Abstract
Protein ubiquitination and the subsequent degradation are important means by which aberrant proteins are removed from cells, a key requirement for long-term survival. In this study, we found that the overall level of ubiquitinated proteins dramatically decreased as yeast cell grew from log to stationary phase. Deletion of SCH9, a gene encoding a key protein kinase for longevity control, decreased the level of ubiquitinated proteins in log phase and this effect could be reversed by restoring Sch9 function. We demonstrate here that the decrease of ubiquitinated proteins in sch9Δ cells in log phase is not caused by changes in ubiquitin expression, proteasome activity, or autophagy, but by enhanced expression of stress response factors and a decreased level of oxidative stress. Our results revealed for the first time how Sch9 regulates the level of ubiquitinated proteins and provides new insight into how Sch9 controls longevity.
Keywords: SCH9, Yeast, Ubiquitination, Oxidation, Hydrogen peroxide
Introduction
The homeostasis of intracellular proteins needs to be precisely regulated to maintain proper cell functions [1]. The deficiency in removal of aberrant or unwanted proteins will result in the accumulation of those deleterious proteins and subsequent malfunction of cell machinery. This process is also referred as proteotoxicity [2]. The ubiquitin proteasome system (UPS) is one of major proteolytic systems which selectively scavenge intracellular proteotoxic proteins. The proteins to be degraded are first recognized and ubiquitinated by a set of enzymes including ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2) and ubiquitin ligase (E3). Ubiquitinated proteins, especially those of polyubiquitinated through K48 linkage, are bound to 26S proteasome and degraded in an ATP dependent manner [3–5]. In addition to proteasomal degradation, recent studies have revealed that ubiquitinated protein aggregates are selectively degraded via autophagy [6,7]. The proteotoxicity caused by the incompetence of removing aberrant proteins by UPS or autophagy is involved in the pathology of many human diseases such as cataracts, macular degeneration, Alzheimer’s, Parkinson’s, and Huntington’s syndromes [2,8–10]. Despite extensive studies have investigated how proteotoxicity contributes to those diseases prevalent in aging human population, relative rare information is acquired regarding the effects of proteotoxicity on longevity. The interventions such as calorie restriction (CR) and rapamycin treatment which extend lifespan from yeast to mammalians have also been demonstrated to decrease the incidences of several age-related diseases in rodents or primates, suggesting those interventions may regulate the proteostasis [11–17]. Indeed, CR modulates redox state, removal of oxidized protein and the UPS in yeast cell [18]. And rapamycin has been well established to be an autophagy inducer [19].
One of the key regulators for longevity and aging in yeast cell is Sch9, a homolog of mammalian protein kinase S6K1 [20–22]. Sch9 senses nutrition and stress signals from Pkh1/2 and TORC1 and is a direct substrate of them [21,23,24]. Pkh1/2 phosphorylate the activation loop of Sch9 to activate the kinase while TORC1 phosphorylates the serine/threonine residues at the hydrophobic motif to fine tune the activity [21,25,26]. Sch9 regulates multiple aspects of cell metabolism to affect the growth and lifespan. It has been demonstrated that Sch9 controls translation by regulating ribosome biogenesis [27,28]. Sch9 also decreases the phosphorylation of translation initiation factor eIF2a to maintain its function [21]. Several stress response regulators, including Rim15, Msn2/4, Gis1 and Hcm1, are downstream effectors of Sch9 [29–33]. Therefore Sch9 plays a key role in stress response. On the other hand the deletion of Sch9 enhances the respiration of mitochondria during growth by increasing the translation of mtDNA-encoded OXPHOS complex subunits, which in turn promotes the generation of superoxide [34–36].
There is only limited information regarding the role of Sch9 in protein ubiquitination. Cdc34, an E2 ubiquitin conjugation enzyme also called Ubc3, is found to be a substrate of Sch9 and the interaction between Cdc34 and Sch9 is proposed to regulate the transition between cell division and cell cycle arrest [37,38]. However, it is unclear if and how Sch9 regulates the protein ubiquitination. Considering the importance of both Sch9 and protein ubiquitination for cell homeostasis, here we studied if Sch9 regulates protein ubiquitination and investigated the mechanism of the regulation.
Results
Sch9 regulates the level of ubiquitinated proteins during cell growth
When yeast cells are cultured in rich medium (YPD), growth progress from log phase, where cells divide every ~90 min, through the diauxic shift to stationary phase where division ceases [39–41]. Dramatic changes of cellular events occur during the transition from log phase to stationary phase, including the dynamics of ubiquitinated proteins (Fig. 1A, B and S1). When wild type (WT) cells entered to diauxic phase 24 h after inoculation, the overall protein ubiquitination level dropped more than 50% and did not decrease further during the stationary phase (Fig. 1A –C).
Fig. 1.
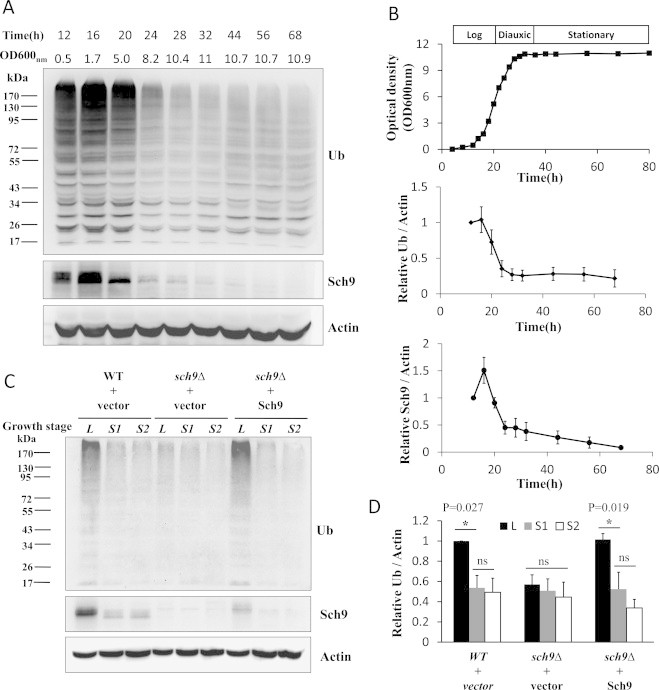
The level of ubiquitinated proteins during cell growth is regulated by Sch9. (A) The levels of ubiquitinated proteins in WT cells (TB50a) at indicated times after inoculation were tested by western blotting with actin as the loading control. The expression of Sch9 was also detected. (B) The quantifications of three repeats from panel A together with the related cell growth curve estimated by the change of cell concentration. (C) The levels of ubiquitinated proteins in WT cells (TB50a) transformed with empty vector (WT+vector) and Sch9 deletion mutant cells (TS120-2d) transformed with empty vector (sch9Δ+vector) or pRS416−SCH9 (sch9Δ+Sch9) at indicated growth stages were tested by western blotting. L: Log phase at 12–16 h after inoculation (OD600 nm=0.5), S1: Stationary phase at 72 h after inoculation (OD600 nm=8–9), S2: Stationary phase at 120 h after inoculation (OD600 nm=8–9). (D) The quantifications of three repeats from panel C. (*P represents<0.05 and “ns” denotes no significance p>0.05 between the indicated comparisons.).
