Abstract
Purpose.
To examine the relationship between proportion of African ancestry (PAA) and proliferative diabetic retinopathy (PDR) and to identify genetic loci associated with PDR using admixture mapping in African Americans with type 2 diabetes (T2D).
Methods.
Between 1993 and 2013, 1440 participants enrolled in four different studies had fundus photographs graded using the Early Treatment Diabetic Retinopathy Study scale. Cases (n = 305) had PDR while controls (n = 1135) had nonproliferative diabetic retinopathy (DR) or no DR. Covariates included diabetes duration, hemoglobin A1C, systolic blood pressure, income, and education. Genotyping was performed on the Affymetrix platform. The association between PAA and PDR was evaluated using logistic regression. Genome-wide admixture scanning was performed using ANCESTRYMAP software.
Results.
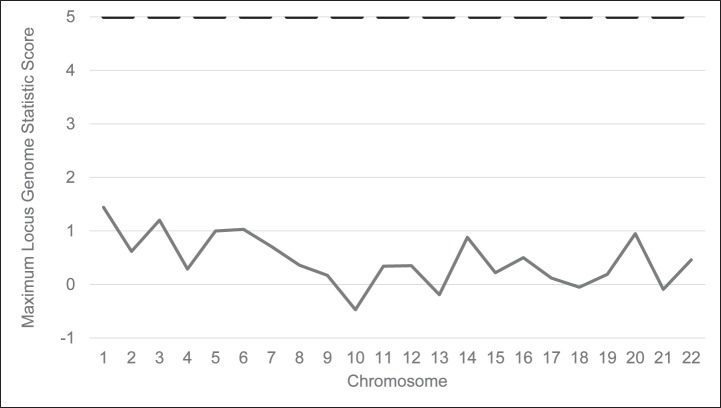
In the univariate analysis, PDR was associated with increased PAA (odds ratio [OR] = 1.36, 95% confidence interval [CI] = 1.16–1.59, P = 0.0002). In multivariate regression adjusting for traditional DR risk factors, income and education, the association between PAA and PDR was attenuated and no longer significant (OR = 1.21, 95% CI = 0.59–2.47, P = 0.61). For the admixture analyses, the maximum genome-wide score was 1.44 on chromosome 1.
Conclusions.
In this largest study of PDR in African Americans with T2D to date, an association between PAA and PDR is not present after adjustment for clinical, demographic, and socioeconomic factors. No genome-wide significant locus (defined as having a locus-genome statistic > 5) was identified with admixture analysis. Further analyses with even larger sample sizes are needed to definitively assess if any admixture signal for DR is present.
Keywords: genetics, diabetic retinopathy, proliferative diabetic retinopathy, African Americans, admixture, ancestry
In African Americans with type 2 diabetes, proportion of African ancestry was associated with proliferative diabetic retinopathy (PDR) but the association was no longer present with adjustment for clinical/socioeconomic variables. No genome-wide significant loci were found with admixture scanning.
Diabetic retinopathy (DR) is a major complication of type 2 diabetes (T2D) and the leading cause of new cases of blindness among adults aged 20 to 74 years in the United States.1 The severity of this disease ranges from mild nonproliferative DR to potentially-blinding proliferative diabetic retinopathy (PDR). However, the development and severity of DR are highly heterogeneous from patient to patient. Duration of diabetes, glycated hemoglobin (HbA1C) levels, and elevated blood pressure (BP) are the most consistently established risk factors for DR progression.2–4 However, these known risk factors explain some, but not all, of the observed heterogeneity. For example, less than 25% of patients with diabetes develop PDR, a blinding advanced form of the disease, despite long-term hyperglycemia.5
Genetic variation may explain some of the remaining heterogeneity in DR development. Heritability estimates for PDR have been found as high as 52%.6 Genetic association studies, including candidate gene studies and genome-wide association studies, are a potentially powerful way to identify genetic variants underlying DR, but most reported associations have not been consistently reproduced.7–14
Admixture mapping is an alternative genetic study design that has proven to be a successful for complex traits in populations of mixed ancestry.15–18 Admixed individuals are those who inherit chromosomal segments of distinct continental ancestry. African Americans are one example of an admixed population; they have an average of 20% European ancestry and 80% African ancestry.19 Admixture mapping is useful when the disease of interest has a significant difference in prevalence in the ancestral populations. Several studies have found that African Americans have a higher risk of developing DR compared with Caucasians,20 even while controlling for the known DR risk factors.21–25 The prevalence of moderate nonproliferative DR was higher for African Americans veterans than for Caucasian veterans in the Veterans Affairs Diabetes Trial of T2D after controlling for traditional risk factors.23 Given these differences in DR prevalence, determining if genetic ancestry is associated with PDR in African Americans may help to explain these disparities.
