Abstract
Adiponectin stimulation of its receptors, AdipoR1 and AdipoR2, increases AMPK and PPAR activities, respectively, thereby contributing to healthy longevity as key anti-diabetic molecules. AdipoR1 and AdipoR2 were predicted to contain seven transmembrane helices with the opposite topology to G protein-coupled receptor (GPCR)s. Here we report the crystal structures of human AdipoR1 and AdipoR2 at 2.9- and 2.4-Å resolution, respectively, which represent a novel class of receptor structure. The seven-transmembrane helices, conformationally distinct from those of GPCRs, enclose a large cavity where three conserved histidine residues coordinate a zinc ion. The zinc-binding structure may play a role in the adiponectin-stimulated AMPK phosphorylation and UCP2 upregulation. Adiponectin may broadly interact with the extracellular face, rather than the C-terminal flexible tail, of the receptors. The present information will facilitate the understanding of novel structure-function relationships and the development and optimization of AdipoR agonists for the treatment of obesity-related diseases, such as type 2 diabetes.
Adiponectin (ADIPOQ)1–4 is an anti-diabetic adipokine. Plasma adiponectin levels are reduced in obesity and type 2 diabetes5, while the replenishment of adiponectin reportedly ameliorated glucose intolerance and dyslipidemia in mice6–8. These beneficial effects of adiponectin are likely to be exerted, at least in part, by the activation of AMP-activated protein kinase (AMPK)9–11 and the peroxisome proliferator-activated receptor (PPAR)-α12,13.
We previously reported the expression cloning of the complementary DNAs encoding adiponectin receptors 1 and 2 (ADIPOR1 and ADIPOR2)14. AdipoR1 and AdipoR2 are predicted to contain a seven-transmembrane domain14, with an internal N-terminus and an external C-terminus, which is the opposite configuration to G-protein-coupled receptors (GPCRs). Therefore, AdipoR1 and AdipoR2 are thought to be structurally and functionally distinct from GPCRs15. AdipoR1 and AdipoR2 serve as the major receptors for adiponectin in vivo, with AdipoR1 activating the AMPK pathways and AdipoR2 activating the PPAR-α pathways such as increased expression of uncoupling protein 2 (UCP2)16. Thereby, they regulate glucose and lipid metabolism, inflammation, and oxidative stress in vivo. Recently, the small-molecule AdipoR agonist AdipoRon was shown to ameliorate diabetes and increase exercise endurance, and at the same time prolong the shortened lifespan in obesity17. It should also be noted that adiponectin receptors are conserved in evolution from mammals to plants and yeasts (http://www.ncbi.nlm.nih.gov/)18, strongly suggesting that they play essential biological roles.
It is extremely difficult to crystallize GPCRs, due to their conformational complexity. By achieving technical breakthroughs, Kobilka et al. successfully crystallized human β2 adrenoceptor (β2AR) and reported the crystal structures19–21. First, the conformational complexity of β2AR was controlled with high-affinity ligands (nanomolar dissociation constants), agonists and inverse agonists, to fix β2AR in the active and inactive forms, respectively20–23. Second, the crystallization was performed with antibody fragments and/or a protein fusion, in the lipidic mesophase. These technical advancements enabled the structure determination of many other GPCRs, and an understanding of their ligand specificities24. Furthermore, Kobilka et al. presented the first crystal structure of the active-state complex of an agonist-occupied β2AR with a nucleotide-free Gs heterotrimer25. Thus, the β2AR structures greatly impacted the fields of GPCR research and drug development26,27.
In contrast to the GPCRs, no information is available about the conformational states of AdipoR1 and AdipoR2 with respect to transmembrane signaling. Although the AdipoR agonist AdipoRon was successfully developed17, further refinement of the AdipoR agonists to achieve nanomolar dissociation constants is still underway. The structural information about AdipoR1 and/or AdipoR2, if available, would be very important for understanding the AdipoR signaling mechanisms, and for developing and optimizing AdipoR agonists.
We optimized the properties of human AdipoR1 and AdipoR2 by deleting their N-terminal tails, and then used the Fv fragment of an anti-AdipoR monoclonal antibody and the lipidic mesophase for crystallization28. In this study, we successfully determined the crystal structures of human AdipoR1 and AdipoR2 at 2.9- and 2.4-Å resolution, respectively. The structures revealed their novel structural and functional properties, including the 7TM architecture, the zinc-binding site, and a putative adiponectin-binding surface, which are completely distinct from those of GPCRs, thus highlighting the uniqueness of the adiponectin receptors. This study should open new avenues toward the elucidation of an unprecedented paradigm of signal transduction and the development and optimization of AdipoR agonists.
Structure determinations of AdipoR1 and AdipoR2 in complexes with an Fv fragment
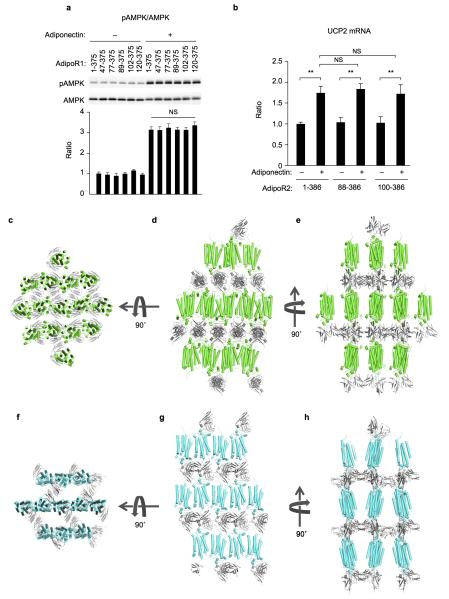
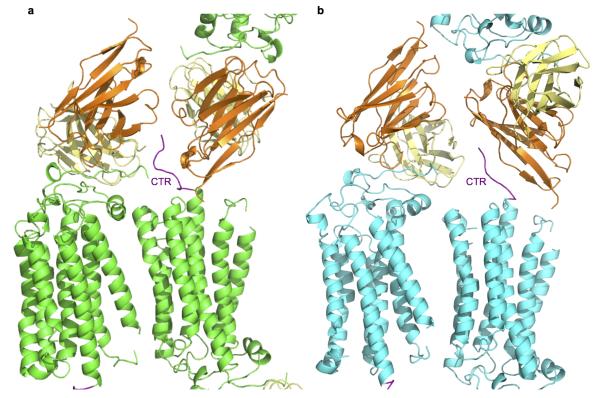
The N-terminally truncated constructs of human AdipoR1 and AdipoR2 (residues 89–375 and 100–386, respectively) exhibited better expression and purification properties than the full-length proteins28. These N-terminally-truncated AdipoR1 and AdipoR2 displayed the same extents of adiponectin-stimulated AMPK phosphorylation9–11 (Extended Data Fig. 1a) and UCP2 upregulation12,13 (Extended Data Fig. 1b), respectively, as those of the full-length proteins. Therefore, the N-terminally-truncated AdipoR proteins were crystallized with the Fv fragment of a monoclonal antibody that recognizes a conformational epitope of both AdipoR1 and AdipoR2 in a cholesterol-doped monoolein lipidic mesophase28. Thus, we determined the crystal structures of AdipoR1 (Fig. 1a, Extended Data Fig. 1c–e) and AdipoR2 (Fig. 1b, Extended Data Fig. 1f–h) at 2.9- and 2.4-Å resolution, respectively. Data collection and refinement statistics are provided in Extended Data Table 1.