To probe the mechanisms by which the dynamics of ubiquitinated proteins is regulated during cell growth, the effect of protein kinase Sch9, a key regulator for cell growth [42], on protein ubiquitination was investigated. The deletion of SCH9 decreased the overall level of protein ubiquitination in log phase cells, but displayed no further decline in stationary phase cells (Figs. S1 and 1C, compare lanes 4–6 to 1–3, and Fig. 1D). Adding SCH9 back to the mutant stain restored the ubiquitination level to that of WT cells, confirming the role of Sch9 in regulating the dynamics of ubiquitinated proteins during cell growth (Figs. S1 and 1C, compare lanes 7–9 to 1–3, and Fig. 1D). The decrease in Sch9 protein level during the transition from log to stationary phase (Fig. 1A, the middle panel, and Fig 1B) when overall protein ubiquitination level went down also suggests the involvement of Sch9 in the regulation of protein ubiquitination.
The alteration of the dynamics of ubiquitinated proteins by Sch9 does not depend on the ubiquitin expression or proteasomal activity
Although overall protein ubiquitination is decreased in sch9Δ cells (Fig. 1), the level of free ubiquitin in these cells was indistinguishable to that in cells expressing Sch9 (Fig. 2A), suggesting that the decrease of ubiquitinated proteins does not result from a shortage of free ubiquitin.The proteasome pathway is one of the major pathways which regulate the dynamics of intracellular polyubiquitinated proteins [1,43]. The decline of ubiquitinated proteins in sch9Δ cells may be caused by the enhanced proteasomal degradation. However, the proteasome activities in WT, sch9Δ, and sch9Δ(Sch9) cells are indistinguishable (Fig. 2B), indicating that the decreased protein ubiquitination in sch9Δ cells is not due to enhanced proteasome activity. In agreement with this result, we found that inhibiting proteasome activity by MG132 on sch9Δ cells could not restore the ubiquitinated proteins to the same level as that in WT and sch9Δ(Sch9) cells (Fig. 2C and D).
Fig. 2.
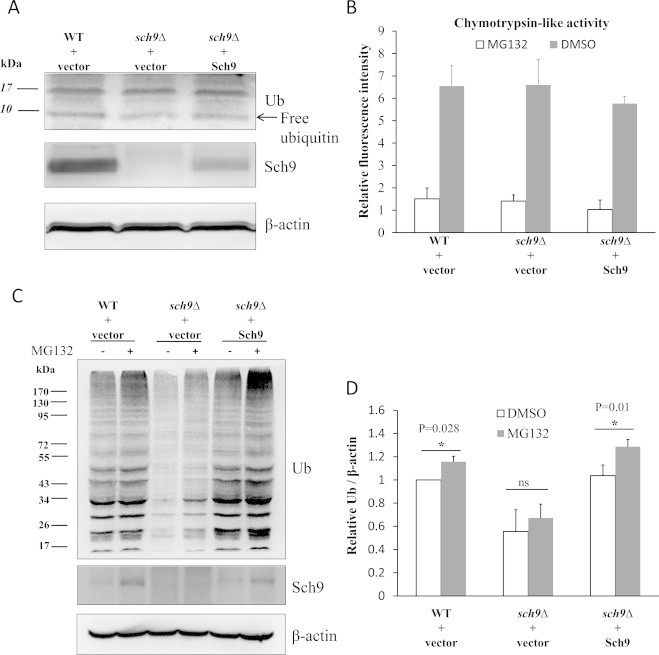
Sch9 does not regulate the level of ubiquitinated proteins by affecting ubiquitin expression or proteasomal activity. (A) Lysates of log phase yeasts (OD600 nm=0.5) were resolved on 15% SDS gels and proteins of interest were detected by Western blotting. (B) Chymotrypsin-like activity of the proteasome was monitored by Suc-Leu-Leu-Val-Tyr-AMC digestion using lysates with equal amounts of total protein from log phase yeasts at OD600nm of 0.5. The proteasome inhibitor MG132 (75 µM) was added to the isopyknic lysates as the blank control. (C) The log phase yeasts (OD600 nm=0.5) were treated with 75 µM MG132 for 1 h and the levels of ubiquitinated protein and Sch9 were tested by western blotting with actin as the loading control. (D) The quantifications of three repeats from panel C. (*represents P<0.05 and “ns” denotes no significance p>0.05 between the indicated comparisons.).
Enhanced autophagy by deletion of SCH9 does not contribute to the decrease of ubiquitinated proteins
Besides proteasome, autophagy is another major intracellular proteolytic system which degrades ubiquitinated proteins [6]. Genome-wide studies have suggested that the deletion of SCH9 enhances the expression of autophagy related genes [36]. We also found that the transcription of Atg8 gene, the homolog of mammalian LC3 which is a key factor of autophagy [44], was upregulated to 3 folds upon SCH9 deletion (Fig. 3A). It has been demonstrated that PKA and Sch9 cooperatively regulate the induction of autophagy [45]. By using the Atg8-GFP protein as an autophagy activity reporter [46], we found that sch9Δ cells generated more degradation product which diminished when autophagy was inhibited by the deletion of Atg1, confirming that sch9Δ cells have higher autophagy activity than WT cells (Fig. 3B, compare lane 3 to lane 1). If the enhanced autophagy contributes to the decreased ubiquitination in sch9Δ cell, shutting down autophagy should be able to restore the ubiquitination level. However, the depletion of ATG1 did not elevate the level of ubiquitinated proteins in sch9Δ cells despite its autophagy blocking effect (Fig. 3C and D), suggesting that the enhanced autophagy in sch9Δ cells was not the direct mechanism by which the ubiquitinated proteins are decreased.
Fig. 3.
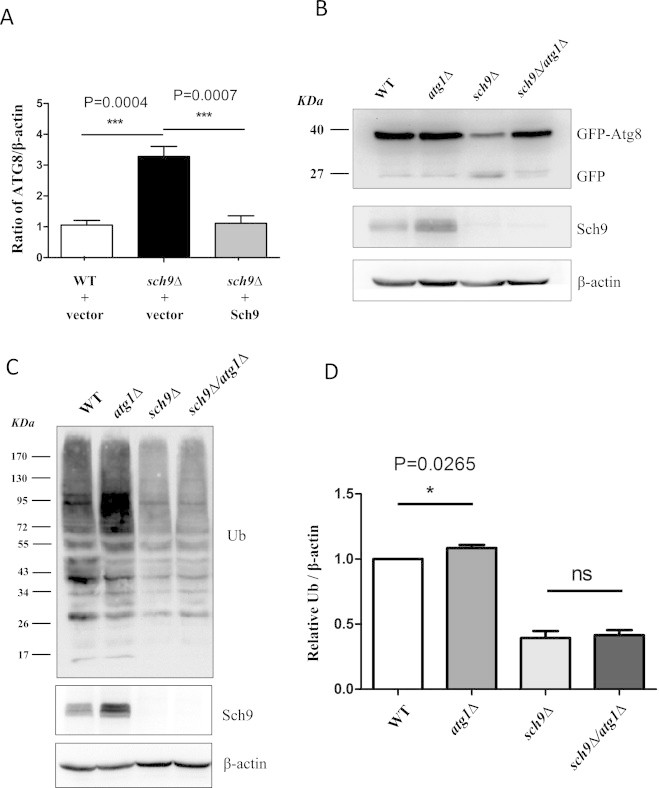
Autophagy is not responsible for the decreased level of ubiquitinated proteins in sch9Δ cells. (A) The mRNA levels of ATG8 were tested in WT cells (DBY746) transformed with empty vector (WT+vector) and SCH9 deletion mutant cells (PF102(1-1)) transformed with empty vector (sch9Δ+vector) or pRS416−SCH9 (sch9Δ+Sch9) at log phase (OD600 nm=0.5). (B) GFP-Atg8 processing is enhanced by the deletion of SCH9. WT (DBY746), atg1Δ, sch9Δ and sch9Δ/atg1Δ strains were transformed with pUG36-Ura/ATG8 and the autophagy activities were detected in log phase cells (OD600 nm=0.5). (C) WT (DBY746), atg1Δ, sch9Δ and sch9Δ/atg1Δ were harvested at OD600 nm of 0.5 and the levels of ubiquitinated protein and Sch9 were tested by western blotting with actin as the loading control. (D) The quantifications of three repeats from panel C (*P<0.05, ***P < 0.001, “ns” no significance).