The concept of whole-genome admixture mapping for PDR in African Americans is to scan the genome in a large number of diabetic individuals looking for regions where the proportion of African ancestry (PAA) of those with PDR is strikingly higher or lower than that seen in those without PDR. This is accomplished by genotyping ancestry informative markers (AIMs), polymorphisms that differ significantly between the two populations. Finding such a region would indicate the presence of at least one genetic risk variant for PDR whose frequency differed between the ancestral populations. The purposes of this study were to examine the relationship between proportion of African ancestry and PDR and to identify genetic loci associated with PDR in African Americans with T2D using whole-genome admixture mapping.
Methods
Participants
This investigation followed the tenets of the Declaration of Helsinki. The institutional review boards of the University of Mississippi Medical Center (UMMC), Massachusetts Eye and Ear Infirmary (MEEI), and Boston Medical Center (BMC) approved this study. All participants gave written, informed consent after explanation of the nature and possible consequences of the study. Samples came from two sources: the African American Proliferative Diabetic Retinopathy (AAPDR) Study and the Candidate-gene Association Resource (CARe). The AAPDR Study was designed specifically for the purposes of the genome-wide admixture scan. The CARe consortium was created to facilitate a wide variety of genetic association studies. The two samples are described in further detail below.
African American Proliferative Diabetic Retinopathy (AAPDR) Study.
Participants who self-identified as African Americans and had T2D were recruited from four clinical sites: UMMC, MEEI, BMC, and Harvard Vanguard Medical Associates. The inclusion criterion was a diagnosis of T2D by the 2003 American Diabetes Association (ADA) criteria.26 Dilated, digital seven-standard field fundus photography of both eyes were obtained using a Topcon TRC 50 DX camera (Topcon, Tokyo, Japan). The photographs were scored for degree of DR by two independent, masked readers using the Early Treatment Diabetic Retinopathy Study (ETDRS) adaptation of the modified Airlie House classification system.27
Covariate data were collected by using uniform methods at the study visit or were retrieved from the medical record as previously described.28 Duration of diabetes was recorded using a standardized questionnaire and verified by review of the medical record. With an appropriately sized arm cuff, sitting BP was measured three times. Mean systolic and diastolic BP was calculated. Standing height, weight, and waist circumference were also recorded. For each participant, two 10-mL tubes of EDTA anticoagulated blood were obtained. One tube was sent immediately for HbA1c measurement at a centralized laboratory. The other tube was transferred into labeled 10-mL plastic tubes, frozen at −80°C, and shipped to the Broad Institute for genotyping.
The Candidate-Gene Association Resource (CARe).
CARe is a collaboration of large epidemiologic studies of heart disease for association analyses of genotypes and cardiovascular disease phenotypes.29 Three CARe cohorts have fundus photography on self-identified African American T2D participants and genome-wide genotyping done through CARe: Atherosclerosis Risk in Communities (ARIC) Study, Jackson Heart Study (JHS), and Multi-Ethnic Study of Atherosclerosis (MESA).30–33 These studies have collected extensive covariate measures using standardized methods.
For the purposes of this investigation, all African Americans participants with T2D who had been genotyped as part of CARe and had graded fundus photographs were eligible for inclusion. Type 2 diabetes was defined according to the 2003 ADA Criteria.26 The fundus photography protocol for each cohort has been previously described.33–36 In all studies, except for the JHS, fundus photographs were graded by masked readers at the University of Wisconsin Ocular Epidemiology Reading Center according to the modified Airlie House Classification system.37 Fundus photographs for the JHS were graded by masked JHS ophthalmologist-investigators according to the same criteria.33
Case-Control Definitions
We defined cases as participants with ETDRS grade greater than or equal to 60, which denotes PDR, in the eye with the higher ETDRS grade or in the only eye photographed, depending on the cohort's study protocol. Controls were T2D participants with an ETDRS grade less than 60, which denotes no DR or any degree of nonproliferative DR
Admixture Mapping
Genotyping.
