Abstract
Intrauterine exposure to gestational diabetes mellitus (GDM) is linked to development of hypertension, obesity, and type 2 diabetes in children. Our previous studies determined that endothelial colony-forming cells (ECFCs) from neonates exposed to GDM exhibit impaired function. The current goals were to identify aberrantly expressed genes that contribute to impaired function of GDM-exposed ECFCs and to evaluate for evidence of altered epigenetic regulation of gene expression. Genome-wide mRNA expression analysis was conducted on ECFCs from control and GDM pregnancies. Candidate genes were validated by quantitative RT-PCR and Western blotting. Bisulfite sequencing evaluated DNA methylation of placenta-specific 8 (PLAC8). Proliferation and senescence assays of ECFCs transfected with siRNA to knockdown PLAC8 were performed to determine functional impact. Thirty-eight genes were differentially expressed between control and GDM-exposed ECFCs. PLAC8 was highly expressed in GDM-exposed ECFCs, and PLAC8 expression correlated with maternal hyperglycemia. Methylation status of 17 CpG sites in PLAC8 negatively correlated with mRNA expression. Knockdown of PLAC8 in GDM-exposed ECFCs improved proliferation and senescence defects. This study provides strong evidence in neonatal endothelial progenitor cells that GDM exposure in utero leads to altered gene expression and DNA methylation, suggesting the possibility of altered epigenetic regulation.
Introduction
The Barker hypothesis postulates that alterations in the intrauterine environment and in fetal and infant nutrition correlate with development of adult diseases (1). This concept of a developmental origin of adult disease was based upon a landmark study linking low birth weight with increased risk of death from ischemic heart disease (2). Several subsequent studies have confirmed that infants born small at birth are at increased risk of developing hypertension, stroke, type 2 diabetes, and obesity (1). Collectively, these observations infer that permanent changes occur during fetal development allowing adaptation and survival in a suboptimal intrauterine environment and that in a postnatal setting these developmental adaptations mechanistically contribute to the pathogenesis of multiple chronic diseases. Similarly, infants born to women with pre–gestational diabetes mellitus and gestational diabetes mellitus (GDM) have an increased risk for developing chronic diseases including hypertension, type 2 diabetes, and obesity (3–7). Numerous animal studies show long-term, harmful effects of fetal overnutrition (8,9). Thus, fetal intrauterine exposure to either undernourishment or diabetes increases disease risk later in life.
Because of the long-lasting nature of an individual’s response to adverse intrauterine environment exposure, dysfunctional stem and progenitor cells are hypothesized to participate in disease pathogenesis. Our previous work evaluating the function of endothelial progenitor cells supports this supposition. Using cord blood endothelial colony-forming cells (ECFCs), a highly proliferative and self-renewing endothelial progenitor population, we identified numerous functional deficits of ECFCs exposed to GDM in utero (10). Importantly, fetal GDM exposure resulted in increased proliferation, reduced vasculogenesis, and resistance to hyperglycemia-induced senescence of ECFCs (10).
A potential mechanism for these fetal adaptations includes epigenetic modifications that lead to aberrant gene expression and subsequent cellular dysfunction (11,12). Epigenetic changes, such as alterations in DNA methylation and histone acetylation, have been reported in animal models of intrauterine growth restriction and diabetes (13,14). However, few studies have been conducted in humans to solidify whether epigenetic changes alter the functional capacity of cells from infants born small for gestational age or to women with GDM. Furthermore, the majority of published data do not address whether molecular adaptations occur in stem and/or progenitor cells. We hypothesized that epigenetic changes are induced in ECFCs during fetal exposure to GDM, resulting in abnormal gene expression and cellular dysfunction. The goals of the current study were to identify candidate genes with altered expression that contribute to the aberrant function of GDM-exposed ECFCs and to determine whether impaired DNA methylation promotes aberrant expression of a candidate gene.
Research Design and Methods
Umbilical Cord Blood Collection
Umbilical cord blood samples were collected from healthy control pregnancies and pregnancies complicated by GDM after obtaining informed consent from the mothers. GDM was defined according to the guidelines of the American College of Obstetricians and Gynecologists (15). All pregnancies were singleton gestations. Women with preeclampsia or hypertension, women with other illnesses known to affect glucose metabolism, and women taking medications known to affect glucose metabolism were excluded. In addition, infants with known chromosomal abnormalities were excluded. The institutional review board at the Indiana University School of Medicine approved this protocol. GDM samples were separated into two groups for initial analyses: conservatively managed (diet and exercise) and insulin treated. Clinical data for mothers (Supplementary Table 1) and infants (Supplementary Table 2) are included for cohorts 1 and 2. Glucose values were obtained for all women from the 50-g, 1-h glucose screening test that is performed between 24 and 28 weeks of gestation during routine obstetric care.
Cell Culture
ECFCs were cultured from umbilical cord blood samples by the Indiana University Simon Cancer Center Angio BioCore (formerly the Angiogenesis, Endothelial and Pro-Angiogenic Cell Core) as previously described (10). HEK/293 cells (American Type Culture Collection, Manassas, VA) were cultured in DMEM (Mediatech; Corning Cellgro, Manassas, VA) containing 10% FCS (Atlanta Biologicals, Flowery Branch, GA) and antibiotic-antimycotic solution (Mediatech). Jurkat cells were the kind gift of Helmut Hanenberg (Indiana University School of Medicine) and were cultured in RPMI-1640 (Invitrogen, Grand Island, NY) containing 10% FCS and antibiotic-antimycotic solution.