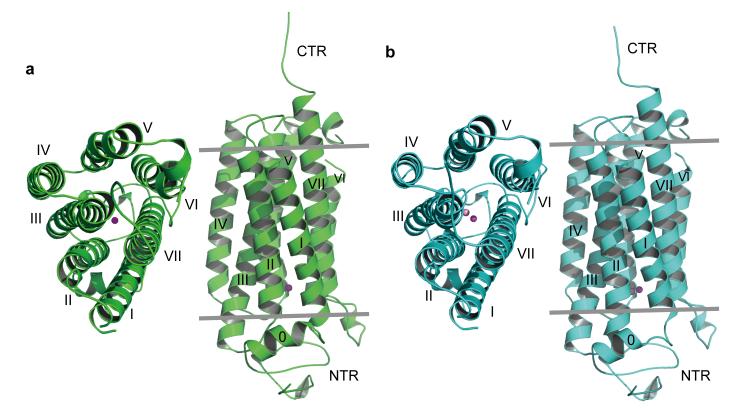
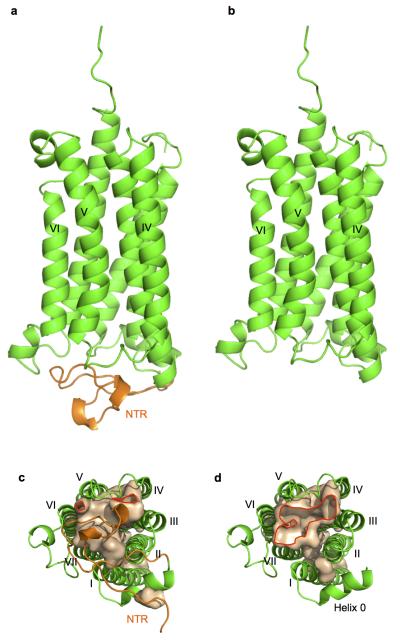
Figure 1. Overall structures of AdipoR1 and AdipoR2.
a, The 2.9-Å resolution structure of AdipoR1. b, The 2.4-Å resolution structure of AdipoR2. The structures were determined for their complexes with an Fv fragment, but the Fv fragments are omitted here for clarity. The structures are viewed from the extracellular side (left) and parallel to the membrane (right). The NTR, helix 0, transmembrane helices I–VII, and the CTR of AdipoR1 (a) and AdipoR2 (b) are indicated.
Overall structures of the AdipoR1•Fv and AdipoR2•Fv complexes
The AdipoR1 protein (residues 89–375) contains the N-terminal intracellular region (residues 89–120, NTR), a short intracellular helix (residues 121–129, helix 0), the 7TM domain (residues 134–364), and the C-terminal extracellular region (residues 365–375, CTR) (Figs. 1a and 2). The Fv fragment is bound to the NTR (Extended Data Fig. 2a). The seven transmembrane helices (I–VII) are formed by residues 135–157, 169–192, 198–227, 232–252, 264–288, 305–319, and 336–364, respectively, and are connected by three intracellular loops (ICL1–3) and three extracellular loops (ECL1–3). ECL3 has a short α helix (residues 291–295, the ECL helix) in its center, while ICL3 has another short α helix (residues 322–325, the ICL helix) just after helix VI. All of the residues are structurally ordered, except for residues 159–160 in ECL1, residues 298–299 in ECL3, and residues 374–375 in the CTR.
Figure 2. Sequence alignment of human AdipoR1 and AdipoR2.
Amino acid residues that are not conserved between these receptors are shown in green (AdipoR1) and cyan (AdipoR2). The deleted residues in the constructs and the disordered residues in the crystal structures are shown in grey and yellow, respectively. The helices in the crystal structures are surrounded with blue squares. The identical and similar residues between the two proteins are indicated with red asterisks and black colons, respectively. The Gly residues in helix V and in the CTR are indicated with red and blue number signs, respectively.
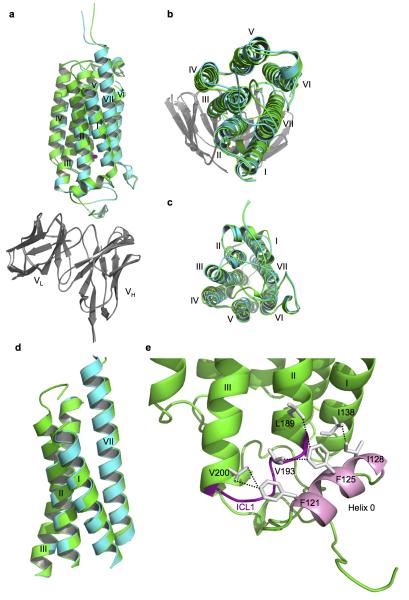
The seven transmembrane helices of AdipoR1 form a helix bundle. As viewed from the outside of the cell, the seven transmembrane helices in the helix bundle are arranged circularly in a clockwise manner, from helix I to helix VII (Fig. 1a). The structure of AdipoR2 is quite similar to that of AdipoR1 (Fig. 1, Extended Data Fig. 2a–c). The r.m.s.d. value for the main-chain Cα atoms between the AdipoR1 and AdipoR2 structures is as small as 0.56 Å.
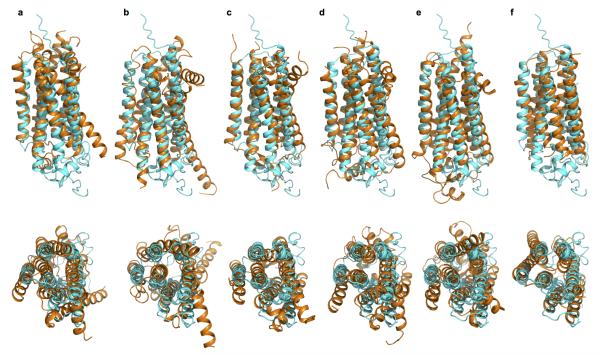
The DALI search29 indicated that the AdipoR1 and AdipoR2 structures share no similarity with other protein structures in the Protein Data Bank. The C-terminus-out topology of the 7TM domain of AdipoR1/AdipoR2, relative to the plasma membrane, is opposite to the N-terminus-out topology of the conventional 7TM proteins, such as GPCRs30 and microbial rhodopsins31. Furthermore, the conformational characteristics, such as the proline-induced kink19–21, of the transmembrane helices of GPCRs in classes A, B, and C20,21,32,33 are not observed for those of AdipoR1/AdipoR2 (Extended Data Fig. 3). In the AdipoR1/AdipoR2 structures, the transmembrane helices are not kinked, while helix V is slightly curved due to three Gly residues (Fig. 2). Consequently, we concluded that the AdipoR1 and AdipoR2 structures are novel.
