Abstract
OprF is the major outer membrane porin in bacteria belonging to the Pseudomonas genus. In previous studies, we have shown that OprF is required for full virulence expression of the opportunistic pathogen Pseudomonas aeruginosa. Here, we describe molecular insights on the nature of this relationship and report that the absence of OprF leads to increased biofilm formation and production of the Pel exopolysaccharide. Accordingly, the level of c-di-GMP, a key second messenger in biofilm control, is elevated in an oprF mutant. By decreasing c-di-GMP levels in this mutant, both biofilm formation and pel gene expression phenotypes were restored to wild-type levels. We further investigated the impact on two small RNAs, which are associated with the biofilm lifestyle, and found that expression of rsmZ but not of rsmY was increased in the oprF mutant and this occurs in a c-di-GMP-dependent manner. Finally, the extracytoplasmic function (ECF) sigma factors AlgU and SigX displayed higher activity levels in the oprF mutant. Two genes of the SigX regulon involved in c-di-GMP metabolism, PA1181 and adcA (PA4843), were up-regulated in the oprF mutant, partly explaining the increased c-di-GMP level. We hypothesized that the absence of OprF leads to a cell envelope stress that activates SigX and results in a c-di-GMP elevated level due to higher expression of adcA and PA1181. The c-di-GMP level can in turn stimulate Pel synthesis via increased rsmZ sRNA levels and pel mRNA, thus affecting Pel-dependent phenotypes such as cell aggregation and biofilm formation. This work highlights the connection between OprF and c-di-GMP regulatory networks, likely via SigX (ECF), on the regulation of biofilm phenotypes.
Keywords: OprF, exopolysaccharides, biofilm, c-di-GMP, SigX, ECF
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium found in almost every wet ecological niches including soils, water, and plants (Spiers et al., 2000). It is also an opportunistic human pathogen, causing a variety of infections including chronic lung infections in cystic fibrosis patients (Ramsey and Wozniak, 2005). This requires a particularly well-developed ability to adapt to changes in environmental conditions, in which outer membrane (OM) proteins may play an important function considering their role in communication with the extracellular medium. The major Pseudomonas sp. OM protein OprF is homologous to OmpA of Enterobacteriaceae, and is the only general P. aeruginosa porin (Tamber and Hancock, 2004). It allows a non-specific diffusion of ionic species (Yoon et al., 2002) and small polar nutrients, including polysaccharides up to 1.5 kDa in size (Nestorovich et al., 2006). OprF anchors the OM to the peptidoglycan layer (Woodruff and Hancock, 1989; Rawling et al., 1998), is involved in host-pathogen interactions, and is required for expression of full virulence (Fito-Boncompte et al., 2011). Binding of human interferon-γ to OprF leads to the expression of the PA-I lectin, a quorum-sensing-dependent virulence determinant, suggesting that OprF acts as a sensor of the host immune system (Wu et al., 2005). OprF has also been proposed to be involved in rhamnolipid production (Bouffartigues et al., 2011), adhesion onto eukaryotic cells (Azghani et al., 2002; Fito-Boncompte et al., 2011), and biofilm development under anaerobic conditions (Hassett et al., 2002; Yoon et al., 2002). OprF expression is controlled by at least two extracytoplasmic function (ECFs) sigma factors, AlgU and SigX (Brinkman et al., 1999; Firoved et al., 2002; Bouffartigues et al., 2012), which are involved in cell envelope homeostasis (Wood et al., 2006; Boechat et al., 2013; Duchesne et al., 2013; Gicquel et al., 2013; Blanka et al., 2014).
Biofilm growth is a bacterial lifestyle of particular importance for P. aeruginosa pathogenesis (Ramsey and Wozniak, 2005). These surface-associated microbial communities of cells are embedded into a matrix of extracellular polymeric substances (Donlan and Costerton, 2002; Allesen-Holm et al., 2006; Ma et al., 2009). In P. aeruginosa, the matrix contains an array of components such as extracellular DNA (Allesen-Holm et al., 2006; Ma et al., 2009), proteinaceous adhesins, vesicles and exopolysaccharides (EPS; Ryder et al., 2007; Gooderham and Hancock, 2009; Mikkelsen et al., 2011). In several non-mucoid strains, such as the PAO1 strain, Psl and Pel are two key EPS that maintain biofilm structure (Friedman and Kolter, 2004a; Ma et al., 2006; Colvin et al., 2012). Pel is a glucose-rich EPS that is required to form pellicles at the air-liquid interface or mature solid surface-associated biofilms (Friedman and Kolter, 2004a,b; Ghafoor et al., 2011). Psl is a repeating penta-saccharide containing D-mannose, D-glucose and L-rhamnose (Byrd et al., 2009), which is of particular importance for the stability (shear resistance) of non-mucoid biofilms (Bazire et al., 2010). Expression of the pel and psl gene clusters is highly complex. It is regulated transcriptionally by the second messenger bis-(3′–5′)-cyclic guanosine monophosphate (c-di-GMP; Lee et al., 2007; Starkey et al., 2009; Borlee et al., 2010), and translationally by the Gac/Rsm signaling network (Brencic and Lory, 2009; Irie et al., 2010).
We report here that the absence of OprF in the OM of P. aeruginosa results in increased biofilm formation. We show that this is concomitant to (i) the overproduction of Pel but not Psl, (ii) the increase in c-di-GMP levels and (iii) the up-regulation of rsmZ, a small non-coding RNA of the Gac/Rsm signaling pathway. The artificial reduction of c-di-GMP in the oprF mutant resulted in the restoration of these phenotypes. Two genes of the SigX regulon that are involved in regulating c-di-GMP levels, adcA (PA4843; Jones et al., 2014) and PA1181, are up-regulated in the oprF mutant, which thus links OprF and c-di-GMP regulatory networks.
Materials and Methods
Bacterial Strains and Growth Conditions
The strains are listed in Table 1. Strains were grown overnight at 37°C on a rotary shaker (180 rpm) in Luria Bertani (LB) broth. Five hundred μg streptomycin ml-1 only or with 300 μg carbenicillin ml-1 were added in H636 and H636O precultures, respectively. Under these conditions, the growth of the three strains was closely similar (Fito-Boncompte et al., 2011). When indicated, 40 μg.ml-1 of congo red (CR) was added to the LB growth medium. Fifty μg.ml-1 of tetracyclin were added to the growth medium for the culture of the H636C complemented strain. The PcdrA-gfp, pJN105 and pJN2133 plasmids were introduced into P. aeruginosa strains by electroporation and transformants were selected and grown in LB containing 100 μg gentamicin ml-1.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains/plasmids | Relevant characteristic(s) | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | Cloning host | Life technology |
| SM10pir | Host strain for mini-CTX1 | de Lorenzo and Timmis (1994) |
| Pseudomonas aeruginosa | ||
| H103 | PAO1 derivative | Hancock and Carey (1979) |
| H636 | PAO1 H103 oprF::Ω | Woodruff and Hancock (1988) |
| H636O | pRW5 containing H636 strain | Woodruff and Hancock (1988) |
| H636C | H636 complemented strain with | This study |
| a copy of oprF downstream | ||
| pBAD promoter | ||
| Plasmids | ||
| pRW5 | OprF cloned in pUCP19 vector, Cbr | Woodruff and Hancock (1988) |
| pJN105 | araC-pBAD cassette cloned in pBBR1MCS-5, | Newman and Fuqua (1999) |
| Gmr | ||
| pJN2133 | PA2133 cloned in pJN105, Gmr | Hickman et al. (2005) |
| mini-CTX1 | Plasmid for the integration of genes into | Hoang et al. (2000) |
| the att site of the P. aeruginosa chromosome, Tcr | ||
| mini-CTX- | araC-pBAD cassette cloned in mini-CTX1, Tcr | This study |
| araC-pBAD | ||
| mini-CTX- | oprF cloned upstream pBAD promoter in | |
| araC-pBAD-oprF | mini-CTX-araC-pBAD, Tcr | This study |
| PcdrA-gfp | Rybtke et al. (2012) |
Cbr, carbenicillin resistance; Gmr, gentamycine resistance; Tcr, tetracycline resistance.
