Abstract
The structure of chromatin, affected by many factors from DNA linker lengths to posttranslational modifications, is crucial to the regulation of eukaryotic cells. Combined experimental and computational methods have led to new insights into its structural and dynamical features, from interactions due to the flexible core histone tails of the nucleosomes to the physical mechanism driving the formation of chromosomal domains. Here we present a perspective of recent advances in chromatin modeling techniques at the atomic, mesoscopic, and chromosomal scales with a view toward developing multiscale computational strategies to integrate such findings. Innovative modeling methods that connect molecular to chromosomal scales are crucial for interpreting experiments and eventually deciphering the complex dynamic organization and function of chromatin in the cell.
Keywords: Chromatin, atomistic simulations, coarse-grained models, polymer and continuum models, multiscale modeling
1. Introduction
Scientists by nature seek order and clarity. In the field of chromatin—the nucleoprotein complex that stores the genetic material in eukaryotes—such order has been difficult to describe. For a long time, a hierarchical helical model of chromatin organization has been envisioned, from the base-pair level of the DNA double helix to the megabase-pair level of states associated with chromosomes and nuclear DNA; the latter higher-order states are also known to alter flexibly throughout the cell cycle—from looser states at interphase (between cell division) to recognizable chromosomes at mitosis or meiosis (metaphase stage) in heterochromatin (see Fig. 1).
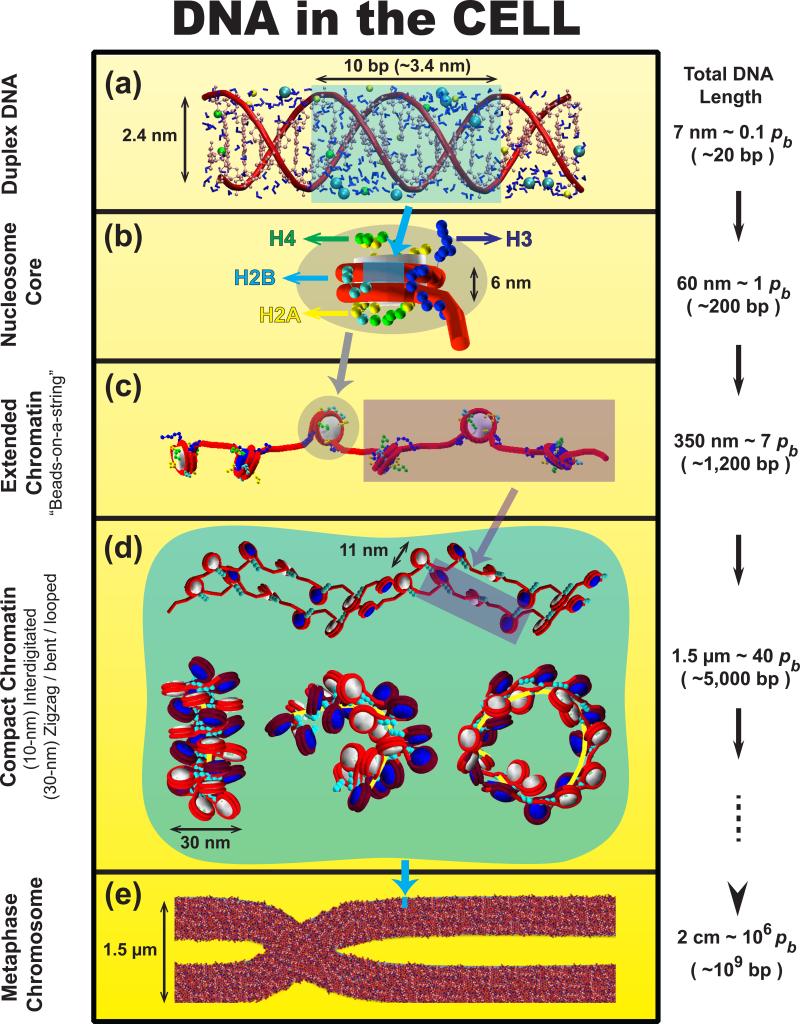
Figure 1.
DNA's many levels of folding [99]. Various colored backgrounds/masks are used to illustrate the extent of DNA compaction from one stage to the next. (a) DNA is relatively straight for length scales smaller than its persistence length (~ 0.15pb ≈ 15 bp). (b) In eukaryotic cells, DNA wraps around a core of eight histone proteins to form the nucleosome, the fundamental unit in chromatin. (c) Linker DNAs connect consecutive nucleosomes to form chromatin fibers, whose structures are unknown. The stretched 10-nm fiber, “beads-on-a-string”, is observed at low salt concentrations or when applying stretching forces to unfold the polymer. (d) At physiological conditions with multivalent ions and binding proteins such as linker histones, chromatin fibers condense. This can lead to side-by-side inter-digitated 10-nm fibers [11] (top), canonical 30-nm zigzag fibers with solenoid bent linker DNA motifs (bottom left), bent and looped fibers (bottom right) [12••]. The precise spontaneous secondary structure of chromatin depends on the cell type and other internal and external factors, and is still under debate [6,49••]. (e) Chromosomes are made up of dense chromatin fibers, shown here in the metaphase stage. Nuclear arrangements of chromatin in interphase stages are thought to be less ordered and more diverse (See polymer model in Fig. 4). The blue strip of size ~80×120 nm on the metaphase chromosome of size ~1.5×10 μm helps illustrate the enormous compaction of DNA in the cell.
The compression involved in this DNA folding problem is enormous. In humans, for instance, the 2 meters of stretched DNA corresponding to 23 pairs of chromosomes must fit into a cell nucleus of ~ 6μm diameter. This translates into a compression ratio of up to 10,000, or 3 and 4 orders of magnitude for mammalian interphase and metaphase chromosomes, respectively.
The first level of packing consists of chains of DNA wrapped around nucleosome protein-octamer cores in ~1.7 left-handed superhelical turns with linker DNA connecting successive nucleosomes (see Fig. 1). Each of the nucleosome core histone proteins (two copies each of H2A, H2B, H3, and H4) has protruding N-terminal tails that are important targets for charge modulation and hence DNA regulation (see below). For a long time, a compact form of this polynucleosome polymer called the ‘30-nm fiber’ lay at the heart of this initial helical coiling of the chromatin fiber. Yet a 30-nm compression represents only one order of magnitude condensation of the genetic material (Fig. 1). The hierarchical helical folding model imagines successive coiling above such coiling to produce much thicker fibers in interphase and metaphase to accomplish the required condensation in eukaryotic cells (Fig. 2).
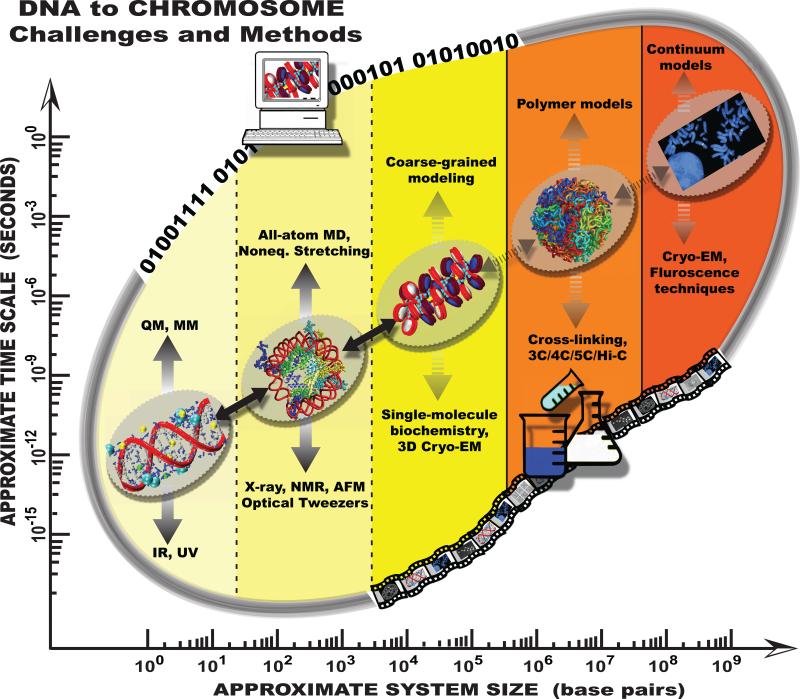
Figure 2.
Chromatin challenges and techniques on multiple spatial (base pair) and temporal (seconds) scales. Along the diagonal, representative systems are illustrated. Major computational (top) and experimental (bottom) techniques are listed, and the strength of connection between them is indicated by the boldness of the vertical arrows. Similarly, the boldness of the arrows along the diagonal reflect the strength of the connection between each pair of scales. Although inevitably losing some details during parameterization, the information transferred from high-resolution models generally enhances the overall validity of the coarser models. For instance, some of the current mesoscale models of the nucleosome are parameterized using all-atom simulations, and shown to produce many experimentally observed features of chromatin (Fig. 3). At the polymer level, existing models have not yet been connected to higher-resolution models and instead used to interpret specific experimental data. Thus, more work is required on higher scales for example to develop a computationally efficient polymer model that can be parameterized with various cell-specific information such as linker DNA length, linker histone presence, or monovalent/divalent salt concentration. See proposed multiscale connections in Fig. 4. Metaphase chromosome image credit: Wessex Reg. Genetics Centre. Wellcome Images.