Both the AAPDR Study and CARe participants separately underwent genome-wide genotyping on the Affymetrix 6.0 platform (Santa Clara, CA, USA) at the Broad Institute. These genotyping data sets were each culled to keep only the AIMs specific to the Affymetrix panel.38 After genotyping, we examined the samples to ensure that (1) all samples had greater than or equal to 95% call rate, and (2) there were no duplicate samples defined as a greater than 75% match in the genotypes between the two samples. All samples met these criteria. The AIMs were used to estimate European/African ancestry proportion in each individual. A list of the AIMs has been previously published.38
SNP Quality Filters.
To decrease the likelihood of false-positives in our scan, we applied a series of filters that had the goal of detecting and removing any SNPs with problematic genotyping. First, SNPs were dropped if there were atypical clustering patterns, ill-defined clusters, or low genotyping success rate (<95%). We eliminated SNPs if they did not meet the requirement for Hardy-Weinberg equilibrium (P > 0.01) in both ancestral West African and European populations. We applied a filter that examined whether the observed frequency of a SNP in African Americans was statistically consistent with being a mixture of the frequencies observed in West Africans and European American samples that we used to represent the ancestral populations. Finally, we applied a filter that for each sample iteratively eliminated SNPs that were less informative (in terms of information content about ancestry) until none were within 200 Kbs of each other or in detectable linkage disequilibrium (LD) with each other in the ancestral West African or European populations.
Estimating Genome-Wide Ancestry.
Using the ANCESTRYMAP software (in the public domain, genepath.med.harvard.edu/∼reich), we estimated the PAA. ANCESTRYMAP uses a Markov Chain Monte Carlo approach to account for uncertainty in the unknown parameters (including SNP allele frequencies in the West African and European ancestral populations, the numbers of generations since mixture, and the average proportion of ancestry inherited from ancestral populations) that emerge from the Hidden Markov Model analysis. For the subsequent analyses, we defined African Americans as those having at least 50% African ancestry. This cut-off of 50% African ancestry was chosen to be consistent with our other ongoing genetic study of DR, a genome-wide association study for DR in African Americans and Caucasians where it is important to distinguish correct ancestry as populations' genetic data are imputed and meta-analyzed separately. Only six samples had less than 50% African ancestry and were eliminated from the downstream analyses. The distribution of cases and controls that were included in the analyses are shown in Table 1.
Table 1.
Distribution of Cases and Controls by Study

Statistical Analysis
Association Between Percentage of African Ancestry and PDR.
We tested the association between PDR and the following variables: sex, age, hemoglobin A1C, duration of diabetes, systolic blood pressure, body mass index (BMI), education level, yearly family income, and percentage of African ancestry. Variables were chosen because they have either been consistently associated with PDR39–41 or because they have been found to be associated with genetic ancestry in prior studies.42 Association testing was conducted using logistic regression in univariate and multivariate analyses. The multivariate model was constructed using the covariates that were significantly associated with PDR in the univariate analyses as well as income and education. This was done because socioeconomic variables have previously been correlated with genetic ancestry in African American and Latino populations.42,43 The multivariate model was adjusted for age and sex. A P value less than 0.05 was considered statistically significant. All analyses were conducted using Stata/IC12.1 (Stata, College Station, TX, USA).
Admixture Mapping.
