Abstract
Enzymatic and non-enzymatic lipid metabolism can give rise to reactive species that may covalently modify cellular or plasma proteins through a process known as lipoxidation. Under basal conditions, protein lipoxidation can contribute to normal cell homeostasis and participate in signaling or adaptive mechanisms, as exemplified by lipoxidation of Ras proteins or of the cytoskeletal protein vimentin, both of which behave as sensors of electrophilic species. Nevertheless, increased lipoxidation under pathological conditions may lead to deleterious effects on protein structure or aggregation. This can result in impaired degradation and accumulation of abnormally folded proteins contributing to pathophysiology, as may occur in neurodegenerative diseases. Identification of the protein targets of lipoxidation and its functional consequences under pathophysiological situations can unveil the modification patterns associated with the various outcomes, as well as preventive strategies or potential therapeutic targets. Given the wide structural variability of lipid moieties involved in lipoxidation, highly sensitive and specific methods for its detection are required. Derivatization of reactive carbonyl species is instrumental in the detection of adducts retaining carbonyl groups. In addition, use of tagged derivatives of electrophilic lipids enables enrichment of lipoxidized proteins or peptides. Ultimate confirmation of lipoxidation requires high resolution mass spectrometry approaches to unequivocally identify the adduct and the targeted residue. Moreover, rigorous validation of the targets identified and assessment of the functional consequences of these modifications are essential. Here we present an update on methods to approach the complex field of lipoxidation along with validation strategies and functional assays illustrated with well-studied lipoxidation targets.
Keywords: Mass spectrometry, Reactive carbonyl species, Electrophilic lipids, Cyclopentenone prostaglandins, Target validation, Vimentin cysteine lipoxidation
Abbreviations: ACR, acrolein; AGE, advanced glycation end product(s); ALE, advanced lipoxidation end product(s); CID, collisional induced dissociation; CHH, 7-(diethylamino)coumarin-3-carbohydrazide; cyPG, cyclopentenone prostaglandin(s); DNPH, 2,4-dinitrophenylhydrazine; ESI, electrospray ionization; GO, glyoxal; 4-HHE, 4-hydroxyhexanal; HNE, 4-hydroxynonenal; HEL, Nε-hexanoyl-lysine; MALDI, matrix assisted laser desorption/ionization; MDA, malondialdehyde; MS, mass spectrometry; NLS, neutral loss scans; ONE, 4-oxononenal; 15d-PGJ2, 15-deoxy-Δ12,14-prostaglandin J2; RCS, reactive carbonyl species.
Graphical abstract
Highlights
-
●
Protein lipoxidation may regulate protein activity in health and disease.
-
●
Lipoxidation by structurally diverse moieties may elicit specific functional outcomes.
-
●
The study of lipoxidation requires derivatization strategies and high resolution MS.
-
●
Lipoxidation functional assessment requires molecular and cellular biology approaches.
-
●
Importance of considering the potential interplay with other oxidative modifications.
1. Introduction
Redox balance is emerging as an important physiological and pathophysiological regulator of cellular behavior and outcome. The functions and activities of a growing number of proteins have been found to be altered by the oxidation state of the protein, especially at redox-sensitive cysteines. Proteins can also be modified by covalent reactions with oxidized sugars, often referred to as advanced glycation end products (AGE), or with oxidized products of lipids (advanced lipoxidation end products, or ALE) [1]. These reactions can occur mainly on the nucleophilic residues cysteine, histidine, arginine and lysine, although reactions with glutamine and asparagine have also been reported; this leads to the formation of a wide variety of adducts, as described previously [1], [2], [3], [4]. While, originally, such modifications were considered solely as detrimental to protein function, more recently there have been some reports of protein lipoxidation increasing the activity or altering the nature of the activity of specific proteins, and therefore lipoxidation is starting to be considered alongside cysteine thiol-sulfenate-disulfide switches as an additional mechanism of protein regulation [3], [5], [6]. The aim of this article is to provide a succinct update on the importance of lipoxidation in vivo and progress in the methods employed for its study.
2. Types of oxidized lipids that generate adducts
Phospholipid peroxidation occurs following radical attack, usually on polyunsaturated fatty acyl chains, and generates many different products including full-chain length oxidized fatty acids or phospholipids, chain-shortened oxidized phospholipids and small fragmentation products from the chain scission reactions. These reactions are now quite well understood and have been described in detail in several recent reviews [7], [8], [9], showing that the structure of the parent lipid and the site of radical damage determine the products. There are also enzymatic pathways for producing oxidized fatty acids and phospholipids, starting with cytochrome P450 enzymes, lipoxygenases and cyclooxygenases; products of the latter are further metabolized by a variety of prostaglandin synthases [10]. Many of the products generated by both enzymatic and non-enzymatic pathways are reactive and electrophilic owing to the presence of carbonyl groups (aldehydes or ketones) or α, β-unsaturated moieties, and can be categorized into five principal groups: alkanals (and hydroxyalkanals), 2-alkenals, 4-hydroxy–2-alkenals, keto-alkenals, and alkanedial (dialdehydes) [3]. The most reactive and commonly studied are malondialdehyde (MDA), acrolein (ACR), 4-hydroxyhexanal (4-HHE) and 4-hydroxynonenal (HNE), which also reflects the fact that these products are produced at higher levels than many other products [7] (please see Fig. 1 for the structures of some electrophilic lipids involved in protein lipoxidation). In addition, compounds with more complex structures, such as oxidized phospholipids, arachidonic acid metabolites and nitrated fatty acids are emerging as important lipid mediators in pathophysiological situations, in some cases associated with the onset and/or the resolution of inflammation. The type of adducts formed depends on the reactivity of the oxidized lipid species. Compounds containing aldehydes or ketones can react with amines (e.g. on lysine) to form Schiff base adducts by loss of water, whereas those containing an α, β-unsaturated moiety form Michael adducts by a nucleophilic addition reaction of the protein sidechain at the β-carbon. Furthermore, some electrophilic lipids have been described to contain epoxide moieties, which also react with nucleophiles giving rise to different structures. It is interesting to note that some bi-functional lipid oxidation products, such as dialdehydes or hydroxyalkenals, do react with proteins and still present free carbonyls, which can be exploited in some detection procedures, as discussed below. Nevertheless, in many cases, the carbonyl group is involved in the reaction and is not available for detection. In addition, bi-functional electrophilic lipids can induce protein cross-linking, as has been shown for HNE, isoketals and cyclopentenone prostaglandins (cyPG) with dienone structure, and this may have important consequences on protein fate [11], [12], [13].
Fig. 1.
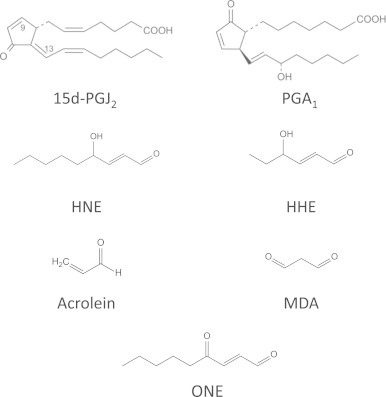
Structure of some of the electrophilic lipids involved in protein lipoxidation.
3. Pathophysiological relevance of lipoxidation adducts
Evidence for occurrence of lipoxidation products in vivo has expanded greatly in the last 10 years, as more sensitive and specific methodology has been developed, and now there are many examples of lipoxidized proteins in both healthy and diseased tissues. Much of the work has focused on HNE, but there are also many examples of adducts formed by other short chain electrophilic products, whereas studies of lipoxidation by long chain and esterified products are rarer.
As discussed below, generation of reactive species is increased in pathological conditions, and, in parallel, levels of protein lipoxidation increase in several diseases, favoring progress in the detection of adducts and identification of the modified proteins. A condition in which protein lipoxidation may have a particularly high relevance is ageing. ALE-adducted proteins have been found to accumulate during ageing and certain age-related diseases (see [14] for review). Both enzymatic and non-enzymatic lipid metabolism can contribute to the generation of increased levels of reactive lipid species in ageing [2]. On one hand, even physiological ageing is associated with a sub-clinical systemic inflammatory state, in which pro-inflammatory cytokines may elicit the enzymatic formation of inflammatory lipid mediators such as prostaglandins. On the other hand, the oxidative state can contribute to the non-enzymatic oxidation of unsaturated lipids. Nevertheless, the potential for oxidized lipid products to cause lipoxidation will depend on a balance of the rate of formation of the product, its reactivity, and the rate of detoxification by glutathione peroxidases [15], glutathione S-transferases [16], or aldoketoreductases [17]. Importantly, protein lipoxidation in ageing and age-related diseases can result in protein dysfunction contributing to the accumulation of damaged proteins. This occurs because the modified proteins can be involved in mitochondrial function, clearance of damaged proteins or in the antioxidant cellular defense; thus, their impairment can give rise to a vicious circle of protein lipoxidation and accumulation.
Therefore, an important issue regarding the pathophysiological relevance of protein lipoxidation is the nature of the adducts formed. Various proteomic studies indicate that protein lipoxidation occurs at restricted sets of proteins within the cellular proteome [18], [19]. Interestingly, there seem to be certain “hot spots” among lipoxidized proteins. Regarding plasma proteins, albumin appears to be the protein most sensitive to lipoxidation, not only because of its abundance but of the high reactivity and accessibility of some nucleophilic sites, such as Cys34 and Lys199 [20]. In the cellular environment, particular pathways or protein classes have been detected as lipoxidation targets in several experimental or clinical settings. Among them, mitochondrial, protein homeostasis-related, such as the chaperones Hsp70 and Hsp90 [21], key regulators of the antioxidant response such as Keap 1 [22], and cytoskeletal proteins emerge as targets for oxidation and/or lipoxidation in oxidative stress, cellular senescence and ageing or age-related diseases [14]. Not only actin, but also tubulin and vimentin are frequently identified cytoskeletal lipoxidation targets [18], [23], [24], [25], [26], [27]. Within proteins, some nucleophilic residues act as “hot spots” towards the electrophilic agents. Usually, the residues that undergo covalent adduction by reactive carbonyl species (RCS) are cysteine, histidine and amino acids bearing a reactive amino group, such as lysine and arginine. In general, the order of reactivity of nucleophilic amino acids towards electrophilic compounds is Cys⪢H is>Lys. However, factors including solvent accessibility and nucleophilicity regulate the reactivity of such hot spots. In addition, the microenvironment around the residue greatly affects the susceptibility to RCS adduction, as in the case of Cys34 and Cys374 in albumin and actin, respectively. The high reactivity of these particular residues is due to their significant accessible surface together with the remarkable acidity of the S–H bond, as indicated both by the polarity of S–H bond and the stability of the corresponding sulfur anion [20], [23].