The zinc-binding sites of AdipoR1 and AdipoR2
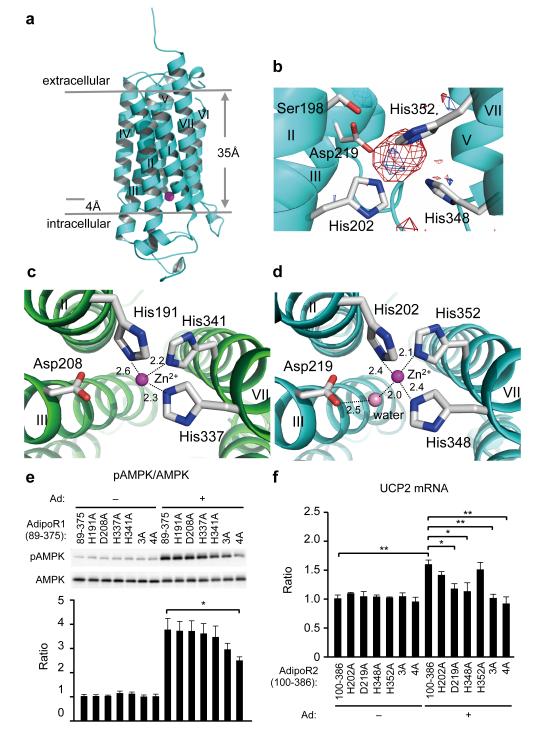
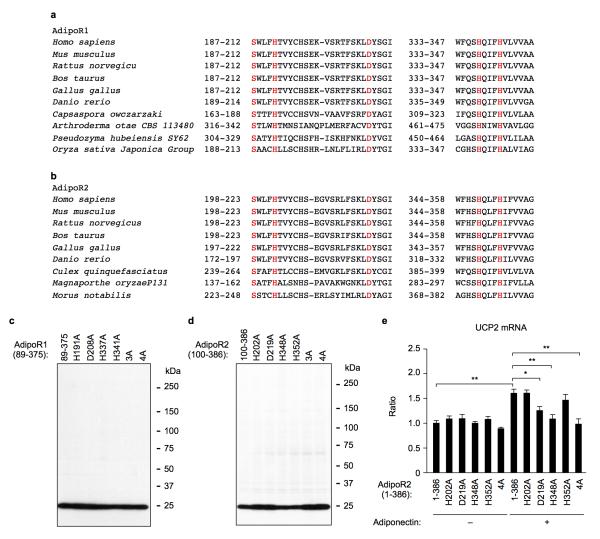
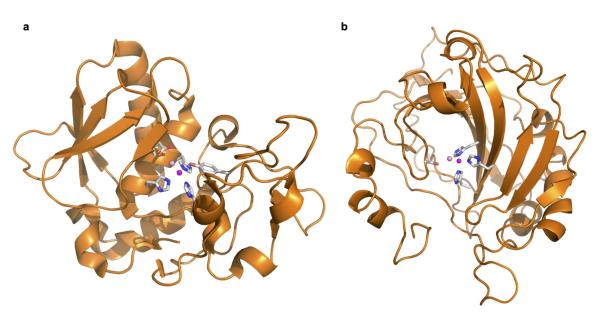
Remarkably, we found a zinc ion bound within the 7TM domain in the AdipoR1 and AdipoR2 structures (Fig. 3a), by X-ray absorption spectroscopy (data not shown) and the anomalous difference Fourier map (Fig. 3b). The zinc-binding site is located in the intracellular layer of the membrane. The zinc ion is coordinated by three His residues, H191 in helix II and H337 and H341 in helix VII of AdipoR1 and H202 in helix II and H348 and H352 in helix VII of AdipoR2, at zinc–nitrogen distances of 2.1–2.6 Å (Fig. 3c, d). The zinc ion is thus located approximately 4 Å deep from the inner surface of the plasma membrane (Fig. 3a). Furthermore, a water molecule is observed between the zinc ion and the side-chain carboxyl group of D219 in helix III of AdipoR2. Thus, the zinc ion has a tetrahedral coordination (Fig. 3d). The zinc ion binds helices II, III, and VII together (Fig. 3c, d), and probably stabilizes the structure of the subdomain consisting of helices I, II, III, and VII (Extended Data Fig. 2). The 3×His and Asp residues of AdipoR1 and AdipoR2 are strictly conserved in the homologues from mammals to plants and bacteria (Extended Data Fig. 4a, b).
Figure 3. The zinc-binding sites of AdipoR1 and AdipoR2.
a, The position of the zinc ion (magenta sphere) in the AdipoR2 structure. b, The anomalous difference maps of AdipoR2 calculated from the peak data set (red, 1.28 Å) and the low remote data set (blue, 1.288 Å), at a resolution of 3.0 Å and contoured at 3.0 σ. Helix I has been omitted for clarity. c, d, Coordination of the zinc ion (magenta sphere) by three His residues of AdipoR1 (c) and AdipoR2 (d), viewed from the cytoplasmic side. A water molecule (pink sphere) is also coordinated to the zinc ion, and is fixed by D219 in AdipoR2. e, Phosphorylation and amounts of AMPK in HEK293 cells transfected with AdipoR1 (residues 89–375) or its mutants (see text), treated for 5 min with adiponectin (15 μg ml−1). f, UCP2 mRNA levels in HEK293 cells transfected with AdipoR2 (residues 100–386) or its mutants (see text), treated for 18 hr with adiponectin (3 μg ml−1). All values are presented as mean ± s.e.m., n = 3–4, *P < 0.05, and **P < 0.01 compared to control cells or as indicated. Ad, adiponectin.
We mutated the zinc-coordinated 3×His and Asp residues of AdipoR1 (residues 89–375) (Fig. 3e). As compared with the parent AdipoR1 molecule (89–375), the adiponectin-stimulated AMPK phosphorylation was reduced by the triple mutant H191A/H337A/H341A (3A) and more seriously by the quadruple mutant H191A/D208A/H337A/H341A (4A), while none of the single H191A, D208A, H337A, and H341A mutations affected it (Fig. 3e, Extended Data Fig. 4c). Therefore, the results suggested that zinc binding is not directly required for the adiponectin-stimulated AMPK phosphorylation, but exerts a putative structure-stabilizing effect.
