Summary
In vitro studies have demonstrated that surface expression of CD49d on chronic lymphocytic leukaemia (CLL) B cells facilitates leukaemic cell– stromal interactions by binding to fibronectin. This interaction reduces both spontaneous and drug-induced apoptosis. The present study measured CD49d expression by flow cytometry in a cohort of untreated CLL patients previously accrued to a prospective observational study and evaluated the relationship with overall survival (OS). Among the 158 CLL patients tested, the percentage of leukaemic B cells expressing CD49d ranged from 0 to 100%. When all risk factors were treated as continuous variables, CD49d expression showed moderate correlation with expression of ZAP-70 (r = 0.54; P < 0.0001) and CD38 (r = 0.58; P < 0.0001) but not % IGHV mutation. As a continuous variable, CD49d expression strongly correlated with OS (P < 0.0001). Recursive partitioning analysis suggested the 45% threshold of CD49d expression best predicted OS. Multivariate analysis, controlling for disease stage, ZAP-70, IGHV status and fluorescent in situ hybridization defects identified CD49d as an independent predictor of OS and was a better predictor of clinical outcome than ZAP-70, IGHV, or cytogenetics. This observational cohort study suggests that CLL B-cell expression of CD49d is an easily measurable and independent predictor of OS and CD49d expression in CLL. Importantly, anti-CD49d antibodies are already approved for treatment of other human diseases. Clinical testing of anti-CD49d therapy in CLL appears warranted.
Keywords: chronic lymphocytic leukaemia, stromal cells, therapy, prognostic factors, CD49d
Chronic lymphocytic leukaemia (CLL) is a common lymphoid malignancy with a variable clinical course. While the majority of patients with CLL are asymptomatic and have early stage disease at the time of diagnosis, some experience an aggressive disease course that leads to premature death while others live for decades and never require therapy. A number of recently identified prognostic parameters, including ZAP-70, IGHV mutation status, CD38 and cytogenetic abnormalities, can predict which early stage patients will experience early disease progression (Shanafelt et al, 2004).
The ideal prognostic parameter would not only identify patients at risk for worse clinical outcome, but also be relevant to disease biology and provide a therapeutic target. Her2-Neu overexpression in breast cancer patients is one example of a prognostic parameter associated with aggressive disease behaviour that can be directly targeted to improve clinical outcomes (Slamon et al, 2001) (Romond et al, 2005). Although the novel prognostic parameters identified for CLL are linked to leukaemic cell biology, the reason these characteristics are associated with worse clinical outcome is poorly understood and none are currently a direct target of available anti-neoplastic therapies.
Although CLL B cells relentlessly accumulate and are resistant to cell death in vivo, these cells rapidly become apoptotic on in vitro culture, suggesting that survival signals in the in vivo environment prevent apoptosis (Kay et al, 2007) (Ghia et al, 2002) (Caligaris-Cappio, 2003). Potentially supportive interactions between CLL B cells and various nurturing environments are complex and involve cytokine-mediated effects, cell–cell interactions, and cell–extracellular matrix interactions. As the bone marrow is invariably infiltrated by CLL B cells and is typically the site of relapse in patients who achieve treatment-induced complete remission, it is believed this tissue site provides a critical nurturing environment for CLL B cells in vivo (Caligaris-Cappio, 2003).
A limited number of studies have begun to explore specific interactions between the CLL B-cell and its environment. A variety of chemokines, such as stromal derived factor-1 (SDF1), are thought to attract leukaemic B cells to the marrow micro-environment by binding to receptors on the CLL B-cell surface (e.g. CXCR4) (Burger et al, 1999, 2000, 2001; Till et al, 2002). Once in the marrow, physical contact between stromal elements and leukaemic cells have been shown to promote CLL B-cell survival (Panayiotidis et al, 1996; Lagneaux et al, 1998; Burger et al, 1999, 2000). This protective effect is mediated in part through integrins on CLL B cells [e.g. alpha4 beta1 integrin (also known as very late antigen-4, VLA-4, CD49d) (Lagneaux et al, 1999)] and ligands [e.g. vascular cell adhesion molecule 1 (VCAM1) and fibronectin] expressed on marrow stromal cells (de la Fuente et al, 1999; Plate et al, 2000; Burger et al, 2001; Pedersen et al, 2002).
CD49d (integrin alpha 4) plays a critical role in leucocyte trafficking, activation, and survival, and also facilitates interactions between leucocytes and stromal cells found in the marrow or germinal center of lymphoid follicles via VCAM-1 and fibronectin (Rose et al, 2002). Notably, in addition to these adhesion functions, CD49d can also serve as a signalling receptor that influences B-cell survival via upregulation of Bcl-2 family members (Koopman et al, 1994; Hayashida et al, 2000). Other studies suggest that CD49d expression on CLL B cells is lower than normal B cells (Baldini et al, 1992; Eksioglu-Demiralp et al, 1996; Lucio et al, 1998), differs from other low grade B-cell malignancies (Baldini et al, 1992; Pinto et al, 1993; Eksioglu-Demiralp et al, 1996; Csanaky et al, 1997; Lucio et al, 1998), demonstrates intra-patient variation (Baldini et al, 1992; Eksioglu-Demiralp et al, 1996; Vincent et al, 1996; Sembries et al, 1999; Zucchetto et al, 2006a), and is associated with disease stage and the presence of lymphadenopathy (Eksioglu-Demiralp et al, 1996; Behr et al, 1998; Lucio et al, 1998; Till et al, 2002).
While several investigators have demonstrated that signalling via CD49d in CLL B cells reduces both spontaneous and drug-induced apoptosis in vitro (de la Fuente et al, 1999, 2002) little is known about the prognostic importance of CD49d in patients with CLL. We and others recently reported that CD49d gene (ITGA4) expression in CLL B cells correlates with CD38 expression (Durig et al, 2003) (Pittner et al, 2005) and have confirmed the correlation between CD49d and CD38 in CLL B cells at the protein level (Pittner et al, 2005). Preliminary studies suggest that CD49d expression in CLL B cells may relate to overall survival (OS) among CLL patients (Sembries et al, 1999) (Zucchetto et al, 2006a) independent of CD38 status (Zucchetto et al, 2006a) or 11q- (Sembries et al, 1999). Others studies have used CD49d as one of six factors used to assign a prognostic ‘surface antigen profiling’ score (Zucchetto et al, 2005)(Zucchetto et al, 2006b). However, there is no published data on the correlation of CD49d expression with a wider range of established prognostic parameters and its value as an independent prognostic factor. We report that the level of expression of CD49d protein on CLL B cells is prognostically important in a cohort of patients with untreated CLL previously enroled on a prospective observational study.