From a biological perspective, the selectivity of protein lipoxidation elicits great interest. According to some hypothesis, selectivity could be related to specific biological pathways whereas others consider that it reflects defense mechanisms. The modification of Keap 1 results in the activation of transcription factor Nrf-2 and induction of antioxidant response element-dependent genes [22]. Actin scavenges reactive electrophilic aldehydes, up to C9, such as ACR and HNE, through its highly reactive Cys374 residue, thus contributing to detoxification mechanisms without undergoing significant polymerization impairment [28]. Similarly, albumin, through its reactive nucleophilic sites, acts as a protein carbonyl scavenger in the extracellular milieu, which contains only low levels of glutathione (1.5–4 µM in human plasma with respect to 1–10 mM in cells) [20]. Hence, actin and albumin can act as carbonyl scavengers, protecting more vulnerable cellular and circulating proteins whose covalent adduction would lead to damage. From an analytical point of view, several limitations should be taken into account when defining the selectivity of protein lipoxidation. Currently, most in vitro studies attempting identification of lipoxidation targets in cells or tissues use non-physiological excess of RCS leading to high noise levels due to off-target effects. To overcome this lack of biochemical specificity, some analytical platforms have been developed that reduce the off-target effects by minimizing the amounts of exogenous RCS [22], [29]. In addition, factors such as protein abundance and the number of studies addressing protein lipoxidation of given targets or performed in particular experimental or pathological conditions, can introduce a bias in these results. For instance, neurodegenerative and cardiovascular diseases have attracted a great deal of attention, and the number of studies addressing protein lipoxidation in these pathological conditions may determine the nature of some of the targets most frequently identified. Extensive work has been carried out on HNE-modified proteins in neurodegenerative diseases [30] and it was recently reported that lysines on neurofilament heavy and medium subunits are major sites of intramolecular cross-linking by HNE [11]. Strong evidence has been accumulating over many years for lipoxidation adducts of aldehydes in atherosclerotic plaques [31], [32], [33], while oxidized phosphatidylcholine adducts of ApoB-100, extensively studied in atherosclerosis [34], [35], appear to be good biomarkers and predictive of disease [36]. More recently, it has been suggested that HNE-adducts on myocardial alcohol dehydrogenase 2 might contribute to cardiomyocyte hypertrophy in a mouse model of disease [37]. Lipoxidation has also been observed in several other inflammatory diseases. For example, HNE-modified catalase has been reported in erythrocytes of patients with systemic lupus erythematosus [38], while Hsp90 modified by HNE was found in chronic alcoholic liver disease [39]. A more recent study identified increased levels of HNE adducts of AMPKα in a murine model of alcoholic liver disease that might contribute to changes in lipid metabolism, and analysis of recombinant AMPKα in vitro demonstrated that several cysteines were susceptible to formation of Michael adducts with HNE [40]. Recent work on chronic periodontitis in humans found increased prevalence HNE-His adducts both in serum and gingival crevicular fluid, which was exacerbated in patients who also had type 2 diabetes [41].
The importance of lipoxidation has been broadly discussed in the context of disease in several recent reviews. Pamplona [2] reviewed several products of lipoxidation (Nε-hexanoyl-lysine or HEL, HNE-lysine, carboxymethyl lysine, glyoxal-lysine dimer) and the proteins bearing these adducts, and discussed evidence for their accumulation in longevity. The role of α, β-unsaturated aldehydes and their adducts in the pathophysiology of vascular diseases (including diabetes and atherosclerosis), Alzheimer’s disease and chronic obstructive pulmonary disorder has been reviewed by Lee and Park [42]. There are recent focused reviews on the occurrence of ACR–protein adducts [32] and the role of HNE–protein adducts [43] in human diseases. Another area of growing interest is the role of ALE end products in age-related ocular diseases and diabetic retinopathy, where evidence for carboxymethyl lysine, Nε(3-formyl-3,4-dehydropiperidino)lysine and other adducts has been collected, as reviewed recently [44], [45].
While it can be seen that there is abundant evidence for the occurrence of lipoxidation and ALE in a range of different diseases, and it is clear that many have value as biomarkers of ageing, damage and inflammation, it is more challenging to demonstrate a physiological role of the modifications. Lipoxidation can be detected in healthy tissue; for example, healthy human plasma has been found to contain HNE adducts on Apolipoprotein B-100 [46] and HNE-adducts were detected in healthy human erythrocyte membranes [47], while protein adducts of HNE, 4-oxononenal (ONE), HHE, ACR and 3-hydroxyacrolein have all been identified in healthy rat heart mitochondria [48]. In many cases, protein modification by reactive electrophilic species has been found to inhibit protein and enzyme function, in some cases leading to beneficial effects. In addition, the possibility that moderate levels of electrophilic species participate in adaptive or preconditioning responses needs to be considered. In these cases, electrophilic species would trigger cell defense mechanisms, such as expression of phase II enzymes or antioxidant defenses, thus protecting cells from further damage. The cyPG 15-deoxy-Δ 12,14-prostaglandin J2 (15d-PGJ2) has been found to inhibit soluble epoxide hydrolase by forming adducts with a catalytic cysteine (Cys521), but this contributes to the coronary vasodilation observed in hypoxia in mice, and can be considered a compensatory effect [49]. Furthermore, a role of 15d-PGJ2 in the resolution of inflammation in relation to its electrophilic nature has been set forth in various experimental systems [50], [51]. A role of electrophilic lipids in cell signaling is exemplified by the fact that the central signaling protein H-Ras is a target for modification and activation by 15d-PGJ2 and other electrophilic prostanoids, which bind to critical cysteine residues in a structure-dependent fashion leading to modulation of downstream cascades [52], [53]. Thus strong support for regulation of physiological processes by lipoxidation is emerging, but more research in the area is required. This in turn depends on the application of robust and specific methods for identifying and quantifying proteins modified by these electrophilic lipid species, which are discussed in the following sections. In addition, lipidomic approaches that shed light on the reactive lipid species generated both under basal and pathological conditions, as well as on their reactivity, will greatly help to understand of the consequences of lipoxidation, and even predict the modification of certain targets. Currently, controversy exists about the levels of some of the species generated and whether they are sufficient to promote changes in protein activity through lipoxidation. Some of the difficulties arise from the high reactivity of these species, which makes it challenging to detect/quantify free and total levels. In addition, analysis of whole cell or tissue extracts can overlook potential compartmentalization of these species, which may be generated at, or preferentially act in, specific subcellular environments.
4. Detection of reactive oxidized lipid species, particularly aldehydes, as precursors of lipoxidation
The electrophilic lipids that have received most attention are those containing carbonyl groups. Analysis of free reactive carbonyl species (RCS) and of electrophilic lipid species in general, especially in biological matrices is very challenging due to their intrinsic physicochemical properties (i.e. high reactivity towards the matrix and low molecular weights), low amounts in biological samples, high hydrophilicity not suitable for their retention on common reversed phase LC columns, and the fact that most of the detectors are unsuitable for direct detection. In addition, most of these reactive species do not have a chromophore or their absorbance is very limited, and they do not possess ionizable functional groups, which makes their ionization yield in electrospray ionization (ESI) very poor and hampers their detection by mass spectrometry (MS). Due to these limitations, most of the analytical approaches for profiling RCS in biological matrices are based on derivatization approaches. The derivatizing agents of RCS are characterized by a functional group that rapidly reacts with the carbonyl group, such as hydralazine, and by a moiety required for their detection. Usually the derivatizing agent is chosen on the basis of the available detector used. Table 1 summarizes the most commonly used derivatizing agents for RCS.
Table 1.
Compounds used in the derivatization of RCS.
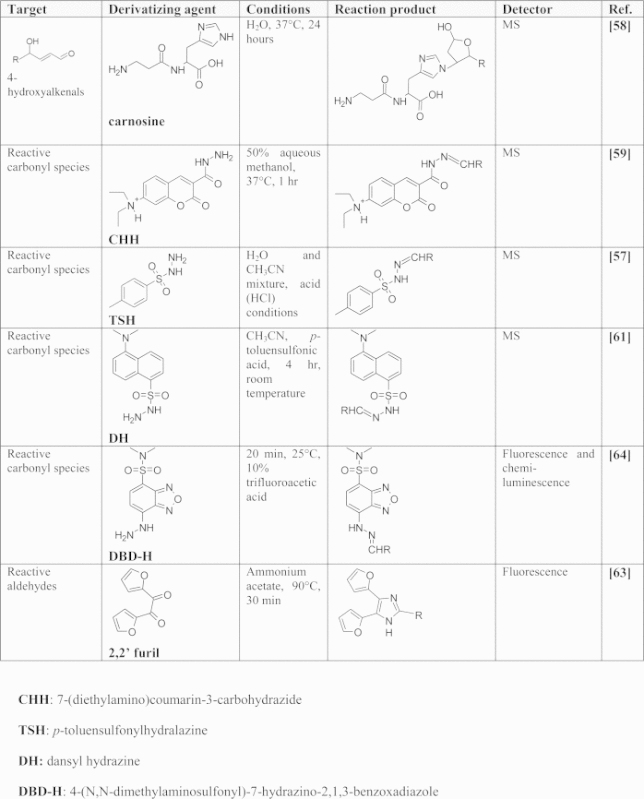 |
As explained below, some of these derivatization methods can be applied to the detection of bifunctional electrophilic lipids bound to proteins, when the adducted lipid still possesses a free carbonyl group. The first popular methods for RCS analysis were based on 2,4-dinitrophenylhydrazine (DNPH) as a derivatizing agent and the corresponding reaction products, the hydrazones, were detected by UV detection for an overall analysis of RCS or by a UV detector coupled to HPLC when each individual aldehyde needed to be identified [54], [55]. This approach was widely used for the analysis of free RCS in vitro, but was inadequate for in vivo conditions due to the lack of sensitivity and selectivity. Later, some methods based on LC–MS and GC–MS analysis of 2,4-dinitrophenylhydrazone derivatives of free aldehydes and ketones were proposed and they were mostly applied to volatile airborne carbonyls or carbonyls contained in food [56]. Use of DNPH as a derivatizing agent is currently limited due to disadvantages such as limited solubility in aqueous solvents, explosiveness, and tendency to deposit in the MS ion-source [57].
In line with these limitations, in recent years some innovative analytical approaches have been reported for RCS profiling and for their quantitative analysis in biological matrices including serum, urine and cells. The proposed methods so far reported can be roughly divided into two groups: (1) untargeted methods able to profile unknown RCS; (2) targeted methods characterized by high sensitivity for an absolute quantitative analysis of a known RCS.
5. Untargeted methods for RCS profiling
In recent years, some oxo-lipidomic approaches have been proposed with the aim of (i) obtaining a comprehensive measurement of the oxidized lipids and of the relative break-down products present in a biological matrix and (ii) profiling the individual analyte responses in various pathophysiological conditions.
One of the first oxo-lipidomic approaches to identify 4-hydroxyalkenal species in lipid extracts used carnosine as a derivatizing agent followed by a direct infusion on a nano ESI source connected to a triple quadrupole [58]. Carnosine rapidly reacts with unsaturated aldehydes and the Michael adducts formed are ionizable and hence, easily detectable using an ESI source. Moreover, collisional induced dissociation (CID) of carnosine adducts leads to diagnostic neutral losses that were used to specifically identify unknown adducts, whereas the structures of 4-hydroxyalkenal species were readily deduced from the detected molecular weight and the knowledge of the existence of naturally occurring polyunsaturated fatty acid structures. Four hydroxyalkenal species were identified in the lipid extracts of various mouse tissue samples, namely 4-hydroxyhexenal, 4-hydroxynondinenal, 4-hydroxynonenal, and 4-hydroxydodecatrienal. Moreover, by using deuterated standards, a quantitative measurement was also performed.