Of course, such condensation is only one half of the mystery surrounding chromatin organization. The highly-compact, transcriptionally silent states of chromatin must unravel through the influence and direct interactions with a host of accessory proteins and posttranslational modifications of the histone tails of the nucleosome-core proteins to allow DNA access to the cellular machinery for template-directed processes. Such covalent modifications—acetylation and methylation of ly-sine residues and phosphorylation of serines—destabilize the charge-screening residues of the DNA polyelectrolyte as well as attract specific regulatory proteins. These structural aspects of chromatin occupy another active branch of biological research surrounding the ‘histone-code’ [1, 2, 3].
Over the past decade, a plethora of experimental studies complemented by computational modeling has managed to puncture several holes in this idealized picture of chromatin's hierarchical helical folding. First, we now realize how the local structure of chromatin is variable and pliant, depending critically on many factors, including the DNA linker length connecting successive nucleosomes, and linker histone concentration, ionic environment [4], not to speak of sequence at the finer levels. Thus, for example, rather than speaking about a ‘zigzag’ versus ‘solenoid’ topologies, a heteromorphic structure—a compact state that blends features of both straight and bent DNA linkers [5•]—is more appropriate. Second, work has shown that besides a more heteromorphic view of the chromatin fiber's ‘secondary’ structure, a more dynamic picture is appropriate at all levels of chromatin structure [6, 7]. This is because many conformational states frequently interchange, as is typical for a thermodynamic ensemble of a flexible polymer in solution. Thus, intra-fiber and inter-fiber interactions can compete to lead to a variety of energetically similar forms. In particular, both coiled 30-nm fibers (dominated by intra-nucleosome interactions) and inter-digitated side-by-side 10-nm fibers (dominated by inter-nucleosome interactions) are viable (Fig. 3) [8, 9, 10, 11]. In addition, variations in internal parameters, such as large differences in successive DNA linker lengths, can promote looping and bending, enhancing distant interactions and favoring certain conformations (Fig. 3) [12••,13•]. Third, we now accept that the lynch pin of the helical chromatin view—existence of the 30-nm fiber, may not be dominant in vivo. Models for higher-order organization of chromatin are therefore undergoing revision and re-examination, remaining largely mysterious [6, 14]. Perhaps this is the chaos that comes before the clarity.
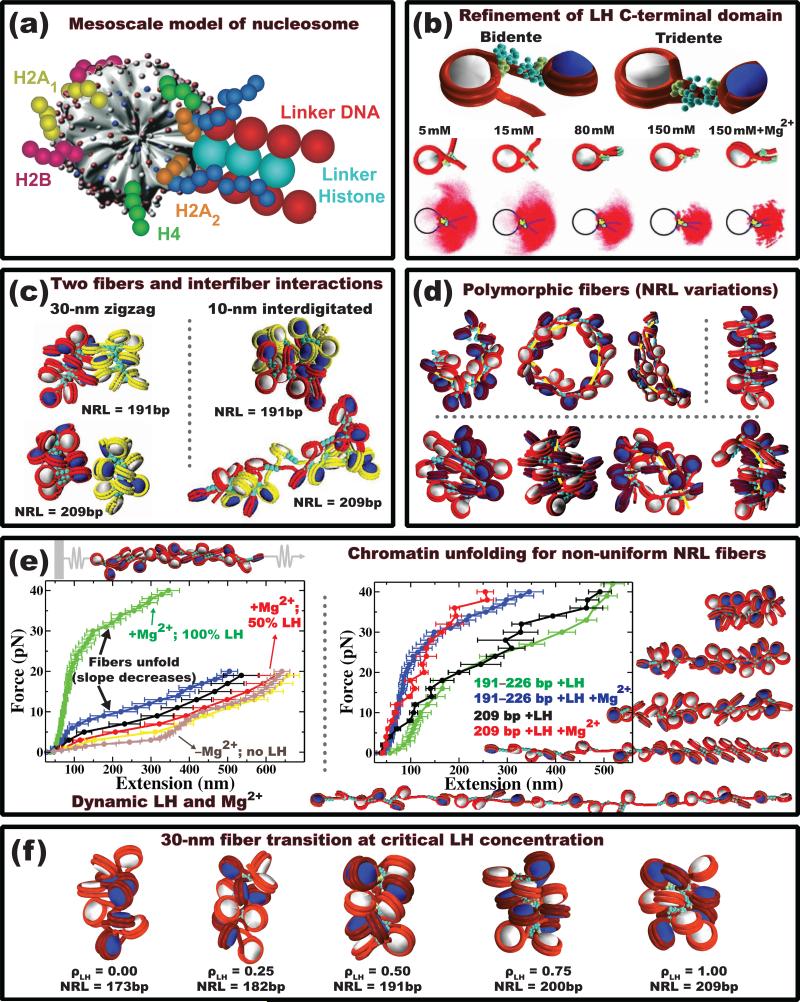
Figure 3.
Recent findings on chromatin fibers using variations of the Schlick group's mesoscale model. (a) The mesoscale model of nucleosome is composed of an irregularly shaped core with uniformly distributed 300 Debye-Hückel charges, 10 flexible core histone tails representing the C-termini of H2A and the N-termini of H2A/H2B/H3/H4, and linker DNA beads described as a worm-like chain; it can also include a linker histone attached to the dyad axis of the nucleosome [68,5•]. (b) A refined model captures the spontaneous condensation of the linker histone C-terminal domain upon nucleosome binding [69••]. The side view trajectories of the nucleosome exiting/entering linker DNA show increasing formation of the nucleosome stem as a function of the monovalent salt concentration and the presence of divalent salts in the environment. At low salt, the nucleosome adopts a bidente configuration (top left), where the linker histone interacts with the nucleosome core and the entry linker DNA, leading to the formation of extended fibers. At high salt, the nucleosome adopts a tridente configuration (top right), where the linker histone interacts with the nucleosome core and both linker DNAs, promoting the stem formation and overall fiber compaction. (c) Two chromatin fibers (red and yellow) can interact in various ways. Low chromatin densities favor the formation of segregated canonical zigzag 30-nm fibers with weak inter-fiber interactions. Denser chromatin environments lead to heteromorphic and metastable 10-nm interdigitated fibers stabilized by stronger inter-fiber interactions [11]. (d) Nonuniform (alternating) DNA linker lengths (measured in units of nucleosome repeat lengths, NRL) increase the structural diversity of chromatin fibers [12••]. Fibers with one short linker DNA length (26 bp) can form bent-ladder like structures (top left); fibers with medium to long linker DNAs (35 to 79 bp) and moderate NRL variations, NRL (see full details in Ref. [12••]), mostly adopt canonical zigzag conformations (top right); long linker length (>44 bp) fibers with large NRL variations are highly polymorphic as to adopt various stable equilibrium conformations ranging from canonical to slightly bent to intra-looped nature. (e) Various chromatin unfolding simulations help interpret force/extension curves measured by single-molecule manipulation experiments. These have helped interpret the effect of linker histones, divalent Mg2+ ions, and non-uniform nucleosome repeat lengths (NRL) on chromatin unfolding. Here, a strong negative correlation between forced extension and linker histone concentration is captured (left) [73]. Fibers with nonuniform NRLs exhibit a smoother structural transition than uniform NRL fibers (right) [75]. (f) The empirical relationship between linker histone concentration and NRL was recently associated with formation of compact 30-nm fibers. The critical linker histone concentration, ρLH (ratio of linker histones per nucleosome), for this transition increases linearly with the NRL [74].
As modern molecular biology textbooks continuously revise the levels and architecture of DNA folding, new questions and challenges emerge on multiple scales. How to link these scales in a comprehensive understanding of genomic organization and function will undoubtedly occupy scientists in the decades to come.
In this perspective, we present key questions on various levels of chromatin organization (Fig. 2) and highlight computational approaches to them, with a focus on methods and models that may extend naturally to other scales. Indeed, multiscale modeling has long been a dream for computational biologists, as described, for example, in a U.S. Department of Energy report [15, Fig. 1.1] and Ref. [16]. Multiscale modeling has also earned the 2013 Nobel Prize in Chemistry for Karplus, Levitt, and Warshel for their pioneering work in linking classical and quantum-mechanical levels for enzymatic reactions [17]. General multiscale computations remain a challenge, though much progress has been made [18•]. There is, however, no general approach for linking chromatin's many scales of spatial as well as temporal scales in analogy to the quantum and classical degrees of freedom for protein catalysis. Ideally, computational tools should make possible a telescoping and zooming from one temporal/spatial scale to another, to focus on the process of interest. Though such an ‘in-silico microscope’ is nowhere in site today, a blending of polymer level studies of nuclear folding of DNA can be imagined with polymer and mesoscale-level modeling of oligonucleosomes and chromosomes, atomistic studies of DNA-protein complexes, and even quantum-resolution studies. By presenting questions and available models for these disparate scales (Fig. 2), we hope to inspire further ideas in this quest.
There are numerous excellent biological reviews on the state of chromatin organization, with many thought-provoking titles indicative of both the excitement and confusion surrounding this fascinating subject of biology [6, 7, 14, 19]; we refer readers to them for a biological perspective. Here we focus on the oligonucleosome level of modeling and discuss ways to approach connections to it at both ends—atomistic simulations of histones and other chromatin components on one hand, and genome-level modeling on the other hand. Establishing these connections with reliable modeling frameworks will define an ambitious goal for current and future generations of computational biologists.