We used ANCESTRYMAP to perform admixture mapping analyses.19 Because ANCESTRYMAP uses Bayesian statistics, a prior distribution of risk models is required. To assess whether a locus showed an unusual association of ancestry with PDR, we calculated the likelihood of the data at each locus assuming a particular disease model, divided by the likelihood with no disease locus. The admixture scan was run using a range of increased risk due to having either one or two population ancestry alleles. The risk models were parameterized as follows: 0.5, 0.6, 0.7, 0.8, 0.9, 1.2, 1.3, 1.45, 1.55, 1.7, and 2.0. By convention, a risk less than 1.0 for inheritance of either one or two copies of a European ancestral allele at a given locus represents a risk model where European ancestry decreases risk relative to African ancestry. This set of models reflects the hypothesis that European ancestral alleles are less likely to confer risk but also tests for the alternative possibilities. To accumulate evidence of association in these models, we averaged the Bayes factors emerging from each model at each point in the genome, taking the log10 of this number to produce a locus-genome statistic (LGS) score. As previously described, we declared genome-wide significance only when the LGS score was greater than 5.19 Based on simulation studies, this threshold for genome-wide significance is conservative (exceeded in < 1 in 100 repetitions of simulated data sets where there is no disease locus).19 A peak with a LGS score greater than 4 was our criterion for genome-wide suggestiveness, and further exploration. The powers of our admixture mapping analysis to detect a disease locus at which a population African ancestry allele confers 1.3-, 1.5-, 1.7- and 2-fold multiplicative increased risk are 65%, 94%, 96%, and 100%, respectively.19
Results
Clinical Characteristics and Univariate Analyses
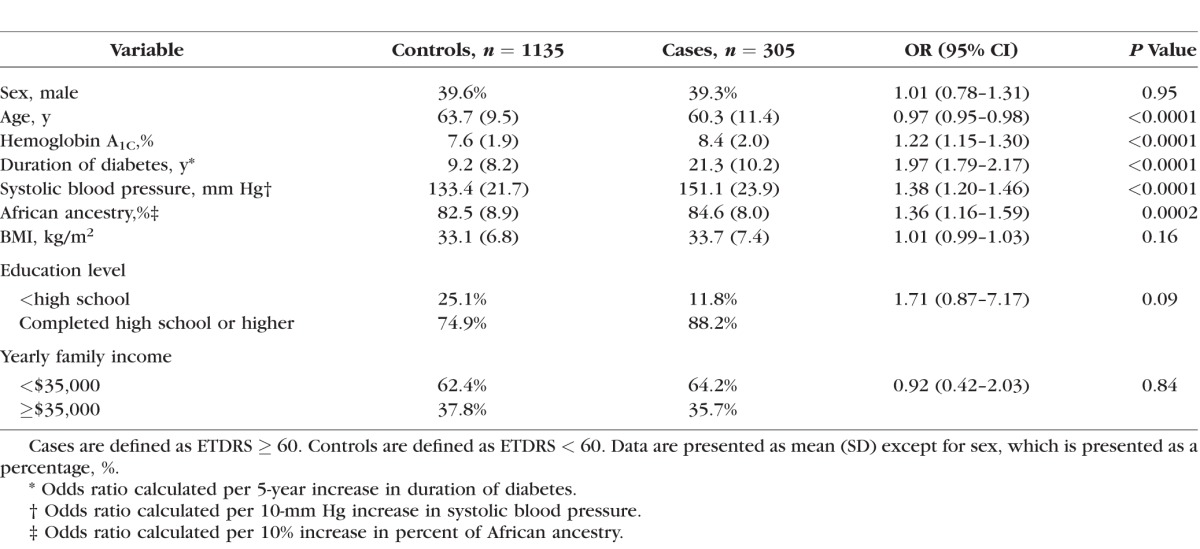
Table 2 shows the clinical characteristics of the study sample and the univariate associations of various candidate risk factors. In the univariate analysis, age (P < 0.0001), HbA1C (P < 0.0001), duration of diabetes (P < 0.0001), systolic BP (P < 0.0001), and PAA (P = 0.0002) were significantly associated with PDR. Compared with controls, cases were younger (60.3 vs. 63.7 years), had a higher HbA1C (8.4% vs. 7.6%), longer duration of diabetes (21.3 years vs. 9.2 years), higher systolic BP (151.1 vs. 133.4 mm Hg), and higher PAA (84.6% vs. 82.5%). Sex, BMI, education level, and income did not have significant associations with PDR. In Supplementary Figure S1, we plotted the first two principal components for cases and controls and for the different cohorts alongside HapMap African and European samples with Eigensoft's smartpca program (in the public domain, http://genetics.med.harvard.edu/reich/Reich_Lab/Software.html). There was no evidence for population stratification between cases and controls or among different cohorts.
Table 2.
Characteristics of the Study Sample and Univariate Association Results
Multivariate Analyses
Table 3 shows the multivariate logistic regression results. The association between PDR and PAA was significantly attenuated and nonsignificant in this multivariate analysis (odds ratio [OR] = 1.21, P = 0.61) when compared with the univariate analysis (OR = 1.36, P = 0.0002). We examined the income and education and their association with PAA separately using Pearson's correlation. Higher PAA was associated with lower education level (correlation coefficient = −0.12, P < 0.0001) and income (correlation coefficient = −0.07, P = 0.03). We note that inclusion of the six individuals with less than 50% African ancestry who were excluded in the analyses above did not change these results materially. When we included these six individuals, the association between PDR and higher PAA was significant in the univariate analysis (P = 0.01) but nonsignificant in the multivariate analysis (P = 0.89).