More comprehensive oxo-lipidomic approaches have taken advantage of the derivatization of RCS with 7-(diethylamino)coumarin-3-carbohydrazide (CHH) followed by ESI-MS analysis in positive ion mode [59]. Identification of CHH-derivatized compounds and elucidation of the cleavage sites and the oxidative modifications were carried out by specific neutral losses or fragment ions. This method allowed the identification of 69 and 122 products of in vitro oxidation of free fatty acids and phosphatidylcholine vesicles, respectively, thus illustrating the complexity of the reactive species generated. More recently this method has been applied to the characterization of lipid extracts from rat primary cardiomyocytes treated with the peroxynitrite donor SIN-1 [60]. Another derivatization method for comprehensive analysis of oxidized lipids uses p-toluenesulfonylhydrazine-(TSH) [57], which has a reactivity similar to DNPH but improved solubility and high volatility, and is suitable for the global derivatization of aldehydes and ketones coupled to a detection based on SWATH (Sequential Window Acquisition of all Theoretical Fragment-Ion Spectra) at high mass resolution. The approach was applied to the comprehensive quantification of known compounds as well as the identification of uncharacterized compounds in biological samples based on the detection of signature fragment ions originated from the derivatization reagent sub-structure. Similarly, another LC–ESI-MS approach uses dansyl hydrazine as a derivatizing agent and a MS analyzer operating in SRM mode, monitoring the ion at m/z 236.1 generated by the CID of the derivatized aldehydes whose [M+H]+ is scanned from m/z 275–949 [61]. A derivatization-free approach based on UHPLC–HRMS has also been proposed as a tool for the identification of aldehydes (2-alkenals, 4-hydroxy-2-alkenals, and 4-hydroxy-2,6-alkadienals). This method was suitable for detecting aldehydes from 6 to 16 carbons, and signals were found for [M+NH4]+ and [M+H]+ adducts in positive-ion mode ESI and for [M+HCOO]− adducts in negative-ion mode [62].
6. Targeted methods
Several methods have been reported for the quantitative analysis of free RCS in biological fluids, including capillary GC–MS, micellar electrokinetic chromatography, LC–MS/MS and various derivatizing procedures. Methods targeting a wide spectrum of RCS, including lipid- and glucose-derived RCS are desirable since, in pathological conditions, there is cross-talk between modification of proteins by the two classes of reactive species.
A method based on derivatization with 2,2′-furyl followed by HPLC coupled to fluorescence detection has been validated for the quantitative analysis of MDA, ACR, glyoxal (GO) and HNE in sera of humans, with detection limits ranging from 0.03 to 0.11 nmol/ml [63]. MDA was found to be the most abundant species in healthy volunteers, with a serum concentration greater than 10 nmol/ml, followed by the other aldehydes at concentration around 1 nmol/ml. Interestingly, this method detected significantly higher concentrations of ACR, MDA, and HNE in sera from diabetic and rheumatoid arthritis patients as compared to healthy controls. Carbonyl groups can also be derivatized with 4-(N,N-dimethylaminosulfonyl)-7-hydrazino-2,1,3-benzoxadiazole to generate per-oxyoxalate chemiluminescence [64]. This method has been applied for the analysis of methylglyoxal, ACR, crotonaldehyde and trans-2-hexenal with results ranging from 4.4 to 6.5 nM.
In spite of the comprehensive oxo-lipidomic methods developed over the last years, several limitations and unanswered questions on RCS analysis in biological matrices still remain. On one hand, the stability of RCS can be widely different, with α, β-unsaturated aldehydes being less stable than saturated aldehydes. This can affect the recovery yield and the accuracy of the method. Hence the recovery of RCS from biological samples should be carefully standardized and made reproducible by suitable sample preparation with addition of the derivatizing agent during sample collection and spiking of deuterated internal standards. On the other hand, it is important to consider the source of RCS detected by derivatization methods, and specifically, what proportion of RCS is in a free or protein-bound form, and what proportion can arise from reversible protein adducts such as Schiff bases or Michael adducts. Furthermore target analysis based on derivatization misses Schiff base protein–lipid adducts.
Notwithstanding these unanswered questions, owing to advances in analytical strategies there is now a better understanding of RCS. In particular, we now know that there are many more RCS besides the well-known RCS species such as HNE, HHE, ACR, GO and methylglyoxal, which have been extensively studied. Indeed, comprehensive lipidomic approaches have identified more than 400 different RCS species in rodent serum [61]. This, together with the knowledge about other lipidic reactive species, such as nitrated fatty acids, which do not possess carbonyl moieties [65], highlights the structural variety of reactive species that can modify proteins. Taking into account that the functional consequences of the modification may depend on the structure of the adducted moiety, a vast number of functionally diverse protein species may arise through lipoxidation. Moreover, significant advances have been made in quantitative detection of RCS in biological samples. From a quantitative point of view it would be important to determine the amount, not only of free RCS but also of protein-bound species. This information would be very valuable for studies in model systems to ascertain the role of lipoxidation using concentrations in the range of those occurring in vivo. A combination of strategies will therefore be needed for understanding the roles of lipoxidation in pathophysiology.
7. Analysis of protein lipoxidation through MS-based methods
7.1. Label-free MS methods
Mass spectrometry (MS)-based analytical approaches are one of the most popular methodologies to study biological systems and are used extensively in the study of protein–lipid adducts. MS technology offers unmatched performance in identification and quantification of these compounds, providing precise, sensitive and high-throughput analysis. It allows detailed structural information to be obtained at a molecular level, providing structural characterization of protein–lipid adducts in vitro and in vivo. Although the ultimate goal is to characterize the adducts formed in real pathophysiological conditions, an initial characterization of potentially important adducts can be obtained from the analysis of peptide–lipid or protein–lipid adducts prepared in vitro under controlled reaction conditions using pure preparations of the candidate peptides/protein and electrophilic lipids. The high reactivity of the electrophilic lipids allows the formation of adducts with the amino (–NH2) or sulfhydryl (–SH) groups almost spontaneously. The new peptide–lipid adducts can be easily detected by MS analysis with matrix assisted laser desorption/ionization (MALDI) or electrospray (ESI), often coupled to liquid chromatography (LC–MS), in positive ion mode. In the case of protein–lipid adducts, characterization by MS can follow the top-down or bottom-up proteomic strategies. Bottom-up approaches are by far the most popular, involving enzymatic hydrolysis of the protein–lipid adducts, usually using trypsin, followed by analysis of tryptic peptides and peptide–lipid adducts by MS (MALDI-MS or LC–MS) and MS/MS [66].
Peptide–lipid adducts are identified in MS as protonated molecules [M+H]+ or multiple charged ions [M+nH]n+ and modified peptides are typically recognized in the MS spectrum based on the mass shift against the unmodified peptide. In the case of Michael adducts, the mass shift caused by the modification is equal to the molecular weight of the RCS, while for the Schiff base it is equal to the molecular weight of the RCS minus 18 amu, owing to loss of H2O during adduct formation. For example, regarding peptide adducts with MDA, the mass deviation is +72 amu for Michael adducts and +54 for Schiff adducts in the case of single charged ions, or +36 amu for Michael adducts and 27 for Schiff adducts in the case of double charged ions, as observed in the study of MDA adduction of beta-lactoglobulin by LC–MS [67], [68]. Tandem mass spectrometry (MS/MS) of peptide–lipid adduct ions is usually performed to confirm the nature of the oxidative modification and to pinpoint its location in the peptide backbone. MS/MS commonly utilizes collision-induced dissociation (CID), and details about the site of modification and type of adducts in peptides are inferred by the deviation of the typical y or b-type fragment ions or by the presence of modified immoniun ions [69]. CID-tandem mass spectra also show typical loss of the carbonyl moiety, leading to the formation of the non-modified b and y-ions, which might prevent the identification of the position of the adduct. It is possible to take advantage of this characteristic loss of the electrophilic lipid and of the modified immonium ions. These typical fragmentation pathways can be used to define specific target reporter ion-based MS approaches, namely neutral loss scans (NLS) or precursor ions scan (PIS). NLS have been widely used for the detection of HNE–peptide adducts. MS3 neutral loss scanning of 156 amu (singly charged), 78 amu (doubly charged) or 52 amu (triply charged) approaches were used to locate HNE–peptide Michael adducts [70], [71], as reported for example for HNE–cytochrome C oxidase adducts [71] or HNE with beta-spectrin in human blood samples [72]. More recently, dissociation techniques that do not induce fragmentation of labile bonds, such as electron capture dissociation (ECD) and electron transfer dissociation (ETD) have been used [4], [73]. MS/MS spectra obtained under these conditions can be more informative, showing abundant c- and z-type fragment ions of modified peptides, since the carbonyl–peptide bond is more stable, preventing the loss of the carbonyl moiety. ETD is also preferred for the analysis of bigger peptides and intact proteins and thus for top-down approaches.
Peptide and protein adducts with small electrophilic lipids, like the α, β-unsaturated aldehydes HNE, MDA and ACR are by far the most studied [3], [66], [74], and protein adducts involving these lipids have been detected and characterized both in vitro and in many pathophysiological situations [30], [32]. Nevertheless, for other lipid classes, the evidence is scarcer. There are numerous reports of the formation of protein adducts of cyPG in several experimental models of pathophysiological situations through various techniques, either involving endogenously generated or exogenously added cyPG. Most studies combine the detection of modified proteins through the use of metabolic labeling (see below) with MS characterization of the candidate cyPG–peptide adducts in the positive mode, using purified protein or peptides incubated in vitro with cyPG (for review see [3], [19]). Nevertheless, at present, the MS detection of prostaglandin–protein adducts formed endogenously is yet to be established. Peptide/protein modifications by electrophilic lipids esterified to phospholipids have rarely been considered [3]. Recently, several groups have identified adducts of RCS derived from phosphatidylcholine with His, Cys and Lys in synthetic peptides of ApoA and ApoB-100 [75], [76]. This is a field of research that should be explored further since adducts of proteins with oxidized phospholipids have been detected in HDL and in human platelets enriched with electrophilic truncated oxidized phosphatidylcholine (KODA-phospahtidylcholine) [77].
Thus, the number of protein–lipid adducts formed under controlled chemical conditions is relatively low, and their spectra are relatively easy to analyze. Data obtained through this approach allows the identification of the type of adducts that can be formed for each electrophilic lipid. Additionally, this approach has the advantage of decoding the reactivity of each specific electrophilic lipid species and specific fragmentation patterns of lipoxidation adducts under tandem MS conditions can be identified, which can be very useful for target analysis in biological samples. However, use of more complex samples from cells, biological fluids and tissues requires more complicated, specific and targeted approaches to identify and characterize the protein adducts. These can take advantage of enrichment procedures, chemical labeling and bottom-up proteomic strategies combining enzymatic digestion, chromatographic methods and analysis by MALDI-MS and/or LC–MS and MS/MS. Enrichment and derivatization methods, followed by MS analysis, are commonly used to improve sensitivity and selectivity, which aids in the structural identification and may be performed before or after protein digestion. In this regard, it should be noted that aldehydes that react with lysines and arginines pose additional difficulties since these modifications may interfere with trypsin digestion, resulting in long peptides that are difficult to sequence. Also, false positives of HNE adducts can be obtained since they would add the same mass as arginine. Stabilization of lipid adducts with NaBH4 prior to enzymatic digestion and MS analysis is usually performed [78]. Targeted analysis based on NLS scans, of either underivatized or derivatized samples is becoming a popular analytical strategy to search for specific protein modifications arising from a particular lipoxidation product. Recently published papers illustrate how combined MS approaches (MALDI-TOF and LC–ESI-MS/MS) can contribute to uncover protein lipid adducts in biological samples. HNE adducts of metalloproteinase rhMMP-13 were identified by bottom-up approaches using HR-LC–MS/MS and target analysis by multiple-reaction monitoring (MRM) based on specific neutral loss of HNE in vitro and in chondrocytes from osteoarthritic patients [79]. Results from proteomic approaches, using MALDI-TOF/TOF combined with SDS-PAGE and enzymatic digestion, showed that liver fatty acid-binding protein (L-FABP) is a target for modification by HNE [80]. Several nuclear proteins including actin, chromodomain-helicase-DNA-binding protein 4, heterogeneous nuclear ribonucleoprotein L(hnRNPs), and neuroblastoma differentiation-associated protein AHNAK, were found to be modified by HNE, HHE, and ONE, among others, using SDS-PAGE and LC–MS in a model of liver steatosis [81].
