 2006; Rämet et al., 2002). Activated Relish, another Rel factor, then migrates into the nucleus to turn on a set of immunity-related genes overlapping with that induced by Dorsal and Dif (Imler and Hoffmann, 2001; Mellroth et al., 2005). Cytokines, growth factors, or stress signals stimulate the MAPK-JNK-p38 pathway to regulate apoptosis, Imd pathway, and cell differentiation (Ragab et al., 2011; Chen et al., 2010; Dong et al., 2002). The JAK-STAT pathway, RNA interference, autophagy and other defense mechanisms are involved in antiviral responses (Kisseleva et al., 2002; Baeg et al., 2005; Kingsolver et al., 2013). Based on the available information, these pathways are mostly conserved among insects but differences do exist. For instance, the honeybee Apis mellifera has considerably fewer immunity-related genes (Evans et al., 2006). A. mellifera has five Toll genes compared with the nine found in D. melanogaster. The pea aphid Acyrthosiphon pisum lacks the entire Imd pathway (Gerardo et al., 2010). With such plasticity observed among the few genomes available in the Insecta, it is therefore critically important to examine and characterize the immune signaling components in different major orders of insects.
Lepidoptera comprises about 160,000 described species of moths and butterflies in 126 families and 46 superfamilies (Kristensen et al., 2007). Larvae of many lepidopterans are serious agricultural pests but they are susceptible and can be controlled by biological agents such as entomopathogens (e.g. viruses, bacteria, fungi) and parasitoid wasps. Studies of lepidopteran immune systems and the associated signaling pathways are extremely important for developing effective biological control methods. Manduca sexta and Bombyx mori have been used as powerful biochemical models to explore various aspects of innate immunity (Jiang et al., 2010). Immunity-related genes in the silkworm were previously compared with those in D. melanogaster, Anopheles gambiae and A. mellifera (Tanaka et al., 2008) and analyses of the M. sexta hemocyte and fat body transcriptomes revealed a set of 232 genes encoding proteins for pathogen recognition, signal transduction, microbe killing (Gunaratna and Jiang, 2013), and modulation of mRNA levels in response to an immune challenge (Zhang et al., 2011). Recently, the M. sexta genome assembly became available along with 52 RNA-Seq datasets of tissues at various life stages (X et al., 2015). To better understand immune signal transduction in this undomesticated pest species, we have annotated genes for the putative pathway members, studied their expression patterns, and proposed a signal transduction network based on 1:1 orthology. The results represent working models for future studies on M. sexta and other lepidopteran pests.
2. Materials and methods
2.1. Gene identification, sequence improvement, and feature prediction
Manduca Genome Assembly 1.0 and gene models in Manduca Official Gene Sets (OGS) 1.0 and 2.0 and Cufflinks Assembly 1.0 (X et al., 2015) were downloaded from Manduca Base (ftp://ftp.bioinformatics.ksu.edu/pub/Manduca/). Protein sequences of the putative signal transducers from M. sexta (Gunaratna and Jiang, 2013) and other insects were used as queries to search Cufflinks 1.0, OGS 1.0 and OGS 2.0 using the TBLASTN algorithm (http://darwin.biochem.okstate.edu/blast/blast_links.html). Hits with aligned regions longer than 30 residues and identity over 40% were retained for retrieving corresponding cDNA sequences. Errors resulting from problematic regions (e.g. NNN…) in the genome assembly were manually corrected after BLASTN search of Manduca Oases and Trinity Assemblies 3.0 of the RNA-Seq data (Cao and Jiang, 2015). The two genome-independent RNA-Seq assemblies were developed to cross gaps between genome scaffolds or contigs and detect errors in the gene models. In some complex cases, all exons of a gene were examined based on the GT-AG rule and sequence alignment to identify the splicing junctions. Correct open reading frames in the improved sequences were identified using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and validated by BLASTP search against GenBank (http://www.ncbi.nlm.nih.gov/) or Uniprot (http://www.uniprot.org/). Signal peptides were predicted using SignalP4.1 (Petersen et al., 2011). Conserved domain structures were identified using SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi) and InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/).
2.2. Sequence alignment and phylogenetic analysis
Multiple sequence alignments of immune signal transducers from M. sexta and other insects were performed using MUSCLE, a module of MEGA 6.0 (Tamura et al., 2013) at the following settings: refining alignment, gap opening penalty (−2.9), gap extension penalty (0), hydrophobicity multiplier (1.2), maximal iterations (100), UPGMB clustering (for iterations 1 and 2) and maximum diagonal length (24). The aligned sequences were used to construct neighbor-joining trees by MEGA 6.0 with bootstrap method for the phylogeny test (1000 replications, Poisson model, uniform rates, and complete deletion of gaps or missing data).
2.3. Gene expression profiling
Coding DNA sequences from the improved gene models were retrieved and employed as templates for mapping reads in the 52 M. sexta RNA-Seq datasets, representing mRNA samples from whole insects, organs or tissues at various developmental stages. Illumina reads (M. sexta genome and transcriptome project; http://www.ncbi.nlm.nih.gov/bioproject/PRJNA81039) were trimmed to 50 bp and mapped to the coding regions using Bowtie (0.12.8) (Langmead et al, 2009). Numbers of the mapped reads were used to calculate FPKM (fragments per kilobase of exon per million fragments mapped) values using RSEM (Li and Dewey, 2011). Hierarchical clustering of the log2(FPKM+1) values was performed using MultiExperiment Viewer (v4.9) (http://www.tm4.org/mev.html) with the Pearson correlation-based metric and average linkage clustering method. To study transcript level changes after immune challenge, the entire CDS set was used to search for corresponding contigs in the CIFH09 database (http://darwin.biochem.okstate.edu/blast/blast_links.html) (Zhang et al., 2011) by TBLASTN. The numbers of CF, CH, IF, and IH reads (C for control, I for induced after injection of bacteria, F for fat body, H for hemocytes) assembled into these contigs were retrieved for normalization and calculation of IF/CF and IH/CH ratios. When a polypeptide sequence corresponded to two or more contigs, sums of the normalized read numbers were used to calculate its relative mRNA abundances in fat body and hemocytes (Gunaratna and Jiang, 2013).