EPS Production Assays
To observe colony morphology, overnight cultures were diluted at an OD580 of 0.08 in LB (8.107 CFU.ml-1) and 5 μl were spotted onto a LB plate containing 40 μg.ml-1 of CR and 20 μg Coomassie brilliant blue as described by Friedman and Kolter (2004a,b). Plates were incubated at 37°C overnight before visual inspection of the colony morphology. To quantify the CR binding on bacterial cells, a CR release assay was achieved as previously described (Lee et al., 2007). Briefly, an aliquot of overnight cultures corresponding to 109 cells, were centrifuged at 7,500 g during 10 min at room temperature. Cells were resuspended in 1 ml of LB containing CR (40 μg.ml-1), and incubated for 15 min at room temperature before centrifugation at 7,500 g during 10 min. The absorbance of the unbound CR was measured in the supernatant at 490 nm.
Total Carbohydrate Content Assay
Sugar composition of the cell-associated carbohydrates was determined by gas chromatography analysis of trimethylsilyl methyl glycoside derivatives using inositol as internal standard for quantification. Bacteria (200 ml), grown overnight in LB medium, were centrifuged at 7,500 g for 5 min. Cells were washed gently with 10 ml of distilled water before centrifugation at 7,500 g for 5 min. EPS were detached from cells as followed. Ten ml of distilled water was added to the cell pellet, and the tubes were vigorously mixed each 2 min for 14 min before centrifugation at 7,500 g for 10 min. This step was repeated five times. The supernatants were collected, pooled, filtered through a 0.2 μm filter and precipitated overnight at -20°C by adding cold ethanol to a final concentration of 70% (v/v). The precipitate was collected by centrifugation at 13,000 g for 1h at 4°C, resuspended in 5 ml of distilled water, dialyzed against distilled water using a 6–8 kDa molecular weight cut-off membrane for 24 h at 4°C, and lyophilisated. Samples (about 2 mg of dried material) and 50 μL of 2.10-3M inositol were hydrolyzed in 0.5 mL of 2 M trifluoroacetic acid for 2 h at 110°C, dried and then heated in 0.250 mL of dry 1 M HCl in methanol at 80°C for 24 h for methanolysis. After evaporation of the methanol, the sample was resuspended in 500 μL of methanol and dried again. The methyl glycosides were then converted into their trimethylsilyl derivatives by heating the samples for 20 min at 80°C in 200 μL of hexamethyldisilazane/trimethylchlorosilane/pyridine: 3/1/9. After evaporation of the reagent, the samples were suspended in 100 μL of cyclohexane before being injected on a FDB-1 column (DB-1 Supelco). Chromatographic data were integrated with gas chromatography Star Workstation software (Varian), each surface being corrected according to its response factor. For comparison of amounts of cell-associated carbohydrates, total monosaccharide contents for each sample were normalized according to the OD measured at 580 nm.
General DNA Procedures
Restriction enzymes, T4 DNA ligase, and alkaline phosphatase were purchased from New England Biolabs (Ipswich, MA, USA) and used accordingly to the manufacturer. PCR assays were carried out with 1 μg of P. aeruginosa chromosomal DNA, 20 pmol of each primer and failsafe Taq polymerase (Epicentre Biotechnologies, Madison, WI, USA). When necessary, PCR products and plasmids were purified with the QIAquick or QIAprep Spin Miniprep kits (QIAGEN), respectively. DNA sequencing was achieved by Genome Express (France). E. coli [commercial electrocompetent DH5α cells (Promega, Madison, WI, USA) or S17.1 cells] and P. aeruginosa cells were transformed by electroporation or by conjugation as previously described (Bouffartigues et al., 2012).
H636C Complemented Strain Construction
To complement the H636 oprF mutant strain with a copy of the oprF gene into the chromosome, the mini-CTX1-araC-pBAD was constructed as follows. The region containing araC and the pBAD promoter was amplified from pJN105 (Table 1) using the primer pair FaraC-pBAD and RaraC-pBAD (Table 2) and cloned into pCR2.1-TA (Invitrogen, Carlsbad, CA, USA). Insert was then cut with SacI and NheI restriction enzymes and inserted into mini-CTX1 using the SacI and SpeI sites. A 1113 bp DNA fragment containing the oprF gene (PA1777) was amplified by PCR using the primer pair FoprFPstI and RoprFHindIII (Table 2). The PCR product was digested with PstI and HindIII and ligated into the PstI-HindIII digested mini-CTX1-araC-pBAD vector to create the mini-CTX1-araC-pBAD-oprF. The sequence of this construct was verified by DNA sequencing. This vector was constructed in DH5α, purified and transferred into E. coli SM10 strain. The mini-CTX1-araC-pBAD-oprF vector was mobilized from E. coli SM10 into H636 by conjugation. Transconjugants were selected onto PIA agar plate containing 250 μg.ml-1 of tetracyclin. The insertion was verified by PCR using the FoprFPstI and RoprFHindIII primers. It is known that the pBAD promoter is not fully locked in P. aeruginosa, we used the basal level of expression from this promoter, without addition of arabinose, to obtain adequate amount of OprF in complementation experiments (Becher and Schweizer, 2000; Hoang et al., 2000).
Table 2.
Primer sequences of the indicated genes used for quantitative RT-PCR reactions and for the construction of the mini-CTX1-araC-pBAD and its derivative vector mini-CTX1-araC-pBAD-oprF.
| PA number | Gene name | Primer name | Sequence (5′-3′) |
|---|---|---|---|
| PA2063 | pelB | FPA3063 | CGGCTACGTGCAGCGTTAT |
| RPA3063 | CACTGCATGCGTTCCTTGAC | ||
| PA2231 | pslB | FPA2231 | ACACCAACGAATCCACCTTCA |
| RPA2231 | CGCTCTGTACCTCGATCATCAC | ||
| PA3621.1 | rsmZ | FPA3621.1 | CGTACAGGGAACACGCAAC |
| RPA3621.1 | ATTACCCCGCCCACTCTTC | ||
| PA0527.1 | rsmY | FPA0527.1 | CGCCAAAGACAATACGGAAAC |
| RPA0527.1 | TTTTGCAGACCTCTATCCTGACATC | ||
| PA1776 | sigX | FPA1776 | AATTGATGCGGCGTTACCA |
| RPA1776 | CCAGGTAGCGGGCACAGA | ||
| PA1775 | cmpX | FPA1775 | GGCAGATCATTGCAGGAATCTAC |
| RPA1775 | TCTCTTCAATAGTGCCTTCAACGT | ||
| PA0762 | algU | FPA0762 | GAAGCCCGAGTCTATCTTGG |
| RPA0762 | GCGATACCTCTCTTGGCATT | ||
| PA3540 | algD | FPA3540 | GGGCTATGTCGGTGCAGTATG |
| RPA3540 | GCGACTTGCCCTGGTTGAT | ||
| PA4843 | PA4843 | FPA4843 | CCTGGGCACCGAATTGG |
| RPA4843 | CGGCGGACAGGTAGATGATC | ||
| PA2072 | PA2072 | FPA2072 | CCAGGCATCAGGACGACAT |
| RPA2072 | CGATTCTGCAGCGCCTTT | ||
| PA1181 | PA1181 | FPA1181 | CCAGATGGAGAAGCGCTACCTCGCTT |
| RPA1181 | CGCTTGCGACTGTCGATATC | ||
| 16sRNA | F16SRNA | AACCTGGGAACTGCATCCAA | |
| R16SRNA | CTTCGCCACTGGTGTTCCTT | ||
| PA1777 | oprF | FoprFPstI | ∗taataactgcagAGATGGGGATTTAACG |
| RoprFHindIII | ∗taataaaagcttTCCTTAGAGGCTCAGCCGATT | ||
| FaraC-PBAD | ∗aacatatgCGTCAATTGTCTGATTCGTTACCAAT | ||
| RaraC-PBAD | ∗aatcgctagcCCAAAAAAACGG |
∗Nucleotides not included in the gene sequence are indicated in lower case.