Suppression of oxidative stress by the deletion of SCH9 results in the decrease of ubiquitinated proteins.
Previous gene chip studies have shown that sch9Δ cells have enhanced expression of stress response genes [36]. By using quantitative PCR we found that the mRNA level of several stress response genes, including GPX1 (Phospholipid hydroperoxide glutathione peroxidase), HSP104 (disaggregase), HSP12 (12 kD small heat shock protein), TSA2 (Stress inducible cytoplasmic thioredoxin peroxidase) and CTT1 (Cytosolic catalase T) increased 2–30 fold in sch9Δ cells (Fig. 4A). Consistently, the catalase activity of Ctt1 elevated ~50% upon SCH9 deletion while adding Sch9 back decreased the catalase activity of Ctt1 back to a level similar to that in WT cells (Fig. S2A and B). These results suggest that sch9Δ cells have stronger capability of ROS scavenging and protein refolding and therefore may generate less damaged proteins as substrates for ubiquitination.
Fig. 4.
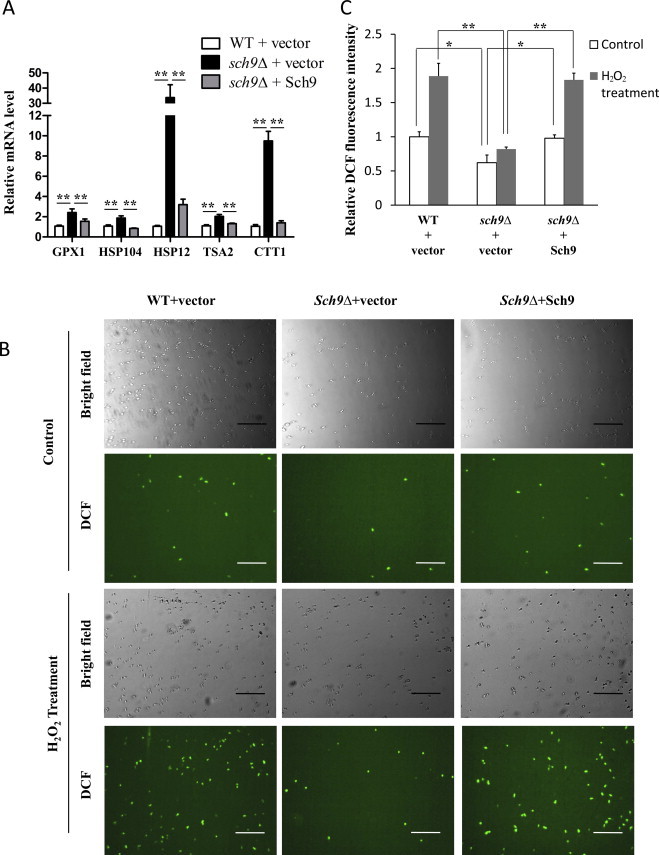
sch9Δ cells are resistant to ubiquitination due to decreased intracellular oxidation. (A) The mRNA levels of stress response genes (GPX1, HSP104, HSP12, TSA2 and CTT1) were tested by qPCR and normalized by the levels of actin (ACT1) mRNA in log phase cells (OD600 nm=0.5). (B) Microscopy of DCF fluorescence on WT cells (TB50a) transformed with empty vector (WT+vector), SCH9 deletion mutant cells (TS120-2d) transformed with empty vector (sch9Δ+vector) or pRS416−SCH9 (sch9Δ+Sch9) at log phase (OD600 nm=0.5) after treated with or without 0.5 mM H2O2 for 1 h. Bars: 100 µm. (C) Spectrofluorimeter analysis of DCF-stained cells (*P <0.05, **P<0.01).
Consistent with the increased expression of peroxidase and catalase in sch9Δ cells, we found that the deletion of SCH9 resulted in ~40% less intracellular DCF staining (Fig. 4B upper panels and C, compare column 3 to 1), a wildly used fluorescent probe for the detection of intracellular ROS and redox signaling [47,48], suggesting that sch9Δ cells are more capable of scavenging intracellular oxidants. This is also confirmed by the results that sch9Δ cells generate ~60% less DCF signals than WT cells in the presence of exogenous H2O2 (Fig. 4B lower panels and C, compare column 4 to 2). Additionally, our parallel experiments of measuring intracellular fluorescence product derived from DHR123, another widely used fluorescent probe for intracellular oxidative stress [49–51], also demonstrated consistent results (Fig. S3A and B).
H2O2 is a major intracellular ROS which emerged as a central hub in redox signaling and oxidative stress [52]. The enhanced CTT1 expression in sch9Δ cells suggest that the lower protein ubiquitination and oxidative stress may be contributed by the lower H2O2 level. To unveil if the difference of intracellular redox statuses between WT and sch9Δ cells is related to the different levels of intracellular H2O2, the oxidation of DCF and DHR123 in both strains was compared in the presence or absence of elevated catalase T activity. By overexpressing Ctt1 in WT cells, the catalase T activity was elevated to a level similar to that in sch9Δ cells (Fig. S4A and B) and the oxidation of DCF and DHR123 was decreased ~20% and ~30% respectively (Figs. 5A and B, and S5A and B). However, enhancing the catalase T activity in sch9Δ cells did not further decrease the oxidation unless exogenous H2O2 was added into the cell culture (Figs. 5A and B, and S5A and B). These results suggest that the enhanced H2O2 removal in sch9Δ cells contribute to their alleviated oxidative stress.
Fig. 5.
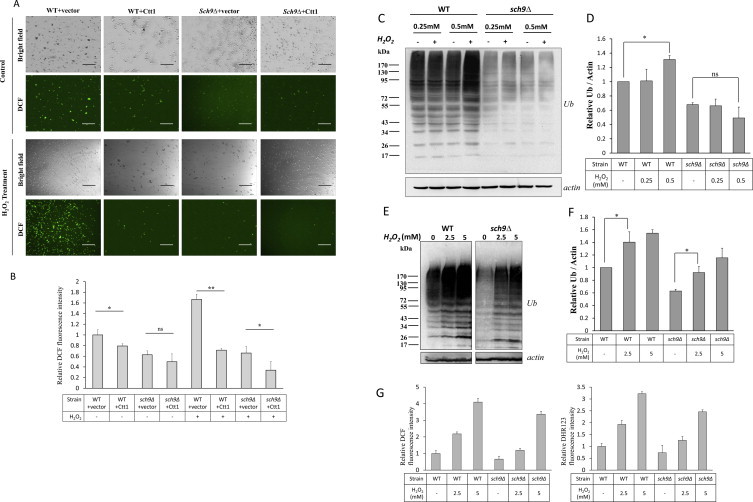
H2O2 contributes to the enhanced intracellular oxidation and ubiquitination of sch9Δ cells. (A, B) Microscopy (A) or spectrofluorimeter analysis (B) of DCF fluorescence on WT (TB50a) and sch9Δ (TS120-2d) cells transformed with pEGH-CTT1 or control vector with or without 1 hour treatment of 0.5 mM H2O2. Bars: 100 µm. (C–F) WT and sch9Δ cells were treated with or without H2O2 at indicated concentrations for 1 h and the total ubiquitination were tested by western blotting (C, E) with respective quantifications (D, F). (G) Spectrofluorimeter analysis of DCF- or DHR123-stained cells with or without treatment of H2O2 at indicated concentrations for 1 hour (All cells were collected at log phase with an OD600 nm of 0.5. *P < 0.05, **p < 0.01, “ns” no significance).