3. Results and discussion
3.1. Spätzle-1–7, cytokines with distinct structures, functions, and expression patterns
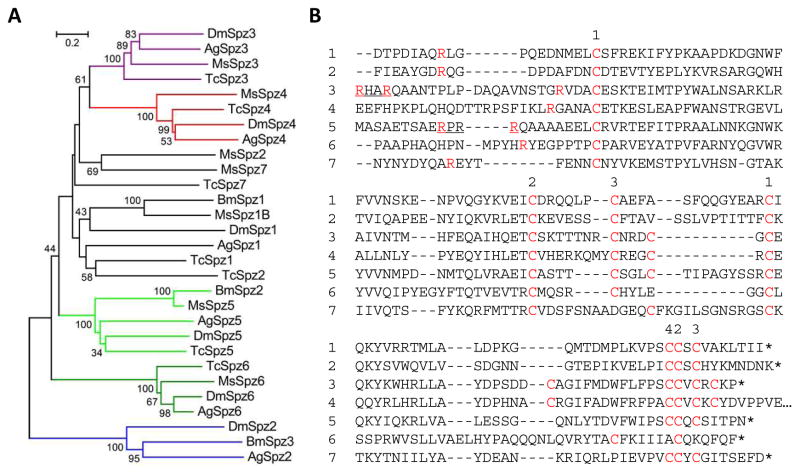
There are seven genes encoding Spätzle-like proteins in M. sexta (Table S1), which differs from the number present in Tribolium castaneum (9), D. melanogaster (6), and A. gambiae (6), B. mori (3) and A. mellifera (2) (Tanaka et al., 2008). The M. sexta proteins contain a signal peptide, a 50 to 360-residue segment with 0 to 4 low complexity regions, and a cystine-knot cytokine domain (Fig. 1). For Spätzle-1, cleavage between QR and LG results in a dimer of the C-terminal fragment that induces antimicrobial peptide synthesis (An et al., 2010), presumably via a Toll receptor. While Spätzle-2–7 may be activated by trypsin-like serine proteases, Spätzle-3 and 5 can also be processed by furin-like enzymes next to their recognition sequences, RHAR and RPRR, respectively. The C-terminal fragments of Spätzle-3–6 contain an even number of Cys residues, which might allow them to possibly dimerize via additional disulfide bonds. Molecular modeling suggests that Spätzle-1–5 and 7 adopt a similar fold with three pairs of antiparallel β-strands stabilized by 3 or 4 intrachain disulfide bonds (data not shown). Phylogenetic analysis of the entire proteins indicates that Spätzle-3–6 each forms a tight group with their orthologs from the other insects (Fig. 1A), suggestive of conserved functions. From parallel studies (Cao et al., 2015; Rao et al., 2015; He et al., 2015), we have noticed that the mRNA levels of many immunity-related genes in fat body and midgut greatly increase at the onset of wandering stage and peak on day 1 of the pupal stage. This infection-independent gene up-regulation during metamorphosis also occurs in other lepidopterans such as Galleria mellonella (Altincicek and Vilcinskas, 2008). Spätzle-1, 2 and 7 transcripts display this pattern with the highest FPKM values of 224, 760 and 564, respectively (Fig. 2A). These three genes are induced upon immune challenge, whereas Spätzle-3–6 mRNAs were detected only at very low levels (Gunaratna and Jiang, 2013; Zhang et al., 2011; Table 1). Spätzle-1B mRNA level is low in ovary, higher in eggs and down-regulated after hatching. In contrast, Spätzle-2 mRNA levels are high in ovary, lower in eggs, and become higher in 1st instar larvae. The expression patterns of Spätzle-3 and 5 are similar to each other. Spätzle-4 and 6 are almost exclusively produced in the midgut of 2nd and 3rd instar larvae. The detection of Spätzle-2, 3, 5 and 7 mRNAs in head is interesting, since Drosophila Toll6, Toll7 and Toll8 act as receptors of neurotrophins (Drosophila Spätzle-2, 3 and 5) (McIlroy et al., 2013; Ballard et al., 2014).
Fig. 1.
Phylogenetic relationships of Spätzles in M. sexta, B. mori, T. castaneum, and D. melanogaster. (A) Tree. Based on the sequence alignment of 29 full-length Spätzles, a tree was generated with branches shown in colors representing closely related groups. (B) Aligned sequences of the cystine-knot cytokine domains in M. sexta Spätzles-1 through 7. Cys residues are indicated in a red font. Some Cys residues may form intra- (1-1, 2-2, 3-3) and inter- (4) chain disulfide bonds. Proteolytic activation sites, known for Spätzle-1, are predicted to be next to the Arg (red) in Spätzle-2 through 6. The putative processing site (RXXR) is underlined in Spätzle-3 and 5.
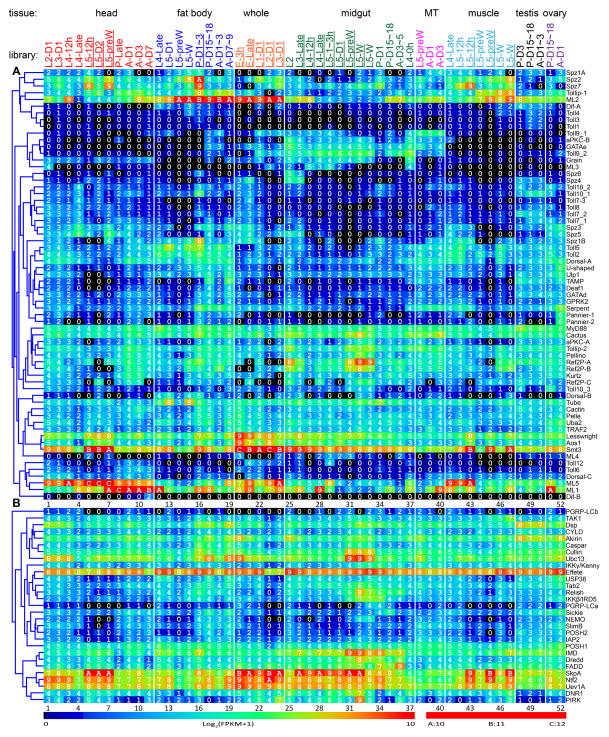
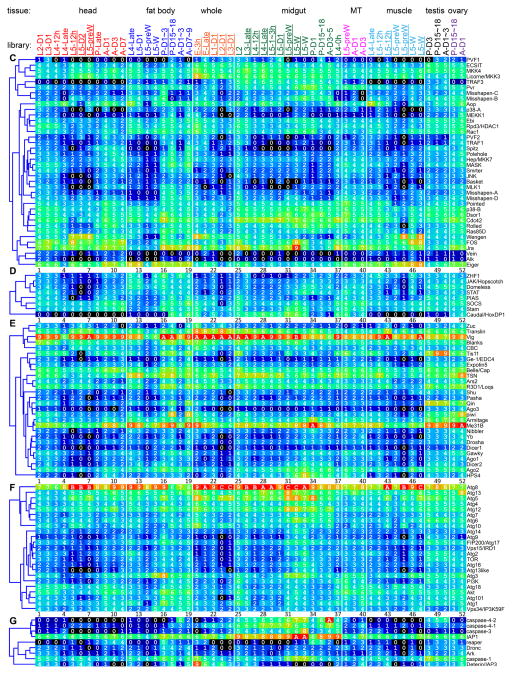
Fig. 2.
Transcript profiles of the putative signaling protein genes in the 52 tissue samples. The mRNA levels, as represented by log2(FPKM+1) values, are shown in the gradient heat map from blue (0) to red (≥10). The values of 0–0.49, 0.50–1.49, 1.50–2.49 ··· 8.50–9.49, 9.50–10.49 10.50–11.49, and 11.50–12.49 are labeled 0, 1, 2 … 9, A, B and C, respectively. The cDNA libraries are constructed from the following tissues and stages: head [2nd (instar) L (larvae), d1 (day 1); 3rd L, d1; 4th L, d0.5; 4th L, late; 5th L, d0.5; 5th L, d2; 5th L, pre-W (pre-wandering); P (pupae), late; A (adults), d1; A, d3; A, d7], fat body (4th L, late; 5th L, d1; 5th L, pre-W; 5th L, W; P, d1-3; P, d15-18; A, d1-3; A, d7-9), whole animals [E (embryos), 3h; E, late; 1st L; 2nd L; 3rd L), midgut (2nd L; 3rd L; 4th L, 12h; 4th L, late; 5th L, 1–3h; 5th L, 24h; 5th L, pre-W; 5th L, W; P, d1; P, d15-18; A, d3-5; 4th L, 0h), Malpighian tubules (MT) (5th L, pre-W; A, d1; A, d3), muscle (4th L, late; 5th L, 12h; 5th L, pre-W; 5th L, W), testis (P, d3; P, d15-18; A, d1-3), and ovary (P, d15-18; A, d1). Some libraries (underlined) are from single-end sequencing; the others are from paired-end sequencing. Note that some synonymous libraries exhibit different FPKMs due to method differences. Panel A, Toll; B, Imd with JNK branch; C, MAPK-JNK-p38; D, JAK-STAT; E, pi- si- and mi-RNA pathways, F, autophagy; G, apoptosis.
Table 1.