Methods
Patient cohort
As previously reported, between January 1994 and October 2002, we enroled 159 patients with untreated B-cell CLL/SLL seen at Mayo Clinic (Rochester or Jacksonville campus) on a prospective study evaluating the prognostic importance of cytogenetic abnormalities and clonal evolution (Shanafelt et al, 2006a). The study was approved by the Mayo Clinic Institutional Review Board and all patients gave written consent to participate. Patients provided blood specimens for research purposes at study entry and follow-up specimens over the ensuing 2–11 years.
Prognostic tests
The CLL B cells were isolated from heparinized venous blood by density gradient centrifugation. Purified peripheral blood mononuclear cells (PBMC) from CLL patients were either used immediately or suspended in RPMI 1640 medium/20% fetal calf serum/10% dimethyl sulphoxide (DMSO) and stored at −80 C until used. IGHV mutation status and cytogenetic abnormalities by fluorescent in situ hybridization (FISH) panel testing were assessed using methods previously described by our group (Jelinek et al, 2001, 2003; Dewald et al, 2003; Shanafelt et al, 2006a,b). CD38 expression in CD19+ cells was measured using antibodies specific for CD38 and CD19 with gates set by technicians with extensive experience assessing CD38 in CLL B cells. Antibodies to CD19 and ZAP-70 were used to detect ZAP-70 expression in CD19+ cells using the gating strategy of Rassenti et al (2004). Where presented as categorically variables (i.e. positive or negative), the traditional thresholds were used to classify CD38 (<30% or ≥30%) and ZAP-70 (<20% or ≥20%) status. For categorical comparison, patients with mutated IGHV genes using the IGHV3-21 rearrangement were grouped with unmutated patients (Tobin et al, 2003).
CD49d analysis
For the present study, we measured CD49d expression by flow cytometry. After purification of PBMC, lymphocytes were gated based on forward and side scatter to exclude debris, monocytes and doublets. CD49d expression in CD19+ cells was then measured using antibodies specific for CD19 (fluorescein isothiocyanate; BD Biosciences, Franklin Lakes, NJ, USA) and CD49d (phycoerythrin; BD Pharmingen, San Diego, CA, USA). As expected (Alon et al, 1995) (Hemler, 1990), where present, CD19 negative lymphocytes (T cells) expressed CD49d and served as positive controls. CD49d was assessed on stored specimens collected within 2 years of study entry for all patients and within 12 months of study entry for 149 of the 158 patients (94%). CD49d, CD38 and ZAP-70 status were all analysed on the same stored specimen and 145 of 158 (92%) patients were untreated at the date of CD49d assessment.
Statistical considerations
CD49d expression was assessed in relation to level of ZAP-70 expression, CD38 expression and degree of IGHV mutation (all treated as continuous variables). Relationships between these continuous variables were explored graphically (scatter-plots) as well as quantitatively (Spearman rank correlation coefficient). In addition, we evaluated the relationship between CD49d and categorical prognostic variables (e.g. mutated/ unmutated IGHV; Zap-70 positive/negative, etc.) using the previously published 30% threshold to classify CD49d expression as ‘high’ or ‘low’ (Zucchetto et al, 2006b). Differences between CD49d and other categorical variables were evaluated using Fisher's exact test. All tests were two-sided and statistical significance was defined as P < 0.05, where multiple comparison corrections were not used.
Overall survival was calculated as the time from date of diagnosis to date of death. Time to treatment (TTT) was defined as the time from diagnosis until treatment or censored at last date of follow-up. Differences in OS and TTT between prognostic groups were evaluated using standard Kaplan– Meier methods and log-rank statistics. Survival status was confirmed for all 158 patients and treatment status could be confirmed for 157 patients. As the interval from diagnosis to study entry was >1 year for some patients, we also calculated OS and TTT from the date of study entry. In addition, we looked at OS and TTT from diagnosis in the subset of patients who enroled on study within 1 year of diagnosis (n = 101). Cox models were used to evaluate the impact of multiple variables simultaneously on OS and TTT from diagnosis. The number of deaths observed provided an adequate sample to evaluate multivariate models including up to five prognostic variables. Based on publications suggesting that CD38 adds little prognostic utility to the combination of IGHV mutation status and FISH (Krober et al, 2002) (Oscier et al, 2002), IGHV, FISH, ZAP-70, Rai stage and CD49d were selected as the five factors for evaluation. The leaps and bounds variable selection approach was used to identify variables for multivariable models.
Recursive partitioning algorithm methods were used in models for OS with CD49d as a continuous variable in order to identify an optimal cut-off point for CD49d expression based on our data. Both this cut-off point and the previously described 30% cut-off point (Zucchetto et al, 2006a,b) were evaluated in our analysis.
Results
Of the 159 untreated patients with CLL/small lymphocytic leukaemia (SLL) accrued to our prospective observational study, 158 had cryopreserved specimens available for CD49d testing. Patient characteristics are described in Table I. The median time from diagnosis to study enrolment was 2.8 months and the median follow-up from diagnosis was 9.9 years (range 0.8–23 years). The majority of patients had Rai stage 0 or 1 disease (83%) at study entry. ZAP-70 status, CD38 status and cytogenetic analysis by FISH were available for all 158 patients. IGHV sequencing was attempted in all 158 patients, with 126 (80%) patients classifiable as mutated or unmutated.
Table I.
Patient characteristics.
| n = 158 | |
|---|---|
| Median age*, years | 64 (range: 37-87) |
| Male | 112 (71) |
| Median ALC*, ×109/l | 17·6 |
| Rai stage* | |
| 0 | 83 (53) |
| I | 48 (30) |
| II | 18 (11) |
| III | 2 (1) |
| IV | 7 (4) |
| ZAP-70 ≥20% | 95 (60) |
| IGHV unmutated† | 55 (44) |
| CD38 ≥30% | 46 (29) |
| FISH | |
| del(13q14·2) | 71 (45) |
| Normal | 39 (25) |
| Trisomy 12 | 23 (15) |
| del (11q22·3) | 12 (8) |
| del (17p131) | 7 (4) |
| Other | 6 (4) |
Values in parenthesis are percentage except where otherwise stated.