Quantitative RT-PCR
Extraction of RNAs, synthesis of cDNAs and real time PCR were achieved as previously described (Gicquel et al., 2013), using primers described in Table 2. PCR reactions were performed in triplicate and the standard deviations were lower than 0.15 CT. The relative quantification of the mRNAs or sRNAs of interest was obtained by the comparative CT (2-ΔΔCt) method (Livak and Schmittgen, 2001), using 16S rRNA as endogenous control (Corbella and Puyet, 2003).
c-di-GMP Quantification
Indirect c-di-GMP quantification was evaluated using PcdrA-gfp plasmid, in which the c-di-GMP depending cdrA promoter is fused to gfp (Rybtke et al., 2012). P. aeruginosa strains containing the PcdrA-gfp plasmid were grown overnight and subcultured in LB with gentamicin to a starting OD600 of 0.1 (108 CFU.ml-1). After 4 h incubation at 37°C in shaking conditions, 1 mL of culture was harvested and re-suspended in the same volume of 1x PBS. OD600 and fluorescence (excitation 485 nm, emission 520 nm) was measured in a black 96-well plate with see-through flat bottom (Falcon) using a FLUOstarOptima plate reader (BMG Labtech). Quantifications were performed in triplicate and data are presented as relative fluorescent units (RFUs) which are arbitrary fluorescent units corrected for cell density.
c-di-GMP was also directly quantified by LC-MS/MS. Extraction and quantification of intracellular c-di-GMP was performed with some modifications as previously described (Spangler et al., 2010; Strehmel et al., 2015). Cultures of P. aeruginosa were grown for 4 h in LB medium at 37°C under shaking conditions to an optical density OD600 of ∼2.0 (2.109 CFU.ml-1). Subsequent aliquots of 10 ml of each culture, adjusted to a cell density OD600 of 0.5 (5.108 CFU.ml-1) by dilution with LB medium, were harvested by centrifugation for 2 min, 8000 × g and 4°C. Cell pellets were resuspended in 600 μl ice-cold extraction buffer [40% (v/v) acetonitrile, 40% (vol/vol) methanol and 20% (vol/vol) ddH2O] by vigorous vortexing with and without 5 μl of a 10 μg.ml-1 solution of xanthosine-(3′,5′)-cyclic monophosphate (cXMP; BioLog Life Science Institute; Bremen, Germany) in ddH2O as an internal standard. After 15 min of incubation on ice, cells were lysed at 95°C for 10 min, followed by centrifugation at 20000 × g and 4°C for 5 min. Supernatants were collected in a new 1.5 ml reaction tube and stored on ice. Remaining pellets were again resuspended in 400 μl extraction buffer, incubated on ice for 15 min and subjected to centrifugation (15000 × g, 4°C, 5 min). This procedure was repeated, and supernatants of all three extraction steps were pooled and subsequently evaporated using a concentrator 5301 (Eppendorf, Hamburg, Germany) at 45°C. The remaining residue were then resuspended in 500 μl ddH2O and c-di-GMP levels were quantified by LC-MS/MS as described previously (Strehmel et al., 2015), with the following modifications. The chromatographic separation was performed on a 1100 Series HPLC system (Agilent, Waldbronn, Germany) using a Multospher AQ RP 18, 5 μm, 250 mm× 4.0 mm HPLC column (CS Chromatography Service GmbH, Langerwehe, Germany) in a gradient mode using 10 mM ammonium acetate with 0.1% acetic acid as eluent A and methanol as eluent B. The injection volume of each sample was set to 40 μl and the flow rate was 0.4 ml/min. The gradient program was as follows: from 0 to 4 min 100% A, followed by a linear gradient from 100 to 80% A in 1 min, held for 2 min at 80% A, followed by a linear gradient from 80 to 60% A in 1 min and held for additional 9 min at 60%. Finally, re-equilibration of the column was obtained by constantly running 100% A for 16 min.
Electrospray ionization (ESI) MS was performed on an API 4000 triple quadrupole mass spectrometer (Applied Biosystems, Toronto, ON, Canada) using a turbo ion spray interface used in positive mode at an ionization potential of 5000 V and a temperature of 400°C using nitrogen as curtain, nebulizer, heater and collision gas. The parameter settings were optimized by infusion experiments. Data were acquired in multiple reaction monitoring (MRM) mode using the Analyst software version 1.6. (Applied Biosystems, Toronto, ON, Canada). Identification and quantification of c-di-GMP was performed by using three specific mass transitions from molecule ion m/z 691 to the product ions m/z 152, m/z 135, and m/z 540. The external calibration was carried out at c-di-GMP concentrations ranging from 10 to 200 ng in 500 μl ddH2O using the internal standard cXMP (50 ng). Obtained concentrations of c-di-GMP were normalized against total protein contents of respective cultures, which were determined by the bicinchoninic acid assay (Smith et al., 1985). All experiments were performed for three independent cultures each analyzed in duplicate.
Bacterial Attachment Assay on Glass and Scanning Electron Microscopy
Bacteria were grown overnight in LB at 37°C, harvested by centrifugation for 10 min at 7,000 g, washed twice, and suspended in 0.9% NaCl to an OD600 of 0.6 (6.108 CFU.ml-1). Twenty-five ml were then poured into a Petri dish containing a glass slide (24 × 50 mm) and stored at 37°C for 2 h. The slide was then washed for 5 min with sterile demineralized water to remove non-adherent cells. The attached cells were observed with a scanning electron microscope (SEM) as follows. Each sample was fixed with 3% glutaraldehyde overnight at 4°C and washed three times with 0.1 M phosphate buffer (pH 7.35). The cells were then sequentially dehydrated with 70, 95, and 100% ethanol (three times for 10 min each). The samples were dried in ethanol in a CPD 030 Critical Point Dryer (Bal-Tec, France), using CO2 as a transitional fluid to reach the critical point. The samples were mounted on aluminum stubs and coated for 120 s at 20 mA with gold/palladium alloy using a 501 sputter coater (Edwards Pirani, UK) and observed with a JEOL 6460LV microscope (JEOL Ltd, Japan) at a magnification of 4,500. The voltage was kept at 10 or 15 kV, at an average distance from the electron gun of about 10 mm.
Biofilm Formation in a Flow Cell System and Confocal Laser Scanning Microscopy
Biofilms were grown at 37°C in a three-channel flow cell with individual channel dimensions of 1 mm× 4 mm × 40 mm (Biocentrum, DTU, Danemark, Pamp et al., 2009), using a microscope coverslip (ST1, VWR) as substratum. The flow system was assembled, prepared and sterilized as described by Tolker-Nielsen and Sternberg (2011). Flow cells were sterilized by flushing 0.5% sodium hypochlorite for 30 min, then rinsed overnight with sterile water using a 250S Watson Marlow peristaltic pump. Bacteria were grown in LB, harvested by centrifugation for 10 min at 7,000 g, washed twice and suspended in 0.9% NaCl to obtain an OD600 of 0.6. Each channel was inoculated with 1 ml of bacterial suspension and left for attachment during 2 h without flow at 37°C. LB medium was then pumped with a 2.5 ml h-1 flow. Biofilms were visualized after 24 and 48 h by confocal laser scanning microscopy (CLSM) with a TCS-SP2 microscope (Leica Microsystems, Heidelberg, Germany), using a 63x oil immersion objective. Image stacks were collected and processed using Leica Confocal Software and Adobe Photoshop. The thicknesses (μm) and biovolumes (in μm3/μm2) of the biofilms were measured using the COMSTAT software (Heydorn et al., 2000). Each experiment was made in triplicate and 10 positions were analyzed per channel.
Biofilm Formation on Polystyrene and Crystal Violet Staining
Quantification of biofilm formation was performed in 12-wells polystyrene microtiter plates as previously described (Bouffartigues et al., 2014). Briefly, LB medium (2 ml per well) supplemented with appropriate antibiotics was inoculated to a final OD580 of 0.08 (8.107 CFU.ml-1) and incubated at 37°C without shaking for 24 h. Biofilms were stained with 3 ml of 0.4% of crystal violet (CV) for 20 min at room temperature, and washed with 3 ml of water until removing the unbound dye. After washing, CV was solubilized in 100% ethanol before measuring the absorbance at 595 nm. For each biofilm assay, three independent experimental repetitions were performed.