2. Atomistic simulations of chromatin components
The nucleosome is the fundamental structural unit of chromatin. Since the early crystal studies in the 1970s [20] and the triumphant atomic structure in 1997 [21], multiple high-resolution experiments have characterized the internal structure of the nucleosome at unprecedented resolution and scope, as recently reviewed [22]. The number of reconstructed crystal structures for canonical nucleosomes and their variants has increased steadily, reaching around 60 structures in the Protein Data Bank [23]. This, however, is still a small number compared to the ~15 million unique nucleosome particles in the nucleus due to variations of core histones, modifications of histone tails, and di erent sequences of wrapped DNA [22]. Nevertheless, the availability of all-atom structures combined with improved simulation algorithms and computing technologies [16] has opened the door to all-atom molecular dynamics (MD) simulations of nucleosomes in explicit solvent and ions. All-atom studies not only provide significant insights into the overall structure of the nucleosome and specific effects of histone tails, DNA variations, and solvent/ion modifications; they also lay critical foundations for coarse-grained investigations. In fact, the finding from such atomistic studies that the canonical nucleosome is relatively rigid [24] provides a fundamental justification for treating a nucleosome as a unit in coarse-grained models focusing on larger chromatin entities (see Section 3). Recent all-atom simulations, however, continuously provide details of the nucleosome's structure as a function of various internal and external factors [25•]; this information in turn helps improve and refine coarse-grained chromatin models.
Each histone in the nucleosome core has highly flexible N-terminal tails that are rich in positively charged residues. Because each such tail has a specific role in the structural stability of the nucleosome and in the compaction of chromatin fibers [26], the tails are important targets for experimental and computational chromatin studies. Due to their intrinsically disordered nature, experimental methods (e.g., X-ray crystallography) cannot determine equilibrium configurations of histone tails in the nucleosome, but all-atom MD simulations help describe the secondary structures adopted by these tails [24], tail-driven intrafiber interactions, and corresponding effects on the overall stability of the nucleosome. Such results from simulations are often obtained from comparisons between wild-type and altered tails (truncated or charge-manipulated tails). For example, the Langowski group found that truncation of the H3 and H2A tails alters the overall nucleosome structure by destabilizing the H2A 3 domain in the histone core, whereas truncation of the H4 or H2B tails does not lead to such nucleosome deformations [27]. Collepardo-Guevara et al. [28•] recently showed that only about 15% of the residues of histone tails are organized into secondary structure, namely β-helices and α-sheets, and thus the tail contour lengths are much longer than corresponding persistence lengths. When the H3 and H4 tails were modified, as by selected lysine acetylation, this percentage of secondary structure increased considerably, and the tails’ ability to form nucleosome-condensing interactions hindered critically. It was suggested that this lack of tail flexibility, rather than charge modulation per se, sacrifices the ability of internucleosomal interactions in oligonucleosomes with acetylated tails to condense chromatin efficiently and hence triggers fiber unfolding [28•].
The positively charged core histone tails interact strongly with both the DNA and the acidic groove on the surface of the nucleosome formed by seven acidic residues from the H2A and H2B his-tones. Interactions between the histone tails—especially the N-terminal domains of H3 and H4—and the “acidic patch” may play a crucial role in regulating both the structure of a single nucleosome and of the chromatin fiber, which ultimately affects transcriptional repression and gene regulation [29]. Thus, understanding the molecular basis for these interactions can shed light into the histone code and epigenetic marks in higher-order chromatin structures. MD simulations have been used to propose several mechanisms for the interactions among the histone tails, the “acidic patch”, and the DNA. For example, recent work has suggested that acetylation of H4 Lysine 16 reduces H4's attraction to the acidic groove on the nucleosome surface, and this in turn decreases the strength of internucleosomal interactions in the organization of chromatin at higher levels [30]. At the mononucleosome level, an attraction between the H3 tail and “acidic patch” can trigger partial unwrapping of the DNA; on the other hand, attraction between the H4 and “acidic patch” was suggested to promote full wrapping of the nucleosome [31••].
MD simulations are also helping develop a fundamental understanding of the DNA interactions with the histone tails as well as structural and dynamical aspects of the DNA constituent of the nucleosome itself. For example, Roccatano et al. showed that positively charged histone tails can neutralize the negatively charged DNA, leading to a slightly more compact nucleosome compared to a nucleosome without tails [24]. In a similar computational experiment, Potoyan and Papoian suggested that acetylation of the H4 tail, despite reducing the positive electrostatic charge, causes a stronger attraction to the DNA [32•]. In a more recent replica exchange MD study using various well-established all-atom force fields, Langowski and coworkers suggested that while, for H4 and H2B there is a single dominant binding configuration to DNA, for H3 and H2B there are multiple stable binding configurations [33].
Interactions between the DNA and core histone tails can also be a factor in triggering the DNA unwrapping in nucleosomes. FRET and stopped flow experiments have suggested that transient DNA unwrapping is crucial in regulating transcription initiation and the access of DNA repair enzymes to the nucleosomal DNA [34]. For example, a pathway for the DNA unwrapping was proposed by force-extension measurements using optical tweezers [35]. MD simulations are contributing to this problem by modeling DNA unwrapping using enhanced sampling nonequilibrium techniques, like steered molecular dynamics (SMD). Such a study by Rippe and Wedemann proposed a mechanism for DNA unwrapping in terms of the broken DNA-histone tail interactions: The DNA–H3 (at the N-terminus) and DNA–H2A (at the C-terminus) interactions are broken initially, and then DNA–H2A, DNA– H2B and DNA–H4 (at the N-termini) tail interactions are successively disrupted [36•]. Although useful in approximating energetic barriers of such events and overall unfolding pathways for DNA unwinding, SMD simulations of a nucleosome in explicit water are limited to stretching speeds (~10 m/s) that are several orders of magnitude higher than AFM experiments (~0.1 μ/s), and this limits the correspondence to experimental manipulations and to in vivo processes. Therefore, modeling cellular-like dynamics of DNA unwrapping, even for a mononucleosome, is still computationally prohibitive at the all-atom level. DNA unwrapping at the fiber scale can only be examined with coarse-graining models, as discussed in the next section.
Although it is not an intrinsic constituent of the nucleosome, the linker histone H1/H5 is an essential molecule that binds to the nucleosome and plays a crucial role in chromatin fiber condensation and cell development [37]. This accessory protein consists of a rigid and well-folded globular head linked to a short N-terminal and a long C-terminal domains, both of which are intrinsically disordered and essential for cell regulation [38]. Although all-atom trajectories of the nucleosome-linker histone complex are too costly, the Wade lab has combined docking, Brownian dynamics, and normal mode analysis to simulate the binding to the H5 globular domain in the nucleosome (without core histone tails) [39•]. Their studies suggest that H5 can adopt various docking positions near the nucleosome's dyad axis rather than a single symmetric position where it interacts with both linker DNAs as previously observed [40]. The positional diversity of H5 binding in the nucleosome is important, as it modulates the entering/exiting angles of linker DNAs, which in turn determine higher-order structure of chromatin fibers [41•].
In addition to the effects of histone tails, wrapped DNA, and linker histones, understanding nucleosome and chromatin structure and dynamics also requires an understanding of their interaction with hundreds of other proteins in the nucleus, which regulate cellular processes. MD studies are helping clarify the mechanisms behind formation of nucleosome and protein complexes related to biochemical reactions in the cell. For instance, recent all-atom MD simulations suggest that the binding of Heterochromatin Protein 1 (HP1) in the nucleosome is critically affected by the chemical modification of H3 tails (phosphorylated S10 and R8) [42]; HP1 binding is believed to be a factor in the formation of hypercondensed, mitotic chromosomes. Dynamics trajectories have also examined the binding of Lysine Specific Demethylase-1 enzyme (LSD1) with its co-repressor protein (CoREST) to chromatin [43]. These studies suggest that LSD1/CoREST binds H3 via an induced-fit mechanism; such knowledge can be useful for designing LSD1 inhibitors and molecular probes for targeting the H3-binding site.
Despite significant improvements in configurational sampling algorithms and long-time approaches for simulating macromolecules [16], atomistic modeling of nucleosomes is limited to small size and short time scales. Coarse-grained models are currently the only general feasible approach to gain insight into chromatin fiber structure and dynamics at the oligonucleosome level. Of course, such models have coarser resolution and necessitate di erent approximations.
3. Simulations of oligonucleosomes
Coarse-grained models can help decipher various experimental findings on chromatin secondary structure. At the secondary level of chromatin organization, nucleosome arrays fold into fibers, whose precise architecture has been a topic of intense debate for several decades (Figs. 1 and 4) [4, 14, 44, 45, 46]. It was long presumed that 30-nm fibers were ubiquitous and that two major models were viable: zigzag and solenoid. Indeed, a recent high-resolution (in vitro) reconstruction of tetranucleosomes (nucleosome repeat length, NRL=167 bp without H1) [47] and 12-oligonucleosomes (NRL=187 bp and one linker histone per nucleosome) [48••] report well-formed 30-nm fibers with zigzag (two-start) organization in which DNA linkers are essentially straight and nucleosomes cross along a global helical axis (Fig. 1). In vivo studies of chromatin of chicken erythrocytes (NRL=210 bp and one H1 per nucleosome) also reveal short, well-formed 30-nm zigzag fibers [49••]. On the other hand, evidence for solenoid fibers with a one-start organization has also been presented [50, 9]. Grigoryev et al. showed that these two forms can arise in one heteromorphic fiber of compact chromatin, especially in divalent ion conditions [5•]. Complicating the picture are recent studies of chromatin in human cells (NRL=181 bp, H1/nucleosome ratio of 0.5, highly dynamic H1) that report a molten polymer state of 10-nm fibers [8, 10]. These diverse findings suggest that chromatin in cell nuclei can organize in di erent secondary configurations depending on the cell type and conditions, not to speak of intrinsic components like linker length variations and the density of linker histones [4]. While experimental techniques cannot fully elucidate the precise instantaneous configuration of di erent chromatin states and the mechanisms underlying the structural transition between 30-and 10-nm fibers, coarse-grained simulations are helping fill the picture by connecting experimental measurements like internucleosome interactions to molecular configurations of fibers (Fig. 3). Yet development of such models is as much an art as it is a science.