At time of study enrolment.
IGHV mutation status classifiable for 126 patients.
Relationship of CD49d expression to other prognostic parameters
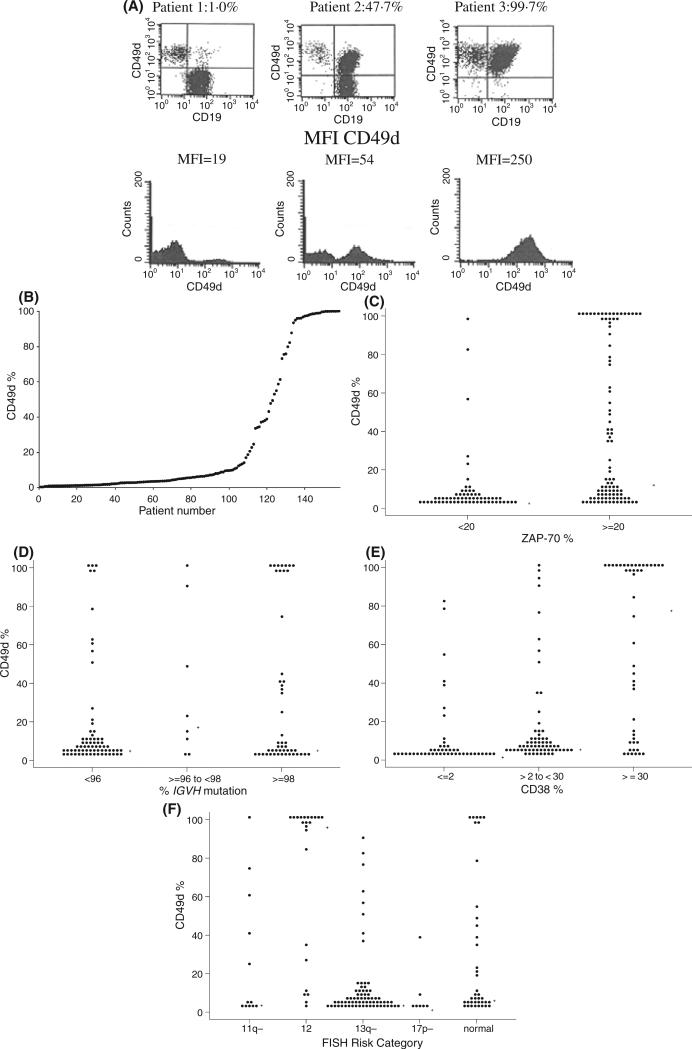
The percentage of B cells expressing CD49d ranged from 0.3 to 100.0% (median expression = 5.5%; n = 158; Fig 1A and B). We next evaluated the relationship between CD49d expression and other prognostic parameters. When ZAP-70 expression, CD38 expression, and the percentage of IGHV mutation were considered as continuous variables, the percentage of CD49d expression correlated with the expression of ZAP-70 (r = 0.54, P < 0.0001) and CD38 (r = 0.58, P < 0.0001) but not the level of IGHV mutation as assessed by the percentage change in mutation sequence (r = )0.10, P = 0.25). To expand on these findings, the relationship between CD49d and these parameters was evaluated using the standard prognostic thresholds to classify ZAP-70 (<20% vs. ≥20%), CD38 (<30% vs. ≥30%), IGHV mutation status (≤2% mutation vs. >2% mutation) and the Dohner classification (Dohner et al, 2000) for FISH abnormalities (Fig 1C–F). Patients with higher CD49d expression were more likely to be ZAP-70 positive, CD38 positive and IGHV unmutated compared to patients with low CD49d expression although substantial discordance was observed. Finally, we evaluated the relationship of CD49d with these parameters using the previously published threshold of 30% to categorize CD49d expression as ‘high’ or ‘low’ (Zucchetto et al, 2006a,b) (Table II). Using this threshold, 113 (72%) patients had low (<30%) CD49d expression and 45 (28%) had high (≥30%) CD49d expression.
Fig 1.
Relationship of CD49d with other prognostic parameters. (A) Examples of dot plots for CLL cells stained with antibodies specific for CD19 (x-axis) and CD49d (y-axis) assessed by flow cytometry. After gating on lymphocytes on forward and side scatter, CD49d expression was measured in CD19+ cells using antibodies specific for CD19 and CD49d. Dots in the upper right quadrant represent leukaemic B cells with higher CD49d expression while the dots in the lower right quadrant represent leukaemic B cells with lower CD49d expression. The percentage of CD19+ cells in the right upper quadrant is given for each patient (i.e. CD19 negative cells are not included in this calculation). Cells in the left upper quadrant represent CD19 negative lymphoid cells (i.e. T cells). (B) Distribution of CD49d expression in B cells of 158 patients. (C) Distribution of CD49d expression by ZAP-70 status (n = 158). (D) Distribution of CD49d expression by IGHV mutation status (n = 126). (E) Distribution of CD49d expression by CD38 status (n = 158). (F) Distribution of CD49d expression by FISH prognostic category (n = 152).
Table II.
Relationship between CD49d expression and other prognostic parameters.
| CD49d <30% (n = 113) | CD49d ≥30% (n = 45) | P-value | |
|---|---|---|---|
| % ZAP-70+ | 46·9 | 93·3 | <0·0001 |
| % IGHV unmutated* | 36·8 | 64·5 | 0·01 |
| % CD38+ | 14·2 | 66·7 | <0·0001 |
| FISH† | <0·0001 | ||
| % with del (17p131) or del (11q22·3) | 12·8 | 11·6 | |
| % with trisomy 12 | 5·5 | 39·5 | |
| % with normal or del(13q14·2) | 81·7 | 48·8 | |
| Current stage | <0·0001 | ||
| 0 | 61·1 | 31·1 | |
| I-II | 38·1 | 51·1 | |
| III-IV | 0·9 | 17·8 |
IGHV genes could be classified for 126 patients (see Methods).
Six patients had other cytogenetic abnormalities that could not be categorized within the Dohner classification.
Relationship of CD49d expression with overall survival and time to treatment
We next evaluated the relationship between CD49d expression and clinical outcomes. As of last follow-up, 72 patients had been treated (46%) and 62 patients (39%) had died. The estimated median OS from time of diagnosis for the entire cohort was 13.2 years [95% confidence interval (CI): 11.1– 20.0 years]. The median TTT was 9.8 years (95% CI: 6.7– 20.2 years).