Relative mRNA abundances of the intracellular signaling pathway members in induced (I) and control (C) fat body (F) and hemocytes from the larvae of M. sexta.
| Name | IF/CF | IH/CH | Name | IF/CF | IH/CH | Name | IF/CF | IH/CH | Name | IF/CF | IH/CH |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spätzle1* | 1.4 | 3.9 | Dredd* | 1.8 | 1.2 | MLK1* | 1 | 2.1 | R3D1/Loqs | 0.8 | 1.3 |
| Spätzle2 | 1.5 | 2.2 | Relish* | 5.2 | 1.5 | MKK4* | 0.8 | 0.5 | Dicer1 | 0 | 0.7 |
| Spätzle7 | 4.1 | 3.6 | NTF2* | 1.2 | 1.7 | JNK* | 1.8 | 1.6 | Ago1 | 1 | 2.6 |
| Toll1* | 2.5 | 6.2 | TAK1* | 0.5 | 3.6 | Basket | 1.3 | 2.7 | Drosha | 1.3 | 1.1 |
| Toll2 | 0.7 | 0.8 | Tab2* | 2.5 | 1 | ECSIT* | 1 | 1.7 | Pasha | - | 1.5 |
| Toll3 | 2.5 | 6.2 | IKKβ* | 0.5 | 0.2 | MEKK1* | 0 | 0.6 | Expotin5 | 4.1 | 1.7 |
| Toll4 | 2.5 | 6.4 | IKKγ* | 2.5 | 1.2 | MKK3* | 1.4 | 1.1 | Nibbler | 0.7 | 1.4 |
| Toll5 | 2.9 | 0.9 | NEMO | 1 | 1.2 | p38* | 2.2 | 1 | Gawky | 0.9 | 0.9 |
| ML1 | 2.2 | 0.6 | Dnr1 | 2.2 | 0.5 | Aop* | 3.1 | 1.2 | Me31B | 1 | 0.8 |
| ML2 | 1.8 | 0.8 | Sickie* | 1 | 33.2 | FOS* | 2 | 1.9 | Ge-1 | 0.5 | 1 |
| MyD88* | 1.5 | 1.5 | Caspar* | 3.1 | 1.6 | Jra* | 1.6 | 0.9 | Atg1 | 2.8 | 0.9 |
| Tube* | 8.4 | 0.8 | IAP2* | 0.3 | 0.9 | Ebi | 1 | 1.4 | Atg13 | 0.4 | 1.3 |
| Pelle* | 5.1 | 2.2 | Bendless* | 1.2 | 2.1 | Smrter | 1.8 | 1.2 | Atg101 | 1.3 | 0.6 |
| Pellino* | 2.3 | 1.3 | Uev1A* | 1.4 | 1.1 | Rpd3/HDAC1 | 0.2 | 1.2 | Atg17 | 0.6 | 0.7 |
| Cactus* | 9.2 | 1.8 | Effete | 0.8 | 1.1 | Domeless* | 2.4 | 0.8 | Vps34 | 0.5 | 1.2 |
| Dorsal* | 1.3 | 1.2 | USP36 | 0.5 | 1.4 | Stam* | 2 | 1.1 | Vps15 | 0.5 | 1.6 |
| Tollip-1* | 1 | - | POSH1* | 1.7 | 0.9 | JAK/Hopscotch* | 1 | 0.5 | Atg6 | 0.5 | 1.2 |
| Tollip-2* | 0.8 | 1.1 | POSH2 | 0.8 | 1.3 | STAT* | 0.4 | 0.7 | Atg18 | 3.6 | 0.8 |
| Ref2P* | 1.5 | 1 | CYLD | 3.6 | 0.8 | PIAS* | 1.6 | 1 | Atg12 | 0.5 | 3 |
| aPKC* | - | 1 | SkpA | 0.5 | 1.7 | SOCS* | 2.5 | 0.8 | Atg7 | 0.5 | 0.5 |
| GPRK2 | 0.1 | 0.7 | Cullin | 0.9 | 1.3 | ZHF1 | 0.7 | 1 | Atg5 | 0.5 | 1 |
| Cactin | 0.5 | 2.5 | SlimB | 1.5 | 0.7 | Piwi | 0.4 | 0.8 | Atg4 | 0.2 | 1.1 |
| Aos1* | 0.7 | 1.7 | Akirin | 9.2 | 1.7 | Armitage | 1.4 | 1 | Atg8 | 3.1 | 0.6 |
| Uba2* | 2.5 | 1.5 | Dsp | 2 | 1.4 | Yb | 0.5 | 0.2 | Atg3 | 0.3 | 1 |
| Lesswright* | 4.6 | 1.2 | Eiger* | 0.8 | 8 | Shu | 0.5 | 5.9 | Atg2 | 1.2 | 0.2 |
| Ulp1 | 5.6 | 0.8 | PVR* | 1 | 1.4 | Qin | 0.7 | 0.9 | Atg9 | 1.5 | 0.9 |
| Kurtz | 1 | 0.7 | Ras85D* | 0.7 | 1.7 | Dicer2 | 1.3 | 1.5 | Akt | 1 | 1 |
| Smt3* | 1.6 | 1.6 | Rac1* | 2.5 | 1.3 | Ago2 | 0.8 | 1 | TOR | 0.2 | 0.8 |
| Deaf1 | - | 0.9 | Cdc42* | 1.3 | 1.4 | Vig | 0.9 | 1.1 | PI3K | 1.5 | 0.9 |
| Serpent* | 0.4 | 0.9 | MASK* | 1.2 | 1.4 | TSN | 0.6 | 1.2 | IAP1 | 1.6 | 1.1 |
| Pannier-1 | 0.5 | 2.4 | Polehole | 0.5 | 1 | Ars2 | 3.1 | 1.7 | Deterin | - | 0.7 |
| Pannier-2 | 1 | 2.4 | Dsor1* | 0.5 | 1.1 | CBC | - | 3.1 | Dronc | 0.5 | 1.2 |
| GATAe | 0.5 | 2.4 | Rolled | 1.9 | 1.1 | Belle/Cap | 1.2 | 1.5 | Ark | 2 | 0.6 |
| U-shaped | 0.3 | 1.1 | Pointed | 0.3 | 0.9 | Blanks | 2.5 | 1.1 | Caspase-1 | 1.5 | 1.4 |
| Imd* | 2.7 | 1 | Misshapen* | 1.6 | 0.8 | Translin | 0.5 | 0.9 | |||
| FADD* | 0.6 | 1.3 | Hep/MKK7* | 1.5 | 1.2 | Tis11 | 1.3 | 0.9 |
As described in Section 2.3, the transcriptome data of larval fat body and hemocytes before and after the immune challenge (Zhang et al., 2011) were processed again according to Gunaratna and Jiang (2013), based on the BLAST search using 196 complete coding sequences as queries. The ones with no hit in the CIFH library are omitted from the table. Note that, due to the increase in query sizes and contig hits, the reported relative abundances (*) (i.e. IF/IH and CF/CH) (Gunaratna and Jiang, 2013) may be different for certain genes. “-”: C and I = 0
3.2. Structure, expression, and evolution of Toll receptors
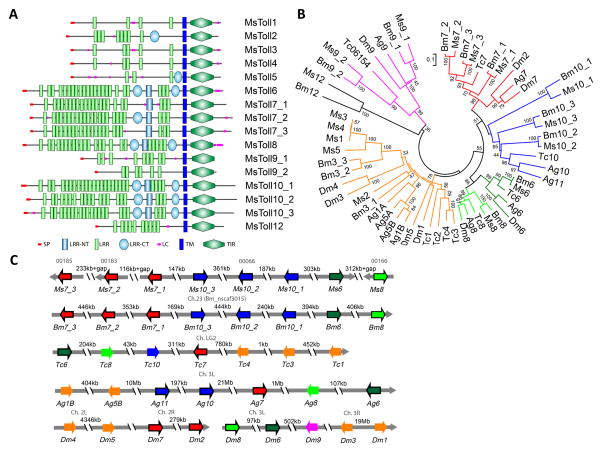
Toll receptors are a group of transmembrane proteins with extracellular Leu-rich repeats (LRRs) and a cytoplasmic Toll/interleukin-2 receptor (TIR) homology domain (Fig. 3A). We have identified sixteen such genes and named them Toll1–6, 7_1–3, 8, 9_1, 9_2, 10_1–3 and 12 (Table S1). These names are based on and mostly consistent with their orthologs in B. mori (Tanaka et al., 2008). Toll1 is reported as an immune-inducible gene that is predominantly expressed in hemocytes (Ao et al., 2008b; Gunaratna and Jiang, 2013) (Table 1). Along with Toll2–5 and B. mori Toll3_1–3, M. sexta Toll1 is grouped with D. melanogaster Toll1, 3–5, A. gambiae Toll1A, 1B, 5A and 5B, and T. castaneum Toll1–4 (Fig. 3B). Nonetheless, M. sexta Toll1, 3 and 4 have only 4 to 5 LRRs (Fig. 3B), instead of the 12 LRRs and 2 Cys-rich C-terminal domains that are present in Drosophila Toll1. The mRNA levels of Toll1, 3 and 4 are very low in the 52 libraries (Fig. 2A). Hence, the putative roles as Spätzle-1 or 2 receptors need validation. In contrast, Toll2 and 5 transcripts are highly abundant in fat body and their profiles of expression are closely similar to those of Dorsal, Serpent and Spätzle-1B. Interestingly, Manduca Dorsal and Serpent may interact with each other to activate moricin gene transcription (Rao et al., 2011). Toll2 and 5 containing a Cys-rich C-terminal domain, are more similar in domain structure to Drosophila Toll1. Based on this and other evidence, we suggest M. sexta Toll2 and 5 are better candidates than Toll1 as receptors of Spätzle-1, 2 and 7. In D. melanogaster, Toll6, 7 and 8 (i.e. Tollo) are involved