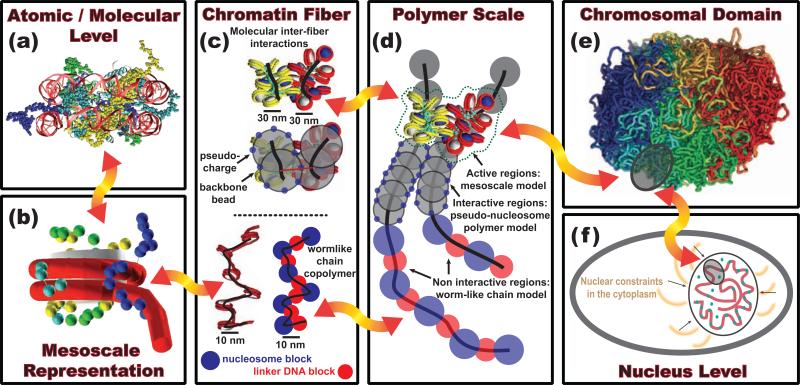
Figure 4.
Proposed dynamic multiscale model connecting atomic to nuclear chromatin levels. (a) All-atom models of nucleosomes provide atomistic details of nucleosome complexes. (b) Such knowledge can be transferred into mesoscale models to build coarse-grained representations of the nucleosome core particle (NCP) interacting with many internal and external elements as found in cellular environment. (c) Because coarse-grained models indicate that chromatin fibers are highly polymorphic— adopting canonical zigzag (30-nm), interdigitated “beads-on-a-string” (10-nm), hairpin-like, bent, and looped conformations [12••]—further coarse graining could describe these various fibers to investigate diverse phase space and structural dynamics of chromosomal and nuclear domains observed in vivo and in vitro experiments. (d) These mesoscale fibers can then be represented as active regions within lower resolution polymer models that capture crucial physical properties of the nuclear environment. (e) By incorporating cell specific information to the all-atom details, different polymer models can be constructed to describe human interphase chromatin [85] (polymer model in Fig. 2) or mitotic chromatin [87••] (Fig. 4e), for example. (f) At the nuclear level, the chromatin polymer organization could be treated as a viscoelastic fluid incorporating passive fluctuations and ATP-active regions [92•]. The red arrows emphasize the dynamic information transfer required, zooming in and out of each level depending on the chromatin properties examined. Such a combined multiscale model could ultimately define an interactive framework to identify the synergy between molecular events and chromatin reorganization, like heterochromatin formation due to specific epigenetic marks.
Over the past decade, several coarse-grained models have investigated different aspects of chromatin fibers, as reviewed in Refs. [4, 51]. Coarse-grained simulations can sample larger chromatin structures than all-atom molecular dynamics, by approximating rather than resolving system components in a selective manner. Initially, geometrical and analytical models were developed to study symmetrical chromatin structures that account for linker DNA length, linker entry-exit angle, and the rotation between consecutive nucleosome core particles (NCPs) [52]; for example, a simple “two-angle” model helped interpret EM chromatin images. Later models incorporated elastic and electro-static interactions and used Monte Carlo and MD simulations to explore more realistic configurations of chromatin fibers. Wedemann and Langowski modeled NCPs as oblate ellipsoids in an elastic chromatin fiber subject to electrostatic repulsion—using a Debye-Hückel approximation—between linker DNA segments. They sampled chromatin chains of 100 NCPs using Monte Carlo and reproduced experimental properties of 30-nm fibers with medium linker DNA lengths (fiber diameter of 32 nm with a mass density of 6.1 NCPs per 11 nm) [53]. Later, the Rippe and Wedemann Labs refined this model by incorporating a nucleosome stem to mimic the presence of linker histones, allowing a comparison to 30-nm chromatin fibers from chicken erythrocytes [54]. This refined model was also used to describe dynamic phase diagrams of the geometric configurational space of chromatin fibers; variant conformations such as 30-nm fibers with interdigidated and cross linked geometries could also be explored [55]. A nucleosome repositioning scheme has recently been incorporated into this model to study irregularly spaced nucleosomes, in particular to suggest how 1–10 bp perturbations of the position of the central nucleosome can alter fiber conformation [56].
Electrostatic interactions between nucleosomes—mostly driven by the flexible core histone tails—are crucial for chromatin compaction. The Nordenskiöld group has developed a coarse-grained electrostatic continuum model that combines a single negatively charged NCP, positively charged beads representing histone tails, and explicit multivalent ions (reviewed in Ref. [57•]). This model was used to study the aggregation of linker-free NCPs [58], condensation of chromatin in the presence of different multivalent ions [59], and cation-induced folding/unfolding of chromatin in response to histone-tail modifications [60]. However, due to the oversimplified model of the nucleosome building block, results with this model may be limited.
With the emergence of single-molecule experiments, additional molecular models and kinetic theoretical models have investigated chromatin unfolding and nucleosome sliding, as reviewed in Refs. [61, 62]. Schiessel et al., for example, studied analytically a “two-angle” model by a mechanical stretching framework and reported that an additional nucleosome-nucleosome attraction term can explain the “swollen” and “condensed” coexistence interpreted from the force-extension diagram [63]. Katritch et al. combined the “two-angle” model with a worm-like chain (WLC) approach to describe the linker DNAs in a series of pulling simulations using a Metropolis-Monte Carlo procedure that reproduced experimental force-extension curves [64]. Wocjan et al. developed a Brownian dynamics coarse-grained model based on the “torus-like” geometry of the nucleosome with uniformly distributed histone/DNA interactions; their simulations reproduced force-extension curves that display distinct regimes observed in experimental nucleosome-stretching studies such as unwrapping of the outer DNA turn at low pulling forces [65]. Drobovolskaia and Arya's Brownian dynamics model accounts for the nonuniform histone/DNA interactions and suggests that nucleosome unraveling occurs in three distinct stages [66•].
Our group's mesoscale chromatin model—shown in Fig. 3a—treats nucleosomes, histone tails, DNA linkers, and linker histones separately according to established theories for each component. The nucleosome with wrapped DNA, but without histone tails, is approximated as an electrostatic object by Debye-Hückel atomistic partial charges; these charges are distributed on an irregularly shaped rigid body using the Discrete Surface Charge Optimization (DiSCO) algorithm developed by Beard and Schlick [67], to reproduce the electric field of the nucleosome robustly at a range of monovalent salt concentrations. This irregular surface is modeled using around 300 point charges and includes the rigid part of the core histones and wrapped DNA. The dynamic and electrostatic properties of the core histone tails are approximated separately, using coarse-grained charged beads with a resolution of five amino-acids per bead using the Warshel-Levitt united-atom protein model [68]. Linker histones were initially incorporated at the entry-exit nucleosome dyad using a 3-rigid-bead model [5•] and have been refined recently as described below to model better the C-terminal domain and globular head components (Fig. 3b) [69••]. Linker DNAs connecting consecutive nucleosomes are also modeled as beads here by a worm-like-chain model that captures the elastic properties of DNA. The negative charge of the DNA beads is approximated at different monovalent salt concentrations using Stigter's procedure, as outlined in Ref. [70]. The combined model (Fig. 3a) defines a complex multiscale but computationally efficient mesoscale model to study oligonucleosomes of up to ~50 nucleosomes [71], which are sampled using Monte Carlo algorithms. Many experimental properties have been reproduced [26, 72], from sedimentation coefficients of fibers to amino-acid distances within the linker-histone C-terminal domain [69••]. Various structural properties of chromatin have been explained, such as the effect of varying linker DNA lengths on the organization of 30-nm fibers [71], the role of dynamic histone tails in bridging inter-nucleosomal interactions [68], the impact of linker histone (LH) binding/unbinding and Mg2+ on chromatin unfolding [72, 73], zigzag/solenoid fiber heteromorphicity in compact chromatin fibers [5•], enhanced fiber bending and looping at large DNA linker length variations (Fig. 3d) [12••], fiber-fiber interactions in interdigitated systems (Fig. 3c) [11], and the relationship between DNA linker lengths and linker histone concentrations (Fig. 3f) [74]. Below we describe some recent findings.
The study of chromatin unfolding is actively being pursued at large by experiment and modeling, as described in the previous section [64,66•]. This is because understanding the chromatin fiber's response to stretching can help decipher its internal structure and also interpret its response to molecular motors. While experiments yield measured force-versus-extension curves, coarse-grained modeling studies can explore the dependence of various factors on the fiber unfolding curves and provide visual descriptions of chromatin conformations at specific points on these force/extension plots. Our recent investigations of the effect of various linker lengths and LH concentrations on fiber unfolding showed that LHs increase fiber resistance to unfolding but that dynamic binding/unbinding of LHs mitigates this effect (Fig. 3e) [72, 73]. Heterogeneous elements promote super beads-on-a-string configurations, which are biologically advantageous due to selective DNA exposure. Interestingly, with their ability to explore diverse conformations, nonuniform linker-length fibers undergo a smoother unfolding process compared to uniform linker-length fibers because of a more continuous range of similar stable conformations (Fig. 3e) [75].