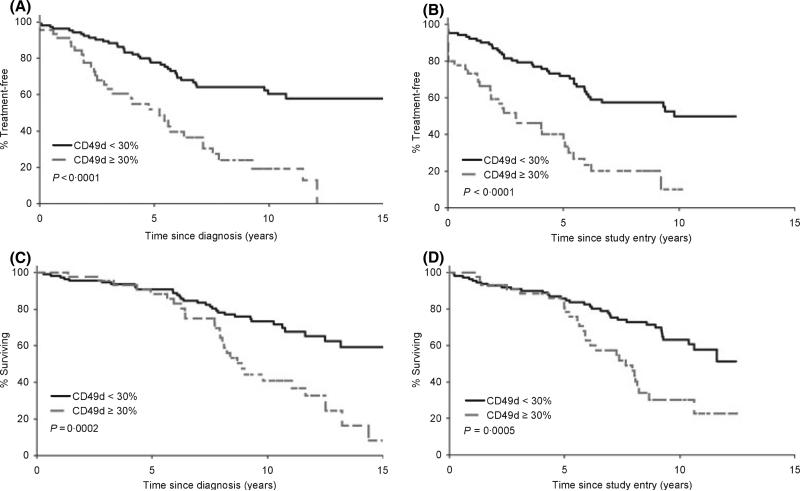
As a continuous variable, CD49d was a significant predictor for both TTT (P < 0.0001) and OS (P < 0.0001) from the date of diagnosis. Similar correlations with TTT and OS were also observed from the date of study entry (TTT P < 0.0001, OS P = 0.0002). Since CD49d was strongly associated with OS as a continuous variable, we next explored the cut-off point for CD49d that best stratified patients with respect to TTT and OS. First, we evaluated differences in OS and TTT using the previously published 30% threshold to classify CD49d expression (Zucchetto et al, 2006a,b). The median TTT from diagnosis for patients with <30% CD49d expression was 18.0 years (95% CI: 10.0–20.5 years), compared to 5.2 years (95% CI: 2.9–7.1 years) for patients with ≥30% CD49d expression (P < 0.0001). There was also a significant difference in OS from diagnosis using the 30% threshold with a median of 20.0 years (95% CI: 13.2 years – not reached) in patients with <30% CD49d expression and a median of 8.9 years (7.9– 11.6 years) in patients with ≥30% CD49d expression, P = 0.0002 (Fig 2A–D).
Fig 2.
Time to treatment and overall survival according to 30% threshold to categorize CD49d. (A) TTT from date of diagnosis based on CD49d expression (n = 156). (B) TTT from on study date based on CD49d expression (n = 156). (C) OS from date of diagnosis based on CD49d expression (n = 158). (D) OS from on study date based on CD49d expression (n = 158).
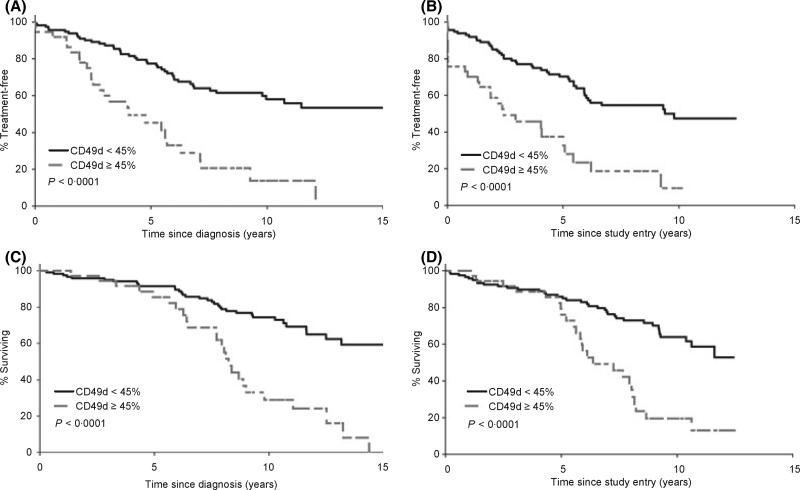
As the optimal threshold to classify CD49d expression is unknown (Zucchetto et al, 2006a,b) recursive partitioning algorithms were used to identify a cut-off point for CD49d expression that best predicted OS. This analysis showed that a cut-off point of 45% CD49d expression best predicted clinical outcome. Using this threshold, 121 (77%) patients had <45% CD49D expression and 37 (23%) had ≥45% CD49D expression. The median TTT from diagnosis for patients with <45% CD49d expression was 18.0 years (95% CI: 9.8– 20.5 years), compared to 4.0 years (95% CI: 2.8–6.3 years) for patients with ≥45% CD49d expression (P < 0.0001). There was also a significant difference in OS from diagnosis using the 45% threshold with a median of 20.0 years (95% CI: 13.2 years – not yet reached), in patients with <45% CD49d expression and of 8.2 years (95% CI: 7.7–9.8 years) in patients with ≥45% CD49d expression (P < 0.0001; Fig 3A–D). TTT (P = 0.046) and OS (P = 0.01) were also both significantly longer in the group with <45% CD49d expression when analysis was restricted to the subset of patients (n = 101) who enroled on study within 1 year of diagnosis.
Fig 3.
Time to treatment and overall survival based on 45% threshold to categorize CD49d. (A) TTT from date of diagnosis based on CD49d expression (n = 156). (B) TTT from on study date based on CD49d expression (n = 156). (C) OS from date of diagnosis based on CD49d expression (n = 158). (D) OS from on study date based on CD49d expression (n = 158).
Multivariate analyses for overall survival
Finally, we evaluated how CD49d (45% cut-off point) was associated with OS when accounting for other relevant prognostic factors, such as Rai stage at study entry, ZAP-70 expression (<20% vs. ≥20%), IGHV mutation status (≤2%; >2%), and cytogenetic abnormalities by FISH. For the purposes of these analyses, a patient with a FISH diagnosis of del(17p13.1) or del(11q22.3) was categorized as having a poor FISH-based prognosis. Similarly, patients with a FISH classification of normal, del(13q14.2), or trisomy 12 were categorized as having a good/intermediate FISH-based prognosis. The analysis considered all possible 1 factor, 2 factor, 3 factor, 4 factor and 5 factor prognostic models and evaluated the goodness of fit of each of these models in predicting OS. CD49d was the best single-variable model to predict OS (Table IIIA). When all factors and all combinations were evaluated, the 2-factor model combining CD49d and FISH could not be significantly improved upon by adding any additional factors. This same model was selected when repeat analysis evaluating various definitions for OS (i.e. OS from diagnosis versus OS from date on study versus OS from diagnosis limited to patients diagnosed within 1 year of study date) was conducted. In the multivariate Cox models, both CD49d status and FISH prognosis group were statistically significant for all definitions of OS (Table IIIB). When analysis was repeated using the 30% threshold to categorize CD49d, the combination of CD49d and FISH was once again selected as the best predictive model. When IGHV, ZAP-70, stage, or FISH were each forced to be one factor in a 2-factor model, CD49d was always the additional factor selected to yield the best predictive model. Overall, CD49d was the single most powerful prognostic parameter.