in neurotrophism (McIlroy et al., 2013; Ballard et al., 2014) and recent studies suggest that Toll7 may also be a pattern recognition receptor for vesicular stomatitis virus, activating cellular autophagy of the virus (Nakamoto et al., 2012). Their orthologous genes (Toll6, 7_1–3 and 8) are expressed in heads at levels higher than other tissues (Fig. 2A) and may play similar roles in M. sexta. The M. sexta Toll9_1 mRNA levels are high in Malpighian tubules of pre-wandering larvae and adults, as well as in midgut of feeding larvae. Human myeloid differentiation factor-2 (MD2) forms a complex with Toll-like receptor-4 to recognize lipopolysaccharide and lead to inflammation and cytokine production. A MD2-like protein (ML1) from A. gambiae specifically regulates the resistance to Plasmodium falciparum (Dong et al., 2006). We have identified five MD2-like proteins (MLs) in M. sexta, which contain a signal peptide and may increase binding specificity of the Toll receptors (Ao et al., 2008a).
Fig. 3.
Domain structures (A), phylogenetic relationships (B), and gene orders (C) of Tolls in M. sexta. (A) Signal peptide (SP), Leu-rich repeat (LRR), amino- and carboxyl-terminal (NT & CT) LRRs, low complexity (LC) region, transmembrane (TM) segment, and TIR (Toll/interleukin-1 receptor) domain are shown in different colors and shapes as indicated. (B) Amino acid sequences of the 58 full-length Toll proteins from M. sexta, B. mori, T. castaneum, A. gambiae, and D. melanogaster are aligned to generate the tree with its branches in different colors for closely related groups. (C) Orientations and orders of the Toll genes in the five insects are schematically shown as arrows in the same colors as in panel B. Arrows for the single exon genes are in black frame.
Despite the fact that the coding regions being 2.2 to 4.0 kb in length, half of the 16 genes (Toll6, 7_1–3, 8, 10_1–3) only contain a single exon (Fig. 3C). They correspond 1:1 with their orthologous genes on chromosome 23 in B. mori. M. sexta Toll7_1, 10_3, 10_2, 10_1 and 6 on Scaffold (S) 00066 have the same orientations as those in the silkworm, flanked by Toll7_3 (S00185), 7_2 (S00183), and 8 (S00166) (Fig. 3C). When we compared the orthologous genes in A. gambiae, D. melanogaster and T. castaneum, similar gene orders were found. These orthologous genes include: Toll7_3 to 1, 10_3 to 1, 6 and 8 in the lepidopterans; Toll11&10, 7, 8 and 6 in the mosquito; Toll2&7, 8 and 6 in the fruit fly; Toll6, 8, 10, and 7 in the beetle. The underlined genes result from lineage-specific gene duplications. Except for AgToll8, TcToll8 and TcToll10 (with 5, 2 and 2 exons, respectively), the remaining genes are intronless. In comparison, MsToll1–5 have 7 or 8 exons, BmToll3_1–3 have 7, 5 and 8 exons, DmToll1, 3–5 have 2 or 4 exons, AgToll1A, 1B, 5A and 5B have 3 exons, and TcToll1–5 have 3 or 4 exons. Together, these observations reveal a dramatic evolutionary history of this ancient family of genes along the lineages of holometabolous insects.
3.3. Intracellular members of the Toll pathway and their regulation
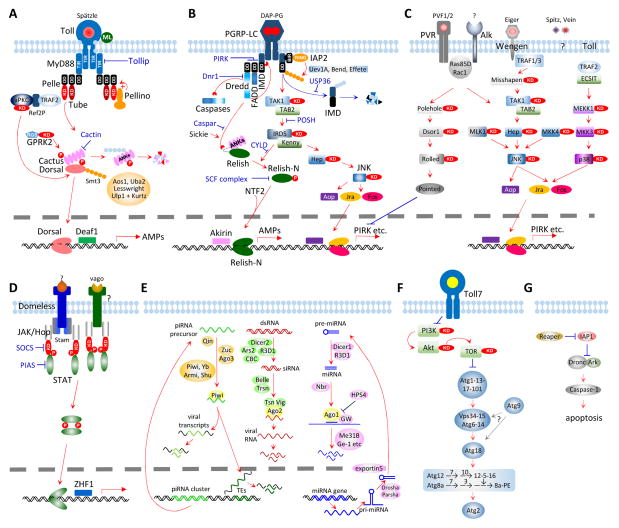
We have identified 1:1 orthologs for most of the intracellular pathway members and modulators known so far. These include MyD88, Tube, Pelle, Pellino, Cactus, G protein-coupled receptor kinase-2 (GPRK2), Tollip-1&2, Cactin, Aos, Uba2, Smt3, Lesswright, and deformed epidermal autoregulatory factor-1 (Deaf1) (Fig. 4A, Table S1) in Drosophila. In the current model, activated Toll receptor associates with its adaptor MyD88 via their TIR domains. MyD88, Tube and Pelle (a kinase-like protein) form a complex via their death domains to phosphorylate Cactus. Pellino, with a RING E3 ubiquitin ligase domain, may ubiquitinate Pelle to enhance the Toll signaling. Unlike its ortholog in the fruit fly, the C-terminal Ser/Thr protein kinase domain of M. sexta Tube is predicted to be active catalytically and thus, actively involved in the pathway activation. The phosphorylation of Cactus by Pelle and perhaps Tube, causes it to dissociate from Dorsal or Dif become polyubiquitinated and degraded by the proteasome. Dorsal and Dif appear to be the products of a lineage-specific gene duplication (data not shown). GPRK2 may interact with Cactus to enhance signaling. Atypical protein kinase C (aPKC), together with its partners Ref2P and TRAF2 (TNF-receptor-associated factor-2), may interact with Pelle and directly activate Dorsal/Dif (Avila et al., 2002). Free, active Dorsal/Dif translocates into the nucleus to activate target gene transcription along with Deaf1 and other transcription factors (e.g. U-shaped and Toll activation mediating protein, TAMP). This pathway is likely regulated at other steps. For instance, Tollips may associate with the Toll receptor and suppress the kinase activity of Pelle (Zhang and Ghosh, 2002). In D. melanogaster, Cactin may bind Cactus to block its function and cause embryonic ventralization (Lin et al., 2000). Conjugation of Dorsal/Dif by Smt3, a small ubiquitin-like modifier (SUMO), may potentiate function of Dorsal/Dif (Bhaskar et al., 2002). Aos1 and Uba2 may form a dimer which acts as an E1 SUMO-activating enzyme (Paddibhatla et al., 2010). The Lesswright homolog of Ubc9, an E2 SUMO-conjugating enzyme, negatively impacts the pathway (Chiu et al., 2005). The E3 SUMO ligase, Ulp1 peptidase and its helper Kurtz, reduces SUMO conjugation and response level of Dorsal/Dif-induced genes (Anjum et al., 2013).