7.2. Label-based MS methods
The most commonly used measurement of protein oxidation is probably the formation of protein carbonyl groups (often referred to as protein carbonylation); this is because robust, simple and economical methods are available for detection of carbonyl groups on proteins [82]. While bi-functional lipid oxidation products, such as dialdehydes or hydroxyalkenals, do react with proteins to yield free carbonyls, a substantial number of other products do not. Moreover, protein carbonyls can also be formed by direct oxidation of the side chains of lysine, proline, arginine and other residues, for example through oxidative deamination. Thus while many studies report on the occurrence of protein carbonyls in disease, this cannot be considered as evidence of lipoxidation unless specific antibodies against lipid oxidation products have been used, or the modifications are analyzed by mass spectrometry, as described in later sections.
To improve sensitivity and selectivity in the detection of carbonylated proteins, some analytical approaches take advantage of chemical labeling of carbonylated lipid–protein adducts prior to MS detection [83]. These derivatization procedures, as discussed in previous sections, usually exploit the high reactivity of the free carbonyl groups of the Michael lipoxidation adducts, which are very prone to react with free amine containing molecules of the derivatizing reagent. Lipid–protein Schiff adducts cannot be derivatized by this approach, unless the electrophilic lipid is a di-aldehyde. Derivatization with DNPH has been used for more than a decade, since DNPH labeled carbonylated peptides showed favorable ionization, providing a sensitive detection method. Nevertheless, this method presents some disadvantages, outlined above for the analysis of free RCS together with a lack of selectivity because DNPH also reacts with sulfenic acid [84]. 7-(Diethylamino)coumarin-3-carbohydrazide has also been used for derivatization of lipid-bound carbonyls [85], although other chemical labels can also be used [86]. One of the most common chemical labeling procedures employs biotin-based hydrazide-functionalized reagents. This approach allows enrichment of biotin-linked adducts by avidin chromatography [4], thus reducing the amount of unmodified peptides in the MS analysis. Two routes can be considered: enrichment of biotinylated proteins followed by enzymatic digestion and MS, or initial digestion of protein and further separation of biotinylated peptides by affinity chromatography [66]. MS based approaches combined with enrichment procedures using biotin are increasingly being used to recognize site-specific modifications of proteins in complex proteomes from biological samples. Recently, Aluise et al. [87] identified a peptidylprolyl cis/trans isomerase A1 (Pin1) adduct with HNE linked to the catalytic cysteine (Cys113) using click chemistry conjugation of biotin to Pin1 and LC–MS/MS analysis. In vitro covalent modification of the mitochondria protein NAD-dependent deacetylase sirtuin-3 (zrSIRT3) by HNE at Cys280 was recognized using biotin hydrazide (BH) treatment and avidin pull-down and further analysis by LC–MS/MS and MALDI-TOF/TOF [88].
As mentioned above, until now most published work has focused on HNE–protein lipid adducts, but many more electrophilic lipid species are generated during lipid oxidation. Further investigation and experimentation in this field will be needed in order to understand the impact of lipoxidation in heath and disease. Future research should therefore concentrate on new methods for in vivo detection of carbonylated proteins and other lipid adducts.
8. Detection of protein lipoxidation through non-MS approaches
Metabolic labeling methods are based on the incubation of biological samples, mostly cells in culture, with labeled analogs of the oxidized or electrophilic species or their precursors. As stated above, some of these methods can be used both in MS and non-MS approaches. Incubation of cells in the presence of radioactive precursors of oxidized lipid species, in particular with radioactive arachidonic acid, which is a precursor of electrophilic prostaglandins and of other reactive lipids, has allowed the detection of incorporation of radioactivity into proteins [89]. These methods have the advantage that the structure of the resulting lipoxidized species is not affected; thus, metabolism and interactions are preserved. As a drawback, this labeling method requires handling of radioactive material and does not allow imaging or enrichment procedures.
Biotinylated derivatives of electrophilic lipid species or their parent compounds have been widely used for the study of protein lipoxidation. Biotinylated analogs of cyPG or their precursors are among the most widely used probes. The fact that some cyPG showed beneficial effects in experimental models of inflammation and tissue injury spurred research in the identification of the proteins modified by these mediators, seeking novel therapeutic targets, even before their real pathophysiological importance had been elucidated. Indeed, the study of protein modification by biotinylated cyPG has paved the way for studies with other lipid species with similar structure or reactivity, including nitrated fatty acids and isoprostanes. In fact, many of the targets of cyPG identified, including NF-κB and transient receptor potential (TRP) channels [90], [91], have later been confirmed to be modified by the other lipids [65], [92]. The progress with cyPG may be due to the fact that these compounds are relatively stable, as are the resulting adducts with proteins, and they are amenable to derivatization through modification of the carboxyl group by various moieties. Therefore, since the first descriptions of biotinylated PGA2 [93] and 15d-PGJ2 [90], many biotinylated derivatives of arachidonic acid-derived electrophilic lipids have been commercialized and are readily available. Advantages of these derivatives include the exploitation of the high affinity avidin–biotin interaction for both detection and enrichment of the samples for purification and identification. Among the drawbacks of this approach is the fact that the biotin moiety imposes structural restrictions that may affect interaction with proteins (see below). Thus, although biotinylated analogs mimic many of the effects of their parent compounds, there are important functional differences [50], [94]. In addition, none of these metabolic labeling procedures are suitable for use in humans.
Analogously, fluorescent labels have been introduced in electrophilic lipid species for the detection and quantitation of lipoxidized proteins either in gels or in cells [95], [96]. In combination with proteomic techniques, these approaches are also suitable for the identification of potential targets of lipoxidation, although it should be taken into account that, as in the case of biotinylated tags, due to the bulky nature of the fluorescent moieties, validation of the identified targets is required and the biological interaction may be affected.
Click chemistry has also been used to monitor the fate of HNE and oxidized phospholipids [87], [97] and presents the advantage of the smaller tagging moiety, which is considered in most instances not to interfere with the metabolism or interactions of the labeled lipids. This type of labeling allows the derivatization of the tag by an alkyne-azide reaction ex vivo, that is, in tissue extracts, partially purified samples or permeabilized cells, thus facilitating detection, enrichment, imaging, etc. This strategy has been successfully used to detect the HNE-induced cross-linking of peptidylprolyl cis/trans isomerase A1 (Pin1) [87].
As discussed above, lipoxidation is a selective process that does not affect cellular proteins randomly but is directed by structural features of the protein and the reactive lipid species [19], [98]. Intracellular redox status and availability of small molecule antioxidants such as glutathione is also an important determinant for this selectivity [98], [99]. In addition, lipoxidation could present a selective compartmentalization depending on the site of generation of electrophilic lipid species, or on the distribution of small molecule thiols that could act as decoys or the presence of oxidized lipid detoxifying enzymes such as GSTs [94], [100]. Therefore, topography of lipoxidation may also be important in its functional outcome. In this context, some probes have been synthesized that combine derivatization of electrophilic lipids with biotin or fluorescent tags, and organelle-specific tags or targeting moieties for selectively detecting lipoxidation at particular subcellular environments [101].
Importantly, all these label-based methods can be used both in “positive” (or direct) and “negative” (or indirect) approaches to detect protein modification. In the positive approaches, the modified targets are identified. In the negative approaches, modification is induced first with an oxidant or electrophilic lipid and the reduction of the signal with the labeled probe is observed [96], [102], or vice versa. These approaches can be used in vitro and in cells or tissues, and are useful to study the competition between the tagged and the parent lipid or the potential interplay between lipoxidation and concurrent modifications, either oxidative, or by endogenous or exogenous electrophiles, such as drugs [53], [103], particularly those affecting the versatile cysteine residues (see below) [104]. Nevertheless, as considered in the following section, especially when using live cells, these approaches need to be complemented with other methods since the modifications occurring in cells treated with oxidized lipids can be structurally quite varied and involve not only lipoxidation but also diverse oxidations related to oxidative stress generated by the reactive species [100].
The ex vivo derivatization of lipoxidized proteins with various reagents has been considered above. In addition to MS procedures, the derivatized moieties can be detected using fluorescence methods, avidin–biotin based approaches or specific antibodies. Nevertheless, some derivatization procedures present limitations. As mentioned above, although lipoxidation can lead to an increase in the amount of carbonyl groups on proteins as a result of the incorporation of some lipid moieties containing these structures, carbonyl groups formed by oxidative modification of certain residues will also be detected by methods based on derivatization of carbonyl groups, such as reaction with DNPH. Moreover, this method will not be specific for lipoxidation, nor will it detect all types of lipoxidation, but only those in which a carbonyl moiety is preserved after lipid addition. In spite of this, the dinitrophenylhydrazone generated after reaction of DNPH with carbonyl groups can be detected spectrophotometrically or by antibodies against the dinitrophenyl group, which has contributed to the wide use of this method. Similar detection can be achieved with biotin hydrazide derivatization, which reacts with carbonyl groups but in this case adds a biotin moiety to the modified residue, enabling avidin-based affinity detection or enrichment strategies. The methods for detecting carbonyl formation on proteins have been reviewed recently [105].
Lastly, there are several antibodies that directly detect adducts of lipid peroxidation products with proteins. These include antibodies against various types of HNE adducts [106], which may be selective for cysteine or histidine adducts, and antibodies against other adducts like ACR–protein, MDA–lysine and oxidized phospholipid–protein (please see [3] for review). In addition, antibodies against 15d-PGJ2 have been used to detect this PG intracellularly by immunohistochemistry [107], as well as protein adducts involving this lipid through ELISA [49]. Kato et al. [108] reviewed antibody-based methods for detection of a range of unusual modifications of proteins with hydroperoxide-derived products that form amide-type lipid–lysine adducts like HEL. In all these cases, assuming an optimal specificity of the antibody, information on the proteins modified can be obtained, but not information on the site of modification, unless the use of antibodies is combined with other strategies, such as the study of site-specific mutants as described below.
9. Validation strategies
The results of the detection and identification of lipoxidation targets may give a general view of the extent of these modifications in a given sample and may also pinpoint targets of potential pathophysiological or therapeutic interest. Lipoxidation is usually not random but occurs at precise residues within proteins [100]. The affected residues are often strong nucleophiles, and may be located in environments that favor the docking or interaction of the reactive lipid species [109], [110]. Frequently, the modified residues are involved in catalysis or interaction with other proteins. Therefore, validation of the modification and assessment of its functional consequences is of special importance in delineating its contribution to the overall effect of electrophilic lipids.
For validation of lipoxidation, usually a combination of strategies is required. In the most favorable scenario, a protein would be identified from a sample of a pathophysiologically relevant model, in which the modification by endogenously generated species would be detected and the sites of addition directly spotted by MS approaches. In this case, functional assessment of the modification can be undertaken. However, in many cases, protein modification is achieved by the use of exogenously added lipid electrophiles, either unmodified or tagged. Above all in this situation, validation strategies are required. In Fig. 2 we present a summary of some of the currently used methods for validation of targets of lipoxidation. If the electrophile was tagged, it is important to confirm the results using the parent compound, since any tag can confer steric hindrance to the interaction either with the targets of interest or with other structures, leading to indirect effects [94]. Nevertheless, the differential behavior of tagged and untagged electrophilic lipids can be exploited to infer functional or structural consequences of the modification [50]. Immunoprecipitation of the modified protein or other affinity-based purification procedures such as those based on avidin–biotin interaction have proved extremely useful for assessing protein lipoxidation in the case of low abundance proteins or of peptides modified in a low proportion [19], [111]. Fig. 3 outlines the identification and characterization approaches that have been used in the case of a thoroughly studied lipoxidation target: the cytoskeletal protein vimentin. In addition, we illustrate several features of the functional characterization of the modification (see below for detailed comments).
