Abstract
Objective
To test the hypothesis that maternal complications significantly affect gut colonization patterns in very low birth weight infants.
Methods
49 serial stool samples were obtained weekly from 9 extremely premature infants enrolled in a prospective longitudinal study. Sequencing of the bacterial 16S rRNA gene from stool samples was performed to approximate the intestinal microbiome. Linear mixed effects models were used to evaluate relationships between perinatal complications and intestinal microbiome development.
Results
Subjects with prenatal exposure to a non-sterile intrauterine environment, i.e. PPPROM and chorioamnionitis exposure, were found to have a relatively higher abundance of potentially pathogenic bacteria in the stool across all time points compared to subjects without those exposures, irrespective of exposure to postnatal antibiotics. Compared with those delivered by Caesarean section, vaginally delivered subjects were found to have significantly lower diversity of stool microbiota across all time points, with lower abundance of many genera, most in the family Enterobacteriaceae.
Conclusions
We identified persistently increased potential pathogen abundance in the developing stool microbiota of subjects exposed to a non-sterile uterine environment. Maternal complications appear to significantly influence the diversity and bacterial composition of the stool microbiota of premature infants, with findings persisting over time.
Keywords: obstetrical complications, obstetrical interventions, prematurity, microbiome, PPPROM, chorioamnionitis, antibiotics
INTRODUCTION
The developing intestinal microbiome, in its symbiotic relationship with the neonatal host, fills critical physiological roles and is an important modulator of immune maturation and metabolic function [1–3]). The nearly blank slate of the intestinal tract beginning at birth has the potential to acquire microbial communities that influence future risk of acute and chronic diseases [2,4]). Recent work comparing the developing gut microbiome among distinct human populations found that the intestinal microbiome exhibits the greatest variation during the period from birth to 3 years, likely due to the influence of environmental factors [5]), and becomes stable by 3 years of age. In healthy neonatal populations, bacterial colonization of the gut begins during the process of delivery [6–8]) and is primarily determined by feeding method, hospitalization, and antibiotic exposure [7–9].
A healthy human gestation involves a primarily sterile environment [10] and birth presents the infant’s first significant encounter with bacteria that rapidly populate the intestines and influence the composition of the gut microbiome [11,12]. Microbial colonization patterns in premature infants requiring intensive care differ from their healthy full term counterparts [8,13]. Premature infants are more often colonized with pathogenic organisms and lack microbial diversity, both of which pose risk for short term complications including sepsis, which is often caused by the predominant intestinal bacteria [4,14,15]. However, less is known about the developing gut microbiome in premature infants who are exposed to a non-sterile intrauterine environment prior to birth. The purpose of this study was to investigate the developing intestinal microbiome in very low birth weight (VLBW) infants using culture-independent deep sequencing of the 16S rRNA gene of bacteria present in stool. We observed the stool microbiota development in VLBW infants at several time points over hospitalization to investigate our hypothesis that maternal complications significantly affect neonatal gut colonization patterns. Our findings point to potential mechanisms of influence of maternal complications on VLBW infant health outcomes, and highlight opportunities to intervene with breast milk feedings, probiotics, and altered antibiotic regimens.
METHODS
We obtained Institutional Review Board approval for this study in April 2011 (CPHS 21761) and renewal in April 2012 from the Center for Protection of Human Subjects at Dartmouth. Parents provided written, informed consent for their infants’ participation. Inclusion criteria, clinical data and sample collection, sample processing, massively parallel sequencing and data analysis are described in SI Methods.