In contrast, the adiponectin-stimulated UCP2 upregulation by AdipoR2 was drastically reduced by each of the single mutations D219A and H348A, and nearly completely eliminated by the triple mutation H202A/H348A/H352A (3A) and the quadruple mutation H202A/D219/H348A/H352A (4A) of AdipoR2 (residues 1–386 and 100–386), as compared with the wild-type AdipoR2 (Fig. 3f, Extended Data Fig. 4d, e). Correspondingly, the single mutations H202A and H352A of AdipoR2 (residues 100–386) did not decrease the amount of bound zinc ion, whereas the single mutations D219A and H348A decreased it moderately, and the multiple mutations 3A and 4A reduced it drastically (data not shown). These results suggested that the zinc ion is directly involved in the adiponectin-stimulated UCP2 upregulation in the case of AdipoR2, in addition to structural stabilization.
An attractive hypothesis is that AdipoR2 has zinc-ion-dependent hydrolytic activity, and uses the water molecule fixed between the zinc ion and the side-chain carboxyl group of D219 of AdipoR2 for the nucleophilic attack on the carbonyl carbon atom of substrates. Free fatty acid (FFA) might be produced from lipid hydrolysis by the adiponectin-stimulated AdipoR2, and PPARα activation by the produced FFA would increase the expression of the target genes, such as UCP2.
The zinc-binding structures in the transmembrane domains of AdipoR1 and AdipoR2 are novel. The only previously reported membrane protein structure with a zinc ion within the transmembrane domain is that of a site-2 protease family intramembrane metalloprotease34. The protease consists of six transmembrane segments, and the catalytic zinc ion is coordinated by two His residues and one Asp residue, and is approximately 14 Å deep from the inner surface of the plasma membrane. Therefore, the structural features of the two proteins are not homologous. By contrast, some globular zinc enzyme structures share architectural similarity, in terms of the coordination of three His residues and a water molecule35,36 (Extended Data Fig. 5). Although the transmembrane alkaline ceramidases share negligible sequence homology with AdipoR, a set of 3×His and Asp residues is conserved in these proteins. However, their crystal structures have not been solved. Therefore, we presently cannot completely exclude the possibility that the AdipoRs have ceramidase activity.
The large internal cavities of AdipoR1 and AdipoR2
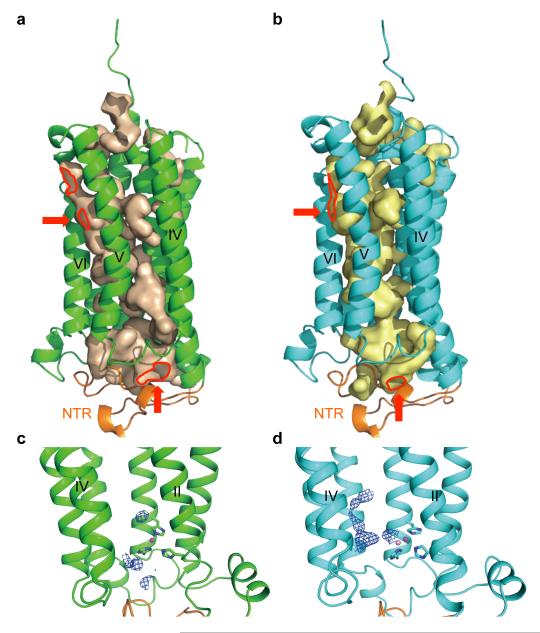
In both the AdipoR1 and AdipoR2 structures, the seven transmembrane helices surround a large internal cavity, including the zinc-binding site (Fig. 4a, b). This large internal cavity is formed between the four- and three-helix subdomains (helices VII-I-II-III and IV-V-VI, respectively) of the 7TM domains of AdipoR1/2 (Extended Data Fig. 2). The cavities extend from the cytoplasmic surface to the middle of the outer lipid layer of the membrane (Fig. 4a, b), and contain unidentified extra electron densities, which are weaker than those of the protein (Fig. 4c, d). In the cavity of AdipoR2, the extra electron densities are observed along with helices III, V, and VI (Fig. 4d). In contrast, in the cavity of AdipoR1, even weaker electron densities are observed on the cytoplasmic side of the cavity (Fig. 4c). These weak electron densities might be relevant to the substrates/products of the hypothesized hydrolytic activities of AdipoR1/AdipoR2.
Figure 4. The large internal cavities in the AdipoR1 and AdipoR2 structures.
a, b, The cavities of AdipoR1 (a) and AdipoR2 (b). The red arrows indicate the openings of the cavities. c, d, The extra electron density maps in the cavities of AdipoR1 (c) and AdipoR2 (d) contoured at 0.5 σ and 1 σ, respectively. The NTR (residues 89–119) is colored orange. The openings of the cavities are bordered in red.
The cavity has small openings between helices V and VI within the outer lipid layer and between helices IV and VI on the cytoplasmic side (Fig. 4a, b). Intriguingly, a much larger opening at helices III–VII would be uncovered on the cytoplasmic side, if the NTR was displaced from its present position (Extended Data Fig. 6). These openings might serve as the entrance/exit for the substrate/product of the hypothesized hydrolytic activity. Intriguingly, the shorter constructs (residues 102–375 and 120–375) of AdipoR1 are also as active as the full-length AdipoR1 with respect to adiponectin-stimulated AMPK phosphorylation (Extended Data Fig. 1a), indicating that the NTR, which covers the large internal cavity, is not required for this activity.
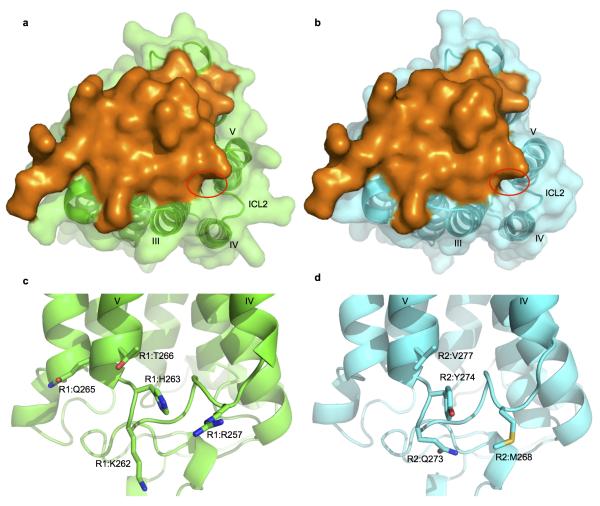
The amino acid sequences of the ICL2 regions are significantly different between AdipoR1 and AdipoR2 (Fig. 2). In particular, AdipoR1 has a cluster of positively charged residues, Arg257, Lys262, and His263, in the ICL2 region (Extended Data Fig. 7), unlike AdipoR2. Consequently, this structural difference in the cytoplasmic face may reflect the distinct signaling pathways downstream of these adiponectin receptors.