Table III.
Uni- and multivariate analyses for overall survival. Multi-variate analysis considered all possible combinations of prognostic factors and evaluated the goodness of fit of each of these models in predicting overall survival (OS). (I) Univariate hazard ratios demonstrated that CD49d was the best single-variable model to predict OS. (II) When all factors and all combinations were evaluated, the 2-factor model combining CD49d and FISH predicted the greatest amount of variation in patient survival and could not be significantly improved upon by adding any additional factors.
| Model | HR | HR 95% CI | P-value |
|---|---|---|---|
| I. Univariate model results for OS | |||
| OS from diagnosis | |||
| CD49d (>45%) | 3·5 | 2·1-5·8 | <0·0001 |
| FISH prognosis group (11q- or 17p-) | 2·6 | 1·4-4·8 | 0·002 |
| IGHV unmutated | 1·6 | 0·9-2·9 | 0·08 |
| Stage | 1·4 | 1·1-1·7 | 0·002 |
| ZAP-70 (≥20%) | 1·8 | 1·0-3·0 | 0·04 |
| II. Multivariate model results for OS | |||
| (A) OS from diagnosis | |||
| CD49d (≥45%) | 4·2 | 2·4-7·3 | <0·0001 |
| FISH prognosis group (11q- or 17p-) | 3·4 | 1·8-6·3 | 0·0001 |
| (B) OS from study entry | |||
| CD49d (≥45%) | 3·7 | 2·2-6·3 | <0·0001 |
| FISH prognosis group (11q- or 17p-) | 3·3 | 1·8-6·2 | 0·0001 |
| (C) OS from diagnosis for patients diagnosed within 1 year of study entry | |||
| CD49d (≥45%) | 2·8 | 1·4-5·6 | 0·0046 |
| FISH prognosis group (11q- or 17p-) | 3·3 | 1·6-7·1 | 0·0018 |
Discussion
In this observational cohort study, there was wide inter-patient variation in CLL B-cell CD49d protein expression (range 0– 100% CD19+ cells). Increased CD49d expression was an independent predictor of OS on multivariate analysis including the other novel biological or molecular prognostic parameters. This is notable since multiple prior studies have suggested that ZAP-70 (Rassenti et al, 2004) (Crespo et al, 2003) (Orchard et al, 2004) (Rassenti et al, 2006), IGHV (Rassenti et al, 2004) (Krober et al, 2002) (Oscier et al, 2002) (Hamblin et al, 2002) and FISH (Shanafelt et al, 2006b) (Krober et al, 2002) (Oscier et al, 2002) (Dohner et al, 2000) are the most powerful molecular prognostic parameters identified to date.
Although a number of studies have looked at the correlation between CD49d expression in CLL B cells and surrogate markers of clinical outcome (i.e. disease stage), only a few have directly evaluated the relationship with OS (Sembries et al, 1999) (Zucchetto et al, 2006a). These small studies did not include comprehensive assessment of the other relevant prognostic parameters and could not evaluate the independent prognostic value of CD49d relative to disease stage, ZAP-70 status, FISH analysis, and IGHV mutation status or provide insight on how to optimally combine CD49d with these factors to predict clinical outcome. In the present study, CD49d was the single strongest prognostic factor in both univariate and MV analysis including these factors and a two factor prognostic model using CD49d and chromosome analysis by FISH could not be significantly improved upon by adding any additional factor(s).
As many prognostic parameters are now available for use in patients with early stage CLL, progress will require identification of molecular features that provide insight into disease biology and have the potential for therapeutic targeting. CD49d mediates interactions between CLL B cells and stromal cells and facilitates in vitro resistance to both spontaneous and drug induced apoptosis (de la Fuente et al, 1999, 2002). The association of CD49d with other poor prognostic parameters (IGHV unmutated, ZAP-70+, CD38+) in our study implies that leukaemic clones with these molecular characteristics may receive enhanced stromal nuturing with resultant effects on leukaemic cell viability, migration and apoptotic resistance (Panayiotidis et al, 1996; Lagneaux et al, 1998, 1999; Burger et al, 1999, 2000, 2001; Plate et al, 2000; Pedersen et al, 2002; Till et al, 2002). CD49d expression/ligation influences a number of factors that relate to the migratory and invasive properties of CLL B cells including adhesion to fibronectin and the endothelium (Eksioglu-Demiralp et al, 1996; Vincent et al, 1996), upregulation of MMP-9 through phosphatidylinositol 3 kinase/Akt signaling (Redondo-Munoz et al, 2006), podosome formation (Redondo-Munoz et al, 2006) and chemokine-dependent motility through the endothelium (Till et al, 2005). Others have demonstrated that the CD49d – fibronectin interaction reduces spontaneous apoptosis in CLL B cells by increasing the Bcl-2/Bax ratio (de la Fuente et al, 1999) and also appears to reduce fludarabine-induced apoptosis by increasing Bcl-XL levels (de la Fuente et al, 2002, 2003).
Perhaps most importantly, our observations have realistic and relatively immediate therapeutic potential since anti-CD49d anti-bodies are available for clinical use, have already been widely studied in humans for treatment of multiple sclerosis (Rudick et al, 2006) (Polman et al, 2006), and are approved for clinical use by the Food and Drug Administration (FDA) for that indication. Combining anti-CD49d antibodies with established chemotherapy or immunotherapy regimens in CLL could impair stromal nuturing of leukaemic cells and overcome apoptotic resistance. Unlike ZAP-70, IGHV mutation status, CD38 and cytogenetic abnormalities, measurement of CD49d could thus be an actionable prognostic test with opportunities for direct therapeutic targeting. Other approaches to therapeutically target CD49d are also in development including small peptide mimetics to block CD49d binding to VCAM-1 or fibronectin in the hopes of enhancing treatment efficacy (Rose et al, 2002).