RESULTS
Patient cohort
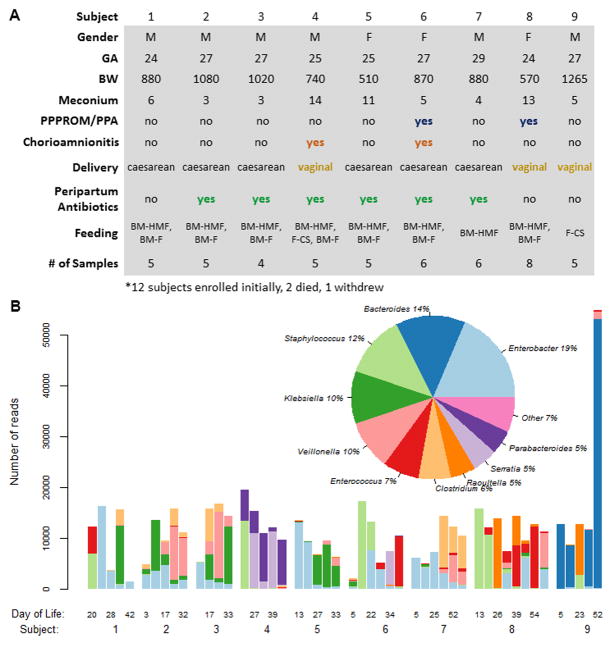
Twelve VLBW infants were enrolled at the time of birth. Two of the 12 died within the first 72 hours of life, and one subject withdrew. The 9 subjects included in the analysis were delivered at 24–29 weeks gestation with birth weights ranging from 510 to 1265 grams (Figure 1a). Three subjects were delivered vaginally and 6 subjects were exposed to peripartum antibiotics. Maternal complications included PPPROM (2 subjects) and chorioamnionitis (2 subjects). Subjects 1–8 were fed breast milk (maternal or donor) mixed with human milk fortifier or formula. Subject 9 was fed only formula with caloric supplementation. Number of stool samples per subject ranged from 4–8. 6 of 9 subjects were previously studied in Madan et al, 2012 [15]. Subjects 2 and 3 are dizygotic twins.
Figure 1.
A) Demographics and clinical variables. Gestational age (GA), birth weight (BW). Meconium represents the day of life at which first stool was passed. PPPROM (prolonged preterm premature rupture of membranes) in conjunction with prolonged prenatal antibiotics (PPA). Feedings include breast milk (BM) and/or formula (F) with or without the addition of human milk fortifier (HMF) or caloric supplementation (CS). Number of stool samples (# of Samples) collected from each subject.
B) Dominant genera among serial samples separated by subject, representing 93% of all raw bacterial reads. The y-axis on the bar graph represents the number of raw reads of the top 10 bacterial genera in a sample, while the x axis shows serial samples in each subject, labeled with the day of life when the sample was collected.
Antibiotic exposure: pre- and postnatal
The mothers of two subjects received prolonged prenatal antibiotics (Figure 1a). Maternal antibiotic exposure included oral antibiotics and broad-spectrum intravenous antibiotics (cephalexin, erythromycin, ampicillin and gentamicin) and exposure occurred 2 hours to 7 days prior to delivery in 6 of 9 subjects. All infant subjects received empiric postnatal antibiotics (ampicillin and gentamicin) at time of birth for a range of 2–8 days. Some subjects received additional postnatal antibiotics (vancomycin, cefotaxime, nafcillin, piperacillin/tazobactam, clindamycin, macrodantin).
Dominant Genera
Ten dominant bacterial genera were identified that represented 93% of the total reads, including Enterobacter (19% of all raw reads), Bacteroides (14%), and Staphylococcus (12%), and others (Figure 1b). Enterobacter and Staphylococcus, which are known to predominate early in VLBW infants [16], were observed as the dominant genera in most early stool samples and are among the most abundantly represented bacteria across all subjects. Bacteroides, an obligate anaerobe and prevalent adult gut commensal with variable colonization of the early infant gut [17,18], was seen in very high numbers in Subject 9 (Figure 1b). Higher numbers of members of the genera Bacteroides in the infant gut have been associated with formula feeding [19,20], and subject 9 was exclusively formula fed (Figure 1a). Bifidobacterium and Escherichia, which normally have an early predominance in term neonates [8,21], were present in low numbers in the cohort, 0.01% and 0.17% of total reads respectively, as is seen in extremely premature infants [14,16,21].