The role of H2O2 in protein ubiquitination was further confirmed by the observation that the ubiquitination in WT cells was stimulated in a medium containing 0.5 mM H2O2 when the ubiquitination of sch9Δ cells was not elevated at the same condition (Fig. 5C and D). Consistently, the oxidation of DCF and DCR123 only increased slightly in sch9Δ cells but dramatically in WT cells in the presence of 0.5 mM H2O2 (Figs. 4B and C, and S3A and B). On the other hand, if the concentration of environmental H2O2 increased to 2.5 or 5 mM, the level of protein ubiquitination in both WT and sch9Δ cells were elevated while it still exhibited lower level in sch9Δ cells at the respective H2O2 concentration (Fig. 5E and F). Consistently, the oxidation of DCF and DHR123 was lower in sch9Δ cells than in WT cells but increased in both strains in the presence of 2.5 or 5 mM H2O2 (Fig. 5G). These results suggest that sch9Δ cells are less sensitive to H2O2 induced ubiquitination.
The level of ubiquitinated proteins is correlated to intracellular protein carbonylation in log phase cells, but not in stationary phase cells
To investigate the correlation between decreased oxidative stress and protein ubiquitination in sch9Δ cells, the concentrations of carbonyl groups in oxidized intracellular proteins in WT and sch9Δ cells were evaluated. In log phase, the level of protein carbonyl in sch9Δ cells was ~30% lower than that in WT cells. Adding Sch9 back restored the level of protein carbonyl to a similar level as that in wild type cells (Fig. 6A).
Fig. 6.
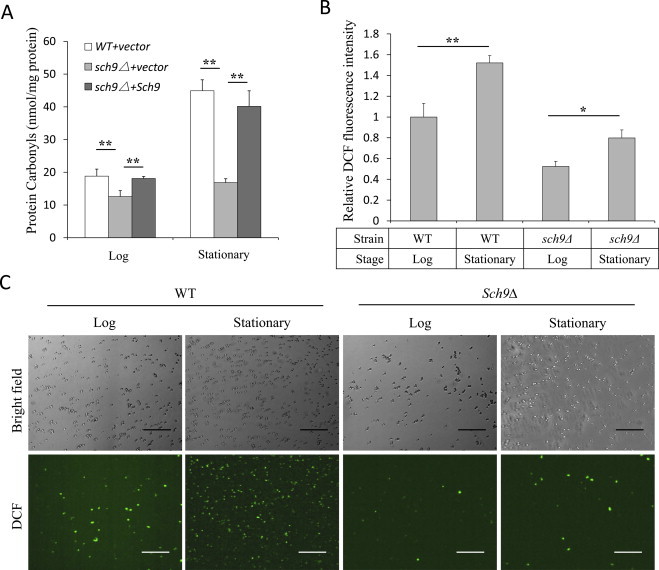
Decreased protein carbonylation by alleviated intracellular oxidation in sch9Δ cells. (A) Measurements of concentration of protein carbonyl groups in WT cells (TB50a) transformed with empty vector (WT + vector), SCH9 deletion mutant cells (TS120-2d) transformed with empty vector (sch9Δ+vector) or pRS416−SCH9 (sch9Δ+Sch9) in log or stationary phase. (B) Fluorescence spectrometer analysis of DCF-stained WT (TB50a) and sch9Δ cells in log or stationary phase. (C) Microscopy of DCF-stained WT (TB50a) and sch9Δ cells in log or stationary phase. Bars: 100 µm (log phase cells were collected 12–16 h after inoculation with an OD600nm of 0.5 while stationary phase cells were collected 72 h after inoculation with an OD600 nm of 8–9. *p < 0.05, **p < 0.01).
When WT cells progressed into stationary phase, the level of protein carbonyl increased more than two folds, while it still exhibited ~70% lower in sch9Δ cells (Fig. 6A). The increased levels of protein carbonyl in both WT and sch9Δ cells in stationary phase are correlated to the enhanced oxidative stress at this stage (Fig. 6B and C), most likely caused by the enhanced respiration [11]. Despite the different levels of redox signaling between wild type and sch9Δ cells during stationary phase, the levels of ubiquitinated proteins were similar in both strains and significantly lower than those in cells of log phase (Fig. 1C and see Discussion).
Discussion
Protein ubiquitination is a key post-translational modification involved in many aspects of cell behavior and malfunctional ubiquitination is related to a broad range of human diseases, including diabetes, neuron degeneration diseases, and several types of cancer [53]. Therefore, it is interesting to note that the level of ubiquitinated proteins dropped to 50% if yeast cells lack a single gene SCH9. The role of Sch9 in regulating protein ubiquitination was verified by the observation that ubiquitinated proteins were recovered after putting Sch9 back into sch9Δ cells. Sch9 is emerged as an important regulator for lifespan of yeast cell [20]. So the understanding about how it affects the protein ubiquitination may lead to a broader perspective into the mechanisms of aging.
To understand how Sch9 is involved in the dynamics of protein ubiquitiniation we first ask if the decreased level of ubiquitinated proteins in sch9Δ cells is due to the enhanced degradation or attenuated formation of them. The proteasome is the major cellular apparatus that digests polyubiquitinated proteins [1,43]. However, we do not find any difference in the proteasome activities between wild type and sch9Δ cells. The fact that proteasome inhibitor does not restore the ubiquitination level in sch9Δ cells also indicates that there is no enhanced proteasomal degradation in sch9Δ cells. Beside proteasomal degradation, recent studies have suggested that ubiquitinated protein can be degraded by autophagy as well [6,7]. Previous studies also indicated that Sch9 is involved in the regulation of autophagy [45]. Our data verify that the deletion of SCH9 promotes the expression of the autophagy-related gene ATG8 and the activity of autophagy. However, when we delete ATG1 in sch9Δ cells to abolish the autophagy, the ubiquitination level remains constant. These data suggest that the decreased level of ubiquitinated proteins in sch9Δ cells is not due to increased degradation. We did not determine if there was enhanced deubiquitination enzyme activity in sch9Δ cells since previous gene chip analysis did not suggest so [27,36].