Fig. 4.
Putative signaling pathways and regulators for antimicrobial immune responses in M. sexta. Panels A, Toll; B, Imd with JNK branch; C, MAPK-JNK-p38; D, JAK-STAT; E, pi- si- and mi-RNA pathways, F, autophagy; G, apoptosis. Panels A through G are described in the text.
Three Dorsal and two Dif variants are generated via alternative splicing (Table S1). The major Dorsal A is widely produced in tissues whereas B- and C-forms are preferentially expressed in fat body and head, respectively (Fig. 2A). Dif mRNA levels are lower compared to Dorsal. Like MyD88, Aos1 and Smt3, Manduca Tube, Pelle, Pellino, Lesswright, Uba2, Ref2Ps, aPKC-A, TRAF2, Cactus, Dorsal-A, ML2, cactin, Tollip-1 and 2 are widely expressed in all the tissues examined. However, mRNA levels of the latter genes (Tube through Tollip-2) increase considerably in fat body during the wandering stage and reach peaks in pupae at day 1. As well, most of these genes are induced by 24 h following an immune challenge (Table 1).
3.4. The Imd pathway, JNK branch, and their regulation
The Imd pathway, considered specific for Gram-negative bacteria, regulates the transcription of a set of immunity-related genes that overlaps with that controlled by the Toll pathway (Kleino and Silverman, 2014). This pathway is also branched to JNK and apoptosis (Fig. 4B). We have identified 1:1 orthologs for nearly all of the pathway components (Table S1) and, therefore, propose that the M. sexta Imd pathway is triggered by DAP-PG, a component of the cell wall in most Gram-negative bacteria as well as Gram-positive Bacillus and Listeria species. Since there is no PGRP-LE ortholog in the moth (Zhang et al., 2015), membrane-bound PGRP-LCa and LCb may work together to detect them. The longer splicing variant LCa contains two transmembrane domains, raising the possibility that it detects intracellular bacteria. Upon DAP-PG binding, a cytosolic portion of these variants may interact with the adaptor Imd and then FADD through their death domains. FADD recruits Dredd, the mammalian caspase-8 homolog, which cleaves Imd and Relish (Ertürk-Hasdemir et al., 2009) or a pro-caspase that leads to apoptosis. Cleaved Imd is susceptible to ubiquitination by IAP2 (inhibitor of apoptosis-2, an E3 ubiquitin ligase), Uev1A, Ubc13/Bendless and Ubc5/Effete (E2 ubiquitin-conjugating enzymes) (Paquette et al., 2010). Following ubiquitination, Imd likely recruits transforming growth factor β-activated kinase-1 (TAK1) and its binding protein TAB2 (Aggarwal, 2003). The dimer of TAB2 and TAK1 may then phosphorylate both Kenny/IKKβ and IRD5/IKKγ/NEMO in a complex, and JNK and Basket through MKK4 or MKK7/hemipterous (Hep) (Geuking et al., 2009). JNK may activate Aop and the AP-1 complex of Jra/Jun and Fos/Kayak to regulate downstream genes (e.g. PIRK). The IKK complex may phosphorylate the cleaved Relish to cause chain separation. While the C-terminal ankrin repeats and death-like domain are destined to be degraded, the N-terminal fragment (Relish-N), assisted by nuclear transport factor 2 (NTF2), could translocate into the nucleus and activate expression of immunity-related genes (e.g. antimicrobial peptides) via its Rel homology domain.
Additional regulatory mechanisms are known for the Imd pathway in Drosophila (Kleino and Silverman, 2014). PIRK interferes with the association of Imd, FADD, and Dredd (Kleino et al., 2008). Dnr1 (defense repressor-1) inhibits the caspase Dredd while Sickie and Caspar have opposite effects on Dredd-induced activation of Relish (Foley and O’Farrell, 2004). USP36 deubiquitinates Imd for its degradation and, thus, represses Imd signaling (Thevenon et al., 2009). Another deubiquitinase, CYLD (for Cylindromatosis), modulates the IKK complex to control Relish phosphorylation (Tsichritzis et al., 2007). POSH controls the complex of TAK1 and TAB2; an SCF complex of Skp1, Cullin and F-box protein regulates the phosphorylated Relish-N; Akirin and Relish-N co-regulate some target genes of the Imd pathway (Tsuda et al., 2005; Cardozo and Pagano, 2004; Bonnay et al., 2014).
Most genes in the putative Imd pathway are widely expressed in different tissues at various life stages (Fig. 2B). The mRNA levels of Imd, FADD, Dredd, Relish, and many other genes are considerably higher in midgut than in fat body. This is consistent with the finding that local immune response of epithelial cells is Imd pathway-dependent, as the Imd pathway is fast and can be activated within minutes following a challenge (Kleino and Silverman, 2014; Paquette et al., 2010). While mRNA levels of a few genes are higher at 24 h after the immune challenge, others are similar to or even lower than the control levels (Table 1). This contrasts drastically with most of the Toll pathway genes, whose induced expression in fat body and hemocytes lasts longer than 24 h. Consistent with their immune inducibility, most Imd pathway members are highly expressed in fat body from the pre-wandering larval stage to the early pupal stage (Fig. 2B). Their up-regulation in midgut is less pronounced and varies among the Imd pathway members during the same period, perhaps due to gut purging.
3.5. MAPK-JNK-p38 pathways
MAPK pathways are responsive to growth factors, cytokines and stress signals, and thereby regulate cell proliferation, differentiation, inflammation, and death. In Drosophila, components of these pathways activate MAPKs (Rolled, JNK and p38), down-regulate the Imd pathway, and stimulate hemocyte proliferation and lamellocyte formation (Fig. 4C) (Ragab et al., 2011; Dong et al., 2002; Lee and Ferrandon, 2011). We have identified homologs of two platelet-derived and vascular endothelial growth factors (PVFs), a PDGF/VEGF receptor (PVR), two small GTPases (Ras85D and Rac1), three kinases (Polehole, Dsor1 and Rolled), and a transcription factor (Pointed) that induces PIRK (poor Imd response upon knock-in) production. By interfering with Imd-FADD-Dredd association, PIRK, a small protein with no known domain structure, may inhibit Imd signaling. JNK may be activated through an Imd branch (Fig. 4B) and perhaps also by MLK1, MKK4, PVR or Alk (PVR and Alk are receptors with a Ser/Thr kinase domain). We have also found putative members of the cytokine-triggered MAPK pathway, namely Eiger, Wengen, TRAF1, and Misshapen that may recruit and sequentially activate TAK1-TAB2 dimer, MKK7/hemipterous, and JNK (Liu et al., 1999; Geuking et al., 2009). A protein called ECSIT (evolutionarily conserved intermediate in Toll pathways) is linked to the Toll receptor through TRAF2, and may activate a kinase cascade of MEKK1, MKK3, and p38 to induce the formation of the AP-1 complex (Kopp et al., 1999). In addition, Spitz and Vein may induce MAPK signaling in the presence of reactive oxygen species but their receptors are unknown in Manduca.