Results
The Lack of OprF Results in Increased EPS Production, Cell Aggregation and Biofilm Formation
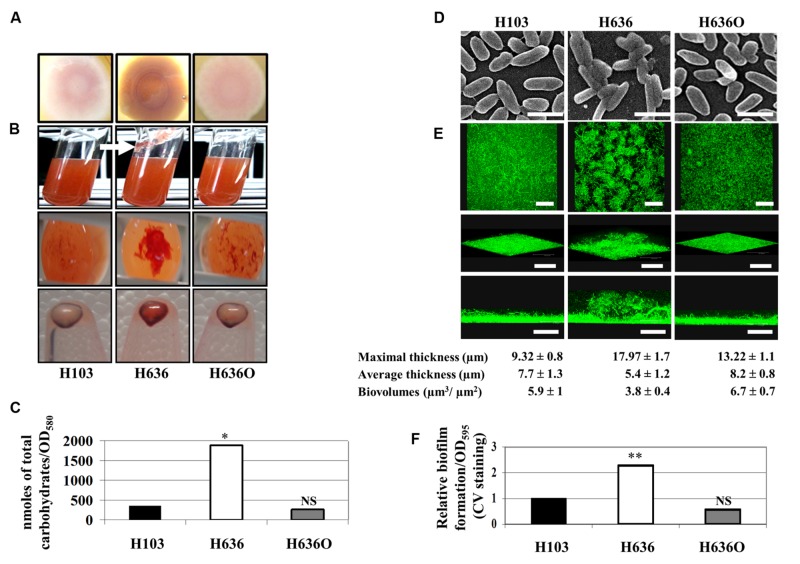
We investigated the role of OprF in biofilm formation and EPS production by comparing three strains, i.e., the P. aeruginosa H103 wild-type strain (a PAO1 derivative, Hancock and Carey, 1979), the isogenic oprF mutant H636 (Woodruff and Hancock, 1988), and the oprF complemented control strain H636O (Woodruff and Hancock, 1988). We first examined colony morphologies on CR plates at 37°C. As shown in Figure 1A, the P. aeruginosa H636 colony, i.e., the oprF mutant, displayed a marked red staining, suggesting higher polysaccharide production when compared to the H103 wild-type strain. Upon complementation of the oprF mutation, a lower polysaccharide production was restored, confirming that the phenotype was due to the oprF deletion. Accordingly, when strains were grown in liquid LB medium, CR staining revealed the presence of cell aggregates and slime attached to the glass in H636, but not in H103 or H636O (Figure 1B). Upon centrifugation of the bacterial cultures, the cell pellets showed higher CR staining for H636 but not for H103 or H636O (Figure 1B). Quantification of the total cell-associated carbohydrates by gas chromatography confirmed that the levels of EPS in the H636 oprF mutant are fivefold higher than in H103 or H636O strains (Figure 1C).
FIGURE 1.
The absence of OprF led to increase EPS production and biofilm formation in P. aeruginosa. (A) Colony morphology observed on Congo red (CR) containing LB agar plates. (B) H103, H636, and H636O were grown in CR containing LB for 24 h at 37°C. Top images: a slime production is indicated by a white arrow in case of H636; middle images: CR colored aggregates were observed at the bottom of the cultures; bottom images: CR binding of pelleted cells (109). (C) Cell-associated carbohydrates were quantified by gas chromatography. Statistics were achieved by unpaired t-test. ∗p < 0.05, NS, not significant. (D) Scanning electron microscopy images of H103, H636, and H636O allowed to attach onto a glass coverslip for 2 h (enlargement: 4,500 x; scales represent 2 μm). (E) Biofilms of H103, H636, and H636O grown in flow cells for 24 h and examined by CLSM. Top images, top views (x, y-plane; scales represent 48 μm); intermediate images, cross section views (scales represent 67 μm); bottom images, 3D-modelizations (x, y, z-axes, scales represent 48 μm). Maximal (μm), average thicknesses (μm), and biovolumes (μm3/μm2) were determined by COMSTAT analyses. The averages and standard deviations were calculated from 10 samples. (F) Microtiter grown biofilm formed by H103, H636, and H636O. Biofilms were quantified by measuring absorbance at 595 nm after crystal violet (CV) staining. At least six assays were performed for each strain. Statistics were achieved by unpaired t-test. ∗∗p < 0.01, NS, not significant.
It is well established that increased EPS production may result in cell aggregation and biofilm formation. Bacteria were thus allowed to attach onto a glass coverslip for 2 h and then observed by scanning electron microscopy (SEM). SEM images showed that H636 cells were slightly thinner than H103 and H636O, and cells aggregates could be observed, suggesting that the absence of OprF led to cell surface alterations that favored interactions between bacteria (Figure 1D). In addition, biofilm formation in flow cells was examined by CLSM after 24 h growth. As shown in Figure 1E, H103 and H636O biofilms were homogeneous and flat, displaying similar thicknesses and biovolumes. Conversely, the H636 oprF mutant formed a biofilm with heterogeneous cell distribution, increased maximal thickness and weaker biovolume. CV staining and quantification of biofilms grown in microtitre plates revealed that the oprF mutant strain yielded a 2.3-fold higher biofilm biomass than the other two strains (Figure 1F).
The OprF-Deficient Strain Displays Increased Levels of pel mRNA and c-di-GMP
Since Psl and Pel are the major EPS of P. aeruginosa PAO1 biofilm matrix (Hickman et al., 2005), we quantified by qRT-PCR the levels of the pelB and pslB transcripts. While pslB mRNA levels were unaffected by the oprF mutation, expression of pelB was 15-fold up-regulated in the H636 mutant strain (Figure 2A). Complementation of the mutant restored the expression of pelB to wild-type level, confirming that the observed variation was OprF-dependent. Since the expression of pel and psl is stimulated by high intracellular c-di-GMP levels (Lee et al., 2007; Starkey et al., 2009; Borlee et al., 2010), the latter was tested using the PcdrA-gfp reporter system in which gfp is fused to the c-di-GMP-dependent cdrA (Rybtke et al., 2012). The reporter plasmid was introduced into the wild-type H103, the H636 mutant strain, and the oprF complemented strain H636C. In this case and in the subsequent experiments, complementation was achieved from a single copy of oprF inserted in the chromosome (see Materials and Methods). The Figure 2B shows that the activity of the cdrA promoter was ca 1.8-fold higher in H636 than in H103, a phenotype that was partly complemented in H636C. This result was confirmed by direct quantification of intracellular c-di-GMP (Figure 2B) using LC-MS/MS (see Materials and Methods).
FIGURE 2.
The c-di-GMP level is increased in the oprF mutant. (A) Relative pel and psl mRNA expression in H636 (white bars) and H636C (gray bars) relatively to H103 (dashed black line). (B) c-di-GMP level evaluation using the PcdrA-gfp reporter fusion (full colored bars), and by LC/MS/MS quantification (dotted colored bars), harbored by H103 (black bar), H636 oprF mutant (white bar) and the complemented H636C strain (gray bar). (C) Exopolysaccharide production by H636 harboring pJN105 (dotted white bar) or pJN2133 (gray bar), using the CR release assay. 100% correspond to the EPS production by P. aeruginosa H103. (D) Biofilms formed by H636 harboring pJN105 (dotted white bar) or pJN2133 (gry bar) were quantified relatively to H103 by CV staining. (E) qRT-PCR assays of pelB mRNA level in H636 harboring pJN105 (dotted white bars) or pJN2133 (gray bars), relatively to H103. Each experiment (A–E) was performed at least three times. Statistics were achieved by unpaired t-test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, NS, not significant, relatively to H103.