Linker histones (LH) are highly flexible and can adopt various conformations upon binding to nucleosomes [39•]. Studies have shown that N-terminal domain of the LH does not have significant effect on the higher-order chromatin organization; however, the intrinsically disordered C-terminal domain can be a crucial factor in chromatin architecture [37]. In this context, our refined linker histone model that treats in detail the non-uniform globular head structure as well as the flexible and intrinsically disordered C-terminal domain has captured the dynamic condensation of the C-terminal domain upon binding to the nucleosome (Fig. 3b) [69••]. Namely, a synergistic interplay can be observed between the linker histone condensation and nucleosome stem formation, with concomitant global chromatin condensation. Our recent work also explains the linear relationship between the linker DNA length and linker histone concentration, ρLH, observed experimentally [76] by a linker-length dependent threshold of LH concentration that corresponds to formation of a compact zigzag organization (Fig. 3f) [74]. Moreover, histone tail interactions within the fiber appear very sensitive to these structural variations, and show a similar pattern at the critical transition to a compact zigzag at different linker lengths. These insights suggest a molecular mechanism that connects the linker length and linker histone concentration in the nucleus.
Intra- and interdigitation of chromatin fibers can play a crucial role in the enormous compaction of DNA into chromosome (Fig. 1) [50]. Computational approaches often utilize a cuto for the long-range interactions that limits modeling of such phenomena. In our recent collaboration with experiment, our cross-linking measurements with modeling suggest that, while zigzag 30-nm fibers are thermodynamically robust structures, the presence of Mg2+, low density of linker histones, and non-uniform linker length all favor both intradigidated (via looping) (Fig. 3d) and interdigitated fibers (Fig. 3c) [11]. All the findings described in this section underscore the fluidity of 10-nm and 30-nm fibers observed in the cell nucleus [46] and help reconcile contradictory experimental findings.
Some of these coarse-graining techniques have already benefitted from multiscale modeling [51]: All-atom and short-to-medium length chromatin fiber scales have already been bridged. For example, the Nordenskiöld group has combined atomic details of ions with coarse grained NCPs and histone tails in Langevin molecular dynamics model, confirming the important role of ions in NCP aggregation [57•]. Wong et al. integrated all-atom linker histone simulations in a rigid fiber with elastic linker DNAs, predicting that the linker histone orientation is linker length-dependent [77]. The Langowski lab has also combined their mesoscale model with all-atom MD to propose a detailed unwrapping mechanism where short DNA ends of about 10 bp spontaneously detach from the histone core and the H3 tails occupy this newly exposed area on the nucleosome preventing the instanta neous rewrapping of the DNA [31••]. More recently, the Orozco and Schlick groups have combined all-atom MD with mesoscale modeling to explore the impact of epigenetic marks in chromatin folding [28•]. In this approach, all-atom replica exchange MD show that lysine acetylations of the H3 and H4 tails increase the order of core histone tail secondary structure, reducing their effective lengths and dramatically altering their ability to form compacting internucleosomal interactions. When this all-atom information is transferred into the mesoscale chromatin model, the chromatin fiber unfolds due to the absence of these crucial internucleosomal interactions. This multiscale study helps clarify the molecular mechanism in the tail-driven regulation of chromatin fibers and emphasizes the importance of histone tail flexibility in folding/unfolding events rather than charge modulation per se. Such attempts to bridge the atomic with oligonucleosome scale are key for increasing our understanding of both chromatin levels. A similar information transfer from atomic and mesoscale models to higher-order chromatin structures can be envisioned to construct polymer-scale models of chromatin (Fig. 4).
4. Polymer and continuum models
Emerging techniques based on molecular biology and light microscopy are providing a quantitative characterization of genome structure in cells. By cross linking DNA sequences, Chromosome Conformation Capture (3C) techniques can estimate the frequency of interactions between genome loci in the nucleus. The resolution of the interacting genome parts depends on the specific 3C technique, named accordingly: kilobasepairs (3C), megabasepairs (5C), megabasepairs to genome-wide (4C), and genome-wide (Hi-C) [78]. The measured contacts are typically reported as a statistical average over a large population of cells, although single-cell Hi-C analysis have been recently developed [79••]. Additionally, light microscopy methods, like fluorescence in situ hybridization (FISH), can resolve interactions between genomic loci in single cells, providing a complementary approach to 3C methods on the megabase to genome-wide range [80].
All these techniques are accumulating a wealth of data about global genome structure. Yet, to interpret these data and relate these distances to DNA architecture, chromatin polymer models have been developed. These models are much coarser than the mesoscale models discussed in Section 3 but can approximate structures of larger chromatin regions. By visualizing the measured contacts, such models can provide configurational information and mechanistic insights into the emergence of chromosome territories, the distribution of interactions between distant genes, and the effects of entropy in chromatin organization, as reviewed in Refs. [81, 82, 83]. Here the important concept of fractal architecture refers to a chromosomal region that consists of globules at different scales, where the scaling of the contact probability between two genome loci as a function of the genome distance, or subchain length, follows the same law. This leads to a non-equilibrium hierarchical structure made of polymer domains that are self-similar and maintain an open-state and untangled topology [84].
These polymer models usually approximate chromatin as a large chain of spherical beads with simple effective energetic terms, like harmonic and Lennard-Jones potentials; chromatin configurations are generally sampled by Monte Carlo algorithms and Langevin dynamics.
A model developed in the Mirny Lab has had a key role in identifying the fractal organization of chromatin in human interphase chromosomes [85, 86]. Each monomer in that model has a diameter of 20 nm, with a volume equivalent to 6 nucleosomes and an effective resolution of 1.2 kbp. The only interaction considered between monomers is excluded volume, and a Monte Carlo algorithm samples the configurations of a polymer of 4,000 beads (~9 Mbp). Using this model, it was found that the contact probability measured as a function of the genome distance, follows a power law with exponent ~−1, and is consistent with a fractal architecture made of non-interpenetrating hierarchical entities of chromatin in the region 500 kb to 7 Mbp. To reproduce such a collapsed and unknotted state, the chromatin polymer was rapidly squeezed using a spherical constraint, and then equilibrated to obtain a uniform density but not an equilibrium globular state, incompatible with results for interphase chromosomes [85].
More recently, the same group has refined the model to investigate the organization of human mitotic chromosomes [87••], using 10 nm monomers (600 bp resolution) in a fiber made of 128,000 monomers ~ 77 Mbp). The monomer interactions are captured with attractive and repulsive Lennard-Jones potentials, softened at short distances to allow chain passing, mimicking topoisomerase II action. The contact probability of metaphase Hi-C experiments was found to follow a power law decay of ~−0.5 for 0.1 Mb to 10 Mb and a sharp decay for larger genomic distances. Several chromatin organizations were investigated in an attempt to reproduce the experimental results; an array of chromatin loops linearly distributed in a cylindrical chromosomal region that resembles the shape of metaphase chromosomes matched the data best. To obtain such a structure, the chromatin polymer was constrained in an effective cylinder and simulated by Langevin dynamics using a two-stage process. First, an effective potential restricted each monomer in a longitudinal region— 600-nm high—of the effective cylinder; the potential varies linearly as a function of the monomer sequence. The arrangement obtained is a linear distribution of chromatin loops through the axis of the effective cylinder. Second, an effective attractive potential applied an axial compression to the cylindrical fiber until reaching a chromosome compaction compatible with chromatin density in metaphase cells.
Alternatively, Barbieri et al. have introduced a chromatin polymer model that includes binding molecules, which can have specific interactions with the chromatin fiber and can bridge different genomic regions [88]. In this model, a chromatin fiber is treated as a chain of self-avoiding beads with effective proteins at a concentration cm that can di use in the medium or bind to chromatin. That is, the fiber contains binding sites with heterogeneous chemical a nity, from general to specific, mimicking the a nity of chromatin to different architectural proteins, responsible for the interaction between genomic loci and chromatin looping. A chromatin fiber of 512 monomers with resolution s0 is sampled using Monte Carlo and, by varying the concentration of binding proteins, the model recovers three main architectures as a function of protein concentration cm: open/randomly folded below a threshold concentration cm < Ctr, fractal at cm = Ctr, and compact/collapsed at cm > Ctr.
Gehlen et al. investigated the nuclear organization of the 16 chromosomes of budding yeast (12,156bp) by combining two polymer models [89]. First, a semi-flexible model made of 30 nm spherical beads (3.9 kbp each) represents compact chromatin fibers in yeast, which account for 70% of the genome. Second, a freely jointed chain made of 60 nm beads (1.3kbp each) accounts for loosely coiled 10nm-chromatin fibers. Molecular dynamics simulations of their model in a spherical nucleus of 2,000 nm in diameter—with additional constraints representing centromers, telomers and the nucleolus—show that chromosomes in yeast have preferred positions, identifying also a dynamic clustering of different functional elements in the yeast genome.
More recently, Jost et al. have introduced a block polymer fiber model where different polymer blocks emulate the effect of epigenetic domains in chromatin organization [90]. Each block is associated with an epigenomic domain whose constituent monomers have a higher a nity for self-interaction than with monomers in other domains. Each monomer resolution is 10 kbp, and attractive short-range interactions determine the compaction of the polymer (non-specific interactions) as well as its epigenetic states (specific interactions). The contact probability between monomers is calculated numerically by applying a Gaussian and self-consistency approximation in the Langevin dynamics equations for the polymer. The resulting domain supports Hi-C data obtained for different chromosomes in Drosophila in 10 Mbp regions, with formation of topological domains within regions that are related epigenetically. The overall organization of the fiber is found to be multistable, combining coiled phases with collapsed microphase regions that segregate the different epigenetic domains.