Table 3.
Multivariate Association Results

Admixture Mapping
The initial SNP quality control steps resulted in ten markers being removed for inappropriate allele frequency when compared with the parental frequencies, and one marker failed Hardy-Weinberg equilibrium. After quality control, 2072 markers remained. The results of the admixture scan are shown in the Figure. The maximum genome-wide score for the LGS was 1.44 for chromosome 1. There were no genome-wide significant or suggestive loci. We also separately analyzed the participants from the AAPDR Study, which was recruited specifically for the purpose of this admixture scan (Supplementary Fig. S2). In the AAPDR Study, controls were required to have at least 10 years of T2D to be enrolled. This enrollment criterion was designed to decrease the misclassification that might occur when controls of short diabetes duration are included (e.g., controls could develop more severe retinopathy with time and become cases). No genome-wide significant loci were identified.
Figure.
Genome-wide admixture mapping scan for loci associated with proliferative diabetic retinopathy (1135 controls, 305 cases). The maximum locus genome statistic score was 1.44 on chromosome 1. The dashed line indicates the level of genome wide significance (locus genome statistic score of 5).
Discussion
To our best knowledge, this is the first study to investigate the contribution of African ancestry to PDR in African Americans. In the univariate analyses, a higher percentage of African ancestry was associated with a higher risk of PDR. However, once we adjusted for the well-established clinical risk factors for PDR as well as education and income, the association between PAA and PDR was no longer significant. This suggests that the association between PAA and PDR may be confounded by clinical and socioeconomic variables. Socioeconomic status has been shown to confound the association between T2D itself and genetic ancestry in Latinos.42
The correlation between education and income variables and African ancestry make it difficult to distinguish the relationship between genetics and social factors in the contribution to DR risk. In a predominantly Mexican American population, Native American ancestry has been associated with severe DR even after adjusting for traditional DR risk factors and socioeconomic variables.44 In that study, there was a wide range of Native American ancestry (from a few percent to almost 100%) with the mean percent of Native American ancestry between 45% and 50%. Self-identified African Americans have a narrower range of African American ancestry with the vast majority having at least 50% African ancestry and the mean of the overall population being about 80%. This limits the separation of ancestry proportion between cases and controls in African Americans. This could make the effects of ancestry more subtle and difficult to detect in this population although this has not been the case for analogous variables examined in simulations of gene-by-environment interaction studies.45 There is also some lack of precision inherent in estimations of socioeconomic status, which rely on information provided by participants who may have secondary motives for reporting inaccurate information. This imprecision further complicates the dissection of socioeconomic and genetic variables in risk of DR Finally, differences in PAA can reflect genetic differences but they could also reflect social differences, including discrimination across ranges of ancestry. Thus, we cannot be sure that education and income are truly confounders of the effect of PAA on PDR rather than mediators of the effect. If they are mediators, we may be underestimating the impact of PAA on PDR by overadjusting the model.
To our best knowledge, we assembled the largest collection of African Americans with T2D and fundus photography grading for DR and used admixture mapping to search for genomic regions that may be associated with PDR in African Americans. Despite this effort, we did not find any evidence for a genetic association to PDR. There are several possible reasons for this. First, the sample size, while the largest to date, may still not be sufficient to detect an admixture mapping signal. Previous studies for other diseases that have used admixture mapping successfully have had sample sizes in the 1500 to 2000 range.16,18,46 Our sample size is less than 1500 participants. Admixture mapping is most likely to succeed when the trait of interest found in the admixed population has a significant difference in prevalence in the ancestral populations. While there is evidence that DR prevalence differs between African Americans and Caucasian Americans,22–25 the difference is likely moderate in size, making a large sample size even more crucial to the success of admixture mapping. The observation from our African ancestry analysis that increased DR prevalence in African Americans may be at least partially explained by environmental variables decreases the likelihood that significant genetic risk factors can be easily found through admixture mapping, but it does not rule out the possibility that the method can work.