To assess whether we could revert the oprF mutant phenotypes by modulating the c-di-GMP level, the pJN2133 plasmid carrying the gene encoding the phosphodiesterase PA2133 under control of the pBAD promoter (Hickman et al., 2005), was introduced in the H636 mutant strain. Basal expression of PA2133 without arabinose induction led to restore CR binding (Figure 2C) and biofilm formation (Figure 2D) to wild-type levels in the pJN2133-carrying strain. In agreement with this observation, we observed that reducing the c-di-GMP level led to a significant decrease in pelB expression (Figure 2E). Noteworthy, the level of pelB was not fully restored, suggesting that additional regulatory mechanisms may account for the up-regulation of the pel genes in the oprF mutant (Figure 2E).
rsmZ is Up-Regulated in the oprF Mutant
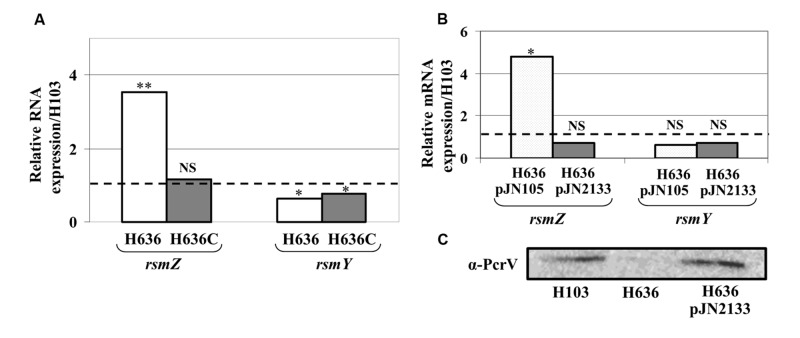
Pel production is known to be post-transcriptionally repressed by the RNA binding protein RsmA, a downstream target of the GacS/GacA two-component system (TCS; Brencic and Lory, 2009; Irie et al., 2010). This regulatory system positively controls factors involved in biofilm formation through the production of two small non-coding RNAs, rsmY and rsmZ that sequester RsmA and thus interfere with its function (Gooderham and Hancock, 2009; Mikkelsen et al., 2011). Using qRT-PCR experiments, we investigated whether the production of rsmY and rsmZ was affected in the H636 oprF mutant. As shown in Figure 3A, rsmZ expression was increased by about 3.5-fold, while that of rsmY was slightly decreased in the oprF mutant. Complementation of the mutant led to restoration of rsmZ levels. Reducing the c-di-GMP level by overexpressing PA2133 led to lower rsmZ expression without affecting that of rsmY, suggesting that rsmZ transcription is connected to the c-di-GMP increase in the oprF mutant (Figure 3B). Finally, we have previously shown that the H636 oprF mutant was impaired in the production of components of the type III secretion system (T3SS; Fito-Boncompte et al., 2011), which is also regulated by the Gac/RsmA signaling network. Consistently, the production of the T3SS needle tip, PcrV, was strongly reduced in H636 compared to H103. The levels of PcrV were restored when the oprF mutation was compensated by reducing the c-di-GMP levels in H636 (Figure 3C).
FIGURE 3.
The lack of OprF led to increase rsmZ, but not rsmY expression. (A) Relative expression of the small RNAs rsmZ and rsmY in H636 mutant (white bars) and the complemented strain H636C (gray bars), relatively to H103 (dashed black line). (B) Relative rsmZ and rsmY sRNA levels in H636 mutant strain harboring pJN105 (dotted white bars) or pJN2133 (gray bars), relatively to H103 (dashed black line). (C) Western blot analyses of T3SS α-PcrV in P. aeruginosa H103, H636, and H636 harboring pJN2133. Each experiment was performed at least three times. Statistics were done by unpaired t-test. ∗p < 0.05, ∗∗p < 0.01, NS, not significant.
The ECF Sigma Factors AlgU and SigX are Up-Regulated in the oprF Mutant
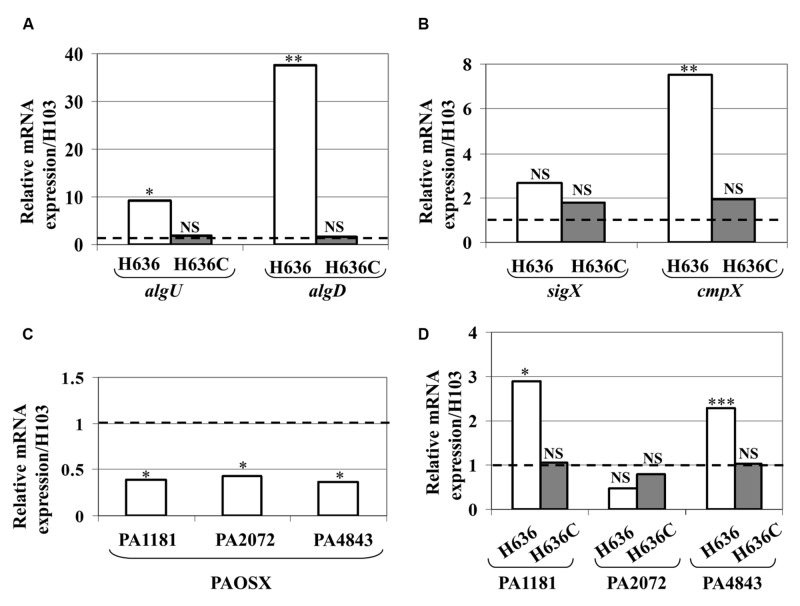
OprF was suspected to be involved in cell shape maintenance, partly through connecting the OM with the peptidoglycan layer. It has been suggested that the absence of OprF leads to alterations of the cell wall integrity (Woodruff and Hancock, 1989; Rawling et al., 1998). P. aeruginosa possesses two extracytoplasmic sigma factors involved in maintaining cell wall integrity, the RpoE homologue AlgU (Wood and Ohman, 2009) and SigX (Boechat et al., 2013; Gicquel et al., 2013; Blanka et al., 2014). As shown in Figure 4A, the expression of algU and algD genes, which are direct AlgU targets (Schurr et al., 1993), was strongly increased in the oprF mutant, a phenotype that is restored in the H636C strain. This strongly supports the idea that the oprF mutant strain encounters cell wall stress. On the contrary, the expression of sigX was not significantly altered in the conditions tested. However, the expression of cmpX, a direct target of SigX (Bouffartigues et al., 2012; Gicquel et al., 2013; Blanka et al., 2014), was significantly increased in the oprF mutant (Figure 4B). We thus hypothesized that SigX activity level is higher in H636 and examined the expression of genes from the SigX regulon that are involved in c-di-GMP metabolism, i.e., PA1181, PA2072 and adcA (PA4843; Gicquel et al., 2013; Blanka et al., 2014). As shown in Figure 4C, PA1181, PA2072 and adcA (PA4843) are down-regulated in the isogenic sigX mutant of P. aeruginosa H103, confirming previous observations made on strain PA14 (Blanka et al., 2014). Using qRT-PCR, we observed that PA1181 and adcA mRNAs are overproduced in H636 (but not in the complemented strain) by 2.9- and 2.1-fold, respectively. In contrast, the expression of PA2072 was unaffected (Figure 4D). Since AdcA has been shown to have diguanylate cyclase activity (Jones et al., 2014), and PA1181 contains both GGDEF and EAL motifs, our data suggest that the two proteins may be responsible for the c-di-GMP increase in the oprF mutant strain.
FIGURE 4.
Increased expression and activity of the extracytoplasmic function sigma factors AlgU and SigX. Relative mRNA levels of algU and algD (A), sigX and cmpX (B) in P. aeruginosa H636 (white bars) and H636C (gray bars) relatively to H103. (C) Relative mRNA levels of PA1181, PA2072 and adcA in PAOSX [sigX mutant strain (Bouffartigues et al., 2012) relatively to H103]. (D) Relative mRNA levels of the SigX-dependent PA1181, PA2072 and adcA (PA4843) gene in P. aeruginosa H636 (white bars) and H636C (gray bars) relatively to H103. Each experiment was performed at least three times. Statistics were achieved by unpaired t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, NS, not significant.