Changes in Bacterial Abundance Over Time
Linear mixed effects modeling revealed statistically significant changes in bacterial abundance over time. Staphylococcus, a genus that includes important pathogens in preterm infants [15], showed the most significant decrease over time (p=0.006), while Veillonella, the genus of a common gut anaerobe in healthy term neonates [8,9], demonstrated the most significant increase over time (p<0.001) (Supplementary Figure 1). Some facultative anaerobes such as those of the genera Escherichia (p=0.030) and Shigella (p=0.027) decreased over time while others such as Streptococcus (p<0.001) and Enterococcus (p=0.004) increased. Facultative anaerobes are generally early colonizers of the neonatal gut, later replaced by obligate anaerobes such as Bacteroides and Clostridia as the gut environment becomes more anaerobic at around 1–2 weeks of life [8,17,22].
Neonatal microbial diversity and perinatal exposures
When comparing microbial diversity across all time points relative to perinatal exposures, we observed that infants delivered by Caesarean section had greater bacterial diversity when compared with those delivered vaginally (p=0.021) (Figure 2a). A significant difference was seen in the microbial diversity trajectories between subjects whose mothers had PPPROM and those whose mothers did not (p=0.032), with the former having a significantly lower initial microbial diversity (p=0.045), and experiencing a strong increase in diversity over time while the latter showed a weak decrease (Supplementary Figure 2).
Figure 2.
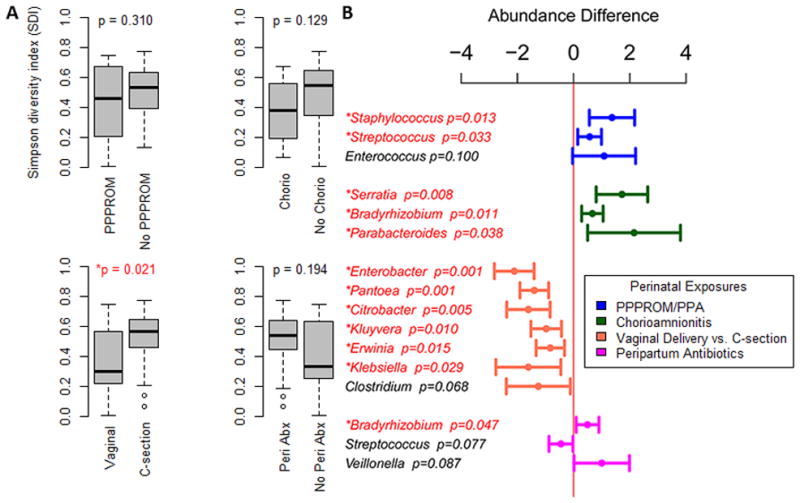
A) Microbial diversity relative to perinatal exposures. X axis depicts box plots of diversity based upon perinatal exposures. PPPROM (prolonged premature preterm rupture of membranes); Chorio (chorioamnionitis); Caesarean section; Peri Abx (Perinatal antibiotics). Y axis depicts Simpson Diversity Index (SDI) 0–1, 0=no diversity, 1=infinite diversity. B) Microbial abundance differences across all time points seen in subjects with and without a perinatal exposure. Microbial genera were evaluated for differences in abundance between groups of subjects with and without exposure to a specific peripartum exposure. A bar to the left of the midline represents a relatively lower abundance of the microbes, and a bar to the right of the midline represents a relatively greater abundance.
Perinatal influences on the developing microbial communities
Nonsterile intrauterine environment results in pathogen predominant infant stool microbiota
Subjects whose mothers had PPPROM (all of whom had prolonged prenatal antibiotics) had significantly greater Staphylococcus (p=0.013) and Streptococcus (p=0.033) abundance across all time points, compared with those who did not (Figure 2b). They also demonstrated a relatively faster increase in abundance over time of normal neonatal gut flora member Enterobacter (p=0.009) [9] and a slower increase in abundance of the genus Clostridium (p=0.020), normally a significant component of the gut flora in both neonates and adults (Supplementary Table 1) [20,23]. Subjects whose mothers had chorioamnionitis demonstrated both a significantly greater abundance across all time points and a faster increase of potential pathogens such as Serratia (p=0.008, p<0.001) [24] and Parabacteroides (p=0.038, p=0.004) (Figure 2b, Supplementary Table 1) [25]. Interestingly, subjects with chorioamnionitis exposure had a greater abundance of the genera Bradyrhizobium, considered a part of the core breast milk microbiome [26]. Additionally, the microbial diversity of subjects exposed to a non-sterile environment (PPPROM and/or chorioamnionitis) was significantly lower across all time points compared to those without the exposures (p=0.045; adjusted for antibiotic administration p=0.090) (Supplementary Figure 3).