Our findings are subject to a number of limitations. Although the patient group studied was a defined cohort of previously untreated patients enroled on a prospective observational trial, larger validation studies are needed before CD49d can be used as a prognostic parameter in clinical practice. Similar to other investigators (Sembries et al, 1999), we used a two-colour (CD19, CD49d) flow cytometry-based strategy analogous to that used for evaluation of CD38 and ZAP-70. Zucchetto et al (2006a) have previously reported a three-colour (CD19, CD5, CD49d) strategy to measure CD49d. The best way to measure CD49d expression in CLL B cells is unknown and will need to be determined. While 45% expression was the best threshold to classify patients at risk of shorter survival in the present cohort, the optimal cut-off point to stratify patient risk will need to be defined in future studies utilizing even larger cohorts. The stability of CD49d expression during the course of the disease is also unknown and clearly requires future evaluation.
Our study also has a number of important strengths. The individuals studied were a well-defined cohort of CLL patients participating in a prospective observational trial. Over 90% of patients had early stage disease at study entry and thus represent the patient group for whom prognostic tools are most needed. CD49d correlated with OS as a continuous variable and was an independent predictor of OS on multivariate analysis that included the other well-established prognostic parameters. Finally, the assay used for CD49d involves a two-colour flow cytometric assay for a cell surface antigen that should be available in nearly every modern clinical and research laboratory.
In conclusion, CLL B-cell membrane expression of CD49d as measured by flow cytometry is a powerful prognostic parameter in patients with CLL. The prognostic utility of CD49d expression is independent of ZAP-70, IGHV mutation status and cytogenetic abnormalities evaluated by FISH. As CD49d is functionally important to CLL B-cell biology and leukaemic cell survival, our findings suggest CD49d may have promise as a therapeutic target in CLL. Anti-CD49d antibodies are already FDA-approved for treatment of other human diseases. Clinical testing of anti-CD49d therapy in CLL appears to be warranted.
Acknowledgements
This study was supported through grants from the National Cancer Institute (NCI CA 113408; NCI CA 94919; NCI CA97274) are gratefully acknowledged.
References
- Alon R, Kassner PD, Carr MW, Finger EB, Hemler ME, Springer TA. The integrin VLA-4 supports tethering and rolling in flow on VCAM-1. Journal of Cell Biology. 1995;128:1243–1253. doi: 10.1083/jcb.128.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini L, Cro L, Calori R, Nobili L, Silvestris I, Maiolo AT. Differential expression of very late activation antigen-3 (VLA-3)/VLA-4 in B-cell non-Hodgkin lymphoma and B-cell chronic lymphocytic leukemia. Blood. 1992;79:2688–2693. [PubMed] [Google Scholar]
- Behr SI, Korinth D, Schriever F. Differential adhesion pattern of B cell chronic lymphocytic leukemia cells. Leukemia. 1998;12:71–77. doi: 10.1038/sj.leu.2400883. [DOI] [PubMed] [Google Scholar]
- Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94:3658–3667. [PubMed] [Google Scholar]
- Burger JA, Tsukada N, Burger M, Zvaifler NJ, Dell'Aquila M, Kipps TJ. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood. 2000;96:2655–2663. [PubMed] [Google Scholar]
- Burger JA, Zvaifler NJ, Tsukada N, Firestein GS, Kipps TJ. Fibroblast-like synoviocytes support B-cell pseudoemperipolesis via a stromal cell-derived factor-1- and CD106 (VCAM-1)-dependent mechanism. Journal of Clinical Investigation. 2001;107:305–315. doi: 10.1172/JCI11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligaris-Cappio F. Role of the microenvironment in chronic lymphocytic leukemia. British Journal of Haematology. 2003;123:380–388. doi: 10.1046/j.1365-2141.2003.04679.x. [DOI] [PubMed] [Google Scholar]
- Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, Marce S, Lopez-Guillermo A, Campo E, Montserrat E. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. New England Journal of Medicine. 2003;348:1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- Csanaky G, Matutes E, Vass JA, Morilla R, Catovsky D. Adhesion receptors on peripheral blood leukemic B cells. A comparative study on B cell chronic lymphocytic leukemia and related lymphoma/leukemias. Leukemia. 1997;11:408–415. doi: 10.1038/sj.leu.2400582. [DOI] [PubMed] [Google Scholar]
- Dewald G, Brockman S, Paternoster S, Bone N, O'Fallon J, allmer C, James C, Jelinek D, Tschumper R, Hanson C, Pruthi R, Witzig T, Call T, Kay N. Chromosome anomalies detected by interphase fluorscence in hybridization: correlation with significant biological features of chronic lymphocytic leukemia. British Journal of Haematology. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. New England Journal of Medicine. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Durig J, Nuckel H, Huttmann A, Kruse E, Holter T, Halfmeyer K, Fuhrer A, Rudolph R, Kalhori N, Nusch A, Deaglio S, Malavasi F, Moroy T, Klein-Hitpass L, Duhrsen U. Expression of ribosomal and translation-associated genes is correlated with a favorable clinical course in chronic lymphocytic leukemia. Blood. 2003;101:2748–2755. doi: 10.1182/blood-2002-09-2683. [DOI] [PubMed] [Google Scholar]
- Eksioglu-Demiralp E, Alpdogan O, Aktan M, Firatli T, Ozturk A, Budak T, Bayik M, Akoglu T. Variable expression of CD49d antigen in B cell chronic lymphocytic leukemia is related to disease stages. Leukemia. 1996;10:1331–1339. [PubMed] [Google Scholar]