Discussion
In a previous study, we have shown that an oprF mutant is altered in the production of many virulence factors, including pyocyanin, elastase and T3SS effectors (Fito-Boncompte et al., 2011). Here, we further show that an oprF mutant overproduces polysaccharides, a phenotype that is mainly linked to pel gene overexpression rather than psl. In PA14, or when overexpressed in PAO1, Pel promotes attachment to abiotic surfaces, aggregation in broth culture, and biofilm formation (Yang et al., 2011; Colvin et al., 2012). Accordingly, the oprF mutant displayed an aggregative phenotype and an increased biofilm formation in our conditions. Interestingly, the biofilm structure produced by the mutant strain was altered. The maximal thickness was increased while the biofilm biovolume was reduced. These data suggest that H636 cells grew slower than both the wild-type and the complemented mutant strains. Otherwise, the lack of OprF may weaken the cell structure leading to increased bacterial death within the biofilm. We also observed increased c-di-GMP level in the oprF mutant. High levels of c-di-GMP are known to (i) impact positively polysaccharide production and biofilm formation (Hickman et al., 2005; Güvener and Harwood, 2007; Starkey et al., 2009; Borlee et al., 2010), and (ii) impact negatively the production of T3SS components (Moscoso et al., 2011), which we were able to confirm in the oprF mutant. Decrease of the c-di-GMP level in the oprF mutant strain upon overproduction of a phosphodiesterase led to the restoration of polysaccharide production, biofilm formation and T3SS activity to wild-type levels. These observations suggest that an elevated c-di-GMP level could be the leading cause of the phenotypic alterations in this strain. Both pel and psl genes are known to be regulated by intracellular c-di-GMP level (Hickman et al., 2005), so the question remains on how c-di-GMP increase in the oprF mutant specifically affects Pel synthesis but not Psl. Recently, another study has shown that c-di-GMP produced by the diguanylate cyclase WspR specifically affects Pel but not Psl synthesis (Huangyutitham et al., 2013). This same study suggests that phenotypes that are very dominant in P. aeruginosa may not be much affected by the small c-di-GMP increase [this could be the case for Psl, which is the EPS that is mostly responsible for attachment and biofilm formation in P. aeruginosa strain PAO1 (Colvin et al., 2012)]. This study also suggests that the differential effects are due to differences in the affinity of the receptor proteins for c-di-GMP. PelD and FleQ are the two proteins that regulate Pel synthesis in response to c-di-GMP and their affinity to c-di-GMP is in the micromolar range (lower affinity; Hickman et al., 2005; Whitney et al., 2012), whereas other known c-di-GMP receptors have affinities in the nanomolar range (higher affinity; Christen et al., 2010). So it is conceivable that a change in the c-di-GMP level like the one observed in the oprF mutant could affect FleQ and/or PelD activity but not other high-affinity receptors which are already saturated for c-di-GMP binding. Finally, the levels of pelB mRNA were not fully reverted to wild-type levels upon artificial reduction of the c-di-GMP level in the oprF mutant, suggesting that factors other than c-di-GMP may contribute to pelB increased expression in the oprF mutant.
It is known that the pel and psl mRNAs are furthermore post-transcriptionally repressed by RsmA, the activity of which is controlled by the two sRNAs rsmZ and rsmY through complex regulatory systems involving several sensor kinases and accessory components (Mikkelsen et al., 2011). Interestingly, we show that rsmZ transcription is increased in response to the higher c-di-GMP level displayed by the oprF mutant. Expression of these sRNAs is positively regulated by the phosphorylated GacA response regulator from the TCS GacS/GacA. GacS is itself regulated by the orphan sensors, LadS and RetS, acting positively and negatively on GacA, respectively. PhoP, the regulator of the PhoPQ TCS that is activated in response to low magnesium concentration, also controls expression of rsmZ but not rsmY (Mulcahy and Lewenza, 2011). Noticeably, the PA1181 gene encoding a c-di-GMP cyclase is located nearby PA1179 and PA1180, encoding the PhoPQ TCS. However, the three genes were not predicted to be part of an operon (http://www.pseudomonas.com). Furthermore, HptB is a negative regulator of rsmY but not of rsmZ (Bordi et al., 2010). Similarly, the BfiSR two components system controls rsmZ but not rsmY via the RNase CafA (Petrova and Sauer, 2010). Remarkably, a mutation in lptA, encoding a lysophosphatidyl transferase, altered membrane fluidity (Baysse et al., 2005), led to increase expression of rsmZ, but not of rsmY (Baysse and O’Gara, 2007). Our study shows a c-di-GMP-dependent control of OprF, on rsmZ expression, but not on rsmY expression, providing a novel example of the dissociated control of the two sRNAs, rsmY and rsmZ.
How the c-di-GMP pool level is increased in the oprF mutant is not a trivial question. OprF is connecting the OM and the peptidoglycan layer and the absence of OprF provokes OM alterations (Rawling et al., 1998; Chevalier et al., 2000). It has been previously shown that H636 cells were shorter than the wild-type as judged by image analysis (Woodruff and Hancock, 1988) and by electron microscopy (Gotoh et al., 1989). Here we observe by SEM analyses that H636 cells were slightly thinner that H103 and H636O suggesting cell wall alterations. Cell morphology is, however, highly versatile and depends on growth conditions, as well as on the physiological state of the bacteria. However, despite suspected, it has never been clearly demonstrated that an oprF mutant encounters cell wall stress. Here, we show that the two ECFs sigma factors AlgU and SigX that are important for peptidoglycan and membrane homeostasis (Wood et al., 2006; Wood and Ohman, 2009; Boechat et al., 2013; Gicquel et al., 2013; Blanka et al., 2014), are active in the oprF mutant. Cell wall stress induced by D-cycloserin treatment (Wood and Ohman, 2009), or low shear modeled microgravity (Crabbé et al., 2010), is known to have similar effects on AlgU and SigX so one can hypothesize that deletion of oprF may result in a cell wall stress that then leads to production or activation of AlgU and SigX. Interestingly, two diguanylate cyclases, PA1181 and adcA were recently reported to belong to the SigX regulon of P. aeruginosa PA14 (Blanka et al., 2014). Accordingly, the two genes have lower expression levels in a sigX-deleted mutant of strain H103. While PA1181 and adcA expression is increased in the oprF mutant strain, the same was not true for PA2072, another c-di-GMP related protein belonging to the SigX regulon. Taken together, these data support the idea that P. aeruginosa oprF mutant displays higher levels of c-di-GMP as a consequence of the cell wall stress that the lack of OprF imposes to the cells. In addition to this, it is known that the two ECFs are involved in P. aeruginosa PAO1 biofilm formation (Bazire et al., 2010; Gicquel et al., 2013), so this study also highlights the possible role of cell envelope stress in biofilm formation. Our observation might in fact consolidate this concept, since an ompA mutant of Escherichia coli was shown to form sticky colonies by overproducing the exopolysaccharide cellulose through activation of the CpxRA TCS that responds to membrane stress (Ma et al., 2009). The bacteria might compensate the loss of the major OM protein OprF and the related membrane stress by producing an EPS that may stabilize the membrane, or protect itself from the environment.
This hypothesis is further strengthened by the fact that upon contact with surfaces, c-di-GMP is stimulated through the chemosensory-like surface-sensing system Wsp (Güvener and Harwood, 2007; O’Connor et al., 2012; Huangyutitham et al., 2013), though the exact nature of the signal remains obscure. Very recently, it was suggested that membrane alterations led to increase biofilm production through WspR activation resulting in c-di-GMP production (Blanka et al., 2015). In Vibrio cholerae, growth at low temperature modulates membrane fluidity and alters c-di-GMP signaling and biofilm formation (Townsley and Yildiz, 2015). Interestingly, Townsley and Yildiz (2015) show that temperature modulates c-di-GMP levels in a similar fashion in P. aeruginosa but not in the Gram-positive pathogen Listeria monocytogenes. Taken together, our study further supports the concept associated with these data and suggests that membrane alterations affect activity of enzymes involved in c-di-GMP metabolism, although the molecular mechanism remains to be elucidated.
The alternative or additional possibility that the effects caused by the absence of OprF result from something else other than OM disorganization cannot be disregarded, especially considering the role of OprF as a host immune system sensor (Wu et al., 2005; Mishra et al., 2015), more generally as an environmental sensor (Wagner et al., 2006; Fito-Boncompte et al., 2011). It is also conceivable that OprF-dependent c-di-GMP modulation is linked to its ability to transmit or to transduce unknown signals through the OM, which would then be detected by a sensor protein in the cytoplasmic membrane. Taken together, our data show for the first time that OprF is linked to c-di-GMP level modulations, through a direct (signaling) and/or indirect (envelope stress) mechanism.
Author Contributions
Conceived and designed experiments: EB, JM, PL, AD, AF, SC.