Certain members of the putative MAPK-JNK-p38 pathways (i.e. Eiger, Rac1, MASK, Rolled, JNK, p38, Aop, Jra, Fos) in M. sexta are transcriptionally activated in larval fat body or hemocytes after an immune challenge (Gunaratna and Jiang, 2013) (Table 1). In addition to these, PVF2, PVR, Wengen, Ras85D, Cdc42, Dsor1, Misshapen, MLK and Pointed show mRNA level increases in fat body from pre-wandering to early pupal stage (Fig. 2C). Transcript levels for most of these genes in midgut are similar to or higher than those in fat body. Levels of PVR, Rac1, Misshapen-B&C, p38B, Ras85D, Jra and Fos mRNAs reach peak levels during pupation. Expression of the pathway members in head, muscles, Malpighian tubules, testis, and ovary clearly indicates that roles of the MAPK-JNK-p38 pathways are beyond immunity.
3.6. JAK-STAT pathway and other antiviral mechanisms
3.6.1. JAK-STAT pathway and its regulation
The JAK-STAT pathway is involved in antiviral immune responses in insects (Dostert et al., 2005; Kingsolver et al., 2013). In Drosophila, an extracellular protein, Unpaired3, binds to Domeless, causes receptor dimerization, and recruits STAM and Hopscotch/JAK, which in turn phosphorylates itself and then STAT (Fig. 4D). We did not find an Unpaired3 ortholog in M. sexta or T. castaneum (Zou et al., 2007). However, the M. sexta ortholog of Vago may bind to an unknown receptor to activate JAK and STAT in a way similar to the unknown ligand of Domeless. After phosphorylation, the STAT dimer translocates into the nucleus to induce antiviral gene expression. SOCS (a JAK inhibitor) and PIAS (protein inhibitor of activated STAT) may down-regulate the pathway. Except for the ligand, orthologs of all the pathway components are present in M. sexta (Table S1). Domeless and SOCS mRNA levels increased 2.6-fold in larval fat body at 24 h after the injection of a mixture of bacteria (Gunaratna and Jiang, 2013) (Table 1). We also found that their mRNA levels became more abundant in fat body and midgut between wandering larval and early pupal stages (Fig. 2D). Similar increases were observed for other members of the predicted pathway, including JAK, STAT and STAM.
3.6.2. RNA interference (RNAi) pathways
RNA interference plays important roles in limiting viral infection in insects (Kingsolver et al., 2013; Fablet, 2014). There are three RNAi pathways (Fig. 4E): 1) small interfering RNAs (siRNAs) are generated from double-stranded RNA (dsRNA) of viruses and siRNAs degrade or inhibit viral RNA and thereby disrupt the viral infection cycle; 2) microRNAs (miRNAs) are produced from cellular gene transcripts and typically function to control the translation or half-life of their target transcripts, including those regulating immune responses; 3) Piwi-interacting RNAs (piRNAs) provide epigenetic control of transposable elements and viral transcripts in germ-line cells in order to prevent genome disruption. The siRNA pathway is mostly responsible for antiviral activity in insects. Viral RNAs may form double stranded RNAs due to innate secondary structures or via replication intermediate, and these dsRNAs are recognized and cleaved by Dicer-2 to generate siRNAs, which are then loaded into RNA-induced silencing complexes consisting of Argonaute-2 and other proteins. Unwinding of the duplex occurs along with guide strand selection. After target RNA recognition by the guide RNA, the targeted viral RNA is degraded by Argonaute-2. We have identified 28 putative pathway members suggesting that these pathways are functional in M. sexta (Fig. 4E, Table S1). Since R2D2 is not found in M. sexta, we suggest that R3D1 (an ortholog of Drosophila Loquacious) acts as a Dicer-1 partner in the miRNA pathway, as well as a Dicer-2 partner in the siRNA pathway. Unlike Drosophila, which has distinct Piwi and Aubergine genes, lepidopteran insects have a single PIWI-clade protein that we refer to as Aub/Piwi. Transcript levels for members of the siRNA pathway are relatively higher than those for either piRNA or miRNA pathways (Fig. 2E), consistent with its greater role in antiviral immunity (Kingsolver et al., 2013). Expression profiles of these pathways do not exhibit fat body- and midgut-specific up-regulation from wandering to early pupal stage, except for Dicer-2 and Argonaute-2. The Argonaute-2 mRNA levels increased moderately in induced fat body and hemocytes (Table 1). Although transcript abundances for piRNA pathway components vary, they are almost always higher in testis and ovary than the other tissues, consistent with their roles in the germline cells.
3.6.3. Autophagy
Autophagy is a cellular process in which dysfunctional or unnecessary cellular materials or components are selectively targeted, then separated from the cytoplasm in double membrane vesicles (autophagosomes), and ultimately degraded by lysosomes (Mulakkal et al., 2014). Some pathogens may also be targeted to autophagosomes. Autophagy recycles the cellular materials and maintains cellular homeostasis under a variety of conditions. It is implicated in cellular responses to stress by nutrient-restriction, developmental changes involving tissue reorganization during metamorphosis, and certain pathological processes. The signaling of autophagy is mediated through the phosphoinositide 3-kinase (PI3K)-Akt pathway (Fig. 4F), which phosphorylates TOR to suppress autophagy. Autophagy itself involves about 20 components conserved throughout eukaryotes from yeast to mammals. In Drosophila, autophagy is induced upon infection by some viruses, intracellular bacteria (e.g. Listeria monocytogenes), and other pathogens (Yano et al., 2008; Kingsolver et al., 2013), suggesting that in addition to other cellular functions, it may also serve as an ancient cellular immune response. We have identified orthologs of all known autophagy pathway members (Fig. 4F, Table S1) and examined their expression profiles (Fig. 2F). As components of a ubiquitination complex, Atg3, 4, 5, 7, 8, 10, 12 and 16 are highly expressed in all the tissue samples used for RNA-Seq analyses. The mRNA levels of these autophagy pathway genes are generally higher in midgut than in fat body, testis and ovary. Since there is no major increase in mRNA levels in the pupal stage, autophagy may be partly supported by pre-existing proteins. Transcript levels of Atg2 through 6, 8, 9, and 16 are up-regulated in fat body and midgut from wandering larvae and young pupae, and decrease in the later stages. These changes may correlate with cellular reorganization in cells undergoing metamorphosis. In contrast, the PI3K, Akt, TOR, Vps34, Atg1, 7, 10, 12, 13, 17, 18, and 101 mRNA levels remain high from pupal to adult stage. Based on our current data (Fig. 2F), expression of autophagy-related genes appears to be a development-regulated process. There is no strong correlation with their immune inducibility, perhaps due to the fact we did not use viruses or intracellular bacteria to challenge the larvae.