Fig. 2.
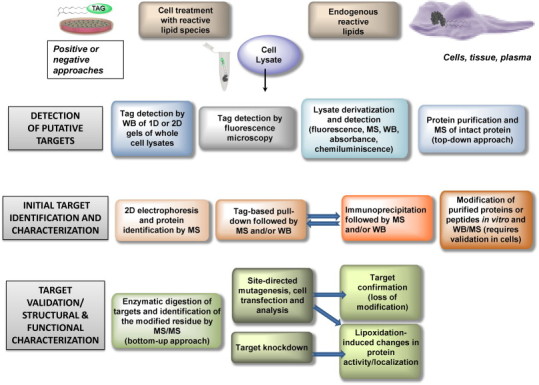
Summary of procedures useful for the identification, characterization and functional assessment of protein lipoxidation. The rows represent increasing complexity and depth of information from top to bottom. At each level of complexity there are several alternative but complementary approaches that can be used. WB, western blot.
Fig. 3.
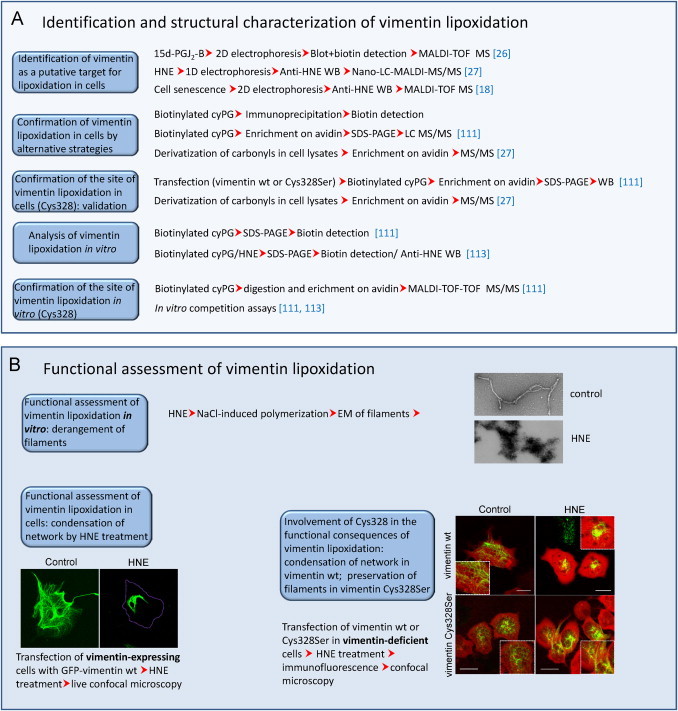
Methods employed in the study of the lipoxidation of vimentin. (A) Combination of strategies employed in the chemical and structural characterization of vimentin lipoxidation. (B) Several approaches used in the assessment of the consequences of vimentin lipoxidation in vitro and in cellular contexts.
From a functional point of view, it is important to take into account that oxidized lipids bind to many targets in the cell proteome and their effects arise from the complex interplay of the diverse modifications. As stated above, induction of oxidative stress may be a source of concomitant modifications that may coexist or compete with lipoxidation, hence complicating the structural and functional outcomes. This is particularly relevant in the case of modifications of cysteine residues, which, in addition to lipoxidation by various reactive species, may be targeted by oxidative and nitrosative modifications of varied structure, including glutathionylation, nitrosylation, cysteinylation, sulfenylation, etc. Some recent comprehensive reviews have addressed the interplay between lipoxidation and oxidative modifications [100], [112]. Therefore, care should be exercised to assign functional roles to lipoxidation of proteins by specific moieties without prior assessment of the overall oxidative state of the protein and its cysteine residues. Moreover, lipoxidation may result in activation or inhibition depending on the target and the structure of the adducted moiety. Modifications of Ras proteins may have different consequences on subcellular localization and activation platforms. Whereas modification by small hydrophobic moieties appears to favor localization at and/or signaling from the Golgi, modification by fatty acids (i.e. palmitoylation) or addition of cyPG or cGMP to the C-terminal cysteines seems to promote localization and/or signaling at the plasma membrane (reviewed in [100]). Use of labeled electrophilic species in combination with markers specific for various subcellular compartments has been used to obtain information on the presence of a particular target at a defined compartment through fluorescence microscopy. Nevertheless, given the high number of cellular targets of lipoxidation, colocalization studies may offer information on the subcellular localization of adducts but is insufficient for target validation.
The proportion of the target modified is also an important issue which may have diverse implications, if we consider a signal versus background effect. Activation of a small proportion, i.e. 1% of an otherwise inactive protein may have significant effects, whereas inhibition of a higher proportion, i.e. 10% of a fully active protein may not be enough to elicit a detectable outcome. In order to establish a correlation between target modification and functional effects, the use of mutant constructs and/or model systems in which the levels of the target can be modulated are needed. In cases in which lipoxidation occurs at a specific residue within one protein, mutation of that residue can sometimes block the modification and completely rescue the functional alteration. Such is the case with the transcription factor NF-κB and the intermediate filament protein vimentin [26], [90] (see Fig. 3B). While the effect of the mutation can be clearly observed in vitro when working with purified proteins, in cellular systems it is usually necessary to work with a model that does not express the wild type form. In the case of vimentin, transfection of cells that do not express the endogenous protein has allowed the response of homogeneous protein constructs/variants to electrophilic lipids to be explored [113]. These studies have shown that mutation of the target residue, in this case Cys328, protects the mutant protein from the morphological derangement induced by electrophilic lipids [113], as illustrated in Fig. 3 for HNE. Thus, this residue behaves as a sensor for this type of stress, either through direct modification by HNE addition or by reactive species generated during HNE treatment. In some cases, mutation of the target residue is deleterious for protein function, making it more difficult to assess the functional consequence of lipoxidation. This may be the case of actin, which is targeted by various electrophiles at Cys374 [23], [24], [25], a residue located at the interface between actin monomers in such a way that its mutation may induce altered microfilament patterns per se [114]. In some cases, although the function of a particular protein may be spared by mutation of the target residue, the general consequences for the cell could even be worsened if the mutated site was acting as a decoy, thus scavenging the reactive species.
It is also important to consider that lipid electrophiles are key regulators of gene expression, thus, the functional effects, above all in the long run can be the result of protein modification plus altered gene expression, which can amplify or counteract the effect of the electrophiles [50], [98].
Ultimately, confirmation of the modification of a given target will be achieved when the adduct is detected on a peptide from the protein, whereas functional confirmation will be achieved by combined strategies, such as those involving mutation of the modified residue or alterations of expression levels.
10. Concluding remarks
Protein lipoxidation is obtaining recognition as a mechanism for regulation of protein function in health and disease. Given the wide structural variability and complexity of these posttranslational modifications, MS-based methods are essential for their characterization. In turn, functional assessment of the modification requires integrated approaches that take into account other potential modifications. Lipoxidation may interact or compete with other modifications, including oxidative modifications or adduct formation with drugs in therapeutic regimes of patients, thus creating complex patterns, the characterization of which requires highly specific and sensitive methods. In vitro studies using either labeled or tag-free oxidized lipids are essential to unveil the potential sites of modification by these compounds, though further work regarding protein modifications in biological samples by endogenously generated species and their functional relevance will be key for a full understanding of the role of these lipids in pathophysiological scenarios.
Acknowledgments
Work at the authors’ laboratories is supported by grants: MINECO SAF2012-36519 and ISCIII RETIC RD12/0013/0008 (Spain) to DPS; PEst-C/QUI/UI0062/2013, FCOMP-01-0124-FEDER-037296 to QOPNA research unit and RNEM-REDE/1504/REM/2005 to Portuguese National Mass Spectrometry Network by Fundação para a Ciência e a Tecnologia (FCT, Portugal), European Union, QREN, FEDER, and COMPETE to MRMD and PD. CMS acknowledges funding from the Engineering and Physical Sciences Research Council, UK, EP/I017887/1 Cross-Disciplinary Research Landscape Award (the Proxomics Project). Collaboration among the authors’ laboratories has been supported by EU COST Action CM1001 on “Chemistry of non-enzymatic protein modification − modulation of protein structure and function”.