There are issues related to DR phenotyping that could have also contributed to a null scan. While the AAPDR Study and JHS in CARe both used gold-standard, seven-field fundus photography through pharmacologically dilated pupils to assess DR in each eye, ARIC took nonstereoscopic 40° film images in one eye centered between the fovea and optic disc in a darkened room though a nonpharmacologically dilated pupil while MESA took two nonpharmacologically dilated digital images centered on the fovea and on the disc of each eye. While limited field photography has been validated in the study of DR,47 there is still potential for misclassification bias, particularly among African Americans whose darker fundus pigmentation and more frequent presence of lens opacities makes it harder to discern retinal lesions especially when limited fields are available.21,48 This could bias the study toward the null. We chose PDR as the case definition because it is more heritable than overall DR, and therefore there is a higher likelihood of detecting modest genetic effects for PDR.6 To maximize the control sample size, we included all other participants as controls. This strategy was previously successful in identifying an association between Native American ancestry and DR.44 However, the optimal case and control definitions for genetic studies of DR are still not clear with even the largest studies in the field having used very different case-control definitions.10–12,14,49 In the present study, controls had a shorter mean duration of diabetes (9.2 years) compared with cases (21.3 years). When using ETDRS grade, there is always the possibility of case-control misclassification as controls could develop more severe retinopathy with time and become cases. Other DR phenotypes such as diabetic macular edema may be better endpoints for genetic studies than grade of retinopathy. There are no heritability estimates available for macular edema but it has been shown to be more prevalent in African Americans than in other US populations.50
In conclusion, in this large study among African-Americans with diabetes, there is no association between PAA and PDR after adjustment for clinical, demographic, and socioeconomic factors. Future efforts will be focused on different strategies to increase the possibility of obtaining a more informative admixture scan. These could include examining different case-control definitions and recruiting additional participants to increase the power to detect a signal and to allow for tests for interactions between genes and environment in DR risk.
Supplementary Material
Acknowledgments
Supported by grants from the American Diabetes Association Clinical Translational Research Award 1-11-CT-51 (Alexandria, VA, USA), National Eye Institute Grant K12-EY16335 (Bethesda, MD, USA), Research to Prevent Blindness Career Development Award (New York City, NY, USA), Harvard Catalyst | The Harvard Clinical and Translations Science Center Faculty Fellowship (Boston, MA, USA), Massachusetts Lions Eye Research Fund (Abington, MA, USA), Eleanor and Miles Shore Fellowship (Boston, MA, USA), Sara Elizabeth O'Brien Trust (Boston, MA, USA). The National Heart, Lung Blood Institute (NHLBI) Grant N01-HC-65226 (Bethesda, MD, USA) has supported genotyping and has created a genotype-phenotype database with data and samples from nine cohorts as part of Candidate Gene Association Resource (CARe). Please see the Supplementary Material for detailed information about the grants supporting three cohorts that were part of this study: Atherosclerosis Risk in Communities (ARIC) Study, Jackson Heart Study (JHS) and Multi-Ethnic Study of Atherosclerosis (MESA).
Disclosure: A. Tandon, None; C.J. Chen, None; A. Penman, None; H. Hancock, None; M. James, None; D. Husain, None; C. Andreoli, None; X. Li, None; J.Z. Kuo, None; O. Idowu, None; D. Riche, None; E. Papavasilieou, None; S. Brauner, None; S.O. Smith, None; S. Hoadley, None; C. Richardson, None; T. Kieser, None; V. Vazquez, None; C. Chi, None; M. Fernandez, None; M. Harden, None; M.F. Cotch, None; D. Siscovick, None; H.A. Taylor, None; J.G. Wilson, None; D. Reich, None; T.Y. Wong, None; R. Klein, None; B.E.K. Klein, None; J.I. Rotter, None; N. Patterson, None; L. Sobrin, None
References
- 1. National Institute of Diabetes and Digestive and Kidney Diseases: National Diabetes Statistics. US Department of Health and Human Services, National Institute of Health, Publication No 11-3892; Bethesda: 2011. [Google Scholar]
- 2. Klein R,, Klein BE,, Moss SE,, Cruickshanks KJ. The Wisconsin Epidemiologic Study of Diabetic Retinopathy: XVII. The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology. 1998; 105: 1801–1815. [DOI] [PubMed] [Google Scholar]