In addition to 3C and fluorescence hybridization methods, other experimental techniques, based on biophysical methods, are helping characterize the organization and dynamics of chromatin. These in turn are triggering other types of theoretical methods based on continuum rather than polymer approaches. Zidovska et al., for example, have recently developed a displacement correlation spectroscopy method for single cells to capture the in vivo dynamics of nucleosomes in human cancer cells at a resolution of 65 nm and 250 ms [91•]. A coherent movement extending beyond single-chromosome territories was observed across large chromatin regions (4-5 μm). In collaboration with Bruinsma et al., a two-fluid analytical model for the cell nucleus (with nucleoplasm as solvent and chromatin as solute) was able to distinguish between the dynamic modes associated with passive thermal fluctuations and active events that depend on ATP hydrolysis [92•]. In this way, the researchers suggested that coherent movements of large chromosomal regions observed in vivo are associated with the synchronized action of ATP-dependent molecular motors.
Other 3D imaging techniques, like soft X-ray tomography [93], have also motivated the development of other continuum models to investigate the effect of chromatin density on the diffusion of binding proteins in the nucleus by Isaacson et al. [94•,95].
While these polymer and continuum models represent a valuable reference to interpret experiments, they rely on ad hoc parameters and non-systematic design protocols that strongly depend on each cell of study. The reliance on measured experimental contacts for fitting the data also reduces the predictive power of these models. These elements make interpretation of the underlying molecular mechanisms that regulate chromatin organization uncertain. To achieve a deeper understanding, a systematic approach to account for the nuclear conditions that impact chromatin and generate chromosomal data is required. As we discuss below, next generation multiscale models could bridge theoretical techniques applied at all required levels: atomic, mesoscopic, and chromosomal, as suggested in Figs. 2 and 4.
5. Perspective
Decades of chromatin research have shown us that computational models at different levels of resolution can help solve fundamental problems of chromatin architecture at each relevant scale (Fig. 2). All-atom simulations, for instance, can probe how posttranslational modifications impact the structure and dynamics of histone tails [28•,31••], touching upon one level of the molecular mechanisms underlying the histone code. Mesoscale models focus on the effect of relevant properties, such as the ionic concentration, linker DNA length distribution, core-histone tail flexibility, and linker histone condensation, on the stabilization and variation of 10-nm and 30-nm fibers [57•,68,71,73]. The connections between experiment and theory are high at these scales (bold vertical arrows in Fig. 2). For higher-levels of chromatin organization, physical polymer models provide connections to measured contact data concerning the distribution of long-range genome interactions, formation of domains in Mb-regions, segregation of chromosomes in territories, and global architecture of chromatin [82]. Of course, while providing significant information at their relevant levels, all these models are far from perfect and certainly open to further refinement to improve our understanding of chromatin and provide more accurate representations of all relevant features [4, 78]. The connections between experiment and theory at these levels are relatively weak (light vertical arrows in Fig. 2).
If achieving a comprehensive understanding of chromatin structure and function at the cellular level is the ultimate goal, current atomistic and mesoscale models are limited by computational resources, while polymer models describing chromatin at the interphase/metaphase level are quite primitive. Novel approaches, however, could be envisioned to bridge the properties of chromatin observed at all these levels—atomic, mesoscale, and chromosomal, as denoted by the diagonal arrows in Fig. 2.
Multiscale method development has been an active area of research (e.g., Refs. [16,18•,51,96]). Yet there is no general framework as for hybrid classical/quantum mechanics methods, and the best techniques are inherently tailored. For chromatin, an integration of the structure and dynamics of different scales can be envisioned in a self-consistent fashion so that the modeling approach can be adjusted to “active-site” regions, similar to the techniques recognized by the 2013 chemistry Nobel Prize [17].
Our vision for how to embed active high-resolution regions within lower-resolution chromatin environments is illustrated in Figure 4. For instance, in a nuclear region where epigenetic marks are incorporated in the nucleosome, high-resolution atomistic models could capture the structural transition of the core histone tails and the subsequent binding of architectural proteins, like linker histones, at all-atom details (Figs. 4a,b). A lower resolution model of chromatin could be embedded to approximate the physical environment surrounding the active region. If the epigenetic marks promote chromatin silencing or cell differentiation, the active region could be remodeled at an intermediate level, by combining mesoscale and pseudo-polymer models (Figs. 4c,d). This level of refinement could capture the transition from a chromatin molten state made of 10-nm inter-digitated fibers to 30-nm heterochromatin fibers. Once the transition is completed, the regional properties could be translated into a polymer or continuum model (Figs. 4e,f), capturing the dynamics and the structure of chromatin at the nuclear and chromosomal level. Such a multiscale approach could rigorously explore how posttranslational modifications of histone tails, transcription factors, sca old proteins, and other internal/external factors activate a cascade of structural events that continuously reshape chromatin at the nuclear level. Accurate computational characterization of the structure and dynamics of chromatin at the polymer level could provide a significant framework to interpret and predict the self-organizing structure of the genome as related to cell function [97].
Since the structure of chromatin strongly depends on the organism and cell-type [98] (Fig. 2), a multiscale approach as outlined above could ultimately help formulate a chromatin atlas to describe genomic interactions as a function of many relevant nuclear factors [78], such as posttranslational modifications and histone variants, linker histone concentration and binding rate, chromosome territory proteins. Taken together, we could attempt to capture how all these and other factors interact as a complex dynamic network. For example, by taking cell-specific features into account, different multiscale models could be developed for yeast, HeLa cells (proliferation cells), or chicken erythrocytes (differentiated cells), to improve our understanding of the diverse range and complex dynamics of chromatin organization. Eventually, accurate and computationally efficient models will shed light into old questions, like the long debated secondary and tertiary structures of chromatin, as well as pose new challenges, which will be probed by an iterative combination of experiment and theory [16]. These combined efforts will undoubtedly lead to new structural tools that are essential to integrate the fast-growing body of genomic, epigenomic, and biochemical data concerning chromatin in the cell nucleus. Continuous refinement of such models will in turn help connect experiment to theory better as well as inspire new experiments concerning the structure and dynamics of chromosomal chromatin.
HIGHLIGHTS.
Review of DNA compaction in silico on multiple spatial and temporal scales
Survey of chromatin models, with emphasis on combining experiment and theory
Perspective on multiscale challenges in chromatin organization
Proposal for novel multiscale approaches connecting atomistic to nuclear scales
Acknowledgments
Computing support from the New York University High Performance Computing Mercer cluster and the CCNI at RPI is acknowledged. This work was supported by the National Institutes of Health (R01 GM55164 to T.S.), Philip Morris USA and Philip Morris International (to T.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;222:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlick T, Hayes J, Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J Biol Chem. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Grigoryev S, Arya G, Correll S, Woodcock C, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [Pioneering cross-linking experiment combined with mesoscale modeling shows that the two different models—zigzag and solenoid fibers—could be merged into one heteromorphic compact fiber with enhanced i±1 internucleosome interactions due to small percentage (Ȉ20%) of linker DNA bending. Such a fiber can be dominant in moderate divalent ion concentrations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Hübner MR, Spector DL. Chromatin dynamics. Annu Rev Biophys. 2010;39:471–489. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eltsov M, MacLellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruithof M, Chien F, Routh A, Rhodes C, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat Struct Mol Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 10.Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoryev S, Luque A, Schubert M, Woodcock C, Schlick T. Persistent zigzag architecture within interphase and metaphase chromatin with enhanced looping and self association in metaphase chromosome. Submitted. [Google Scholar]
- 12••.Collepardo-Guevara R, Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc Natl Acad Sci USA. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [This Ishikawa the first detailed study of chromatin fibers containing variable nucleosome repeat lengths of the linker DNAs using a mesoscale model. Alternating linker lengths are found to promote polymorphic forms of chromatin, such as bent, looped, and hairpin-like conformations. The polymorphic behavior of fibers emphasizes the heterogeneity of chromatin structure and suggest connections between chromatin structure and specific biological functions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Kulaeva OI, Zheng G, Polikanov YS, Colasanti AV, Clauvelin N, Mukhopadhyay S, Sengupta AM, Studitsky VM, Olson WK. Internucleosomal interactions mediated by histone tails allow distant communication in chromatin. J Biol Chem. 2012;287:20248–20257. doi: 10.1074/jbc.M111.333104. [A combination of in vitro transcription and mesoscale modeling reveals that long-range histone tail interactions play a crucial role in distant communication within chromatin fibers over large distances (i.e., more than 1.5 kb). The findings suggest that chromatin fibers of 10-25 nucleosomes undergo significant bending/looping transformations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Holde K, Zlatanova J. Chromatin fiber structure: Where is the problem now? Semin Cell Dev Biol. 2007;18:651–658. doi: 10.1016/j.semcdb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 15.DOE Workshop Report. Chicago, IL: Aug 17-19, Scientific grand challenges: Opportunities in biology at the extreme scale of computing. http://science.energy.gov/~/media/ascr/pdf/program-documents/docs/Biology_report.pdf. [Google Scholar]