References
- 1.Vistoli G., De Maddis D., Cipak A., Zarkovic N., Carini M., Aldini G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): an overview of their mechanisms of formation. Free Radical Research. 2013;47:3–27. doi: 10.3109/10715762.2013.815348. 23767955 [DOI] [PubMed] [Google Scholar]
- 2.Pamplona R. Advanced lipoxidation end-products. Chemico-Biological Interactions. 2011;192(1–2):14–20. doi: 10.1016/j.cbi.2011.01.007. 21238437 [DOI] [PubMed] [Google Scholar]
- 3.Domingues R.M., Domingues P., Melo T., Pérez-Sala D., Reis A., Spickett C.M. Lipoxidation adducts with peptides and proteins: deleterious modifications or signaling mechanisms? Journal of Proteomics. 2013;92:110–131. doi: 10.1016/j.jprot.2013.06.004. 23770299 [DOI] [PubMed] [Google Scholar]
- 4.Vasil’ev Y.V., Tzeng S.C., Huang L., Maier C.S. Protein modifications by electrophilic lipoxidation products: adduct formation, chemical strategies and tandem mass spectrometry for their detection and identification. Mass Spectrometry Reviews. 2014;33(3):157–182. doi: 10.1002/mas.21389. 24818247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ullery J.C., Marnett L.J. Protein modification by oxidized phospholipids and hydrolytically released lipid electrophiles: investigating cellular responses. Biochimica Biophysica Acta. 2012;1818(10):2424–2435. doi: 10.1016/j.bbamem.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stemmer U., Hermetter A. Protein modification by aldehydophospholipids and its functional consequences. Biochimica Biophysica Acta. 2012;1818(10):2436–2445. doi: 10.1016/j.bbamem.2012.03.006. 22450235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niki E. Lipid peroxidation: physiological levels and dual biological effects. Free Radical Biology and Medicine. 2009;47(5):469–484. doi: 10.1016/j.freeradbiomed.2009.05.032. 19500666 [DOI] [PubMed] [Google Scholar]
- 8.Guéraud F., Atalay M., Bresgen N., Cipak A., Eckl P.M., Huc L., et al. Chemistry and biochemistry of lipid peroxidation products. Free Radical Research. 2010;44(10):1098–1124. doi: 10.3109/10715762.2010.498477. 20836659 [DOI] [PubMed] [Google Scholar]
- 9.Reis A., Spickett C.M. Chemistry of phospholipid oxidation. Biochimica Biophysica Acta. 2012;1818(10):2374–2387. doi: 10.1016/j.bbamem.2012.02.002. 22342938 [DOI] [PubMed] [Google Scholar]
- 10.Murphy S.A., Al-Aaswad N.M., Nicolaou A. In: Lipid Oxidation in Health and Disease. Spickett C.M., Forman H.J., editors. CRC Press; London, UK: 2015. Enzymatic oxidation of polyunsaturated fatty acids. [Google Scholar]
- 11.Perry E.A., Castellani R.J., Moreira P.I., Nunomura A., Lui Q., Harris P.L., et al. Neurofilaments are the major neuronal target of hydroxynonenal-mediated protein cross-links. Free Radical Research. 2013;47(6–7):507–510. doi: 10.3109/10715762.2013.794265. 23566300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts L.J., 2nd, Morrow J.D. Products of the isoprostane pathway: unique bioactive compounds and markers of lipid peroxidation. Cellular and Molecular Life Sciences. 2002;59(5):808–820. doi: 10.1007/s00018-002-8469-8. 12088281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sánchez-Gómez F.J., Díez-Dacal B., Pajares M.A., Llorca O., Pérez-Sala D. Cyclopentenone prostaglandins with dienone structure promote cross-linking of the chemoresistance-inducing enzyme glutathione transferase P1-1. Molecular Pharmacology. 2010;78(4):723–733. doi: 10.1124/mol.110.065391. 20631055 [DOI] [PubMed] [Google Scholar]
- 14.Baraibar M.A., Friguet B. Oxidative proteome modifications target specific cellular pathways during oxidative stress, cellular senescence and aging. Experimental Gerontology. 2013;48(7):620–625. doi: 10.1016/j.exger.2012.10.007. 23127722 [DOI] [PubMed] [Google Scholar]
- 15.Thomas J.P., Maiorino M., Ursini F., Girotti A.W. Protective action of phospholipid hydroperoxide glutathione peroxidase against membrane-damaging lipid peroxidation. in situ reduction of phospholipid and cholesterol hydroperoxides. Journal of Biological Chemistry. 1990;265(1):454–461. 2294113 [PubMed] [Google Scholar]
- 16.Alary J., Guéraud F., Cravedi J.P. Fate of 4-hydroxynonenal in vivo: disposition and metabolic pathways. Molecular Aspects of Medicine. 2003;24(4–5):177–187. doi: 10.1016/S0098-2997(03)00012-8. 12892995 [DOI] [PubMed] [Google Scholar]
- 17.Spite M., Baba S.P., Ahmed Y., Barski O.A., Nijhawan K., Petrash J.M., et al. Substrate specificity and catalytic efficiency of aldo–keto reductases with phospholipid aldehydes. Biochemical Journal. 2007;405(1):95–105. doi: 10.1042/BJ20061743. 17381426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed E.K., Rogowska-Wrzesinska A., Roepstorff P., Bulteau A.L., Friguet B. Protein modification and replicative senescence of WI-38 human embryonic fibroblasts. Aging Cell. 2010;9(2):252–272. doi: 10.1111/j.1474-9726.2010.00555.x. 20102351 [DOI] [PubMed] [Google Scholar]
- 19.Garzón B., Oeste C.L., Díez-Dacal B., Pérez-Sala D. Proteomic studies on protein modification by cyclopentenone prostaglandins: expanding our view on electrophile actions. Journal of Proteomics. 2011;74(11):2243–2263. doi: 10.1016/j.jprot.2011.03.028. 21459170 [DOI] [PubMed] [Google Scholar]
- 20.Aldini G., Vistoli G., Regazzoni L., Gamberoni L., Facino R.M., Yamaguchi S., et al. Albumin is the main nucleophilic target of human plasma: a protective role against pro-atherogenic electrophilic reactive carbonyl species? Chemical Research in Toxicology. 2008;21(4):824–835. doi: 10.1021/tx700349r. 18324789 [DOI] [PubMed] [Google Scholar]
- 21.Jacobs A.T., Marnett L.J. Systems analysis of protein modification and cellular responses induced by electrophile stress. Accounts of Chemical Research. 2010;43(5):673–683. doi: 10.1021/ar900286y. 20218676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang X., Fu Y., Long M.J., Haegele J.A., Ge E.J., Parvez S., et al. Temporally controlled targeting of 4-hydroxynonenal to specific proteins in living cells. Journal of the American Chemical Society. 2013;135(39):14496–14499. doi: 10.1021/ja405400k. 24015839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldini G., Dalle-Donne I., Vistoli G., Maffei Facino R., Carini M. Covalent modification of actin by 4-hydroxy-trans-2-nonenal (HNE): LC–ESI-MS/MS evidence for Cys374 Michael adduction. Journal of Mass Spectrometry. 2005;40(7):946–954. doi: 10.1002/jms.872. 15934040 [DOI] [PubMed] [Google Scholar]
- 24.Gayarre J., Sánchez D., Sánchez-Gómez F.J., Terrón M.C., Llorca O., Pérez-Sala D. Addition of electrophilic lipids to actin alters filament structure. Biochemical and Biophysical Research Communications. 2006;349(4):1387–1393. doi: 10.1016/j.bbrc.2006.09.005. 16979589 [DOI] [PubMed] [Google Scholar]
- 25.Aldini G., Carini M., Vistoli G., Shibata T., Kusano Y., Gamberoni L., et al. Identification of actin as a 15-deoxy-Delta12,14-prostaglandin J2 target in neuroblastoma cells: mass spectrometric, computational, and functional approaches to investigate the effect on cytoskeletal derangement. Biochemistry. 2007;46(10):2707–2718. doi: 10.1021/bi0618565. 17297918 [DOI] [PubMed] [Google Scholar]
- 26.Stamatakis K., Sánchez-Gómez F.J., Pérez-Sala D. Identification of novel protein targets for modification by 15-deoxy-Delta12,14-prostaglandin J2 in mesangial cells reveals multiple interactions with the cytoskeleton. Journal of the American Society Nephrology. 2006;17(1):89–98. doi: 10.1681/ASN.2005030329. 16291835 [DOI] [PubMed] [Google Scholar]
- 27.Chavez J., Chung W.G., Miranda C.L., Singhal M., Stevens J.F., Maier C.S. Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP-1 monocytic cells: protein carbonylation is diminished by ascorbic acid. Chemical Research in Toxicology. 2010;23(1):37–47. doi: 10.1021/tx9002462. 20043646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aldini G., Granata P., Orioli M., Santaniello E., Carini M. Detoxification of 4-hydroxynonenal (HNE) in keratinocytes: characterization of conjugated metabolites by liquid chromatography/electrospray ionization tandem mass spectrometry. Journal of Mass Spectrometry. 2003;38(11):1160–1168. doi: 10.1002/jms.533. 14648823 [DOI] [PubMed] [Google Scholar]
- 29.Yang J., Tallman K.A., Porter N.A., Liebler D.C. Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Analytical Chemistry. 2015;87(5):2535–2541. doi: 10.1021/ac504685y. 25654326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butterfield D.A., Reed T., Sultana R. Roles of 3-nitrotyrosine- and 4-hydroxynonenal-modified brain proteins in the progression and pathogenesis of Alzheimer’s disease. Free Radical Research. 2011;45(1):59–72. doi: 10.3109/10715762.2010.520014. 20942567 [DOI] [PubMed] [Google Scholar]
- 31.Leonarduzzi G., Chiarpotto E., Biasi F., Poli G. 4-hydroxynonenal and cholesterol oxidation products in atherosclerosis. Molecular Nutrition and Food Research. 2005;49(11):1044–1049. doi: 10.1002/mnfr.200500090. 16270277 [DOI] [PubMed] [Google Scholar]
- 32.Aldini G., Orioli M., Carini M. Protein modification by acrolein: relevance to pathological conditions and inhibition by aldehyde sequestering agents. Molecular Nutrition and Food Research. 2011;55(9):1301–1319. doi: 10.1002/mnfr.201100182. 21805620 [DOI] [PubMed] [Google Scholar]
- 33.Dunér P., Gonçalves I., Grufman H., Edsfeldt A., To F., Nitulescu M., et al. Increased aldehyde-modification of collagen type IV in symptomatic plaques − a possible cause of endothelial dysfunction. Atherosclerosis. 2015;240(1):26–32. doi: 10.1016/j.atherosclerosis.2015.02.043. 25746374 [DOI] [PubMed] [Google Scholar]
- 34.Tsimikas S., Witztum J.L. The role of oxidized phospholipids in mediating lipoprotein(a) atherogenicity. Current Opinion in Lipidology. 2008;19(4):369–377. doi: 10.1097/MOL.0b013e328308b622. 18607184 [DOI] [PubMed] [Google Scholar]
- 35.Zhang W., Salomon R.G. Oxidized phospholipids, isolevuglandins, and atherosclerosis. Molecular Nutrition and Food Research. 2005;49(11):1050–1062. doi: 10.1002/mnfr.200500056. 16270278 [DOI] [PubMed] [Google Scholar]
- 36.Taleb A., Witztum J.L., Tsimikas S. Oxidized phospholipids on apoB-100-containing lipoproteins: a biomarker predicting cardiovascular disease and cardiovascular events. Biomarkers in Medicine. 2011;5(5):673–694. doi: 10.2217/bmm.11.60. 22003918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mali V.R., Ning R., Chen J., Yang X.P., Xu J., Palaniyandi S.S. Impairment of aldehyde dehydrogenase-2 by 4-hydroxy-2-nonenal adduct formation and cardiomyocyte hypertrophy in mice fed a high-fat diet and injected with low-dose streptozotocin. Experimental Biology and Medicine. 2014;239(5):610–618. doi: 10.1177/1535370213520109. 24651616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Souza A., Kurien B.T., Rodgers R., Shenoi J., Kurono S., Matsumoto H., et al. Detection of catalase as a major protein target of the lipid peroxidation product 4-HNE and the lack of its genetic association as a risk factor in SLE. BMC Medical Genetics. 2008;9:62. doi: 10.1186/1471-2350-9-62. 18606005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carbone D.L., Doorn J.A., Kiebler Z., Petersen D.R. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chemical Research in Toxicology. 2005;18(8):1324–1331. doi: 10.1021/tx050078z. 16097806 [DOI] [PubMed] [Google Scholar]