The extracellular faces of AdipoR1 and AdipoR2
The ECL1–3 and the CTR are exposed on the extracellular faces of AdipoR1 and AdipoR2. The three extracellular loops exhibit high conservation between AdipoR1 and AdipoR2 (Fig. 5a–d). Helices VII and III are longer than the others, and the C-terminal two turns of helix VII protrude from the extracellular face. The CTR, which follows helix VII, seems to be independent of the other extracellular structural elements, the ECL1–3 and helix VII (Fig. 1). The very C-terminal L374–L375 (AdipoR1) and A385–L386 (AdipoR2) residues are disordered, and the crystal packing fixed the tail conformations differently (Extended Data Fig. 8). Therefore, the entire CTRs of AdipoR1 and AdipoR2 are likely to be flexible and unstructured (Figs. 1 and 2).
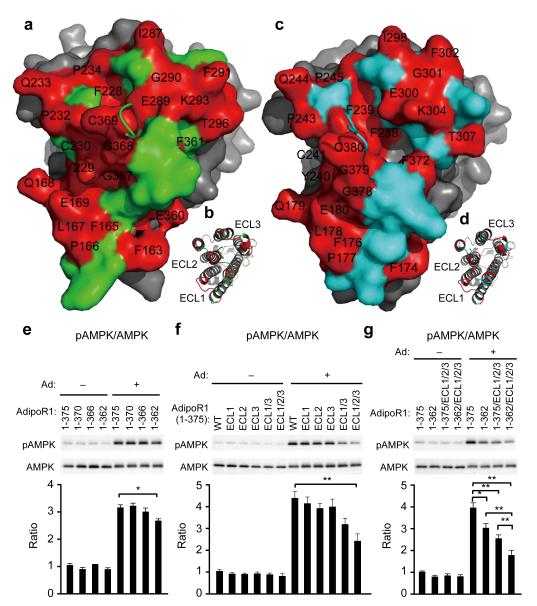
Figure 5. The extracellular faces of AdipoR1 and AdipoR2.
a–d, The extracellular faces of AdipoR1 (a, b) and AdipoR2 (c, d). AdipoR1 and AdipoR2 are shown by surface (a, c) and cartoon (b, d) representations. The residues conserved between AdipoR1 and AdipoR2 are shown in red and labeled in black. The AdipoR1- and AdipoR2-specific residues are shown in green and cyan, respectively. The 7TM domains of AdipoR1 and AdipoR2 are shown in grey. The CTR residues 370TDD372 and 381EED383 of AdipoR1 and AdipoR2, respectively, were removed for clarity. e–g, Phosphorylation and amounts of AMPK in HEK293 cells transfected with full-length AdipoR1 (residues 1–375) or a variety of mutants of AdipoR1, treated for 5 min with adiponectin (15 μg ml−1). All values are presented as mean ± s.e.m., n = 3–4, *P < 0.05, and **P < 0.01 compared to control cells or as indicated. Ad, adiponectin; ECL1, 161MYFMAPL167→SGSSGGS; ECL2, 229YCS231→GGG; ECL3, 291FVKATTV297→SSSGGGS; ECL1/3, ECL1+ECL3; ECL1/2/3, ECL1+ECL2+ECL3.
Adiponectin should bind to the extracellular face of the receptor, and the adiponectin-binding site seems to be shared by AdipoR1 and AdipoR2. A yeast two-hybrid analysis revealed that the C-terminal fragment, which extends from the middle of helix VI to the very C-terminus, as determined by the present structures, interacts with adiponectin37. Therefore, adiponectin might bind to the CTR of AdipoR1/2. However, in this study, the deletion of the CTR (AdipoR1 residues 1–370 and 1–366) did not affect the adiponectin-stimulated AMPK phosphorylation via AdipoR1 (Fig. 5e), indicating that the flexible CTR is not necessarily required for AMPK phosphorylation by adiponectin. By contrast, the deletion of the C-terminal thirteen residues, which include the last two residues of helix VII, Y363–G364, reduced the adiponectin-stimulated AMPK phosphorylation via AdipoR1 (residues 1–362, Fig. 5e, Extended Data Fig. 9a), indicating that the protruding C-terminal turn of helix VII may be involved in adiponectin signaling. Furthermore, the extracellular loop residues conserved between AdipoR1 and AdipoR2 were mutated to Gly/Ser (Fig. 5f, g): the three-loop mutation (ECL1/2/3) combined with the C-terminal thirteen-residue deletion (1–362), the 1–362/ECL1/2/3 mutation, remarkably decreased adiponectin-stimulated AMPK phosphorylation via AdipoR1 (Fig. 5g, Extended Data Fig. 9b). The other mutants with fewer Gly/Ser mutations (ECL1, ECL2, ECL3, and ECL1/3) or with no C-terminal deletion showed correspondingly smaller decreased (Fig. 5f, g, Extended Data Fig. 9a, b). These data raised the possibility that AdipoR1 may recognize adiponectin by the extensive use of its extracellular face, including the three extracellular loops and the C-terminal turns of helix VII.
Conclusions
The structural and functional characteristics of AdipoR1 and AdipoR2 revealed by this study are completely different from those of GPCRs, and therefore the AdipoRs represent an entirely novel class of receptor. The present crystal structures are expected to provide a strong basis for the development and optimization of adiponectin receptor agonists, such as AdipoRon17, as well as for understanding the roles and mechanisms of the AdipoR1/AdipoR2 homologues from animals and plants in putative signaling, such as in defense systems and lipid metabolism (Extended Data Fig. 4a, b).
METHODS
Preparation of the AdipoR1•Fv and AdipoR2•Fv crystals
The human AdipoR1 and AdipoR2 proteins and the Fv fragment of an anti-AdipoR1 monoclonal antibody were prepared as described28. In brief, human AdipoR1 and AdipoR2 (residues 89–375 and 100–386, respectively) were expressed in High Five insect cells. The proteins were purified by Flag antibody affinity chromatography followed by anion exchange chromatography, metal ion affinity chromatography after cleaving the N-terminal Flag tag by His-tagged tobacco etch virus (TEV) protease, and size-exclusion chromatography. The Fv fragment was cloned from hybridoma cells. The Fv fragment was synthesized by the E. coli cell-free protein synthesis method, and purified by Ni-affinity chromatography followed by size-exclusion chromatography. The purified AdipoR1 and AdipoR2 proteins were mixed with the Fv fragment, and the AdipoR1•Fv and AdipoR2•Fv complexes were purified by size-exclusion chromatography, and were crystallized by the lipidic mesophase method38.