Since our results indicate that neither the supply of free ubiquitin nor the degradation of ubiquitinated proteins is related to the decline of ubiquitinated proteins in sch9Δ cells, it seems that this phenomenon is caused by the attenuated ubiquitination. By reviewing the gene expression profile of sch9Δ cells, we do not find any signs suggesting that the ubiquitin conjugating system is altered [27,36]. Previous studies have revealed that Sch9 phosphorylates ubiquitin conjugating enzyme Cdc34 at S97 [54]. However many other kinases including PKA, Gcn2, Mkk2, Snf1, and Vps15/Vps34 may also contribute to the phosphorylation of S97 [54]. Additionally, since both phosphorylated and dephosphorylated forms of S97 in CDC34 are required for the activity of the enzyme [54], the ubiquitination activity of Cdc34 has little chance to be altered largely by the single deletion of SCH9.
It is more likely that sch9Δ cells produce less aberrant protein for ubiquitination. Consistent with this hypothesis and previous gene chip analysis [27,36], our quantitative mRNA measurements and catalase activity analysis indicate a much stronger oxidative stress response with higher expression of heat shock proteins and antioxidant enzymes in sch9Δ cells. We verify that sch9Δ cells exhibit less intracellular oxidation using fluorescent probes DCF and DHR123 which have been wildly used as probes for ROS. The observation that overexpressing catalase Ctt1 decrease the fluorescent signals of DCF and DHR123 (Fig. 5A and B) also supports that both chemicals are effective in probing ROS. However, the oxidation of DCF or DHR123 may also be affected by hemeproteins and iron mobilization in some systems [55]. Therefore more evidences are required to validate the contribution of oxidative stress on protein ubiquitination regulated by Sch9. We then verify that less protein oxidation is produced in sch9Δ cells, likely due to the higher expression of peroxidases and other antioxidant. The effect of redox signaling on protein ubiquitination is further tested by monitoring the levels of ubiquitinated proteins in the presence of exogenous H2O2. 2.5 and 5 mM of environmental H2O2 are able to stimulate the oxidation of ROS probes and increase protein ubiquitination in both WT and sch9Δ cells, while 0.5 mM H2O2 only affects that in WT cells. Our data here suggest that the enhanced oxidative stress response in sch9Δ cells sets a higher threshold for H2O2-induced ubiquitination.
Although the deletion of SCH9 enhances mitochondrial respiration by activating Hcm1 and probably Hap4 to promote the superoxide production [32,33], it may also stimulate the stress response via Rim15 and its downstream factors [30]. The increased level of superoxide may also enhance the oxidative resistance as an adaptive ROS signaling [11]. Together, the deletion of SCH9 causes the up-regulation of the intracellular antioxidants and the decrease of H2O2, which damages intracellular proteins and subsequently causes ubiquitination. The plausible mechanisms by which Sch9 regulates protein ubiquitination are summarized in Fig. 7.
Fig. 7.
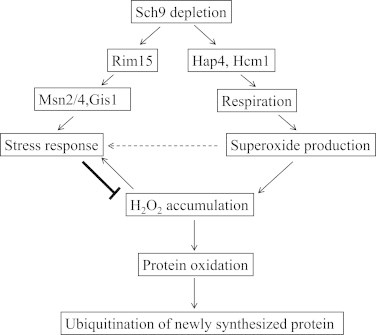
The Regulation of Protein ubiquitination through Sch9 signaling during growth. A speculative model of how reduced Sch9 signaling down-regulated ubiquitinated proteins by Rim15-dependent stress-resistance pathways (left arm) and adaptive mitochondrial ROS signaling (right arm). Elevated respiration in sch9∆ cells is mediated by upregulation of the transcription factors Hap4 or Hcm1 which gives rise to increased respiration and intracellular superoxide during growth [32,33]. Superoxide may serve as an adaptive signal to activate stress response genes (horizontal arrow). Therefore, although elevated cellular superoxide leads to accumulated H2O2, enhanced stress Resistance via Rim15 pathway and adaptive superoxide signal [30], on the other hand, diminishes H2O2 more effectively (bold arrow). Elimination of H2O2 in sch9∆ cells in turn reduces the oxidation of intracellular proteins including those of newly synthesized and their subsequent ubiquitination.
The effects of either H2O2 or Sch9 on protein ubiquitination during cell growth do not extend to stationary phase. Both WT and sch9Δ strains maintain a relative lower level of ubiquitination despite different levels of oxidative stress during stationary phase. A plausible explanation for this phenomenon is that the oxidation-induced ubiquitination preferentially happens during protein synthesis when proteins are folding to native conformation and is therefore more susceptible to oxidative stress [56]. Additionally, during stationary phase although WT cells do not express detectable Sch9 (Fig. 1A), they still present more intracellular oxidation than sch9Δ cells (Fig. 6B and C). It is likely that sch9Δ cells at stationary phase have taken advantage of the elevated antioxidant system during the prior growth phases which induces less protein ubiquitination and benefits the survival.
Materials and methods
Strains, plasmids, and media
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Plasmids pRS316 and pRS416-SCH9 were kind gifts from Dr. Robbie Loewith (University of Geneva, Geneva, Switzerland) [21]. Plasmid pUG36-Ura/ATG8 was used to detect autophagy as described by us previously [57]. Galactose-inducible multi-copy plasmids expressing CTT1 (pEGH-CTT1) was purchased from GE Healthcare (GE Healthcare, Huntsville, AL). Plasmid pEGH was a gift from Dr. Guozhen LIU (University Agriculture of Hebei, China) [58]. Cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or synthetic dextrose complete medium (SDC) medium as described previously [30,59]. Cells transformed with plasmids carrying URA3 were grown in the SDC medium lacking uracil [30,59]. For inducing expression of galactose-inducible multi-copy plasmids, pEGH and pEGH-CTT1, carbon source in routine SDC medium was replaced by 1% galactose and 1% sucrose. For MG132 treatments, cells were grown in SDC medium using l-proline instead of ammonium sulfate as the sole nitrogen source and a small amount of sodium dodecyl sulfate (SDS; 0.003%) was added to enhance the permeability of yeast cells [60].
Table 1.
S. cerevisiae strains.
| Strain | Genotype | Source |
|---|---|---|
| DBY746 | MAT alpha leu2-3.112 his3Δ1 trp1-289 ura3-52 GAL+ | [67] |
| atg1∆ | DBY746 with atg1∆::KanMX | [57] |
| PF102 (1-1) | DBY746 with sch9∆::URA (URA3 silencing) | Gift from Dr. Robert C. Dickson. Made from PF102 [68] by selecting for Ura- phenotype. |
| atg1∆ sch9∆ | DBY746 with atg1∆::KanMX, sch9∆: :HIS3 | This study |
| TB50a | MATa trp1 his3 ura3 leu2 rme1 | [21] |
| TS120-2d | TB50a with sch9∆::KanMX | [21] |
Construction of yeast mutant strains
The atg1∆ sch9∆ (DBY746 with atg1∆::KanMX, sch9∆::HIS3) strain was generated by the transformation of atg1∆ (DBY746 with atg1∆::KanMX) strain with a PCR product generated by amplification of the HIS3 cassette from the pRS303 plasmid using oligonucleotides designed to add homologous bases to regions directly flanking the SCH9-coding sequence (Tables 2, 15 and 16). The mutant was confirmed by PCR using 5′ UTR forward primer and HIS3-ORF-specific reverse primer [61].
Table 2.