- de la Fuente MT, Casanova B, Garcia-Gila M, Silva A, Garcia-Pardo A. Fibronectin interaction with alpha4beta1 integrin prevents apoptosis in B cell chronic lymphocytic leukemia: correlation with Bcl-2 and Bax. Leukemia. 1999;13:266–274. doi: 10.1038/sj.leu.2401275. [DOI] [PubMed] [Google Scholar]
- de la Fuente MT, Casanova B, Moyano JV, Garcia-Gila M, Sanz L, Garcia-Marco J, Silva A, Garcia-Pardo A. Engagement of alpha4beta1 integrin by fibronectin induces in vitro resistance of B chronic lymphocytic leukemia cells to fludarabine. Journal of Leukocyte Biology. 2002;71:495–502. [PubMed] [Google Scholar]
- de la Fuente MT, Casanova B, Cantero E, Hernandez del Cerro M, Garcia-Marco J, Silva A, Garcia-Pardo A. Involvement of p53 in alpha4beta1 integrin-mediated resistance of B-CLL cells to fludarabine. Biochemical and Biophysical Research Communications. 2003;311:708–712. doi: 10.1016/j.bbrc.2003.10.054. [DOI] [PubMed] [Google Scholar]
- Ghia P, Granziero L, Chilosi M, Caligaris-Cappio F. Chronic B cell malignancies and bone marrow microenvironment. Seminars in Cancer Biology. 2002;12:149–155. doi: 10.1006/scbi.2001.0423. [DOI] [PubMed] [Google Scholar]
- Hamblin TJ, Orchard JA, Ibbotson RE, Davis Z, Thomas PW, Stevenson FK, Oscier DG. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002;99:1023–1029. doi: 10.1182/blood.v99.3.1023. [DOI] [PubMed] [Google Scholar]
- Hayashida K, Shimaoka Y, Ochi T, Lipsky PE. Rheumatoid arthritis synovial stromal cells inhibit apoptosis and up-regulate Bcl-xL expression by B cells in a CD49/CD29-CD106-dependent mechanism. Journal of Immunology. 2000;164:1110–1116. doi: 10.4049/jimmunol.164.2.1110. [DOI] [PubMed] [Google Scholar]
- Hemler ME. VLA proteins in the integrin family: structures, functions, and their role on leukocytes. Annual Review of Immunology. 1990;8:365–400. doi: 10.1146/annurev.iy.08.040190.002053. [DOI] [PubMed] [Google Scholar]
- Jelinek DF, Tschumper RC, Geyer SM, Bone ND, Dewald GW, Hanson CA, Stenson MJ, Witzig TE, Tefferi A, Kay NE. Analysis of clonal B-cell CD38 and immunoglobulin variable region sequence status in relation to clinical outcome for B-chronic lymphocytic leukaemia. British Journal of Haematology. 2001;115:854–861. doi: 10.1046/j.1365-2141.2001.03149.x. [DOI] [PubMed] [Google Scholar]
- Jelinek DF, Tschumper RC, Stolovitzky GA, Iturria SJ, Tu Y, Lepre J, Shah N, Kay NE. Identification of a global gene expression signature of B-chronic lymphocytic leukemia. Molecular Cancer Research. 2003;1:346–361. [PubMed] [Google Scholar]
- Kay NE, Shanafelt TD, Strege AK, Lee YK, Bone ND, Raza A. Bone biopsy derived marrow stromal elements rescue chronic lymphocytic leukemia B-cells from spontaneous and drug induced cell death and facilitates an “angiogenic switch”. Leukemia Research. 2007;31:899–906. doi: 10.1016/j.leukres.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman G, Keehnen RM, Lindhout E, Newman W, Shimizu Y, van Seventer GA, de Groot C, Pals ST. Adhesion through the LFA-1 (CD11a/CD18)-ICAM-1 (CD54) and the VLA-4 (CD49d)-VCAM-1 (CD106) pathways prevents apoptosis of germinal center B cells. Journal of Immunology. 1994;152:3760–3767. [PubMed] [Google Scholar]
- Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P, Dohner H, Stilgenbauer S. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- Lagneaux L, Delforge A, Bron D, Bruyn CD, Stryckmans P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood. 1998;91:2387–2396. [PubMed] [Google Scholar]
- Lagneaux L, Delforge A, De Bruyn C, Bernier M, Bron D. Adhesion to bone marrow stroma inhibits apoptosis of chronic lymphocytic leukemia cells. Leukemia & Lymphoma. 1999;35:445–453. doi: 10.1080/10428199909169609. [DOI] [PubMed] [Google Scholar]
- Lucio PJ, Faria MT, Pinto AM, da Silva MR, Correia Junior ME, da Costa RJ, Parreira AB. Expression of adhesion molecules in chronic B-cell lymphoproliferative disorders. Haematologica. 1998;83:104–111. [PubMed] [Google Scholar]
- Orchard J, Ibbotson R, Davis Z, Wiestner A, Rosenwald A, Thomas P, Hamblin T, Staudt L, Oscier D. ZAP-70 expression and prognosis in chronic lymphocytic leukemia. Lancet. 2004;363:105–111. doi: 10.1016/S0140-6736(03)15260-9. [DOI] [PubMed] [Google Scholar]
- Oscier DG, Gardiner AC, Mould SJ, Glide S, Davis ZA, Ibbotson RE, Corcoran MM, Chapman RM, Thomas PW, Copplestone JA, Orchard JA, Hamblin TJ. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–1184. [PubMed] [Google Scholar]
- Panayiotidis P, Jones D, Ganeshaguru K, Foroni L, Hoffbrand A. Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocytic leukaemia cells in vitro. British Journal of Haematology. 1996;92:97–103. doi: 10.1046/j.1365-2141.1996.00305.x. [DOI] [PubMed] [Google Scholar]
- Pedersen IM, Kitada S, Leoni LM, Zapata JM, Karras JG, Tsukada N, Kipps TJ, Choi YS, Bennett F, Reed JC. Protection of CLL B cells by a follicular dendritic cell line is dependent on induction of Mcl-1. Blood. 2002;100:1795–1801. [PubMed] [Google Scholar]
- Pinto A, Carbone A, Gloghini A, Marotta G, Volpe R, Zagonel V. Differential expression of cell adhesion molecules in B-zone small lymphocytic lymphoma and other well-differentiated lymphocytic disorders. Cancer. 1993;72:894–904. doi: 10.1002/1097-0142(19930801)72:3<894::aid-cncr2820720339>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Pittner BT, Shanafelt TD, Kay NE, Jelinek DF. CD38 expression levels in chronic lymphocytic leukemia B cells are associated with activation marker expression and differential responses to interferon stimulation. Leukemia. 2005;19:2264–2272. doi: 10.1038/sj.leu.2403975. [DOI] [PubMed] [Google Scholar]
- Plate JM, Long BW, Kelkar SB. Role of beta2 integrins in the prevention of apoptosis induction in chronic lymphocytic leukemia B cells. Leukemia. 2000;14:34–39. doi: 10.1038/sj.leu.2401621. [DOI] [PubMed] [Google Scholar]