X-ray data collection
Data collection was performed on beamline BL32XU at SPring-8, using an MX225HE CCD detector39–41. X-ray diffraction data were collected at 100 K by the helical scan method, with a beam size of 1 × 10 μm (horizontal × vertical) using 1° oscillation. The AdipoR1 and AdipoR2 crystals diffracted up to 2.8-Å and 2.2-Å resolution, respectively28. Data collection from the AdipoR1 crystals was limited to 10–30 images per crystal, due to radiation damage in the microcrystals, and data from 5 crystals were merged to complete the data set. For AdipoR2, diffraction data were collected from a single crystal. The data from the AdipoR1 crystals and the AdipoR2 crystal were indexed, scaled, and merged with the HKL2000 program suite42 and the XDS package43, respectively. The data collection statistics are shown in Extended Data Table 1. The AdipoR1 crystals belonged to the space group C2221, with unit cell parameters a = 92.3, b = 194.1, c = 74.3 Å, and the AdipoR2 crystal belonged to the space group P21212, with unit cell parameters a = 74.6, b = 108.6, c = 101.0 Å.
Structure solution and refinement
The initial phases for the AdipoR2•Fv complex were obtained by molecular replacement, using Fv (the VH and VL fragments from PDB IDs 1E6J and 1FDL, respectively) in Phaser44 as a search model. The resulting phases were improved by density modification using the program RESOLVE45, and thereby the electron density map around the helix bundle region of AdipoR2 became clearly visible. The initial model (all of Fv and about 80% of AdipoR2) was automatically built using the program AutoBuild46, and the rest of the model (the loops connecting the transmembrane helices) was built manually using COOT47. Refinement was performed with phenix.refine48, and the refined coordinates were rebuilt with COOT. The structure of the AdipoR2•Fv complex was refined with final R/Rfree values of 0.25/0.29. The structure of the AdipoR1•Fv complex was determined by molecular replacement, using that of the AdipoR2•Fv complex as a search model, and was refined with the secondary structure restraints in phenix.refine. Refinement of the AdipoR1•Fv complex was performed similarly to that of the AdipoR2•Fv complex. The structure of the AdipoR1•Fv complex was refined with final R/Rfree values of 0.24/0.30. Each of the final models of the AdipoR1•Fv and AdipoR2•Fv complexes includes 281 residues of the receptor, 119 residues of VH, and 107 residues of VL. The data collection and refinement statistics are summarized in Extended Data Table 1. Structural illustrations were generated using PyMol49.
Cell culture
HEK293T cells (ATCC) were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum. Cells were transfected by using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. The cDNAs encoding the AdipoR mutants were introduced into the pOriP vector, for the expression of proteins tagged with a Flag epitope.
Generation of recombinant adiponectin
Recombinant mouse full-length adiponectin was generated as previously described6,9,13,14,17,50. The expression of histidine-tagged adiponectin was induced by the addition of isopropyl β-D-1-thiogalactopyranoside to the growth medium. Bacterial extracts were prepared using standard methods, and the fusion proteins were purified by elution through a nickel-ion agarose column.
Western blot analysis and measurement of AMPK activities
Phosphorylation and protein levels of αAMPK were determined as described51–54. Western blot analyses were performed with anti-phosphorylated-AMPK (Cell Signaling Technology #2535) and anti-αAMPK (Cell Signaling Technology #2532) antibodies. Protein levels of AdipoR were analyzed by western blotting, using an anti-Flag antibody (Sigma-Aldrich F1804).
Real-time PCR
Real-time PCR was performed according to the method described previously16,50. Total RNA was prepared from cells with Trizol (Invitrogen), according to the manufacturer’s instructions. We used the real-time PCR method to quantify the mRNAs14, with slight modifications.
Extended Data
Extended Data Figure 1. Analysis of N-terminal deletion mutants of AdipoR1 and AdipoR2 and lattice packing of the AdipoR1•Fv and AdipoR2•Fv crystals.
a, Phosphorylation and amounts of AMPK in HEK293 cells transfected with full-length AdipoR1 (residues 1–375) or N-terminally truncated mutants (residues 47–375, 77–375, 89–375, 102–375, and 120–375), treated for 5 min with adiponectin (15 μg ml−1). b, UCP2 mRNA levels in HEK293 cells transfected with full-length AdipoR2 (residues 1–386) or N-terminally truncated mutants (residues 88–386 and 100–386), treated for 18 hr with adiponectin (3 μg ml−1). All values are presented as mean ± s.e.m., n = 3–4, and **P < 0.01 compared to control cells or as indicated. NS, not significant. c–h, Lattice packing of the AdipoR1•Fv crystals (c–e) and the AdipoR2•Fv crystals (f–h). AdipoR1, AdipoR2, and Fv are colored green, cyan, and grey, respectively. The AdipoR1•Fv and AdipoR2•Fv complexes crystallized with an anti-parallel arrangement of the receptor molecules.
Extended Data Figure 2. Comparison of the AdipoR1•Fv and AdipoR2•Fv structures.
a–c, Superimposition of the AdipoR1•Fv and AdipoR2•Fv complexes: side view (a), extracellular view (b), intracellular view (c). Fv was omitted from the intracellular view for clarity. d, Superimposition of the subdomains consisting of helices I, II, III, and VII between AdipoR1 and AdipoR2. The Cα r.m.s.d. value is 0.34 Å. e, Helix 0 (pink) interacts hydrophobically with the cytoplasmic ends of helices I–III, and the ICL1 (purple), as represented by those of AdipoR1. In addition, the zinc ion firmly connects helices VII, II, and III (Fig. 3). Therefore, helices VII, I, II, and III (d) constitute a rigid subdomain in the 7TM-domain structures. In contrast, helices IV, V, and VI are superimposed between AdipoR1 and AdipoR2 with a Cα r.m.s.d. value of 0.73 Å, and are likely to constitute the other subdomain, with some conformational differences in helix V between AdipoR1 and AdipoR2.
Extended Data Figure 3. Comparison of the AdipoR2 structure with other 7TM proteins.
Superimpositions of AdipoR2 (cyan) with the β2AR (PDB ID 2RH1) (r.m.s.d. 3.9 Å) (a), the glucagon receptor (PDB ID 4L6R) (r.m.s.d. 3.8 Å) (b), the metabotropic glutamate receptor 1 (PDB ID 4OR2) (r.m.s.d. 2.9 Å) (c), the sphingosine 1-phosphate receptor 1 (PDB ID 3V2Y)55 (r.m.s.d. 3.0 Å) (d), the A2A adenosine receptor (PDB ID 2YDV)56 (r.m.s.d. 3.4 Å) (e), and sensory rhodopsin (PDB ID 1XIO)57 (r.m.s.d. 3.1 Å) (f) in orange. The AdipoR2 and other 7TM protein structures are viewed parallel to the membrane (top) and from the extracellular and intracellular sides, respectively (bottom).