Delivery mode affects microbial abundances
Vaginally delivered subjects were found to have a significantly lower relative abundance of many genera, most belonging to the family Enterobacteriaeceae, as compared to subjects delivered by Caesarean section (Figure 2a).
Effect of Postnatal Antibiotics on the Developing Stool Microbiota
There was a significant difference between the microbial diversity of the subjects with the most minimal postnatal antibiotic exposure (total of 2 days of antibiotic exposure) and all other subjects, with the minimal antibiotics group demonstrating a much higher overall diversity across all time points (p<0.001, Supplementary Figure 4). Microbial diversity exhibited a downward linear trend with increasing number of days on antibiotic treatment (p=0.191, Supplementary Figure 5a). After administration of antibiotics ceased, an upward linear trend (p=0.073) in microbial diversity over time was demonstrated (Supplementary Figure 5b). There was a significant difference in overall diversity between samples collected on day of antibiotic administration compared to those collected on days when there was no administration of antibiotics (p=0.014) (Supplementary Figure 6). The potentially confounding effect of antibiotic administration was analyzed and while antibiotics contributed to a decrease in microbial diversity irrespective of perinatal exposure (Supplementary Tables 2a and 2b), the effects of perinatal exposures on the infant stool microbiota were generally found to be even more significant after adjustment for postnatal antibiotic exposure, both with regards to microbial diversity and abundance (Supplementary Tables 2a, 2b, 2c and 2d). Stool samples collected on day of antibiotic administration were found to have greater abundances of the genera Ralstonia and Propionibacterium (p=0.009, p=0.026), which are considered be part of the core breast milk microbiome [26]. These samples also had a marginally significant greater abundance of the genera Bradyrhizobium, Serratia and Bacteroides (p=0.051, p=0.077, p=0.088, respectively). The genera Bradyrhizobium and Serratia are a part of the core breast milk microbiome [26], while Bacteroides, as well as Serratia, are potential pathogens [24,25].
Bacterial Clustering
Hierarchical clustering highlights the relationship between groups of bacteria at the genus level from individual patient samples (Figure 3, Supplementary Figure 7). Three discrete clusters emerged (Figure 3; A, B, and C), and each cluster was associated with different clinical characteristics. The first cluster, (A), represents samples from patients with exposure to PPPROM and/or chorioamnionitis and a patient who was exclusively formula fed from birth and who exhibited very high Bacteroides predominance (Subject 9). Additionally, most samples from Group A were from subjects delivered vaginally. Microbial diversity levels (SDI scores) were significantly lower in Cluster A than Cluster B (p≪0.001). There was an enrichment of the potential pathogens Staphylococcus and Serratia as well as an increase in Bacteroides and Parabacteroides in Cluster A. Cluster B represents samples from subjects almost all without PPPROM or chorioamnionitis exposure, all delivered by Caesarean section, with almost all having had exposure to peripartum antibiotics. Unlike Cluster A, Cluster B samples showed high microbial diversity levels, as well as an enrichment of the commensals Klebsiella, Clostridium, Veillonella and Enterobacter, genera thought to be important normal commensals in neonatal life [6–8]. Cluster C had an average SDI of 0.53, intermediary between those of Clusters A (SDI 0.30) and B (SDI 0.63), and contains mostly samples from subjects delivered operatively and with exposure to maternal antibiotic treatment, as well as some samples from subjects exposed to a non-sterile uterine environment. There was a statistically significant difference in SDI scores between Clusters A and C (p=0.001), but not B and C (p=0.062). Clustering analysis shows that Clusters B and C together make up a significant cluster (p=0.047).
Figure 3. Hierarchical clustering of patient samples and bacterial genera.