3.6.4. Apoptosis
Apoptosis, the best characterized mechanism of programmed cell death, is a part of normal developmental processes such as tissue modeling and homeostasis, but apoptosis can also participate in pathological processes including cancer and defense against pathogens (Opferman and Korsmeyer, 2003). In Drosophila, the initiator caspase Dronc and an adaptor protein (Ark) form a large protein complex (apoptosome) in response to intrinsic signals (Hay and Guo, 2006). It is not clear how the other Drosophila initiator caspases, Dredd and Strica, are activated. Once Dronc is activated, it cleaves and activates effector caspases such as Drice and Dcp1 to cleave other protein substrates that lead to the downstream events of programmed cell death. Negative regulators of caspases (e.g. IAPs, Dnr1) control the pathway by inhibiting the activation of initiator caspases through either direct binding or by ubiquitination-induced degradation (Orme and Meier, 2009). Likewise, IAP antagonists (e.g. Reaper, Hid, Grim and Sickle) inactivate IAPs and, thus induce apoptosis. We have identified 12 members of the core apoptosis pathway in M. sexta, including Reaper, IAP1, IAP2, Deterin/IAP3, Dnr1, Ark, Dredd/caspase-6, Dronc/caspase-5, caspase-1, -3, and -4 (Fig. 4G) (Courtiade et al., 2011). While Dnr1, Dredd, and IAP2 are likely involved in the balance between the Imd and apoptosis pathways, the other proteins may be devoted to programmed cell death. Reaper, an indirect pathway activator, is produced in the embryo, pupal fat body and midgut, as well as adult head, Malpighian tubules, testis and ovary (Fig. 2G), suggesting a possible role of apoptosis in tissue remodeling. The IAP3 mRNA, which is related to Survivin, a mitotic spindle-associated protein, is strikingly high and may perhaps regulate embryonic development. With a similar expression profile, IAP1 may block caspase-3 and -4 in cells of midgut, fat body, and other tissues. The high transcript abundances in midgut of feeding and wandering larvae, pupae and adults could indicate that the tissue is poised to undergo or carefully regulate active programmed cell death and regeneration. In addition, the caspase-1 and IAP1 mRNA peaks in fat body and midgut from wandering to early pupal stage correlate with their immune inducibility (Table 1).
3.7. Concluding remarks
Our search of the M. sexta genome has yielded 187 genes encoding 198 putative members of the immunity-related signal transduction pathways, namely Toll, Imd, MAPK-JNK-p38, JAK-STAT, piRNA, siRNA, miRNA, autophagy and apoptosis. Analysis of the expression profiles reveals differences among the proposed pathways (e.g. Toll, Imd, and MAPK-JNK-p38) and among some of the components (e.g. Spätzles, Tolls). These results suggest that the intracellular signaling system is functional in this undomesticated insect, and thus pave the way for understanding and potentially modulating similar pathways in pest lepidopteran species. The proposed signaling network needs experimental validation using biochemical, molecular and cellular biological methods.
Supplementary Material
Highlights.
Identified 186 genes for intracellular immune signaling and responses
Proposed based on 1:1 orthology nine pathways of the cellular processes
Examined the expression profiles of these genes in different tissues and stages
Established an integrated knowledge base for future works on insect immune signaling
Acknowledgments
This work was supported by NIH grants GM58634 (to H. Jiang) and a DARPA/NSF (IOS-1354421) grants (to G. Blissard). Computation for this project was performed at OSU High Performance Computing Center at Oklahoma State University supported in part through NSF grant OCI-1126330. This work was approved for publication by the Director of Oklahoma Agricultural Experimental Station, and supported in part under project OKLO2450.
Abbreviations
- Atg
autophagy-related protein
- aPKC
atypical protein kinase C
- CF
control fat body
- CH
control hemocytes
- IF
induced fat body
- IH
induced hemocytes
- Deaf
deformed epidermal autoregulatory factor
- Dnr1
defense repressor-1
- Dsp1
Dorsal switch protein-1
- ECSIT
evolutionarily conserved intermediate in Toll pathways
- FPKM
fragments per kilobase of exon per million fragments mapped
- GPRK
G protein-coupled receptor kinase
- IAP
inhibitor of apoptosis
- Imd
immunodeficiency
- IKK
IκB kinase
- JNK
Jun N-terminal kinase
- Jra
Jun-related antigen
- MAPK
mitogen-activated protein kinase
- MASK
multiple ankyrin repeats single KH domain
- ML
MD2-like
- MLK
mixed-linage kinase
- NFκB and IκB
nuclear factor-κB and its inhibitor
- NTF
nuclear transport factor
- PIAS
protein inhibitor of activated STAT
- PIRK
poor Imd response upon knock-in
- PVF
platelet-derived and vascular endothelial growth factor
- PVR
PDGF/VEGF receptor
- STAT
signal transducer and activator of transcription
- PG and PGRP
peptidoglycan and peptidoglycan recognition protein
- SUMO
small ubiquitin-like modifier
- TAK
transforming growth factor β activated kinase
- TAMP
Toll activation mediating protein
- TIR
Toll/interleukin-1 receptor
- TRAF
tumor necrosis factor receptor-associated factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- Altincicek B, Vilcinskas A. Identification of a lepidopteran matrix metalloproteinase with dual roles in metamorphosis and innate immunity. Dev Comp Immunol. 2008;32:400–409. doi: 10.1016/j.dci.2007.08.001. [DOI] [PubMed] [Google Scholar]
- An C, Jiang H, Kanost MR. Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta. FEBS J. 2010;277:148–162. doi: 10.1111/j.1742-4658.2009.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum SG, Xu W, Nikkholgh N, Basu S, Nie Y. Regulation of Toll signaling and inflammation by β-arrestin and the SUMO protease Ulp1. Genetics. 2013;195:1307–1317. doi: 10.1534/genetics.113.157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao J-Q, Ling E, Rao XJ, Yu XQ. A novel ML protein from Manduca sexta may function as a key accessory protein for lipopolysaccharide signaling. Mol Immunol. 2008a;45:2772–2781. doi: 10.1016/j.molimm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao J-Q, Ling E, Yu X-Q. A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol Immunol. 2008b;45:543–552. doi: 10.1016/j.molimm.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Avila A, Silverman N, Diaz-Meco MT, Moscat J. The Drosophila atypical protein kinase C-ref(2)p complex constitutes a conserved module for signaling in the toll pathway. Mol Cell Biol. 2002;22:8787–8795. doi: 10.1128/MCB.22.24.8787-8795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard SL, Miller DL, Ganetzky B. Retrograde neurotrophin signaling through Tollo regulates synaptic growth in Drosophila. J Cell Biol. 2014;204:1157–1172. doi: 10.1083/jcb.201308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Smith M, Courey AJ. Conjugation of Smt3 to Dorsal may potentiate the Drosophila immune response. Mol Cell Biol. 2002;22:492–504. doi: 10.1128/MCB.22.2.492-504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnay F, Nguyen XH, Cohen-Berros E, Troxler L, Batsche E, Camonis J, Takeuchi O, Reichhart JM, Matt N. Akirin specifies NF-κB selectivity of Drosophila innate immune response via chromatin remodeling. EMBO J. 2014;33:2349–2362. doi: 10.15252/embj.201488456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster - from microbial recognition to whole-organism physiology. Nat Rev Immunol. 2014;14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, He Y, Hu Y, Zhang X, Wang Y, Zou Z, Chen Y, Blissard G, Kanost MR, Jiang H. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem Mol Biol. 2015 doi: 10.1016/j.ibmb.2014.10.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jiang H. Integrated modeling of protein-coding genes in the Manduca sexta genome using RNA-Seq data from the biochemical model insect. Insect Biochem Mol Biol. 2015 doi: 10.1016/j.ibmb.2015.01.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chen J, Xie C, Tian L, Hong L, Wu X, Han J. Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc Natl Acad Sci USA. 2010;107:20774–20779. doi: 10.1073/pnas.1009223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H, Ring BC, Sorrentino RP, Kalamarz M, Garza D, Govind S. dUbc9 negatively regulates the Toll-NF-kappa B pathways in larval hematopoiesis and drosomycin activation in Drosophila. Dev Biol. 2005;288:60–72. doi: 10.1016/j.ydbio.