Extended Data Figure 4. The zinc-binding sites of AdipoR1 and AdipoR2.
a, b, The zinc-binding sites of AdipoR1 (a) and AdipoR2 (b) are conserved from mammals to plants. The conserved residues, the 3×His and Asp residues and a Ser residue, are shown in red. The side chains of S187 (AdipoR1) and S198 (AdipoR2) in helix II are located 3.7 and 3.8 Å, respectively, away from the zinc ion (data not shown and Fig. 3b). c, d, The amounts of AdipoR1 (c) and AdipoR2 (d) in HEK293 cells transfected with AdipoR1 (residues 89–375), AdipoR2 (residues 100–386) or a variety of mutants of AdipoR1 and AdipoR2 were analyzed by western blotting, using an anti-Flag antibody. The label 89–375 indicates no mutation, and the other labels, such as H191A and 4A, indicate the single and multiple mutations (see text). The label 100–386 indicates no mutation, and the other labels, such as H202A and 4A, indicate the single and multiple mutations (see text). e, UCP2 mRNA levels in HEK293 cells transfected with full-length AdipoR2 (residues 1–386) or a zinc-binding site mutant. All values are presented as mean ± s.e.m., n = 3–4, *P < 0.05, and **P < 0.01 compared to control cells or as indicated.
Extended Data Figure 5. The zinc-binding sites of soluble proteins.
a, b, The zinc-binding sites of Astacus astacus L. astacin (PDB ID 1AST) (a) and human carbonic anhydrase II (PDB ID 1CA2) (b). The zinc ion (magenta) is coordinated by three His residues and a water molecule (pink sphere).
Extended Data Figure 6. The cytoplasmic side of AdipoR1.
a, b, Structures of AdipoR1 residues 89–375 (a) and 120–375 (b) viewed parallel to the membrane. c, d, The cavity of AdipoR1 [residues 89–375 (c) and 120–375 (d)] viewed from the cytoplasmic side. Residues 120–375, including helix 0, the 7TM domain, and the CTR, are colored green. The NTR (residues 89–119) is colored orange.
Extended Data Figure 7. The cytoplasmic faces of AdipoR2 and AdipoR1.
a, b, Intracellular views of AdipoR1 (a) and AdipoR2 (b). The openings of the cavities are circled in red. The N-terminal regions of the AdipoRs are represented as surface models (orange). c, d, The ICL2s of AdipoR1 (c) and AdipoR2 (d).
Extended Data Figure 8. Crystal packing of the CTRs of AdipoR1 and AdipoR2.
Crystal packing of the CTR of AdipoR1 with Fv (a) and the CTR of AdipoR2 with Fv (b). The CTR of AdipoR1 is tucked between the two Fv fragments, whereas the C-terminal tail of AdipoR2 contacts the framework region 1 of VH (orange). The CTRs are colored purple.
Extended Data Figure 9. Expression of the AdipoR1 mutant proteins.
The amounts of AdipoR1 in HEK293 cells transfected with full-length AdipoR1 (residues 1–375) or a variety of mutants of AdipoR1 were analyzed by western blotting, using an anti-Flag antibody. Full-length AdipoR1 (residues 1–375) and the C-terminally truncated mutant (residues 1–370, 1–366, and 1–362) were used in (a). AdipoR1 residues 1–375, MYFMAPL (residues 161–167) changed to SGSSGGS (ECL1); residues 1–375, YCS (residues 229–231) changed to GGG (ECL2); residues 1–375, FVKATTV (residues 291–297) changed to SSSGGGS (ECL3); residues 1–375, ECL1 and ECL3 (ECL1/3); and residues 1–375, ECL1, ECL2 and ECL3 (ECL1/2/3) were used in (b).
Extended Data Table 1. X-ray data collection and refinement statistics.
| Structure | AdipoR1•Fv complex | AdipoR2•Fv complex |
|---|---|---|
| Data collection | ||
| No. of crystals | 5 | 1 |
| X-ray source | BL32XU, SPring-8 | BL32XU, SPring-8 |
| Wavelength (Å) | 1 | 1 |
| Space group | C2221 | P21212 |
| Cell dimensions | ||
| a, b, c (Å) | 92.3, 194.1, 74.3 | 74.6, 108.6, 101.0 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| No. of reflections measured | 112167 | 145165 |
| No. of unique reflections | 15105 | 32174 |
| Resolution (Å) | 20.0–2.9 (3.0–2.9)* | 19.5–2.4 (2.5–2.4) |
| R merge | 0.192 (0.930) | 0.115 (1.297) |
| Mean I/s(I) | 6.73 (2.03) | 8.55 (1.19) |
| Completeness (%) | 99.7 (100.0) | 98.3 (99.4) |
| Redundancy | 7.4 (7.5) | 4.5 (4.5) |
| Refinement | ||
| Resolution (Å) | 19.9–2.9 | 19.5–2.4 |
| No. of unique reflections | 15098 | 32141 |
| Rwork / Rfree (%) | 23.9 / 30.0 | 24.8 / 29.0 |
| CC1/2 | 0.976 (0.517) | 0.966 (0.506) |
| CC* | 0.994 (0.826) | 0.999 (0.820) |
| Number of atoms | ||
| AdipoR | 2294 | 2286 |
| Fv | 1747 | 1747 |
| Zn | 1 | 1 |
| water | - | 52 |
| Average B-factors (Å2) | ||
| overall | 62.1 | 66.3 |
| Protein | 62.1 | 66.4 |
| Zn | 49.5 | 56.3 |
| water | - | 61.5 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.004 | 0.003 |
| Bond angles (°) | 0.81 | 0.76 |
| Ramachandran plot (%) | ||
| Favored region | 95.8 | 95.6 |
| Allowed region | 4.2 | 4.4 |
| Outlier region | 0 | 0 |
Highest resolution shell is shown in parentheses
Acknowledgements
We are grateful to the staffs of BL32XU at SPring-8 (Proposal Nos. 2012A1332, 2012B1453, 2013A1008, 2013A1008, 2013B1034, 2013B1007, 2014A1007, 2014A1008, and 2014A1186), beam line I24 at Diamond Light Source, and beam line X06SA at the Swiss Light Source for their assistance in data collection. We thank R. Akasaka for protein analysis, M. Toyama, M. Inoue, M. Goto, M. Aoki, and K. Ishii for expression plasmid preparation, M. Nishimoto, Y. Tomabechi, and Y. Terazawa for technical assistance with protein expression and purification, and Y. Nishibaba, M. Yuasa, and A. Hayashi for technical assistance and support with the activity assays of the mutants. This work was supported by grants from the Targeted Proteins Research Program (S.Y., T.K., S.I., and M.Y.), the Platform for Drug Discovery, Informatics and Structural Life Science (S.Y. and M.Y.), a Grant-in-Aid for Specially Promoted Research (26000012) (T.K.), Grants-in-Aid for Scientific Research (S) (20229008, 25221307) (T.K.), a Grant-in-Aid for Scientific Research (B) (26293216) (M.O.-I.), a Grant-in-Aid for Young Scientists (A) (30557236) (M. Iwabu), and the Translational Research Network Program (M.O.-I.), from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by the research acceleration program of the Japan Science and Technology Agency (S.I.), and by the BBSRC (BB/G02325/1) (S.I.). The authors are grateful for the use of the Membrane Protein Laboratory funded by the Wellcome Trust (grant 062164/Z/00/Z) (S.I.) at the Diamond Light Source Limited.