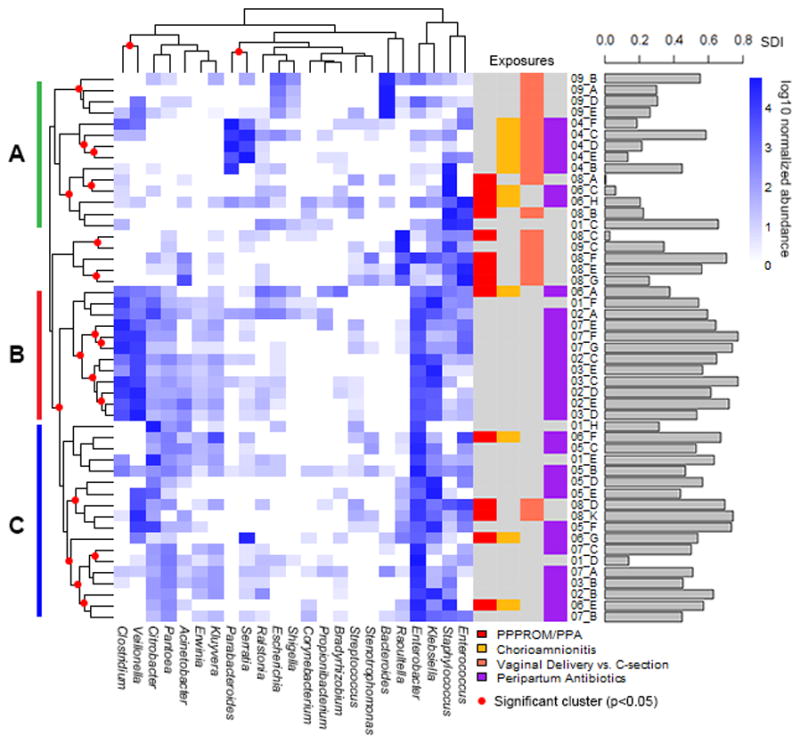
Dark blue squares represent an enrichment of a bacterial genera, while a white square shows a depletion of the bacterial genera. On the right are color-coded bars depicting perinatal exposures, as well as a bar graph showing the microbial diversity (SDI) of each sample. On the left and top, clades with red circles indicate significant clusters (p<0.05). On the left are bars representing discrete clusters, which are defined in the text.
DISCUSSION
We have described our detailed longitudinal investigation of the effects of prenatal and perinatal maternal complications on the developing premature infant gut microbiome and have identified important relationships between a non-sterile intrauterine environment and microbial acquisition patterns that persist throughout hospitalization despite postnatal exposures. Maternal complications leading to preterm delivery frequently involve exposure of the fetus to bacteria secondary to preterm labor, premature rupture of membranes or maternal infection [27]. In our small cohort with frequent longitudinal sampling, we identified that fetal exposure to PPPROM or chorioamnionitis increases the abundance of potential pathogens in the stool, a pattern which persists throughout hospitalization, irrespective of postnatal antibiotic exposure. Clustering analyses revealed that subjects who were exposed to a non-sterile uterine environment shared similar communities of bacterial genera.
Limitations of the study include a small number of subjects and a lack of sample replicates, which make it impossible to determine the variability in microbiota levels within a given stool sample. Though we do not know what the normal variation in bacterial reads is in any one stool sample, we did identify a high degree of inter-subject and intra-subject variability in bacterial reads (Figure 1b). In addition to measuring bacterial abundance, we used Simpson’s Diversity Index to measure the microbial diversity for each sample, a measure of both the number and relative abundance of genera in a sample, to better compare samples exhibiting high degrees of inter- and intra-subject variability. While there appears to be significant variability in the bacterial reads even among the samples from subjects exposed to PPPROM and/or chorioamnionitis (Subjects 4, 6, and 8, Figure 1b), we were able to identify differences between those exposed to a non-sterile environment and those without the exposure by performing additional analyses, such as calculating microbial diversity and bacterial clustering. Additionally, those exposed to a non-sterile environment all appear to have high counts of Staphylococcus in their early stool samples, potentially reflecting similar effects of the lack of sterility in the uterine environment (Figure 1b).