- 16.Schlick T, Collepardo-Guevara R, Halvorsen LA, Jung S, Xiao X. Biomolecular modeling and simulation: a field coming of age. Q Rev Biophys. 2011;44:191–228. doi: 10.1017/S0033583510000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlick T. The 2013 Nobel prize in chemistry celebrates computations in chemistry and biology. SIAM News. 2013;46:20–21. [Google Scholar]
- 18•.Zhou HX. Theoretical frameworks for multiscale modeling and simulation. Curr Opin Struct Biol. 2014;25:67–76. doi: 10.1016/j.sbi.2014.01.004. [This work presents a comprehensive survey of the multiscale strategies used to investigate various chemical and biological processes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson PJ, Rhodes D. Structure of the 30 nm chromatin fibre: A key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 21.Luger K, Mäder A, Richmond R, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 22.Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nuc Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roccatano D, Barthel A, Zacharias M. Structural flexibility of the nucleosome core particle at atomic resolution studied by molecular dynamics simulation. Biopolymers. 2007;85:407–421. doi: 10.1002/bip.20690. [DOI] [PubMed] [Google Scholar]
- 25•.Biswas M, Langowski J, Bishop TC. Atomistic simulations of nucleosomes. Wiley Interdisciplinary Reviews: Computational Molecular Science. 2013;3:378–392. [This work presents a comprehensive review of atomistic simulations of nucleosomes.] [Google Scholar]
- 26.Arya G, Schlick T. A tale of tails: how histone tails mediate chromatin compaction in di erent salt and linker histone environments. J Phys Chem A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biswas M, Voltz K, Smith JC, Langowski J. Role of histone tails in structural stability of the nucleosome. PLoS Comp Biol. 2011;7:e1002279. doi: 10.1371/journal.pcbi.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Collepardo-Guevara R, Portella G, Vendruscolo M, Frenkel D, Schlick T, Orozco M. An epigenetic switch unravels chromatin by inhibiting histone tail flexibility and hence crucial internuclesome interactions. Submitted. [This pioneering multiscale simulation combines long-timescale molecular dynamics simulations with coarse-grained mesoscale modeling of the chromatin fibers to explain the unfolding of chromatin fibers due to lysine acetylation by a critical reduction of compacting internucleosomal interactions.] [Google Scholar]
- 29.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 30.Yang D, Arya G. Structure and binding of the H4 histone tail and the effects of lysine 16 acetylation. Phys Chem Chem Phys. 2011;13:2911–2921. doi: 10.1039/c0cp01487g. [DOI] [PubMed] [Google Scholar]
- 31••.Voltz K, Trylska J, Calimet N, Smith JC, Langowski J. Unwrapping of nucleosomal DNA ends: a multiscale molecular dynamics study. Biophys J. 2012;102:849–858. doi: 10.1016/j.bpj.2011.11.4028. [This work introduces a multiscale methodology that combines all-atom molecular dynamics and coarse grained modeling to study nucleosome unwrapping on the microsecond timescale. A mechanism for the detachment of DNA from the nucleosome is suggested, where H3 tails occupy DNA-freed regions upon unwrapping, thereby preventing instantaneous rewrapping of the DNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Potoyan DA, Papoian GA. Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. Proc Natl Acad Sci USA. 2012;109:17857–62. doi: 10.1073/pnas.1201805109. [All-atom molecular dynamics simulations are used to investigate how H4 Lysine 16 acetylation—a common histone modification—impacts the secondary structure of the H4 tail and its interaction with the DNA. The study predicts that acetylation increases the order of the H4 tail structure and enhances its binding to DNA through nonelectrostatic interactions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erler J, Zhang R, Petridis L, Cheng X, Smith JC, Langowski J. The role of histone tails in the nucleosome: a computational study. Biophys J. 2014;107:2911–2922. doi: 10.1016/j.bpj.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.North JA, Shimko JC, Javaid S, Mooney AM, Sho ner MA, Rose SD, Bundschuh R, Fishel R, Ottesen JJ, Poirier MG. Regulation of the nucleosome unwrapping rate controls DNA accessibility. Nucl Acids Res. 2012;40:10215–10227. doi: 10.1093/nar/gks747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chien FT, van der Heijden T. Characterization of nucleosome unwrapping within chromatin fibers using magnetic tweezers. Biophys J. 2014;107:373–383. doi: 10.1016/j.bpj.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Ettig R, Kepper N, Stehr R, Wedeman G, Rippe K. Dissecting DNA-histone interactions in the nucleosome by molecular dynamics simulations of DNA unwrapping. Biophys J. 2011;101:1999–2008. doi: 10.1016/j.bpj.2011.07.057. [DNA unwrapping in a complete nucleosome and a tailless nucleosome is investigated using steered molecular dynamics. The results show that interactions among the DNA and di erent core histone tails can play crucial roles in the unwrapping process.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalashnikova A, Winkler DD, McBryant SJ, Henderson RK, Herman J, DeLuca JG, Luger K, Prenni JE, Hansen JC. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nuc Acids Res. 2013;41:4026–35. doi: 10.1093/nar/gkt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Pachov GV, Gabdoulline RR, Wade RC. On the structure and dynamics of the complex of the nucleosome and the linker histone. Nuc Acids Res. 2011;39:5255–5263. doi: 10.1093/nar/gkr101. [The binding sites and positions of the linker histone in the nucleosome were investigated by Brownian dynamics-based docking and normal mode analysis. Multiple binding modes of the H5 globular domain were identified, including the dominant mode observed in fluorescence recovery after photobleaching experiments.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer S, Becker NB, Syed SH, Goutte-gattat D, Shukla MS, Hayes JJ, Angelov D, Bednar J, Dimitrov S, Everaers R, Lyon D. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem. Nuc Acids Res. 2011;39:9139–9154. doi: 10.1093/nar/gkr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Zhou BR, Feng H, Kato H, Dai L, Yang Y, Zhou Y, Bai Y. Structural insights into the histone H1-nucleosome complex. Proc Natl Acad Sci USA. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [This experimental study combines NMR with other biophysical methods to characterize an asymmetric configuration for the H1-nucleosome complex in which the globular head interacts with the nucleosome core and 10-bp linker DNA asymmetrically. This conformation is di erent from previous observations and suggests that individual H1 variants might have distinct binding geometries, contributing to chromatin heterogeneity in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papamokos GV, Tziatzos G, Papageorgiou DG, Georgatos SD, Politou AS, Kaxiras E. Structural role of RKS motifs in chromatin interactions: A molecular dynamics study of HP1 bound to a variably modified histone tail. Biophys J. 2012;18:1926–1933. doi: 10.1016/j.bpj.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vellore NA, Baron R. Molecular dynamics simulations indicate an induced-fit mechanism for LSD1/CoREST-H3-histone molecular recognition. BMC Biophysics. 2013;6:15. doi: 10.1186/2046-1682-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grigoryev S, Woodcock CL. Chromatin organization - the 30 nm fiber. Exp Cell Res. 2012;318:1448–1455. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Quénet D, McNally JG, Dalal Y. Through thick and thin: the conundrum of chromatin fibre folding in vivo. EMBO Rep. 2012;13:943–944. doi: 10.1038/embor.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maeshima K, Imai R, Tamura S, Nozaki T. Chromatin as dynamic 10-nm fibers. Chromosoma. 2014;123:225–237. doi: 10.1007/s00412-014-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 48••.Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [In this high-resolution reconstruction of a long chromatin fiber in vitro, two sets of 12 nucleosomes fibers (nucleosome repeat length, NRL=177 bp and NRL=187 bp) with H1 were investigated using cryogenic electorn microscopy, reaching a resolution of 11 Å. Both systems form zigzag two-start 30-nm fibers that repeat the X-ray structure of the tetranucleosome (NRL=167 bp without H1) reported in Ref. [47].] [DOI] [PubMed] [Google Scholar]
- 49••.Sche er MP, Eltsov M, Frangakis AS. Evidence for short-range helical order in the 30-nm chromatin fibers of erythrocyte nuclei. Proc Natl Acad Sci USA. 2011;108:16992–16997. doi: 10.1073/pnas.1108268108. [In this study, cryolectron tomography confirms the presence of short well-formed zigzag 30-nm fibers in chicken erythrocyte chromatin, a model system for di erentiated cells that contains a high concentration of linker histones and an average NRL=211 bp. The 30-nm fibers are left-handed two-start helix with 6.5 nucleosomes per 11 nm.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson P, Fairall L, Huynh V, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korolev N, Fan Y, Lyubartsev AP, Nordenskiöld L. Modelling chromatin structure and dynamics: status and prospects. Curr Opin Struct Biol. 2012;22:151–159. doi: 10.1016/j.sbi.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Woodcock C, Grigoryev S, Horowitz R, Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci USA. 1993;90:9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wedemann G, Langowski J. Computer simulation of the 30-nanometer chromatin fiber. Biophys J. 2002;82:2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stehr R, Kepper N, Rippe K, Wedemann G. The effect of internucleosomal interaction on folding of the chromatin fiber. Biophys J. 2008;95:3677–3691. doi: 10.1529/biophysj.107.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stehr R, Schöpflin R, Ettig R, Kepper N, Rippe K, Wedemann G. Exploring the conformational space of chromatin fibers and their stability by numerical dynamic phase diagrams. Biophys J. 2010;98:1028–1037. doi: 10.1016/j.bpj.2009.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Müller O, Kepper N, Schöpflin R, Ettig R, Rippe K, Wedemann G. Changing chromatin fiber conformation by nucleosome repositioning. Biophys J. 2014;107:2141–2150. doi: 10.1016/j.bpj.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Fan Y, Korolev N, Lyubartsev AP, Nordenskiöld L. An advanced coarse-grained nucleosome core particle model for computer simulations of nucleosome-nucleosome interactions under varying ionic conditions. PloS One. 2013;8:e54228. doi: 10.1371/journal.pone.0054228. [This work develops a coarse-grained model of the nucleosome with flexible histone tails in the presence of K+, Mg2+, and Co(NH3) 3+6 ions to investigate intra- and internucleosome interactions Langevin molecular dynamics simulations of a single nucleosome particle and 10-nucleosome systems show how cation screening, ion-ion correlations, and tail-tail interactions modulate the intraand internucleosomal interactions in the presence of multivalent, and especially, trivalent cations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korolev N, Lyubartsev AP, Nordenskiöld L. Computer modeling demonstrates that electrostatic attraction of nucleosomal DNA is mediated by histone tails. Biophys J. 2006;90:4305–4316. doi: 10.1529/biophysj.105.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korolev N, Allahverdi A, Yang Y, Fan Y, Lyubartsev AP, Nordenskiöld L. Electrostatic origin of salt-induced nucleosome array compaction. Biophys J. 2010;99:1896–905. doi: 10.1016/j.bpj.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y, Lyubartsev A, Korolev N, Nordenskiöld L. Computer modeling reveals that modifications of the histone tail charges define salt-dependent interaction of the nucleosome core particles. Biophys J. 2009;96:2082–2094. doi: 10.1016/j.bpj.2008.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blossey R, Schiessel H. The dynamics of the nucleosome: thermal effects, external forces and ATP. FEBS J. 2011;278:3619–3632. doi: 10.1111/j.1742-4658.2011.08283.x. [DOI] [PubMed] [Google Scholar]