- 40.Shearn C.T., Backos D.S., Orlicky D.J., Smathers-McCullough R.L., Petersen D.R. Identification of 5′ AMP-activated kinase as a target of reactive aldehydes during chronic ingestion of high concentrations of ethanol. Journal of Biological Chemistry. 2014;289(22):15449–15462. doi: 10.1074/jbc.M113.543942. 24722988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pradeep A.R., Agarwal E., Bajaj P., Rao N.S. 4-hydroxy-2-nonenal, an oxidative stress marker in crevicular fluid and serum in type 2 diabetes with chronic periodontitis. Contemporary Clinical Dentistry. 2013;4(3):281–285. doi: 10.4103/0976-237X.118342. 24124291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S.E., Park Y.S. Role of lipid peroxidation-derived alpha, beta-unsaturated aldehydes in vascular dysfunction. Oxidative Medicine and Cell Longevity. 2013;2013:629028. doi: 10.1155/2013/629028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrera G., Pizzimenti S., Ciamporcero E.S., Daga M., Ullio C., Arcaro A., et al. Role of 4-hydroxynonenal-protein adducts in human diseases. Antioxidants and Redox Signaling. 2015 doi: 10.1089/ars.2014.6166. , in press. [DOI] [PubMed] [Google Scholar]
- 44.McDowell R.E., McGeown J.G., Stitt A.W., Curtis T.M. Therapeutic potential of targeting lipid aldehydes and lipoxidation end-products in the treatment of ocular disease. Future Medicinal Chemistry. 2013;5(2):189–211. doi: 10.4155/fmc.12.202. 23360143 [DOI] [PubMed] [Google Scholar]
- 45.Chen M., Curtis T.M., Stitt A.W. Advanced glycation end products and diabetic retinopathy. Current Medicinal Chemistry. 2013;20(26):3234–3240. doi: 10.2174/09298673113209990025. 23745547 [DOI] [PubMed] [Google Scholar]
- 46.Madian A.G., Regnier F.E. Proteomic identification of carbonylated proteins and their oxidation sites. Journal of Proteome Research. 2010;9(8):3766–3780. doi: 10.1021/pr1002609. 20521848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendez D., Hernaez M.L., Diez A., Puyet A., Bautista J.M. Combined proteomic approaches for the identification of specific amino acid residues modified by 4-hydroxy-2-nonenal under physiological conditions. Journal of Proteome Research. 2010;9(11):5770–5781. doi: 10.1021/pr100555v. 20818828 [DOI] [PubMed] [Google Scholar]
- 48.Chavez J.D., Wu J., Bisson W., Maier C.S. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. Journal of Proteomics. 2011;74(11):2417–2429. doi: 10.1016/j.jprot.2011.03.031. 21513823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charles R.L., Burgoyne J.R., Mayr M., Weldon S.M., Hubner N., Dong H., et al. Redox regulation of soluble epoxide hydrolase by 15-deoxy-delta-prostaglandin J2 controls coronary hypoxic vasodilation. Circulation Research. 2011;108(3):324–334. doi: 10.1161/CIRCRESAHA.110.235879. 21164107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Díez-Dacal B., Pérez-Sala D. Anti-inflammatory prostanoids: focus on the interactions between electrophile signalling and resolution of inflammation. Scientific World Journal. 2010;10:655–675. doi: 10.1100/tsw.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez A.E., Sánchez-Gómez F.J., Díez-Dacal B., Oeste C.L., Pérez-Sala D. 15-Deoxy-Δ(12,14)-prostaglandin J2 exerts pro- and anti-inflammatory effects in mesangial cells in a concentration-dependent manner. Inflammation and Allergy Drug Targets. 2012;11(1):58–65. doi: 10.2174/187152812798889349. [DOI] [PubMed] [Google Scholar]
- 52.Renedo M., Gayarre J., García-Domínguez C.A., Pérez-Rodríguez A., Prieto A., Cañada F.J., et al. Modification and activation of Ras proteins by electrophilic prostanoids with different structure are site-selective. Biochemistry. 2007;46(22):6607–6616. doi: 10.1021/bi602389p. 17489560 [DOI] [PubMed] [Google Scholar]
- 53.Oeste C.L., Díez-Dacal B., Bray F., García de Lacoba M., de la Torre B.G., Andreu D., et al. The C-terminus of H-Ras as a target for the covalent binding of reactive compounds modulating Ras-dependent pathways. PLOS One. 2011;6(1):e15866. doi: 10.1371/journal.pone.0015866. 21253588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cordis G.A., Das D.K., Riedel W. High-performance liquid chromatographic peak identification of 2,4-dinitrophenylhydrazine derivatives of lipid peroxidation aldehydes by photodiode array detection. Journal of Chromatography A. 1998;798(1–2):117–123. doi: 10.1016/S0021-9673(97)01161-8. 9542133 [DOI] [PubMed] [Google Scholar]
- 55.Mateos R., Lecumberri E., Ramos S., Goya L., Bravo L. Determination of malondialdehyde (MDA) by high-performance liquid chromatography in serum and liver as a biomarker for oxidative stress. application to a rat model for hypercholesterolemia and evaluation of the effect of diets rich in phenolic antioxidants from fruits. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Science. 2005;827(1):76–82. doi: 10.1016/j.jchromb.2005.06.035. 16009604 [DOI] [PubMed] [Google Scholar]
- 56.Zhu H., Li X., Shoemaker C.F., Wang S.C. Ultrahigh performance liquid chromatography analysis of volatile carbonyl compounds in virgin olive oils. Journal of Agricultural and Food Chemistry. 2013;61(50):12253–12259. doi: 10.1021/jf404368m. 24279346 [DOI] [PubMed] [Google Scholar]
- 57.Siegel D., Meinema A.C., Permentier H., Hopfgartner G., Bischoff R. Integrated quantification and identification of aldehydes and ketones in biological samples. Analytical Chemistry. 2014;86(10):5089–5100. doi: 10.1021/ac500810r. 24745975 [DOI] [PubMed] [Google Scholar]
- 58.Wang M., Fang H., Han X. Shotgun lipidomics analysis of 4-hydroxyalkenal species directly from lipid extracts after one-step in situ derivatization. Analytical Chemistry. 2012;84(10):4580–4586. doi: 10.1021/ac300695p. 22500579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milic I., Hoffmann R., Fedorova M. Simultaneous detection of low and high molecular weight carbonylated compounds derived from lipid peroxidation by electrospray ionization-tandem mass spectrometry. Analytical Chemistry. 2013;85(1):156–162. doi: 10.1021/ac302356z. 23186270 [DOI] [PubMed] [Google Scholar]
- 60.Ni Z., Milic I., Fedorova M. Identification of carbonylated lipids from different phospholipid classes by shotgun and LC–MS lipidomics. Analytical and Bioanalytical Chemistry. 2015 doi: 10.1007/s00216-015-8536-2. 25701423, in press. [DOI] [PubMed] [Google Scholar]
- 61.Tomono S., Miyoshi N., Ohshima H. Comprehensive analysis of the lipophilic reactive carbonyls present in biological specimens by LC/ESI-MS/MS. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2015;988:149–156. doi: 10.1016/j.jchromb.2015.02.036. 25777478 [DOI] [PubMed] [Google Scholar]
- 62.García-Gómez D., Martínez-Lozano Sinues P., Barrios-Collado C., Vidal-de-Miguel G., Gaugg M., Zenobi R. Identification of 2-alkenals, 4-hydroxy-2-alkenals, and 4-hydroxy-2,6-alkadienals in exhaled breath condensate by UHPLC–HRMS and in breath by real-Time HRMS. Analytical Chemistry. 2015;87(5):3087–3093. doi: 10.1021/ac504796p. 25646646 [DOI] [PubMed] [Google Scholar]
- 63.El-Maghrabey M.H., Kishikawa N., Ohyama K., Kuroda N. Analytical method for lipoperoxidation relevant reactive aldehydes in human sera by high-performance liquid chromatography-fluorescence detection. Analytical Biochemistry. 2014;464:36–42. doi: 10.1016/j.ab.2014.07.002. 25017470 [DOI] [PubMed] [Google Scholar]
- 64.Ali M.F., Kishikawa N., Ohyama K., Mohamed H.A., Abdel-Wadood H.M., Mahmoud A.M., et al. Chromatographic determination of low-molecular mass unsaturated aliphatic aldehydes with peroxyoxalate chemiluminescence detection after fluorescence labeling with 4-(N,N-dimethylaminosulfonyl)-7-hydrazino-2,1,3-benzoxadiazole. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2014;953–954:147–152. doi: 10.1016/j.jchromb.2014.02.009. 24614624 [DOI] [PubMed] [Google Scholar]
- 65.Cui T., Schopfer F.J., Zhang J., Chen K., Ichikawa T., Baker P.R., et al. Nitrated fatty acids: endogenous anti-inflammatory signaling mediators. Journal of Biological Chemistry. 2006;281(47):35686–35698. doi: 10.1074/jbc.M603357200. 16887803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colzani M., Aldini G., Carini M. Mass spectrometric approaches for the identification and quantification of reactive carbonyl species protein adducts. Journal of Proteomics. 2013;92:28–50. doi: 10.1016/j.jprot.2013.03.030. 23597925 [DOI] [PubMed] [Google Scholar]
- 67.Zhao J., Chen J., Zhu H., Xiong Y.L. Mass spectrometric evidence of malonaldehyde and 4-hydroxynonenal adductions to radical-scavenging soy peptides. Journal of Agricultural and Food Chemistry. 2012;60(38):9727–9736. doi: 10.1021/jf3026277. 22946674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gürbüz G., Heinonen M. LC–MS investigations on interactions between isolated beta-lactoglobulin peptides and lipid oxidation product malondialdehyde. Food Chemistry. 2015;175:300–305. doi: 10.1016/j.foodchem.2014.11.154. 25577084 [DOI] [PubMed] [Google Scholar]
- 69.Steen H., Mann M. The ABC’s (and XYZ’s) of peptide sequencing. Nature Reviews Molecular Cell Biology. 2004;5(9):699–711. doi: 10.1038/nrm1468. 15340378 [DOI] [PubMed] [Google Scholar]
- 70.Stevens S.M., Jr., Rauniyar N., Prokai L. Rapid characterization of covalent modifications to rat brain mitochondrial proteins after ex vivo exposure to 4-hydroxy-2-nonenal by liquid chromatography–tandem mass spectrometry using data-dependent and neutral loss-driven MS3 acquisition. Journal of Mass Spectrometry. 2007;42(12):1599–1605. doi: 10.1002/jms.1349. 18085542 [DOI] [PubMed] [Google Scholar]
- 71.Rauniyar N., Stevens S.M., Prokai-Tatrai K., Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Analytical Chemistry. 2009;81(2):782–789. doi: 10.1021/ac802015m. 19072288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Méndez D., Hernáez M.L., Kamali A.N., Diez A., Puyet A., Bautista J.M. Differential carbonylation of cytoskeletal proteins in blood group O erythrocytes: potential role in protection against severe malaria. Infection, Genetics and Evolution. 2012;12(8):1780–1787. doi: 10.1016/j.meegid.2012.06.013. 22771625 [DOI] [PubMed] [Google Scholar]
- 73.Fritz K.S., Kellersberger K.A., Gomez J.D., Petersen D.R. 4-HNE adduct stability characterized by collision-induced dissociation and electron transfer dissociation mass spectrometry. Chemical Research in Toxicology. 2012;25(4):965–970. doi: 10.1021/tx300100w. 22404378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carini M., Aldini G., Facino R.M. Mass spectrometry for detection of 4-hydroxy-trans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spectrometry Reviews. 2004;23(4):281–305. doi: 10.1002/mas.10076. 15133838 [DOI] [PubMed] [Google Scholar]
- 75.Spickett C.M., Reis A., Pitt A.R. Use of narrow mass-window, high-resolution extracted product ion chromatograms for the sensitive and selective identification of protein modifications. Analytical Chemistry. 2013;85(9):4621–4627. doi: 10.1021/ac400131f. 23534669 [DOI] [PubMed] [Google Scholar]
- 76.Milic I., Melo T., Domingues M.R., Domingues P., Fedorova M. Heterogeneity of peptide adducts with carbonylated lipid peroxidation products. Journal of Mass Spectrometry. 2015;50(3):603–612. doi: 10.1002/jms.3568. 25800198 [DOI] [PubMed] [Google Scholar]