Primers for PCR.
| No. | Primer name | Oligonucleotide sequence |
|---|---|---|
| 1 | CTT1-F | 5′-tgcaagacttccatctgctg-3′ |
| 2 | CTT1-R | 5′-acggtggaaaaacgaacaag-3′ |
| 3 | GPX1-F | 5′-GATTGTGGCCTTTCCCTGTG-3′ |
| 4 | GPX1-R | 5′-CATTCCAGACTTCCCGCTTAC-3′ |
| 5 | TSA2-F | 5′-caagccccaccatttaagaa-3′ |
| 6 | TSA2-R | 5′-tcggtggaggcaaataaaac-3′ |
| 7 | HSP104-F | 5′-ggatggtgcctttgaaagaa-3′ |
| 8 | HSP104-R | 5′-ccaaagcagaatctggcaat-3′ |
| 9 | ACT1-F | 5′-cgttccaatttacgctggtt-3′ |
| 10 | ACT1-R | 5′-agcggtttgcatttcttgtt-3′ |
| 11 | ATG8-F | 5′-tgtgatttgcgaaaaagctg-3′ |
| 12 | ATG8-R | 5′-agacatcaacgccgcagtag-3′ |
| 13 | HSP12-F | 5′-cgcaggtagaaaaggattcg-3′ |
| 14 | HSP12-R | 5′-tcagcgttatccttgccttt-3′ |
| 15 | sch9-UP35-pRS303(HIS3)-F | 5′-CGTATAAGCAAGAAATAAAGATACGAATATACAATATGACAGAGCAGAAAGCCCTAGTA-3′ |
| 16 | sch9-DW34-pRS303(HIS3)-R | 5′-AAGAAGAGGAAGGGCAAGAGGAGCGATTGAGAAACTACATAAGAACACCTTTGGTGGAG-3′ |
Protein extraction and Western blotting analysis
Yeast cells grown overnight at 30 °C in SDC medium (Figs. 1C, 2A and C, and 3C) or YPD medium (Figs. 1A, and 5C and E) were diluted into 20 ml of medium in 100 ml flask to give an initial OD600 nm of 0.005. Cultures were incubated at 30 °C in an air thermostatic shaker (200 rpm) until desired OD600nm was achieved. Cells at their logarithmic growth phase were collected when the OD600 nm reached 0.5. Cell-free yeast extracts were prepared by using a modified published procedure [59,62]. 10 ml cells at OD600 nm of 0.5 were treated with cold TCA as described above, then suspended in 500 µl of water followed by the addition of 500 µl of 0.2 M NaOH. Thoroughly mixed samples were incubated for 30 min at room temperature, concentrated by centrifugation and suspended in 200 µl of lysis buffer (0.5 M Tris–HCl, pH 6.8, 5% glycerol, 2% SDS, 1% β-mercaptoethanol). After heating at 95 °C for 5 min, samples were centrifuged and the supernatant was mixed with 4× Laemmli Buffer (44.4% glycerol, 4.4% SDS, 10% β-mercapto-ethanol, 0.02% bromophenol blue and 277.8 mM Tris, pH 6.8). For most Western blotting assays protein was loaded in equal amounts onto a 10% SDS-PAGE gel, except 15% for detecting free ubiquitin. Separated proteins were transferred onto a PVDF membrane (Millipore, MA) followed by standard Western blotting protocols with primary antibodies including polyclonal rabbit anti-Sch9 antibodies (1:2000, a gift from Dr. Robert C. Dickson), anti-GFP antibodies (1:5000, Zen BioScience, China), monoclonal mouse anti-Ub antibodies (1:2000, Santa Cruz biotech, MA) or monoclonal mouse anti-actin (1:5000, Zen BioScience, China). Secondary antibodies include alkaline phosphatase-linked anti-rabbit or anti-mouse IgG (1:2000, Zen BioScience, China).
RNA extraction and RT-qPCR analysis
Total RNA was extracted from cells with RNAiso Plus (TaKaRa Bio, China). Before the extraction, 20 ml cells (OD600nm of 0.5) were pretreated with 50 U of lyticase at 30 °C for 30 min to increase extraction efficiency. For each reverse transcription reaction 2.0 µg of total RNA was reverse transcribed to cDNA using a PrimeScript RT reagent kit with gDNA eraser (Takara Bio, China). The primers for real-time quantitative PCR (RT-qPCR) were designed using Primer Premier 5.0 and synthesized commercially (BGI, China). The primer sequences are indicated in Table 2. PCR reaction was performed in a total volume of 25 µl, containing 100 ng cDNA, 12.5 µl SYBR Premix Ex TaqII (TaKaRa Bio, China), 9.5 µl dH2O and 0.2 µM specific primers using Bio-Rad CFX manager real-time PCR system with 1 cycle at 95 °C for 30 s followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s. Data were collected and analyzed by Bio-Rad CFX manager software using the 2−ΔΔCT method [63]. The expression of each mRNA was normalized to that of Actin (ACT1) and data are representative of results from three separate experiments.
Measurement of intracellular reactive oxygen species (ROS) by DCFH-DA and DHR123 staining
Detection of ROS in cells was based on the oxidation of DCFH-DA by intracellular ROS, resulting in the formation of the fluorescent compound 2′,7′-dichlorodihydrofluorescein (DCF) [47]. Cells (OD600 nm of 0.5) were harvested and washed with PBS, then incubated with 10 µM H2DCF−DA (Fanbo Biochemicals, China) for 1 h at 30 °C in dark. After washed three times with PBS (2.7 mM KCl, 1.75 mM KH2PO4, 10 mM Na2HPO4 and 136.75 mM NaCl, pH 7.4), the pellet was resuspended in 1 ml PBS. 10 µl of cells was added to a microscope slide, and DCF fluorescence was monitored using CEWEI LWD200-37FT fluorescence microscope (CEWEI, China). Alternatively, the prepared cells were lysed by glass beads and the fluorescence was measured at 488/525 nm excitation/emission using an f4500 fluorescence spectrometer (Hitachi, Japan). The fluorescence intensity was normalized according to the protein concentration measured by Bradford method.
Detection of intracellular ROS is also achieved by dihydrorhodamine 123 (DHR123) staining, based on the oxidative conversion of DHR123 to its corresponding two-electron oxidized fluorescent product, rhodamine 123 [51]. Cells (OD600 nm of 0.5) were harvested and washed with PBS, then incubated with 10 µM DHR123 (Keygen Biotech, China) for 1 h at 30 °C in dark. The procedures of monitoring DHR123 fluorescence using fluorescence microscope or fluorescence spectrometer were same as the DCF fluorescence measurements, except that 507 and 529 nm were used as excitation and emission wavelengths, respectively.
Catalase activity assay
The catalase activity was measured by native gel electrophoresis as described previously [64]. Briefly, the native gel consisting of 4% stacking gel and 8% separating gel was pre-electrophoresed for 1 h at 40 mA, 4 °C. 107–108 cells were washed twice in 5 ml PBS and resuspended in 50 mM phosphate buffer (pH 7.8) before they were lysed by ten cycles of vortexing with 0.5 mm acid-washed glass beads (Sigma-Aldrich, USA) for 30 s followed by 30 s of cooling on ice. After centrifugation (6000×g, 4 °C, 5 min), the supernatants containing 200 µg of protein per sample was loaded with the sample loading buffer (50% glycerol, 0.05% bromophenol blue, 0.25 M Tris and 4 mM EDTA, pH 6.8) and 10 U of bovine catalase (Solarbio, China) was loaded as a standard. Samples were electrophoresed for 3 h at 40 mA, 4 °C in the pre-electrophoresis buffer (0.2 M Tris, 1 mM EDTA, pH 8.8) followed by another 2 h of electrophoresis in the electrophoresis buffer (0.05 M Tris, 1.8 mM EDTA, 0.3 M glycine, pH 8.3). After electrophoresis, the gel was incubated in 0.003% H2O2 (vol/vol) for 10 min. After rinsing twice gently with ddH2O, the gel was stained with 2% ferric chloride and 2% potassium ferricyanide at room temperature. The stained gels were scanned with the scanner (Microtek Scanmaker i800). The intensities of bands were quantified by Image J and the catalase activities of samples were standardized by comparing to the band of standard catalase with known activity on the same gel.