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Courtiade J, Pauchet Y, Vogel H, Heckel DG. A comprehensive characterization of the caspase gene family in insects from the order Lepidoptera. BMC Genomics. 2011;12:357. doi: 10.1186/1471-2164-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Ann Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler J-L. The JAK-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- Ertürk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stöven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, Kanost M, Thompson GJ, Zou Z, Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fablet M. Host control of insect endogenous retroviruses: small RNA silencing and immune response. Viruses. 2014;6:4447–4464. doi: 10.3390/v6114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, O’Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:e203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, de Vos M, Duncan EJ, Evans JD, Gabaldón T, Ghanim M, Heddi A, Kaloshian I, Latorre A, Moya A, Nakabachi A, Parker BJ, Pérez-Brocal V, Pignatelli M, Rahbé Y, Ramsey JS, Spragg CJ, Tamames J, Tamarit D, Tamborindeguy C, Vincent-Monegat C, Vilcinskas A. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking P, Narasimamurthy R, Lemaitre B, Basler K, Leulier F. A non-redundant role for Drosophila MKK4 and hemipterous/MKK7 in TAK1-mediated activation of JNK. PLoS One. 2009;4:e7709. doi: 10.1371/journal.pone.0007709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JP, Kanost MR, Trenczek T. Biological mediators of insect immunity. Ann Rev Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- Gunaratna RT, Jiang H. A comprehensive analysis of the Manduca sexta immunotranscriptome. Dev Comp Immunol. 2013;39:388–398. doi: 10.1016/j.dci.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Ann Rev Cell Dev Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- He Y, Cao X, Li K, Hu Y, Chen Y, Blissard G, Kanost MR, Jiang H. A genome-wide analysis of antimicrobial effector genes and their transcription patterns in Manduca sexta. Insect Biochem Mol Biol. 2015 doi: 10.1016/j.ibmb.2015.01.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler JL, Hoffmann JA. Toll receptors in innate immunity. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Yano T, Aggarwal K, Lim J-H, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh B-H. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Kingsolver MB, Huang Z, Hardy RW. Insect antiviral innate immunity: pathways, effectors, and connections. J Mol Biol. 2013;425:4921–4936. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- Kleino A, Myllymäki H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, Hultmark D, Valanne S, Rämet M. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180:5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- Kleino A, Silverman N. The Drosophila Imd pathway in the activation of the humoral immune response. Dev Comp Immunol. 2014;42:25–35. doi: 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp E, Medzhitov R, Carothers J, Xiao C. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen NP, Scoble MJ, Karsholt O. Lepidoptera phylogeny and systematics: the state of inventorying moth and butterfly diversity. Zootaxa. 2007;1668:699–747. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KZ, Ferrandon D. Negative regulation of immune responses on the fly. EMBO J. 2011;30:988–990. doi: 10.1038/emboj.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey C. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Huang LH, Steward R. Cactin, a conserved protein that interacts with the Drosophila IκB protein cactus and modulates its function. Mech Dev. 2000;94:57–65. doi: 10.1016/s0925-4773(00)00314-2. [DOI] [PubMed] [Google Scholar]
- Liu H, Su YC, Becker E, Treisman J, Skolnik EY. A Drosophila TNF-receptor-associated factor (TRAF) binds the Ste20 kinase Misshapen and activates Jun kinase. Curr Biol. 1999;9:101–104. doi: 10.1016/s0960-9822(99)80023-2. [DOI] [PubMed] [Google Scholar]
- McIlroy G, Foldi I, Aurikko J, Wentzell JS, Lim MA, Fenton JC, Gay NJ, Hidalgo A. Toll-6 and Toll-7 function as neurotrophin receptors in the Drosophila melanogaster CNS. Nat Neurosci. 2013;16:1248–1256. doi: 10.1038/nn.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Håkansson J. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc Natl Acad Sci USA. 2005;102:6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulakkal NC, Nagy P, Takats S, Tusco R, Juhász G, Nezis IP. Autophagy in Drosophila: from historical studies to current knowledge. BioMed Res Int. 2014;2014:273473. doi: 10.1155/2014/273473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36:658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Korsmeyer SJ. Apoptosis in the development and maintenance of the immune system. Nat Immunol. 2003;4:410–415. doi: 10.1038/ni0503-410. [DOI] [PubMed] [Google Scholar]
- Orme M, Meier P. Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis. 2009;14:950–960. doi: 10.1007/s10495-009-0358-2. [DOI] [PubMed] [Google Scholar]
- Paddibhatla I, Lee MJ, Kalamarz ME, Ferrarese R, Govind S. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS Pathog. 2010;6:e1001234. doi: 10.1371/journal.ppat.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Ertürk-Hasdemir D, Reichhart JM, Meier P, Silverman N. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-κB signaling. Mol. Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Ragab A, Buechling T, Gesellchen V, Spirohn K, Boettcher AL, Boutros M. Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J. 2011;30:1123–1136. doi: 10.1038/emboj.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Rao X-J, Cao X, He Y, Hu Y, Zhang X, Chen Y, Blissard G, Kanost MR, Yu X-Q, Jiang H. Structural features, evolutionary relationships, and transcriptional regulation of C-type lectin-domain proteins in Manduca sexta. Insect Biochem Mol Biol. 2015 doi: 10.1016/j.ibmb.2014.12.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X-J, Xu X-X, Yu X-Q. Manduca sexta moricin promoter elements can increase promoter activities of Drosophila melanogaster antimicrobial peptide genes. Insect Biochem Mol Biol. 2011;41:982–992. doi: 10.1016/j.ibmb.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino RP, Melk JP, Govind S. Genetic analysis of contributions of dorsal group and JAK-Stat92E pathway genes to larval hemocyte concentration and the egg encapsulation response in Drosophila. Genetics. 2004;166:1343–1356. doi: 10.1534/genetics.166.3.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand MR. The insect cellular immune response. Insect Sci. 2008;15:1–15. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evo. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem Mol Biol. 2008;38:1087–1110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, Benaud C, Baudier J, Taillebourg E, Fauvarque MO. The Drosophila ubiquitin specific protease dUSP36/Scny targets Imd to prevent constitutive immune signaling. Cell Host Microbe. 2009;6:309–320. doi: 10.1016/j.chom.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Tsichritzis T, Gaentzsch PC, Kosmidis S, Brown AE, Skoulakis EM, Ligoxygakis P, Mosialos G. A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development. 2007;134:2605–2614. doi: 10.1242/dev.02859. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Langmann C, Harden N, Aigaki T. The RING-finger scaffold protein Plenty of SH3s targets TAK1 to control immunity signaling in Drosophila. EMBO Rep. 2005;6:1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J Biol Chem. 2002;277:7059–7065. doi: 10.1074/jbc.M109537200. [DOI] [PubMed] [Google Scholar]
- Zhang S, Gunaratna R, Zhang X, Najar F, Wang Y, Roe B, Jiang H. Pyro sequencing-based expression profiling and identification of differentially regulated genes from Manduca sexta, a lepidopteran model insect. Insect Biochem Mol Biol. 2011;41:733–746. doi: 10.1016/j.ibmb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, He Y, Cao X, Gunaratna RT, Chen Y, Blissard GW, Kanost MR, Jiang H. Phylogenetic analysis and expression profiling of the pattern recognition receptors: insights into molecular recognition of invading pathogens in Manduca sexta. Insect Biochem Mol Biol. 2015 doi: 10.1016/j.ibmb.2015.02.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Evans J, Lu Z, Zhao P, Williams M, Sumathipara N, Hetru C, Hultmark D, Jiang H. Comparative genome analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.