- Polman CH, O'Connor PW, Havrdova E, Hutchinson M, Kappos L, Miller DH, Phillips JT, Lublin FD, Giovannoni G, Wajgt A, Toal M, Lynn F, Panzara MA, Sandrock AW. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. New England Journal of Medicine. 2006;354:899–910. doi: 10.1056/NEJMoa044397. [DOI] [PubMed] [Google Scholar]
- Rassenti LZ, Huynh L, Toy TL, Chen L, Keating MJ, Gribben JG, Neuberg DS, Flinn IW, Rai KR, Byrd JC, Kay NE, Greaves A, Weiss A, Kipps TJ. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. New England Journal of Medicine. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- Rassenti LZ, Neuberg D, Huynh L, Wierda W, Gribben JG, Brown JR, Flinn IW, Rai KR, Byrd JC, Greaves AW, Kipps TJ. Relative value of CD38 and ZAP-70 vesrsus immunoglobulin mutation status in predicting early disease progression in chronic lymphocytic leukemia. Blood. 2006;108 Abstract no. 2778. [Google Scholar]
- Redondo-Munoz J, Escobar-Diaz E, Samaniego R, Terol MJ, Garcia-Marco JA, Garcia-Pardo A. MMP-9 in B-cell chronic lymphocytic leukemia is upregulated by {alpha}4{beta}1 integrin or CXCR4 engagement via distinct signaling pathways, localizes to podosomes, and is involved in cell invasion and migration. Blood. 2006;108:3143–3151. doi: 10.1182/blood-2006-03-007294. [DOI] [PubMed] [Google Scholar]
- Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG, Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas EP, Lingle WL, Klein PM, Ingle JN, Wolmark N. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. New England Journal of Medicine. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- Rose DM, Han J, Ginsberg MH. Alpha4 integrins and the immune response. Immunological Reviews. 2002;186:118–124. doi: 10.1034/j.1600-065x.2002.18611.x. [DOI] [PubMed] [Google Scholar]
- Rudick RA, Stuart WH, Calabresi PA, Confavreux C, Galetta SL, Radue EW, Lublin FD, Weinstock-Guttman B, Wynn DR, Lynn F, Panzara MA, Sandrock AW. Natalizumab plus interferon beta-1a for relapsing multiple sclerosis. New England Journal of Medicine. 2006;354:911–923. doi: 10.1056/NEJMoa044396. [DOI] [PubMed] [Google Scholar]
- Sembries S, Pahl H, Stilgenbauer S, Dohner H, Schriever F. Reduced expression of adhesion molecules and cell signaling receptors by chronic lymphocytic leukemia cells with 11q deletion. Blood. 1999;93:624–631. [PubMed] [Google Scholar]
- Shanafelt TD, Geyer S, Kay N. Prognosis at diagnosis: integrating molecular biologic insights into clinical practice for patients with CLL. Blood. 2004;103:1202–1210. doi: 10.1182/blood-2003-07-2281. [DOI] [PubMed] [Google Scholar]
- Shanafelt TD, Witzig TE, Fink SR, Jenkins RB, Paternoster SF, Smoley SA, Stockero KJ, Nast DM, Flynn HC, Tschumper RC, Geyer S, Zent CS, Call TG, Jelinek DF, Kay NE, Dewald GW. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. Journal of Clinical Oncology. 2006a;24:4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- Shanafelt TD, Jelinek D, Tschumper R, Schwager S, Nowakowski G, DeWald GW, Kay NE. Cytogenetic abnormalities can change during the course of the disease process in chronic lymphocytic leukemia. Journal of Clinical Oncology. 2006b;24:3218–3219. doi: 10.1200/JCO.2006.06.1077. author reply 3219–3220. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Baja-monde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. New England Journal of Medicine. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Till KJ, Lin K, Zuzel M, Cawley JC. The chemokine receptor CCR7 and alpha4 integrin are important for migration of chronic lymphocytic leukemia cells into lymph nodes. Blood. 2002;99:2977–2984. doi: 10.1182/blood.v99.8.2977. [DOI] [PubMed] [Google Scholar]
- Till KJ, Spiller DG, Harris RJ, Chen H, Zuzel M, Cawley JC. CLL, but not normal, B cells are dependent on autocrine VEGF and alpha4beta1 integrin for chemokine-induced motility on and through endothelium. Blood. 2005;105:4813–4819. doi: 10.1182/blood-2004-10-4054. [DOI] [PubMed] [Google Scholar]
- Tobin G, Thunberg U, Johnson A, Eriksson I, Soderberg O, Karlsson K, Merup M, Juliusson G, Vilpo J, Enblad G, Sundstrom C, Roos G, Rosenquist R. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted V{lambda}2-14 gene usage and homologous CDR3s: implicating recognition of a common antigen epitope. Blood. 2003;13:13. doi: 10.1182/blood-2002-11-3485. [DOI] [PubMed] [Google Scholar]
- Vincent AM, Cawley JC, Burthem J. Integrin function in chronic lymphocytic leukemia. Blood. 1996;87:4780–4788. [PubMed] [Google Scholar]
- Zucchetto A, Sonego P, Degan M, Bomben R, Dal Bo M, Russo S, Attadia V, Rupolo M, Buccisano F, Del Principe MI, Del Poeta G, Pucillo C, Colombatti A, Campanini R, Gattei V. Signature of B-CLL with different prognosis by Shrunken centroids of surface antigen expression profiling. Journal of Cellular Physiology. 2005;204:113–123. doi: 10.1002/jcp.20269. [DOI] [PubMed] [Google Scholar]
- Zucchetto A, Bomben R, Dal Bo M, Bulian P, Benedetti D, Nanni P, Del Poeta G, Degan M, Gattei V. CD49d in B-cell chronic lymphocytic leukemia: correlated expression with CD38 and prognostic relevance. Leukemia. 2006a;20:523–525. doi: 10.1038/sj.leu.2404087. [DOI] [PubMed] [Google Scholar]
- Zucchetto A, Bomben R, Dal Bo M, Sonego P, Nanni P, Rupolo M, Bulian P, Dal Maso L, Del Poeta G, Del Principe MI, Degan M, Gattei V. A scoring system based on the expression of six surface molecules allows the identification of three prognostic risk groups in B-cell chronic lymphocytic leukemia. Journal of Cellular Physiology. 2006b;207:354–363. doi: 10.1002/jcp.20570. [DOI] [PubMed] [Google Scholar]