Proteasome chymotrypsin-like activity assay
20 ml cells were harvested at OD600 nm of 0.5 and resuspended in lysis buffer (50 mM Tris pH 8.0, 5 mM MgCl2, 0.5 mM EDTA, and 1 mM ATP) at a ratio of 1.5 ml buffer per gram of wet cell mass. Cells were lysed by vortexing for 5 min at 4 °C with 0.5 mm acid-washed glass beads (Sigma-Aldrich, USA). Chymotrypsin-like activity of cell lysate was determined by measuring the release of fluorophore 7-amido-4-methylcoumarin (AMC) from 100 µM of N-succinyl-Leu-Val-Tyr-7 (LLVY) amido-4-methylcoumarin (Sigma-Aldrich, USA). Fluorescence was measured by an f4500 fluorescence Spectrometer (Hitachi, Japan) at excitation/emission wavelengths of 380 nm/440 nm. To deduct the influence of non-specific release of AMC, the proteasome inhibitor MG132 (75 µM, Selleck Chemicals, USA) was added to the isopyknic lysate as the control [65].
Protein carbonylation detection
Cells were lysed as described above for a catalase activity assay and the protein carbonylation was quantified by DNPH alkaline method as described [66]. Briefly, 400 µl of DNPH (10 mM in 0.5 M H3PO4) (KeLong Chemical, China) was mixed with 400 µl of protein solution. The mixture was incubated in the dark at room temperature for 10 min and followed by the incubation with 200 µl of NaOH (6 M) for 10 min. The absorbance was read at 450 nm with the UV spectrophotometer (APL Instruments, China). The concentration of carbonyl groups was normalized by the protein concentration of respective sample measured by Bradford method.
Statistical analysis
All data from at least three independent experiments are presented as averages±SD. Statistical analysis and comparisons were performed using two-tailed, unpaired Student t tests. Western blot images were plot and analyzed using ImageJ software. The anti-ubiquitin immunoblots were quantified by the whole lanes.
Acknowledgments
We thank Dr. Robert C. Dickson (University of Kentucky, Lexington, KY) and Dr. Robbie Loewith (University of Geneva, Geneva, Switzerland) for kindly gifting us strains, plasmids and antibodies. We also appreciate comments from Dr. Robert C. Dickson in review of this manuscript. This work was supported by NSFC (30671181/C0603).
Contributor Information
Nianhui Zhang, Email: zhangnianhui@scu.edu.cn.
Linfang Du, Email: dulinfang@scu.edu.cn.
Ke Liu, Email: kliu@scu.edu.cn.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at 10.1016/j.redox.2015.06.002.
Appendix A. Supplementary material
Supplementary material
References
- 1.Breusing N., Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol. Chem. 2008;389(3):203–209. doi: 10.1515/BC.2008.029. 18208355 [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. 17051204 [DOI] [PubMed] [Google Scholar]
- 3.Rechsteiner M. Ubiquitin-mediated pathways for intracellular proteolysis. Annu. Rev. Cell Biol. 1987;3:1–30. doi: 10.1146/annurev.cb.03.110187.000245. 2825735 [DOI] [PubMed] [Google Scholar]
- 4.Su V., Lau A.F. Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell. Mol. Life Sci. 2009;66(17):2819–2833. doi: 10.1007/s00018-009-0048-9. 19468686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. 1323239 [DOI] [PubMed] [Google Scholar]
- 6.Ding W.X., Yin X.M. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4(2):141–150. doi: 10.4161/auto.5190. 17986870 [DOI] [PubMed] [Google Scholar]
- 7.Shaid S. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20(1):21–30. doi: 10.1038/cdd.2012.72. 22722335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor A. Mechanistically linking age-related diseases and dietary carbohydrate via autophagy and the ubiquitin proteolytic systems. Autophagy. 2012;8(9):1404–1406. doi: 10.4161/auto.21150. 22906982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen E., Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat. Rev. Neurosci. 2008;9(10):759–767. doi: 10.1038/nrn2474. 18769445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K. Altered ubiquitin causes perturbed calcium homeostasis, hyperactivation of calpain, dysregulated differentiation, and cataract. Proc. Natl. Acad. Sci. U.S.A. 2015;112(4):1071–1076. doi: 10.1073/pnas.1404059112. 25583491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Y. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13(6):668–678. doi: 10.1016/j.cmet.2011.03.018. 21641548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longo V.D., Finch C.E. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299(5611):1342–1346. doi: 10.1126/science.1077991. 12610293 [DOI] [PubMed] [Google Scholar]
- 13.Clancy D.J. Dietary restriction in long-lived dwarf flies. Science. 2002;296(5566):319. doi: 10.1126/science.1069366. 11951037 [DOI] [PubMed] [Google Scholar]
- 14.Bjedov I. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. 20074526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontana L., Partridge L., Longo V.D. Extending healthy life span – from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. 20395504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattison J.A. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489(7415):318–321. doi: 10.1038/nature11432. 22932268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison D.E. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. 19587680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Cunha F.M., Demasi M., Kowaltowski A.J. Aging and calorie restriction modulate yeast redox state, oxidized protein removal, and the ubiquitin-proteasome system. Free Radic. Biol. Med. 2011;51(3):664–670. doi: 10.1016/j.freeradbiomed.2011.05.035. 21684330 [DOI] [PubMed] [Google Scholar]
- 19.Jung C.H. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. 20083114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabrizio P. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292(5515):288–290. doi: 10.1126/science.1059497. 11292860 [DOI] [PubMed] [Google Scholar]
- 21.Urban J. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cells. 2007;26(5):663–674. doi: 10.1016/j.molcel.2007.04.020. 17560372 [DOI] [PubMed] [Google Scholar]
- 22.Selman C. Ribosomal protein s6 kinase 1 signaling regulates mammalian life span. Science. 2009;326(5949):140–144. doi: 10.1126/science.1177221. 19797661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J. Regulation of the phosphorylation of yeast protein kinase Sch9 under environmental changes and during chronological aging. Prog. Biochem. Biophys. 2014;41(2):192–201. [Google Scholar]
- 24.Liu K. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 2005;280(24):22679–22687. doi: 10.1074/jbc.M502972200. 15840588 [DOI] [PubMed] [Google Scholar]
- 25.Liu K. Signalling functions for sphingolipid long-chain bases in Saccharomyces cerevisiae. Biochem. Soc. Trans. 2005;33(5):1170–1173. doi: 10.1042/BST20051170. 16246074 [DOI] [PubMed] [Google Scholar]
- 26.Biondi R.M. Phosphoinositide-dependent protein kinase 1, a sensor of protein conformation. Trends Biochem. Sci. 2004;29(3):136–142. doi: 10.1016/j.tibs.2004.01.005. 15003271 [DOI] [PubMed] [Google Scholar]