- 3. Klein R,, Klein BE,, Moss SE,, Cruickshanks KJ. Relationship of hyperglycemia to the long-term incidence and progression of diabetic retinopathy. Arch Intern Med. 1994; 154: 2169–2178. [PubMed] [Google Scholar]
- 4. Klein R,, Klein BE,, Moss SE,, Linton KL. The Beaver Dam Eye Study. Retinopathy in adults with newly discovered and previously diagnosed diabetes mellitus. Ophthalmology. 1992; 99: 58–62. [DOI] [PubMed] [Google Scholar]
- 5. Liu Y,, Wang M,, Morris AD,, et al. Glycemic exposure and blood pressure influencing progression and remission of diabetic retinopathy: a longitudinal cohort study in GoDARTS. Diabetes Care. 2013; 36: 3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hietala K,, Forsblom C,, Summanen P,, Groop PH. Heritability of proliferative diabetic retinopathy. Diabetes. 2008; 57: 2176–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lohmueller KE,, Pearce CL,, Pike M,, Lander ES,, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003; 33: 177–182. [DOI] [PubMed] [Google Scholar]
- 8. Abhary S,, Hewitt AW,, Burdon KP,, Craig JE. A systematic meta-analysis of genetic association studies for diabetic retinopathy. Diabetes. 2009; 58: 2137–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirschhorn JN,, Lohmueller K,, Byrne E,, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002; 4: 45–61. [DOI] [PubMed] [Google Scholar]
- 10. Grassi MA,, Tikhomirov A,, Ramalingam S,, Below JE,, Cox NJ,, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011; 20: 2472–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu YP,, Hallman DM,, Gonzalez VH,, et al. identification of diabetic retinopathy genes through a Genome-Wide Association Study among Mexican-Americans from Starr County, Texas. J Ophthalmol. 2010; 2010. [DOI] [PMC free article] [PubMed]
- 12. Sheu WH,, Kuo JZ,, Lee IT,, et al. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum Mol Genet. 2013; 22: 3165–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang YC,, Lin JM,, Lin HJ,, et al. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011; 118: 642–648. [DOI] [PubMed] [Google Scholar]
- 14. Hosseini SM,, Boright AP,, Sun L,, et al. The association of previously reported polymorphisms for microvascular complications in a meta-analysis of diabetic retinopathy. Hum Genet. 2015; 134: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reich D,, Nalls MA,, Kao WH,, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009; 5: e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reich D,, Patterson N,, De Jager PL,, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005; 37: 1113–1118. [DOI] [PubMed] [Google Scholar]
- 17. Reich D,, Patterson N,, Ramesh V,, et al. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am J Hum Genet. 2007; 80: 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freedman ML,, Haiman CA,, Patterson N,, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006; 103: 14068–14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patterson N,, Hattangadi N,, Lane B,, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004; 74: 979–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yau JW,, Rogers SL,, Kawasaki R,, et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. 2012; 35: 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klein R,, Marino EK,, Kuller LH,, et al. The relation of atherosclerotic cardiovascular disease to retinopathy in people with diabetes in the Cardiovascular Health Study. Br J Ophthalmol. 2002; 86: 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leske MC,, Wu SY,, Hennis A,, et al. Nine-year incidence of diabetic retinopathy in the Barbados Eye Studies. Arch Ophthalmol. 2006; 124: 250–255. [DOI] [PubMed] [Google Scholar]
- 23. Emanuele N,, Sacks J,, Klein R,, et al. Ethnicity, race, and baseline retinopathy correlates in the veterans affairs diabetes trial. Diabetes Care. 2005; 28: 1954–1958. [DOI] [PubMed] [Google Scholar]
- 24. Harris EL,, Sherman SH,, Georgopoulos A. Black-white differences in risk of developing retinopathy among individuals with type 2 diabetes. Diabetes Care. 1999; 22: 779–783. [DOI] [PubMed] [Google Scholar]
- 25. Emanuele N,, Moritz T,, Klein R,, et al. Ethnicity, race, and clinically significant macular edema in the Veterans Affairs Diabetes Trial (VADT). Diabetes Res Clin Pract. 2009; 86: 104–110. [DOI] [PubMed] [Google Scholar]
- 26. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 2003; 26 (Suppl 1): S5–S20. [DOI] [PubMed] [Google Scholar]
- 27. Early Treatment Diabetic Retinopathy Study Research Group. Grading diabetic retinopathy from stereoscopic color fundus photographs–an extension of the modified Airlie House classification. ETDRS report number 10. Ophthalmology. 1991; 98: 786–806. [PubMed] [Google Scholar]