In full term neonates, delivery mode is an important factor shaping the initial neonatal microbiome [7,8,11]. Term infants delivered by Caesarean section have primary assemblages of bacteria from mother’s skin and the hospital environment [11,12]. In contrast, infants delivered vaginally have initial colonization from the vaginal microbiome which is less diverse [28], creating patterns in the infant microbiome which persist beyond weaning [19]. Premature infants have been studied, but the impact of delivery mode and neonatal microbial acquisition has not been outlined previously [4,13–16,21]. In our study, we demonstrate that mode of delivery affected gut diversity across all time points. Vaginal delivery appeared to lead to decreased diversity, a finding which mirrors studies in term populations [11,12]. Despite the hospital environment, significant antibiotic exposure, and dietary challenges in this preterm cohort, the mode of delivery had a lasting impact on overall diversity of the stool microbiota.
Our results in a premature population indicated that peripartum administration of antibiotics results in a significantly greater abundance of the genera Bradyrhizobium, a part of the core breast milk microbiome, and a marginally greater abundance across all time points of Veillonella, a bacterium thought to be health-promoting. The genus Streptococcus was marginally less abundant, potentially owing to treatment for Group B streptococcus (GBS) with penicillin [16]. In a separate study that investigated the development of the term neonatal intestinal microbiome in relationship to maternal Caesarean section with its associated maternal antibiotic exposure, the gut microbiota of infants exposed to perinatal antibiotics mirrored the aberrant microflora patterns of infants treated with 4 days of postpartum broad-spectrum antibiotics [29]. The infants exposed to these perinatal antibiotics demonstrated a paucity of Bifidobacterium species and a bloom of Enterobacteriaceae, similar to the colonization patterns in our group of premature infants, particularly those delivered by Caesarean section. These few studies outline the importance of larger epidemiologic studies of the impact of peripartum antibiotics, antibiotic protocols for maternal GBS colonization, and their implications for maternal and neonatal health [30].
In the presence of preterm labor, culture-independent techniques have identified intra-amniotic diversity of microbes that were greater than identified using culture-dependent techniques alone, and these findings associated closely with risk of preterm delivery [31]. A recent study identified a much higher bacterial presence in fetal membranes of PPROM subjects, suggesting a route for pathogenic bacterial colonization in these infants [10]. In our cohort, we identified that fetal exposure to PPPROM or chorioamnionitis increases the abundance of potentially pathogenic bacteria, a pattern which persists throughout hospitalization, and appears to be unrelated to postnatal antibiotic exposure. These findings highlight potential opportunities to intervene in this subgroup of premature infants with breast milk feedings, probiotic regimens, or altered antibiotic regimens, to promote a more diverse and health promoting microbiota.
Conclusion
In very low birth weight infants, fetal exposure to a non-sterile uterine environment secondary to maternal complications (PPPROM and chorioamnionitis) resulted in neonatal stool microbiota with increased potentially pathogenic communities, including Staphylococcus, Streptococcus, Serratia, and Parabacteroides, which persisted throughout months of hospitalization, irrespective of exposure to postnatal antibiotics. Overall, high-risk maternal complications appear to significantly impact the diversity and bacterial composition of the stool microbiota of premature infants. These findings offer opportunities to investigate in larger cohorts the relationship between pregnancy complications and their associated interventions with the initial development of the neonatal intestinal microbiome that has important effects on the burgeoning immune system of infancy.
Supplementary Material
Acknowledgments
The authors are deeply grateful to the children and families that made this study possible. We would also like to acknowledge the efforts of the nurses and staff in the Dartmouth-Hitchcock intensive care nursery. Special thanks to Dr. Olga Zhaxybayeva for helpful discussions. We also thank Dr. Tom Caldwell for database creation and management.