- 62.Collepardo-Guevara R, Schlick T. Insights into chromatin fibre structure by in vitro and in silico single-molecule stretching experiments. Biochem Soc T. 2013;41:494–500. doi: 10.1042/BST20120349. URL: http: //www.ncbi.nlm.nih.gov/pubmed/23514142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiessel H, Gelbart WM, Bruinsma R. DNA folding: structural and mechanical properties of the two-angle model for chromatin. Biophys J. 2001;80:1940–1956. doi: 10.1016/S0006-3495(01)76164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katritch V, Bustamante C, Olson W. Pulling chromatin fibers: computer simulations of direct physical micromanipulations. J Mol Biol. 2000;295:29–40. doi: 10.1006/jmbi.1999.3021. [DOI] [PubMed] [Google Scholar]
- 65.Wocjan T, Klenin K, Langowski J. Brownian dynamics simulation of DNA unrolling from the nucleosome. J Phys Chem B. 2009;113:2639–2646. doi: 10.1021/jp806137e. [DOI] [PubMed] [Google Scholar]
- 66•.Dobrovolskaia IV, Arya G. Dynamics of forced nucleosome unraveling and role of nonuniform histone-DNA interactions. Biophys J. 2012;103:989–998. doi: 10.1016/j.bpj.2012.07.043. [Brownian dynamics simulations using a coarse-grained model of nucleosome that treats the histone octamer and DNA as separate entities are used to study DNA unwrapping. A three-stage unwrapping mechanism is suggested: Rapid unwrapping and rewrapping of the flanking DNA at small extensions, gradual unwrapping of the outer DNA turn at medium extensions, and partial unwrapping of the inner DNA turn at large extensions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Q, Beard D, Schlick T. Constructing irregular surfaces to enclose macromolecular complexes for mesoscale modeling using the discrete surface charge optimization (DISCO) algorithm. J Comput Chem. 2003;24:2063–2074. doi: 10.1002/jcc.10337. [DOI] [PubMed] [Google Scholar]
- 68.Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a new mesoscopic oligonucleosome model. Proc Natl Acad Sci USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69••.Luque A, Collepardo-Guevara R, Grigoryev S, Schlick T. Dynamic condensation of linker his-tone C-terminal domain regulates chromatin structure. Nucl Acids Res. 2014;42:7553–7560. doi: 10.1093/nar/gku491. [This mesoscale chromatin simulation investigates the nucleosome and chromatin fiber configu-ration using a refined H1 model for the linker histone that models the non-uniform structure of the H1 globular head and the flexibility of the H1 C-terminal domain (CTD). The study unveils a synergistic H1 C-terminal domain condensation mechnaism in the nucleosome, whereby the H1 CTD and fiber condense simultaneously as nucleosome stems form with an asymmetric interaction of H1 in the nucleosome, confirmed experimentally in Ref. 41.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schlick T, Li B, Olson WK. The influence of salt on the structure and energetics of supercoiled DNA. Biophys J. 1994;67:2146–2166. doi: 10.1016/S0006-3495(94)80732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perišić O. Collepardo-Guevara R, Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J Mol Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collepardo-Guevara R, Schlick T. The effect of linker histone’s nucleosome binding affinity on chromatin unfolding mechanisms. Biophys J. 2011;111:1670–1680. doi: 10.1016/j.bpj.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collepardo-Guevara R, Schlick T. Crucial role of dynamic linker histone binding and divalent ions for DNA accessibility and gene regulation revealed by mesoscale modeling of oligonucleosomes. Nucl Acids Res. 2012;40:8803–8817. doi: 10.1093/nar/gks600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luque A, Schlick T. A sensitive interplay between nucleosome repeat length, linker histone density, and histone tail interactions. Preparation [Google Scholar]
- 75.Ozer G, Collepardo-Guevara R, Schlick T. Forced unraveling of chromatin fibers with nonuniform linker DNA lengths. J Phys: Condens Matter. 2015;27:064113. doi: 10.1088/0953-8984/27/6/064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woodcock C, Skoultchi A, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 77.Wong H, Victor JM, Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS ONE. 2007;2:e877. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marti-Renom MA, Mirny LA. Bridging the resolution gap in structural modeling of 3D genome organization. PLoS Comput Biol. 2011;7:e1002125. doi: 10.1371/journal.pcbi.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79••.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [A chromosome conformation capture (3C) technique that allows the analysis of chromatin in single cells helps bridge 3C-like studies with single cell light microscopy data. In combination with genome-wide statistical analysis and structural modeling, this pioneering single-cell Hi-C study shows that the domain organization of individual chromosomes is maintained at the megabase scale, though, its chromosome structure is cell-dependent on a larger scale.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rouquette J, Cremer C, Cremer T, Fakan S. Functional nuclear architecture studied by microscopy: present and future. Int Rev Cell Mol Biol. 2010;282:1–90. doi: 10.1016/S1937-6448(10)82001-5. [DOI] [PubMed] [Google Scholar]
- 81.Tark-Dame M, van Driel R, Heermann DW. Chromatin folding–from biology to polymer models and back. J Cell Sci. 2011;124:839–45. doi: 10.1242/jcs.077628. [DOI] [PubMed] [Google Scholar]
- 82.Fudenberg G, Mirny LA. Higher-order chromatin structure: bridging physics and biology. Curr Opin Gen Dev. 2012;22:115–24. doi: 10.1016/j.gde.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nicodemi M, Pombo A. Models of chromosome structure. Curr Opin Cell Biol. 2014;28:90–5. doi: 10.1016/j.ceb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 84.Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bancaud A, Lavelle C, Huet S, Ellenberg J. A fractal model for nuclear organization: current evidence and biological implications. Nuc Acids Res. 2012;40:8783–92. doi: 10.1093/nar/gks586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87••.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–53. doi: 10.1126/science.1236083. [This work combines 5C and Hi-C experiments with chromatin polymer simulations to suggest a linear architecture, enriched with chromatin loops, for mitotic chromosomes. Unlike interphase chromosome, the architecture of which is cell-specific, the proposed mitotic organization of chromatin polymers is common to all chromosomes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barbieri M, Chotalia M, Fraser J, Lavitas LM, Dostie J, Pombo A, Nicodemi M. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci USA. 2012;109:16173–8. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gehlen LR, Gruenert G, Jones MB, Rodley CD, Langowski J, O'Sullivan J. Chromosome positioning and the clustering of functionally related loci in yeast is driven by chromosomal interactions. Nucleus. 2012;3(4):370–383. doi: 10.4161/nucl.20971. [DOI] [PubMed] [Google Scholar]
- 90.Jost D, Carrivain P, Cavalli G, Vaillant C. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nuc Acids Res. 2014;42:9553–9561. doi: 10.1093/nar/gku698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91•.Zidovska A, Weitz DA, Mitchison TJ. Micron-scale coherence in interphase chromatin dynamics. Proc Natl Acad Sci USA. 2013;110:15555–15560. doi: 10.1073/pnas.1220313110. [This work applies displacement correlation spectroscopy to capture the dynamics of nucleosomes in a single cell. Their finding that chromatin motion is coherent and transient suggests mechanical coupling between different chromatin regions on different chromosomes. This communication may have a crucial role in cellular processes such as nuclear response to damaged DNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92•.Bruinsma R, Grosberg AY, Rabin Y, Zidovska A. Chromatin hydrodynamics. Biophys J. 2014;106:1871–81. doi: 10.1016/j.bpj.2014.03.038. [This theoretical framework models chromatin in the nucleus as a viscoelastic fluid to analyze recent experiments that capture nucleosome dynamics correlations [91]. It is found that a simple two-fluid model—with nucleoplasm as the solvent and chromatin fiber as the solute—can describe the experimental data distinguishing between ATP-depleted and ATP-active nuclei.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–37. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94•.Isaacson SA, McQueen DM, Peskin CS. The influence of volume exclusion by chromatin on the time required to find specific DNA binding sites by diffusion. Proc Natl Acad Sci USA. 2011;108:3815–20. doi: 10.1073/pnas.1018821108. [A novel mathematical framework that models chromatin in terms of chromatin density on the microscopic images of the cell nucleus is developed to investigate different penetrability regimes of chromatin. It is shown that an optimal value of chromatin density favors the diffusivity and binding or proteins in the nucleus.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Isaacson SA, Larabell CA, Le Gros MA, McQueen DM, Peskin CS. The influence of spatial variation in chromatin density determined by X-ray tomograms on the time to find DNA binding sites. Bull Math Biol. 2013;75:2093–117. doi: 10.1007/s11538-013-9883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tjioe E, Lasker K, Webb B, Velàzquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A, Sali A. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 98.van Holde K. Chromatin. Second ed. Springer-Verlag; Heidelberg: 1989. [Google Scholar]
- 99.Schlick T. Molecular modeling: an interdisciplinary guide. Second ed. Springer Verlag; New York, NY: 2010. [Google Scholar]