- 27.Huber A. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011;30(15):3052–3064. doi: 10.1038/emboj.2011.221. 21730963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber A. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23(16):1929–1943. doi: 10.1101/gad.532109. 19684113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swinnen E. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006;1:3. doi: 10.1186/1747-1028-1-3. 16759348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei M. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, tor, and Sch9. PLOS Genet. 2008;4(1):e13. doi: 10.1371/journal.pgen.0040013. 18225956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Colman M.J. The forkhead transcription factor Hcm1 promotes mitochondrial biogenesis and stress resistance in yeast. J. Biol. Chem. 2010;285(47):37092–37101. doi: 10.1074/jbc.M110.174763. 20847055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Colman MJ, Sorolla MA, Vall-Llaura N, Tamarit J, Ros J, Cabiscol E. The FOX transcription factor Hcm1 regulates oxidative metabolism in response to early nutrient limitation in yeast. Role of Snf1 and Tor1/Sch9 kinases. Biochimica et biophysica acta. 2013;1833(8):2004–2015. doi: 10.1016/j.bbamcr.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Swinnen E. Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14(1):17–32. doi: 10.1111/1567-1364.12097. 24102693 [DOI] [PubMed] [Google Scholar]
- 34.Bonawitz N.D. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5(4):265–277. doi: 10.1016/j.cmet.2007.02.009. 17403371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y., Shadel G.S. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging. 2009;1(1):131–145. doi: 10.18632/aging.100016. 20157595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavoie H., Whiteway M. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot. Cell. 2008;7(7):1127–1135. doi: 10.1128/EC.00330-07. 18469137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cocklin R., Goebl M. Nutrient sensing kinases PKA and Sch9 phosphorylate the catalytic domain of the ubiquitin-conjugating enzyme Cdc34. PLOS One. 2011;6(11):e27099. doi: 10.1371/journal.pone.0027099. 22087249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goebl M.G. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241(4871):1331–1335. doi: 10.1126/science.2842867. 2842867 [DOI] [PubMed] [Google Scholar]
- 39.Manukyan A. Synchronization of yeast. Methods Mol. Biol. 2011;761:173–200. doi: 10.1007/978-1-61779-182-6_12. 21755449 [DOI] [PubMed] [Google Scholar]
- 40.Werner-Washburne M. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 1993;57(2):383–401. doi: 10.1128/mr.57.2.383-401.1993. 8393130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breeden L.L. Alpha-factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–341. doi: 10.1016/s0076-6879(97)83027-3. 9251031 [DOI] [PubMed] [Google Scholar]
- 42.Roosen J. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol. Microbiol. 2005;55(3):862–880. doi: 10.1111/j.1365-2958.2004.04429.x. 15661010 [DOI] [PubMed] [Google Scholar]
- 43.Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. 10872471 [DOI] [PubMed] [Google Scholar]
- 44.Kabeya Y. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. 11060023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yorimitsu T. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18(10):4180–4189. doi: 10.1091/mbc.E07-05-0485. 17699586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheong H., Klionsky D.J. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. 19185709 [DOI] [PubMed] [Google Scholar]
- 47.Wang H., Joseph J.A. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 1999;27(5–6):612–616. doi: 10.1016/s0891-5849(99)00107-0. 10490282 [DOI] [PubMed] [Google Scholar]
- 48.Owusu-Ansah E., Utpal Banerjee A.Y. A protocol for in vivo detection of reactive oxygen species. Nat. Protoc. 2008 [Google Scholar]
- 49.Patterson E.K. Carbon monoxide-releasing molecule 3 inhibits myeloperoxidase (MPO) and protects against MPO-induced vascular endothelial cell activation/dysfunction. Free Radic. Biol. Med. 2014;70:167–173. doi: 10.1016/j.freeradbiomed.2014.02.020. 24583458 [DOI] [PubMed] [Google Scholar]
- 50.Martins D., Kathiresan M., English A.M. Cytochrome c peroxidase is a mitochondrial heme-based H2O2 sensor that modulates antioxidant defense. Free Radic. Biol. Med. 2013;65:541–551. doi: 10.1016/j.freeradbiomed.2013.06.037. 23831190 [DOI] [PubMed] [Google Scholar]
- 51.Qin Y., Lu M., Gong X. Dihydrorhodamine 123 is superior to 2,7-dichlorodihydrofluorescein diacetate and dihydrorhodamine 6G in detecting intracellular hydrogen peroxide in tumor cells. Cell Biol. Int. 2008;32(2):224–228. doi: 10.1016/j.cellbi.2007.08.028. 17920943 [DOI] [PubMed] [Google Scholar]
- 52.Sies H. Role of metabolic H2O2 generation: redox signaling and oxidative stress. J. Biol. Chem. 2014;289(13):8735–8741. doi: 10.1074/jbc.R113.544635. 24515117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciechanover A., Schwartz A.L. Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology. 2002;35(1):3–6. doi: 10.1053/jhep.2002.30316. 11786953 [DOI] [PubMed] [Google Scholar]
- 54.Cocklin R., Goebl M. Nutrient sensing kinases PKA and Sch9 phosphorylate the catalytic domain of the ubiquitin-conjugating enzyme Cdc34. PLOS One. 2011;6(11):e27099. doi: 10.1371/journal.pone.0027099. 22087249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalyanaraman B. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. 22027063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medicherla B., Goldberg A.L. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 2008;182(4):663–673. doi: 10.1083/jcb.200803022. 18725537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J. Reducing sphingolipid synthesis orchestrates global changes to extend yeast lifespan. Aging Cell. 2013;12(5):833–841. doi: 10.1111/acel.12107. 23725375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu S. A protein chip approach for high-throughput antigen identification and characterization. Proteomics. 2007;7(13):2151–2161. doi: 10.1002/pmic.200600923. 17549792 [DOI] [PubMed] [Google Scholar]
- 59.Huang X., Liu J., Dickson R.C. Down-regulating sphingolipid synthesis increases yeast lifespan. PLOS Genet. 2012;8(2) doi: 10.1371/journal.pgen.1002493. 22319457 p. e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu C. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. BioTechniques. 2007;42(2):158. doi: 10.2144/000112389. 17373478 [DOI] [PubMed] [Google Scholar]
- 61.Brachmann C.B. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. 9483801 [DOI] [PubMed] [Google Scholar]
- 62.Kushnirov V.V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16(9):857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. 10861908 [DOI] [PubMed] [Google Scholar]
- 63.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. 11846609 [DOI] [PubMed] [Google Scholar]
- 64.Weydert C.J., Cullen J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010;5(1):51–66. doi: 10.1038/nprot.2009.197. 20057381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kruegel U. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLOS Genet. 2011;7(9) doi: 10.1371/journal.pgen.1002253. 21931558 p. e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mesquita C.S. Simplified 2,4-dinitrophenylhydrazine spectrophotometric assay for quantification of carbonyls in oxidized proteins. Anal. Biochem. 2014;458:69–71. doi: 10.1016/j.ab.2014.04.034. 24814294 [DOI] [PubMed] [Google Scholar]
- 67.Mikus M.D., Petes T.D. Recombination between genes located on nonhomologous chromosomes in Saccharomyces cerevisiae. Genetics. 1982;101(3–4):369–404. doi: 10.1093/genetics/101.3-4.369. 6757052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fabrizio P. SOD2 functions downstream of Sch9 to extend longevity in y east. Genetics. 2003;163(1):35–46. doi: 10.1093/genetics/163.1.35. 12586694 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material