- 28. Penman A,, Hancock H,, Papavasileiou E,, et al. Risk factors for proliferative diabetic retinopathy in African Americans with type 2 diabetes. Ophthalmic Epidemiol. In press. [DOI] [PMC free article] [PubMed]
- 29. Musunuru K,, Lettre G,, Young T,, et al. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010; 3: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bild DE,, Bluemke DA,, Burke GL,, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002; 156: 871–881. [DOI] [PubMed] [Google Scholar]
- 31. The ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989; 129: 687–702. [PubMed] [Google Scholar]
- 32. Wilson JG,, Rotimi CN,, Ekunwe L,, et al. Study design for genetic analysis in the Jackson Heart Study. Ethn Dis. 2005; 15: S6-30–S6-37. [PubMed] [Google Scholar]
- 33. Penman A,, Hoadley S,, Wilson JG,, Taylor HA,, Chen CJ,, Sobrin L. P-selectin plasma levels and genetic variant associated with diabetic retinopathy in African Americans. Am J Ophthalmol. 2015; 159: 1152–1160 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong TY,, Klein R,, Islam FM,, et al. Diabetic retinopathy in a multi-ethnic cohort in the United States. Am J Ophthalmol. 2006; 141: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheung N,, Wang JJ,, Rogers SL,, et al. Diabetic retinopathy and risk of heart failure. J Am Coll Cardiol. 2008; 51: 1573–1578. [DOI] [PubMed] [Google Scholar]
- 36. Sobrin L,, Green T,, Sim X,, et al. Candidate gene association study for diabetic retinopathy in persons with type 2 diabetes: the Candidate gene Association Resource (CARe). Invest Ophthalmol Vis Sci. 2011; 52: 7593–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diabetic retinopathy study. Report Number 6. Design, methods, and baseline results. Report Number 7. A modification of the Airlie House classification of diabetic retinopathy. Prepared by the Diabetic Retinopathy. Invest Ophthalmol Vis Sci. 1981; 21: 1–226. [PubMed] [Google Scholar]
- 38. Smith MW,, Patterson N,, Lautenberger JA,, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004; 74: 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. West SK,, Munoz B,, Klein R,, et al. Risk factors for Type II diabetes and diabetic retinopathy in a Mexican-American population: Proyecto VER. Am J Ophthalmol. 2002; 134: 390–398. [DOI] [PubMed] [Google Scholar]
- 40. Davis MD,, Fisher MR,, Gangnon RE,, et al. Risk factors for high-risk proliferative diabetic retinopathy and severe visual loss: Early Treatment Diabetic Retinopathy Study Report #18. Invest Ophthalmol Vis Sci. 1998; 39: 233–252. [PubMed] [Google Scholar]
- 41. Porta M,, Sjoelie AK,, Chaturvedi N,, et al. Risk factors for progression to proliferative diabetic retinopathy in the EURODIAB Prospective Complications Study. Diabetologia. 2001; 44: 2203–2209. [DOI] [PubMed] [Google Scholar]
- 42. Florez JC,, Price AL,, Campbell D,, et al. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia. 2009; 52: 1528–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reiner AP,, Ziv E,, Lind DL,, et al. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005; 76: 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao X,, Gauderman WJ,, Marjoram P,, et al. Native American ancestry is associated with severe diabetic retinopathy in Latinos. Invest Ophthalmol Vis Sci. 2014; 55: 6041–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marigorta UM,, Gibson G. A simulation study of gene-by-environment interactions in GWAS implies ample hidden effects. Front Genet. 2014; 5: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng CY,, Reich D,, Wong TY,, et al. Admixture mapping scans identify a locus affecting retinal vascular caliber in hypertensive African Americans: the Atherosclerosis Risk in Communities (ARIC) study. PLoS Genet. 2010; 6: e1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bursell SE,, Cavallerano JD,, Cavallerano AA,, et al. Stereo nonmydriatic digital-video color retinal imaging compared with Early Treatment Diabetic Retinopathy Study seven standard field 35-mm stereo color photos for determining level of diabetic retinopathy. Ophthalmology. 2001; 108: 572–585. [DOI] [PubMed] [Google Scholar]
- 48. Klein R,, Sharrett AR,, Klein BE,, et al. The association of atherosclerosis, vascular risk factors, and retinopathy in adults with diabetes: the Atherosclerosis Risk in Communities Study. Ophthalmology. 2002; 109: 1225–1234. [DOI] [PubMed] [Google Scholar]
- 49. Awata T,, Yamashita H,, Kurihara S,, et al. A genome-wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non-coding RNA. PLoS One. 2014; 9: e111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Varma R,, Bressler NM,, Doan QV,, et al. Prevalence of and risk factors for diabetic macular edema in the United States. JAMA Ophthalmol. 2014; 132: 1334–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.