- 77.Gao D., Willard B., Podrez E.A. Analysis of covalent modifications of proteins by oxidized phospholipids using a novel method of peptide enrichment. Analytical Chemistry. 2014;86(2):1254–1262. doi: 10.1021/ac4035949. 24350680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uchida K., Stadtman E.R. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra- and intermolecular cross-linking reaction. Journal of Biological Chemistry. 1993;268(9):6388–6393. 8454610 [PubMed] [Google Scholar]
- 79.Golizeh M., Abusarah J., Benderdour M., Sleno L. Covalent binding of 4-hydroxynonenal to matrix metalloproteinase 13 studied by liquid chromatography–mass spectrometry. Chemical Research in Toxicology. 2014;27(9):1556–1565. doi: 10.1021/tx5002095. 25116078 [DOI] [PubMed] [Google Scholar]
- 80.Smathers R.L., Fritz K.S., Galligan J.J., Shearn C.T., Reigan P., Marks M.J., et al. Characterization of 4-HNE modified L-FABP reveals alterations in structural and functional dynamics. PLOS One. 2012;7(6):e38459. doi: 10.1371/journal.pone.0038459. 22701647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anavi S., Ni Z., Tirosh O., Fedorova M. Steatosis-induced proteins adducts with lipid peroxidation products and nuclear electrophilic stress in hepatocytes. Redox Biology. 2015;4:158–168. doi: 10.1016/j.redox.2014.12.009. 25560244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rogowska-Wrzesinska A., Wojdyla K., Nedić O., Baron C.P., Griffiths H.R. Analysis of protein carbonylation − pitfalls and promise in commonly used methods. Free Radical Research. 2014;48(10):1145–1162. doi: 10.3109/10715762.2014.944868. 25072785 [DOI] [PubMed] [Google Scholar]
- 83.Fedorova M., Bollineni R.C., Hoffmann R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrometry Reviews. 2014;33(2):79–97. doi: 10.1002/mas.21381. 23832618 [DOI] [PubMed] [Google Scholar]
- 84.Dalle-Donne I., Carini M., Orioli M., Vistoli G., Regazzoni L., Colombo G., et al. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radical Biology and Medicine. 2009;46(10):1411–1419. doi: 10.1016/j.freeradbiomed.2009.02.024. 19268703 [DOI] [PubMed] [Google Scholar]
- 85.Bollineni R.C., Fedorova M., Hoffmann R. Qualitative and quantitative evaluation of derivatization reagents for different types of protein-bound carbonyl groups. Analyst. 2013;138(17):5081–5088. doi: 10.1039/c3an00724c. 23833766 [DOI] [PubMed] [Google Scholar]
- 86.Bollineni R.C., Hoffmann R., Fedorova M. Proteome-wide profiling of carbonylated proteins and carbonylation sites in HeLa cells under mild oxidative stress conditions. Free Radical Biology and Medicine. 2014;68:186–195. doi: 10.1016/j.freeradbiomed.2013.11.030. 24321318 [DOI] [PubMed] [Google Scholar]
- 87.Aluise C.D., Camarillo J.M., Shimozu Y., Galligan J.J., Rose K.L., Tallman K.A., et al. Site-specific, intramolecular cross-linking of Pin1 active Site residues by the lipid electrophile 4-oxo-2-nonenal. Chemical Research in Toxicology. 2015;28(4):817–827. doi: 10.1021/acs.chemrestox.5b00038. 25739016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fritz K.S., Galligan J.J., Smathers R.L., Roede J.R., Shearn C.T., Reigan P., et al. 4-hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chemical Research in Toxicology. 2011;24(5):651–662. doi: 10.1021/tx100355a. 21449565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vunta H., Davis F., Palempalli U.D., Bhat D., Arner R.J., Thompson J.T., et al. The anti-inflammatory effects of selenium are mediated through 15-deoxy-delta12,14-prostaglandin J2 in macrophages. Journal of Biological Chemistry. 2007;282(25):17964–17973. doi: 10.1074/jbc.M703075200. 17439952 [DOI] [PubMed] [Google Scholar]
- 90.Cernuda-Morollón E., Pineda-Molina E., Cañada F.J., Pérez-Sala D. 15-Deoxy-delta 12,14-prostaglandin J2 inhibition of NF-kappaB-DNA binding through covalent modification of the p50 subunit. Journal of Biological Chemistry. 2001;276(38):35530–35536. doi: 10.1074/jbc.M104518200. 11466314 [DOI] [PubMed] [Google Scholar]
- 91.Takahashi N., Mizuno Y., Kozai D., Yamamoto S., Kiyonaka S., Shibata T., et al. Molecular characterization of TRPA1 channel activation by cysteine-reactive inflammatory mediators. Channels. 2008;2(4):287–298. doi: 10.4161/chan.2.4.6745. 18769139 [DOI] [PubMed] [Google Scholar]
- 92.Artim D.E., Bazely F., Daugherty S.L., Sculptoreanu A., Koronowski K.B., Schopfer F.J., et al. Nitro-oleic acid targets transient receptor potential (TRP) channels in capsaicin sensitive afferent nerves of rat urinary bladder. Experimental Neurology. 2011;232(1):90–99. doi: 10.1016/j.expneurol.2011.08.007. 21867704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parker J. Prostaglandin A2 protein interactions and inhibition of cellular proliferation. Prostaglandins. 1995;50(5–6):359–375. doi: 10.1016/0090-6980(95)00136-0. 8838245 [DOI] [PubMed] [Google Scholar]
- 94.Garzón B., Gayarre J., Gharbi S., Díez-Dacal B., Sánchez-Gómez F.J., Timms J.F., et al. A biotinylated analog of the anti-proliferative prostaglandin A1 allows assessment of PPAR-independent effects and identification of novel cellular targets for covalent modification. Chemico-Biological Interactions. 2010;183(1):212–221. doi: 10.1016/j.cbi.2009.09.019. 19800325 [DOI] [PubMed] [Google Scholar]
- 95.Higdon A.N., Dranka B.P., Hill B.G., Oh J.Y., Johnson M.S., Landar A., et al. Methods for imaging and detecting modification of proteins by reactive lipid species. Free Radical Biology and Medicine. 2009;47(3):201–212. doi: 10.1016/j.freeradbiomed.2009.05.009. 19446632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cummins T.D., Higdon A.N., Kramer P.A., Chacko B.K., Riggs D.W., Salabei J.K., Dell’Italia L.J., Zhang J., Darley-Usmar V.M., Hill B.G. Utilization of fluorescent probes for the quantification and identification of subcellular proteomes and biological processes regulated by lipid peroxidation products. Free Radical Biology and Medicine. 2013;59:56–68. doi: 10.1016/j.freeradbiomed.2012.08.014. 22954622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vila A., Tallman K.A., Jacobs A.T., Liebler D.C., Porter N.A., Marnett L.J. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chemical Research in Toxicology. 2008;21(2):432–444. doi: 10.1021/tx700347w. 18232660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pérez-Sala D. Electrophilic eicosanoids: signaling and targets. Chemico-Biological Interactions. 2011;192(1–2):96–100. doi: 10.1016/j.cbi.2010.10.003. 20970408 [DOI] [PubMed] [Google Scholar]
- 99.Gayarre J., Stamatakis K., Renedo M., Pérez-Sala D. Differential selectivity of protein modification by the cyclopentenone prostaglandins PGA1 and 15-deoxy-delta12,14-PGJ2: role of glutathione. FEBS Letters. 2005;579(25):5803–5808. doi: 10.1016/j.febslet.2005.09.069. 16223487 [DOI] [PubMed] [Google Scholar]
- 100.Oeste C.L., Pérez-Sala D. Modification of cysteine residues by cyclopentenone prostaglandins: interplay with redox regulation of protein function. Mass Spectrometry Reviews. 2014;33(2):110–125. doi: 10.1002/mas.21383. 23818260 [DOI] [PubMed] [Google Scholar]
- 101.Diers A.R., Higdon A.N., Ricart K.C., Johnson M.S., Agarwal A., Kalyanaraman B., et al. Mitochondrial targeting of the electrophilic lipid 15-deoxy-delta12,14-prostaglandin J2 increases apoptotic efficacy via redox cell signalling mechanisms. Biochemical Journal. 2010;426(1):31–41. doi: 10.1042/BJ20091293. 19916962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sánchez-Gómez F.J., Gayarre J., Avellano M.I., Pérez-Sala D. Direct evidence for the covalent modification of glutathione-S-transferase P1-1 by electrophilic prostaglandins: implications for enzyme inactivation and cell survival. Archives of Biochemistry and Biophysics. 2007;457(2):150–159. doi: 10.1016/j.abb.2006.10.032. 17169324 [DOI] [PubMed] [Google Scholar]
- 103.Sánchez-Gómez F.J., Dorado C.G., Ayuso P., Agúndez J.A., Pajares M.A., Pérez-Sala D. Modulation of GSTP1-1 oligomerization by electrophilic inflammatory mediators and reactive drugs. Inflammation and Allergy Drug Targets. 2013;12(3):162–171. doi: 10.2174/1871528111312030002. [DOI] [PubMed] [Google Scholar]
- 104.Sánchez-Gómez F.J., Cernuda-Morollón E., Stamatakis K., Pérez-Sala D. Protein thiol modification by 15-deoxy-delta12,14-prostaglandin J2 addition in mesangial cells: role in the inhibition of pro-inflammatory genes. Molecular Pharmacology. 2004;66(5):1349–1358. doi: 10.1124/mol.104.002824. 15317873 [DOI] [PubMed] [Google Scholar]
- 105.Baraibar M.A., Ladouce R., Friguet B. Proteomic quantification and identification of carbonylated proteins upon oxidative stress and during cellular aging. Journal of Proteomics. 2013;92:63–70. doi: 10.1016/j.jprot.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 106.Uchida K., Szweda L.I., Chae H.Z., Stadtman E.R. Immunochemical detection of 4-hydroxynonenal protein adducts in oxidized hepatocytes. Proceedings of National Academy of Sciences of the United States of America. 1993;90(18):8742–8746. doi: 10.1073/pnas.90.18.8742. 8378358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shibata T., Kondo M., Osawa T., Shibata N., Kobayashi M., Uchida K. 15-deoxy-Delta 12,14-prostaglandin J2. A prostaglandin D2 metabolite generated during inflammatory processes. Journal of Biological Chemistry. 2002;277(12):10459–10466. doi: 10.1074/jbc.M110314200. 11786541 [DOI] [PubMed] [Google Scholar]
- 108.Kato Y. The formation of lipid hydroperoxide-derived amide-type lysine adducts on proteins: a review of current knowledge. Subcellular Biochemistry. 2014;77:21–39. doi: 10.1007/978-94-007-7920-4_2. 24374915 [DOI] [PubMed] [Google Scholar]
- 109.Shiraki T., Kamiya N., Shiki S., Kodama T.S., Kakizuka A., Jingami H. Alpha,beta-unsaturated ketone is a core moiety of natural ligands for covalent binding to peroxisome proliferator-activated receptor gamma. Journal of Biological Chemistry. 2005;280(14):14145–14153. doi: 10.1074/jbc.M500901200. 15695504 [DOI] [PubMed] [Google Scholar]
- 110.Díez-Dacal B., Gayarre J., Gharbi S., Timms J.F., Coderch C., Gago F., et al. Identification of aldo-keto reductase AKR1B10 as a selective target for modification and inhibition by prostaglandin A(1): implications for antitumoral activity. Cancer Research. 2011;71(12):4161–4171. doi: 10.1158/0008-5472.CAN-10-3816. 21507934 [DOI] [PubMed] [Google Scholar]
- 111.Gharbi S., Garzón B., Gayarre J., Timms J., Pérez-Sala D. Study of protein targets for covalent modification by the antitumoral and anti-inflammatory prostaglandin PGA1: focus on vimentin. Journal of Mass Spectrometry. 2007;42(11):1474–1484. doi: 10.1002/jms.1291. 17960581 [DOI] [PubMed] [Google Scholar]
- 112.Kim H.J., Ha S., Lee H.Y., Lee K.J. Rosics: chemistry and proteomics of cysteine modifications in redox biology. Mass Spectrometry Reviews. 2015;34(2):184–208. doi: 10.1002/mas.21430. 24916017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pérez-Sala D., Oeste C.L., Martínez A.E., Garzón B., Carrasco M.J., Cañada F.J. Vimentin filament organization and stress sensing depend on its single cysteine residue and zinc binding. Nature Communications. 2015;6:7287. doi: 10.1038/ncomms8287. , [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsapara A., Kardassis D., Moustakas A., Gravanis A., Stournaras C. Expression and characterization of Cys374 mutated human beta-actin in two different mammalian cell lines: impaired microfilament organization and stability. FEBS Letters. 1999;455(1–2):117–122. doi: 10.1016/S0014-5793(99)00848-0. 10428484 [DOI] [PubMed] [Google Scholar]