JCM is a member of the Children’s Environmental Health and Disease Prevention Research Center at Dartmouth (P20 ES018175 from NIEHS and RD-83459901 from the EPA) as well as the Molecular Epidemiology Center at Dartmouth COBRE (NIGMS P20 GM104416). JCM also received funds from the Hearst Foundation, the Joshua Burnett Career Development Award through the Hitchcock Foundation (Dartmouth), the CF Foundation Harry Shwachman Clinical Investigator Award. Additionally, this work was supported by: the Neukom Institute (JHM); NIH grants K01LM011985 (AGH); R01AI59694, GM103534, GM103506 (JHM); 5T32DK007301-35 (DAC); P20RR16448, 4UH3DK083993 (MLS and HGM); K24AT003683 (PLH).
Footnotes
Declaration of Interests
The authors report no declarations of interest.
References
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3(3):14–14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, Stokholm J, Smith B, Krogfelt KA. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128(3):646–52. e1–5. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 4.Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N, Mai V. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 2010;156(1):20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eggesbo M, Moen B, Peddada S, Baird D, Rugtveit J, Midtvedt T, Bushel PR, Sekelja M, Rudi K. Development of gut microbiota in infants not exposed to medical interventions. APMIS. 2011;119(1):17–35. doi: 10.1111/j.1600-0463.2010.02688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98(2):229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 9.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:13–33. doi: 10.1159/000146245. [DOI] [PubMed] [Google Scholar]
- 10.Fortner KB, Grotegut CA, Ransom CE, Bentley RC, Feng L, Lan L, Heine RP, Seed PC, Murtha AP. Bacteria localization and chorion thinning among preterm premature rupture of membranes. PLoS One. 2014;9(1):e83338. doi: 10.1371/journal.pone.0083338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140(6):1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM. The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin Nutr. 2006;25(3):361–368. doi: 10.1016/j.clnu.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P. Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1999;80(3):F167–73. doi: 10.1136/fn.80.3.f167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madan JC, Salari RC, Saxena D, Davidson L, O’Toole GA, Moore JH, Sogin ML, Foster JA, Edwards WH, Palumbo P, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012 doi: 10.1136/fetalneonatal-2011-301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacquot A, Neveu D, Aujoulat F, Mercier G, Marchandin H, Jumas-Bilak E, Picaud JC. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J Pediatr. 2011;158(3):390–396. doi: 10.1016/j.jpeds.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177–e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157(Pt 5):1385–92. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 20.Le Huerou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23(1):23–36. doi: 10.1017/S0954422410000065. [DOI] [PubMed] [Google Scholar]
- 21.Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA. Microbiome assembly across multiple body sites in low-birthweight infants. MBio. 2013;4(6):e00782–13. doi: 10.1128/mBio.00782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15(2):189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- 23.Ringel-Kulka T, Cheng J, Ringel Y, Salojarvi J, Carroll I, Palva A, de Vos WM, Satokari R. Intestinal microbiota in healthy U.S. young children and adults--a high throughput microarray analysis. PLoS One. 2013;8(5):e64315. doi: 10.1371/journal.pone.0064315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahlen SD. Serratia Infections: from Military Experiments to Current Practice. Clinical Microbiology Reviews. 2011;24(4):755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boente RF, Ferreira LQ, Falcao LS, Miranda KR, Guimaraes PL, Santos-Filho J, Vieira JM, Barroso DE, Emond JP, Ferreira EO, et al. Detection of resistance genes and susceptibility patterns in Bacteroides and Parabacteroides strains. Anaerobe. 2010;16(3):190–4. doi: 10.1016/j.anaerobe.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Hunt KM, Foster Ja, Forney LJ, Schütte UME, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire Ma. Characterization of the diversity and temporal stability of bacterial communities in human milk. PloS one. 2011;6:e21313. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tita AT, Andrews WW. Diagnosis and management of clinical chorioamnionitis. Clin Perinatol. 2010;37(2):339–54. doi: 10.1016/j.clp.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, Van den Veyver I, Milosavljevic A, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, Shirakawa T, Sonomoto K, Nakayama J. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. 2009;56(1):80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 30.Bedford Russell AR, Murch SH. Could peripartum antibiotics have delayed health consequences for the infant? BJOG. 2006;113(7):758–765. doi: 10.1111/j.1471-0528.2006.00952.x. [DOI] [PubMed] [Google Scholar]
- 31.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3(8):e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.