Performed the experiments: EB, JM, RD, OM, LF-B, GG, AB, GB-W.
Analyzed the data: EB, JM, RD, PL, AD, AF, SC, JO, GB-W.
Contributed reagents/materials/analysis tools: OM, MB.
Wrote the paper: EB, JM, PL, AD, NO, MF, AF, GB-W, JO, SC.
All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Bob Hancock and Manjeet Bains for the gift of P. aeruginosa H103 and H636, Matt Parsek for the reporter system PcdrA-gfp, Anke Neidig and Michael Nusser for technical assistance. The LMSM is supported by the Région Haute-Normandie (France, GRRs Sésa and IRIB), the Grand Evreux Agglomeration and European FEDER funds. The LBCM is supported by the Région Bretagne (France) and European FEDER funds.
References
- Allesen-Holm M., Barken K. B., Yang L., Klausen M., Webb J. S., Kjelleberg S., et al. (2006). A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59 1114–1128. 10.1111/j.1365-2958.2005.05008.x [DOI] [PubMed] [Google Scholar]
- Azghani A. O., Idell S., Bains M., Hancock R. E. W. (2002). Pseudomonas aeruginosa outer membrane protein F is an adhesin in bacterial binding to lung epithelial cells in culture. Microb. Pathog. 33 109–114. 10.1006/mpat.2002.0514 [DOI] [PubMed] [Google Scholar]
- Baysse C., Cullinane M., Denervaud V., Burrowes E., Dow J. M., Morrissey J. P., et al. (2005). Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology 151 2529–2542. 10.1099/mic.0.28185-0 [DOI] [PubMed] [Google Scholar]
- Baysse C., O’Gara F. (2007). “Role of membrane structure during stress signalling and adaptation in Pseudomonas,” in Pseudomonas: A Model System in Biology Vol. V eds Ramos J., Filloux A. (New York, NY: Springer-Verlag; ) 193–224. [Google Scholar]
- Bazire A., Shioya K., Soum-Soutera E., Bouffartigues E., Ryder C., Guentas-Dombrowsky L., et al. (2010). The sigma factor AlgU plays a key role in formation of robust biofilms by nonmucoid Pseudomonas aeruginosa. J. Bacteriol. 192 3001–3010. 10.1128/JB.01633-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher A., Schweizer H. P. (2000). Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29 948–950. [DOI] [PubMed] [Google Scholar]
- Blanka A., Düvel J., Dötsch A., Klinkert B., Abraham W. R., Kaever V., et al. (2015). Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Sci. Signal. 8 ra36 10.1126/scisignal.2005943 [DOI] [PubMed] [Google Scholar]
- Blanka A., Schulz S., Eckweiler D., Franke R., Bielecka A., Nicolai T., et al. (2014). Identification of the alternative sigma factor SigX regulon and its implications for Pseudomonas aeruginosa pathogenicity. J. Bacteriol. 196 345–356. 10.1128/JB.01034-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boechat A. L., Kaihami G. H., Politi M. J., Lepine F., Baldini R. L. (2013). A novel role for an ECF sigma factor in fatty acid biosynthesis and membrane fluidity in Pseudomonas aeruginosa. PLoS ONE 8:e84775 10.1371/journal.pone.0084775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi C., Lamy M.-C., Ventre I., Termine E., Hachani A., Fillet S., et al. (2010). Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 76 1427–1443. 10.1111/j.1365-2958.2010.07146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlee B. R., Goldman A. D., Murakami K., Samudrala R., Wozniak D. J., Parsek M. R. (2010). Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75 827–842. 10.1111/j.1365-2958.2009.06991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffartigues E., Duchesne R., Bazire A., Simon M., Maillot O., Dufour A., et al. (2014). Sucrose favors Pseudomonas aeruginosa pellicle production through the extracytoplasmic function sigma factor SigX. FEMS Microbiol. Lett. 356 193–200. 10.1111/1574-6968.12482 [DOI] [PubMed] [Google Scholar]
- Bouffartigues E., Gicquel G., Bazire A., Bains M., Maillot O., Vieillard J., et al. (2012). Transcription of the oprF gene of Pseudomonas aeruginosa is dependent mainly on the SigX sigma factor and is sucrose induced. J. Bacteriol. 194 4301–4311. 10.1128/JB.00509-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffartigues E., Gicquel G., Bazire A., Fito-Boncompte L., Taupin L., Maillot O., et al. (2011). The major outer membrane protein oprf is required for rhamnolipid production in Pseudomonas aeruginosa. J. Bacteriol. Parasitol. 2 118 10.4172/2155-9597.1000118 [DOI] [Google Scholar]
- Brencic A., Lory S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72 612–632. 10.1111/j.1365-2958.2009.06670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman F. S., Schoofs G., Hancock R. E., De Mot R. (1999). Influence of a putative ECF sigma factor on expression of the major outer membrane protein, OprF, in Pseudomonas aeruginosa and Pseudomonas fluorescens. J. Bacteriol. 181 4746–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd M. S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A. B., Richardson S. H., et al. (2009). Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73 622–638. 10.1111/j.1365-2958.2009.06795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier S., Burini J. F., Freulet-Marriere M. A., Regeard C., Schoofs G., Guespin-Michel J., et al. (2000). Characterization of an OprF-deficient mutant suggests that OprF is an essential protein for Pseudomonas fluorescens strain MF0. Res. Microbiol. 151 619–627. 10.1016/S0923-2508(00)90128-1 [DOI] [PubMed] [Google Scholar]
- Christen M., Kulasekara H. D., Christen B., Kulasekara B. R., Hoffman L. R., Miller S. I. (2010). Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328 1295–1297. 10.1126/science.1188658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin K. M., Irie Y., Tart C. S., Urbano R., Whitney J. C., Ryder C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14 1913–1928. 10.1111/j.1462-2920.2011.02657.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbella M. E., Puyet A. (2003). Real-time reverse transcription-PCR analysis of expression of halobenzoate and salicylate catabolism-associated operons in two strains of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69 2269–2275. 10.1128/AEM.69.4.2269-2275.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbé A., Pycke B., Van Houdt R., Monsieurs P., Nickerson C., Leys N., et al. (2010). Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 12 1545–1564. 10.1111/j.1462-2920.2010.02184.x [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Timmis K. N. (1994). Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994 386–405. 10.1016/0076-6879(94)35157-0 [DOI] [PubMed] [Google Scholar]
- Donlan R. M., Costerton J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15 167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesne R., Bouffartigues E., Oxaran V., Maillot O., Benard M., Feuilloley M. G. J., et al. (2013). A proteomic approach of SigX function in Pseudomonas aeruginosa outer membrane composition. J. Proteomics 94 451–459. 10.1016/j.jprot.2013.10.022 [DOI] [PubMed] [Google Scholar]
- Firoved A. M., Boucher J. C., Deretic V. (2002). Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184 1057–1064. 10.1128/jb.184.4.1057-1064.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fito-Boncompte L., Chapalain A., Bouffartigues E., Chaker H., Lesouhaitier O., Gicquel G., et al. (2011). Full virulence of Pseudomonas aeruginosa requires OprF. Infect. Immun. 79 1176–1186. 10.1128/IAI.00850-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L., Kolter R. (2004a). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51 675–690. 10.1046/j.1365-2958.2003.03877.x [DOI] [PubMed] [Google Scholar]
- Friedman L., Kolter R. (2004b). Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186 4457–4465. 10.1128/JB.186.14.4457-4465.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafoor A., Hay I. D., Rehm B. H. A. (2011). Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77 5238–5246. 10.1128/AEM.00637-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel G., Bouffartigues E., Bains M., Oxaran V., Rosay T., Lesouhaitier O., et al. (2013). The extra-cytoplasmic function sigma factor sigX modulates biofilm and virulence-related properties in Pseudomonas aeruginosa. PLoS ONE 8:e80407 10.1371/journal.pone.0080407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooderham W. J., Hancock R. E. W. (2009). Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33 279–294. 10.1111/j.1574-6976.2008.00135.x [DOI] [PubMed] [Google Scholar]