Footnotes
Author Information The coordinates and structure factors for the AdipoR1•Fv and AdipoR2•Fv structures have been deposited in the Protein Data Bank, under the accession codes 3WXV and 3WXW, respectively.
The authors declare no competing financial interests.
REFERENCES
- 1.Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 2.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 1996;271:10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 3.Maeda K, et al. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose Most abundant Gene transcript 1) Biochem. Biophys. Res. Commun. 1996;221:286–289. doi: 10.1006/bbrc.1996.0587. [DOI] [PubMed] [Google Scholar]
- 4.Nakano Y, Tobe T, Choi-Miura NH, Mazda T, Tomita M. Isolation and characterization of GBP28, a novel gelatin-binding protein purified from human plasma. J. Biochem. 1996;120:803–812. doi: 10.1093/oxfordjournals.jbchem.a021483. [DOI] [PubMed] [Google Scholar]
- 5.Hotta K, et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler. Thromb. Vasc. Biol. 2000;20:1595–1599. doi: 10.1161/01.atv.20.6.1595. [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi T, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 7.Berg AH, Combs TP, Du X, Brownlee M, Scherer PE. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 8.Fruebis J, et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2005–2010. doi: 10.1073/pnas.041591798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamauchi T, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002;8:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 10.Tomas E, et al. Enhanced muscle fat oxidation and glucose transport by ACRP30 globular domain: acetyl-CoA carboxylase inhibition and AMP-activated protein kinase activation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16309–16313. doi: 10.1073/pnas.222657499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi T, et al. Globular adiponectin protected ob/ob mice from diabetes and ApoE-deficient mice from atherosclerosis. J. Biol. Chem. 2003;278:2461–2468. doi: 10.1074/jbc.M209033200. [DOI] [PubMed] [Google Scholar]
- 14.Yamauchi T, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 15.Wess J. G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 1997;11:346–354. [PubMed] [Google Scholar]
- 16.Yamauchi T, et al. Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat. Med. 2007;13:332–339. doi: 10.1038/nm1557. [DOI] [PubMed] [Google Scholar]
- 17.Okada-Iwabu M, et al. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature. 2013;503:493–499. doi: 10.1038/nature12656. [DOI] [PubMed] [Google Scholar]
- 18.Lyons TJ, et al. Metalloregulation of yeast membrane steroid receptor homologs. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5506–5511. doi: 10.1073/pnas.0306324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen SG, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 20.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbaum DM, et al. Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Graaf C, et al. Crystal structure-based virtual screening for fragment-like ligands of the human histamine H1 receptor. J. Med. Chem. 2011;54:8195–8206. doi: 10.1021/jm2011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanabe H, et al. Expression, purification, crystallization, and preliminary X-ray crystallographic studies of the human adiponectin receptors, AdipoR1 and AdipoR2. J. Struct. Funct. Genomics. 2015 doi: 10.1007/s10969-014-9192-z. DOI:10.1007/s10969-014-9192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 31.Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 32.Siu FY, et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499:444–449. doi: 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 35.Bode W, Gomis-Ruth FX, Huber R, Zwilling R, Stocker W. Structure of astacin and implications for activation of astacins and zinc-ligation of collagenases. Nature. 1992;358:164–167. doi: 10.1038/358164a0. [DOI] [PubMed] [Google Scholar]
- 36.Eriksson AE, Jones TA, Liljas A. Refined structure of human carbonic anhydrase II at 2.0 Å resolution. Proteins. 1988;4:274–282. doi: 10.1002/prot.340040406. [DOI] [PubMed] [Google Scholar]
- 37.Mao X, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat. Cell Biol. 2006;8:516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
References: Methods
- 38.Hato M, Hosaka T, Tanabe H, Kitsunai T, Yokoyama S. A new manual dispensing system for in meso membrane protein crystallization with using a stepping motor-based dispenser. J. Struct. Funct. Genomics. 2014;15:165–171. doi: 10.1007/s10969-014-9187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirata K, et al. Achievement of protein micro-crystallography at SPring-8 beamline BL32XU. J. Phys. Conf. Ser. 2013;425:012002. [Google Scholar]
- 40.Murakami I, et al. Tumor volume and lymphovascular space invasion as a prognostic factor in early invasive adenocarcinoma of the cervix. J. Gynecol. Oncol. 2012;23 doi: 10.3802/jgo.2012.23.3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueno G, Kanda H, Kumasaka T, Yamamoto M. Beamline Scheduling Software: administration software for automatic operation of the RIKEN structural genomics beamlines at SPring-8. J. Synchrotron Radiat. 2005;12 doi: 10.1107/S0909049505004735. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–327. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56 doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terwilliger TC. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D Biol. Crystallogr. 2003;59:45–49. doi: 10.1107/S0907444902018048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano WL. The PyMOL molecular graphics system. DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
- 50.Iwabu M, et al. Adioponectin and AdipoR1 regulate PGC-1α and mitochondria by Ca2+ and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 51.Minokoshi Y, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415:339–343. doi: 10.1038/415339a. [DOI] [PubMed] [Google Scholar]
- 52.Tsao T.S, Murrey, H.E., Hug C, Lee DH, Lodish HF. Oligomerization state-dependent activation of NF-κB signaling pathway by adipocyte complement-related protein of 30 kDa (Acrp30) J. Biol. Chem. 2002;277:29359–29362. doi: 10.1074/jbc.C200312200. [DOI] [PubMed] [Google Scholar]
- 53.Woods A, Salt I, Scott J, Hardie DG, Carling D. The α1 and α2 isoforms of the AMP-activated protein kinase have similar activities in rat liver but exhibit differences in substrate specificity in vitro. FEBS Lett. 1996;397:347–351. doi: 10.1016/s0014-5793(96)01209-4. [DOI] [PubMed] [Google Scholar]
- 54.Hayashi T, et al. Metabolic stress and altered glucose transport. Activation of AMP-activated protein kinase as a unifying coupling mechanism. Diabetes. 2000;49:527–531. doi: 10.2337/diabetes.49.4.527. [DOI] [PubMed] [Google Scholar]
References: Extended Data Figures
- 55.Hanson MA, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogeley L, et al. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 Å. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]