- Gotoh N., Wakebe H., Yoshihara E., Nakae T., Nishino T. (1989). Role of protein F in maintaining structural integrity of the Pseudomonas aeruginosa outer membrane. J. Bacteriol. 171 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güvener Z. T., Harwood C. S. (2007). Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66 1459–1473. 10.1111/j.1365-2958.2007.06008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Carey A. M. (1979). Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140 902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett D. J., Cuppoletti J., Trapnell B., Lymar S. V., Rowe J. J., Yoon S. S., et al. (2002). Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 54 1425–1443. 10.1016/S0169-409X(02)00152-7 [DOI] [PubMed] [Google Scholar]
- Heydorn A., Nielsen A. T., Hentzer M., Sternberg C., Givskov M., Ersbøll B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiol. Read. Engl. 146 2395–2407. [DOI] [PubMed] [Google Scholar]
- Hickman J. W., Tifrea D. F., Harwood C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102 14422–14427. 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Kutchma A. J., Becher A., Schweizer H. P. (2000). Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43 59–72. 10.1006/plas.1999.1441 [DOI] [PubMed] [Google Scholar]
- Huangyutitham V., Güvener Z. T., Harwood C. S. (2013). Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. MBio 4 e00213–e00242. 10.1128/mBio.00242-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y., Starkey M., Edwards A. N., Wozniak D. J., Romeo T., Parsek M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78 158–172. 10.1111/j.1365-2958.2010.07320.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Newsom D., Kelly B., Irie Y., Jennings L. K., Xu B., et al. (2014). ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10:e1003984 10.1371/journal.ppat.1003984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V. T., Matewish J. M., Kessler J. L., Hyodo M., Hayakawa Y., Lory S. (2007). A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65 1474–1484. 10.1111/j.1365-2958.2007.05879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma L., Conover M., Lu H., Parsek M. R., Bayles K., Wozniak D. J. (2009). Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354 10.1371/journal.ppat.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Jackson K. D., Landry R. M., Parsek M. R., Wozniak D. J. (2006). Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188 8213–8221. 10.1128/JB.01202-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H., Sivaneson M., Filloux A. (2011). Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13 1666–1681. 10.1111/j.1462-2920.2011.02495.x [DOI] [PubMed] [Google Scholar]
- Mishra M., Ressler A., Schlesinger L. S., Wozniak D. J. (2015). Identification of OprF as a complement component C3 binding acceptor molecule on the surface of Pseudomonas aeruginosa. Infect. Immun. 10.1128/IAI.00081-15 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso J. A., Mikkelsen H., Heeb S., Williams P., Filloux A. (2011). The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13 3128–3138. 10.1111/j.1462-2920.2011.02595.x [DOI] [PubMed] [Google Scholar]
- Mulcahy H., Lewenza S. (2011). Magnesium limitation is an environmental trigger of the Pseudomonas aeruginosa biofilm lifestyle. PLoS ONE 6:e23307 10.1371/journal.pone.0023307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorovich E. M., Sugawara E., Nikaido H., Bezrukov S. M. (2006). Pseudomonas aeruginosa porin OprF: properties of the channel. J. Biol. Chem. 281 16230–16237. 10.1074/jbc.M600650200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. R., Fuqua C. (1999). Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227 197–203. 10.1016/S0378-1119(98)00601-5 [DOI] [PubMed] [Google Scholar]
- O’Connor J. R., Kuwada N. J., Huangyutitham V., Wiggins P. A., Harwood C. S. (2012). Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol. Microbiol. 86 720–729. 10.1111/mmi.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp S. J., Sternberg C., Tolker-Nielsen T. (2009). Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry A 75 90–103. 10.1002/cyto.a.20685 [DOI] [PubMed] [Google Scholar]
- Petrova O. E., Sauer K. (2010). The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 192 5275–5288. 10.1128/JB.00387-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey D. M., Wozniak D. J. (2005). Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56 309–322. 10.1111/j.1365-2958.2005.04552.x [DOI] [PubMed] [Google Scholar]
- Rawling E. G., Brinkman F. S., Hancock R. E. (1998). Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J. Bacteriol. 180 3556–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtke M. T., Borlee B. R., Murakami K., Irie Y., Hentzer M., Nielsen T. E., et al. (2012). Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78 5060–5069. 10.1128/AEM.00414-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder C., Byrd M., Wozniak D. J. (2007). Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10 644–648. 10.1016/j.mib.2007.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr M. J., Martin D. W., Mudd M. H., Hibler N. S., Boucher J. C., Deretic V. (1993). The algD promoter: regulation of alginate production by Pseudomonas aeruginosa in cystic fibrosis. Cell. Mol. Biol. Res. 39 371–376. [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., et al. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150 76–85. 10.1016/0003-2697(85)90442-7 [DOI] [PubMed] [Google Scholar]
- Spangler C., Böhm A., Jenal U., Seifert R., Kaever V. (2010). A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81 226–231. 10.1016/j.mimet.2010.03.020 [DOI] [PubMed] [Google Scholar]
- Spiers A. J., Buckling A., Rainey P. B. (2000). The causes of Pseudomonas diversity. Microbiol. Read. Engl. 146 2345–2350. [DOI] [PubMed] [Google Scholar]
- Starkey M., Hickman J. H., Ma L., Zhang N., De Long S., Hinz A., et al. (2009). Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191 3492–3503. 10.1128/JB.00119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehmel J., Neidig A., Nusser M., Geffers R., Brenner-Weiss G., Overhage J. (2015). The sensor kinase PA4398 modulates swarming motility and biofilm formation in Pseudomonas aeruginosa PA14. Appl. Environ. Microbiol. 81 1274–1285. 10.1128/AEM.02832-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamber S., Hancock R. E. W. (2004). “The outer membranes of Pseudomonas,” in Pseudomonas Vol. 1 Genomics, Life Style and Molecular Architecture ed. Ramos J.-L. (New York, NY: Springer US; ) 575–601. Available at: http://link.springer.com/chapter/10.1007/978-1-4419-9086-0_19 [Google Scholar]
- Tolker-Nielsen T., Sternberg C. (2011). Growing and analyzing biofilms in flow chambers. Curr. Protoc. Microbiol. Chapter 1 Unit 1B.2 10.1002/9780471729259.mc01b02s21 [DOI] [PubMed] [Google Scholar]
- Townsley L., Yildiz F. H. (2015). Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ. Microbiol. 10.1111/1462-920.12799 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V. E., Frelinger J. G., Barth R. K., Iglewski B. H. (2006). Quorum sensing: dynamic response of Pseudomonas aeruginosa to external signals. Trends Microbiol. 14 55–58. 10.1016/j.tim.2005.12.002 [DOI] [PubMed] [Google Scholar]
- Whitney J. C., Colvin K. M., Marmont L. S., Robinson H., Parsek M. R., Howell P. L. (2012). Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 287 23582–23593. 10.1074/jbc.M112.375378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L. F., Leech A. J., Ohman D. E. (2006). Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 62 412–426. 10.1111/j.1365-2958.2006.05390.x [DOI] [PubMed] [Google Scholar]
- Wood L. F., Ohman D. E. (2009). Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72 183–201. 10.1111/j.1365-2958.2009.06635.x [DOI] [PubMed] [Google Scholar]
- Woodruff W. A., Hancock R. E. (1988). Construction and characterization of Pseudomonas aeruginosa protein F-deficient mutants after in vitro and in vivo insertion mutagenesis of the cloned gene. J. Bacteriol. 170 2592–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff W. A., Hancock R. E. (1989). Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J. Bacteriol. 171 3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Estrada O., Zaborina O., Bains M., Shen L., Kohler J. E., et al. (2005). Recognition of host immune activation by Pseudomonas aeruginosa. Science 309 774–777. 10.1126/science.1112422 [DOI] [PubMed] [Google Scholar]
- Yang L., Hu Y., Liu Y., Zhang J., Ulstrup J., Molin S. (2011). Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 13 1705–1717. 10.1111/j.1462-2920.2011.02503.x [DOI] [PubMed] [Google Scholar]
- Yoon S. S., Hennigan R. F., Hilliard G. M., Ochsner U. A., Parvatiyar K., Kamani M. C., et al. (2002). Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3 593–603. 10.1016/S1534-5807(02)00295-2 [DOI] [PubMed] [Google Scholar]