RNA and DNA Isolation
Total RNA and genomic DNA were isolated from ECFCs during log phase growth at passage 3 or 4. RNA was extracted using an miRNeasy kit (Qiagen, Valencia, CA). RNA concentration was determined by Nanodrop (Wilmington, DE), and RNA quality was examined by either electrophoresis or Bioanalyzer (Agilent Technologies, Santa Clara, CA). DNA was isolated using a QIAamp DNA Mini Kit (Qiagen) per the manufacturer’s instructions.
Affymetrix Microarray
The Center for Medical Genomics (Indiana University School of Medicine) conducted these studies. Total RNA samples were labeled using the standard protocol for the Ambion WT Expression kit (Life Technologies, Grand Island, NY) combined with the Affymetrix GeneChip WT Terminal Labeling and Controls kit (Affymetrix, Santa Clara, CA). Individual labeled samples were hybridized to the Human Gene 1.0 ST GeneChips for 17 h and then washed, stained, and scanned with the standard protocol using Affymetrix GeneChip Command Console Software to generate data (CEL files). Arrays were visually scanned for abnormalities or defects. CEL files were imported into Partek Genomics Suite (Partek, Inc., St. Louis, MO). Robust Multi-Array Average (RMA) signals were generated for the core probe sets using the RMA background correction, quantile normalization, and summarization by Median Polish. Summarized signals for each probe set were log2 transformed. These log-transformed signals were used for principal components analysis, hierarchical clustering, and signal histograms to determine whether there were any outlier arrays; none were found. Untransformed RMA signals were used for fold change calculations. Data were analyzed using a one-way ANOVA using log2-transformed signals with phenotype (control, GDM conservatively managed, and GDM insulin treated) as a factor and all possible contrasts made. Fold changes were calculated using the untransformed RMA signals. Probe sets whose expression level was <4.0 for all phenotypes were removed. False discovery rates were calculated using the Qvalue program in R.
Quantitative RT-PCR
RNA was reverse transcribed using Transcriptor Universal Master cDNA kit (Roche, Indianapolis, IN). Quantitative RT-PCR (qRT-PCR) was performed using Lightcycler 480 SYBR Green I Master Mix (Roche) and gene-specific intron-spanning primers (Supplementary Table 3) as previously described (10).
Rapid Amplification of cDNA Ends
cDNA from control ECFCs was amplified by PCR using the 5′/3′ RACE kit, 2nd generation (Roche). Reverse primers complementary to placenta-specific 8 (PLAC8) exon 3 (Supplementary Table 3) were used to amplify the PLAC8 5′ end. PCR products were cloned into the pCR4-TOPO vector and transformed into One Shot TOP10 Escherichia coli using the TOPO TA Cloning kit (Life Technologies). DNA was isolated using PureYield Plasmid Miniprep System (Promega, Madison, WI), sequenced by the DNA Sequencing Core (Indiana University School of Medicine) and ProteinCT Biotechnologies (Madison, WI), and analyzed using MacVector software (Cary, NC).
Bisulfite Sequencing
DNA was bisulfite treated using EZ DNA Methylation-Direct kit (Zymo Research, Irvine, CA) per the manufacturer’s instructions. Bisulfite-treated DNA was amplified by PCR using ZymoTaq DNA Polymerase and primers listed in Supplementary Table 3. PCR products were cloned as described above. DNA was isolated from 8–12 clones and analyzed by sequencing. The nonconversion rate of cytosines to uracils was determined to be 0.5% (6 of 1,277 cytosines) using individual non-CpG cytosines. This nonconversion rate was representative of 21 clones from 10 different bisulfite conversions.
Western Blotting
Cells were lysed in radioimmunoprecipitation assay buffer containing mammalian protease inhibitor cocktail (Sigma). Equal protein amounts were separated by gel electrophoresis on precast gels (Life Technologies), transferred to nitrocellulose, and immunoblotted with antibodies to PLAC8 (ab122652, Abcam, Cambridge, MA, and HPA040465, Sigma), ALX1 (ab181101, Abcam), NOS3 (610296, BD Biosciences, San Jose, CA), or vinculin (VIN11-5, Sigma). Secondary antibodies conjugated to horseradish peroxidase were from Biorad (Hercules, CA). Blots were developed with Pierce Supersignal West Pico (ThermoFisher, Hanover Park, IL), exposed to film, scanned, and compiled in Photoshop CS5.1 (Adobe, San Jose, CA).
siRNA Transfection
Low-passage GDM-exposed ECFCs were transfected with short-interfering RNAs (siRNAs) using lipofectamine RNAiMAX reagent (Life Technologies) following the manufacturer’s instructions. Cells were transfected with either a nontargeting smart-pool siRNA (siControl) (cat. no. D-001810-10-05; ON-TARGETplus) or human PLAC8 siRNA (siPLAC8) (cat. no. J-020311-10; ON-TARGETplus). All siRNAs were purchased from GE Dharmacon (Lafayette, CO). Media was changed after 18–24 h, and cells were passaged 24 h later for proliferation, apoptosis, and senescence assays. PLAC8 expression was examined 3 days after transfection to confirm knockdown by Western blotting.
BrDU and 7-AAD Proliferation Assays
GDM-exposed ECFCs that were previously transfected with siControl or siPLAC8 were incubated with BrDU labeling reagent (Invitrogen) for 1 h. Cells were then trypsinized and stained using standard ethanol fixation and acid-denaturation protocols for BrDU (anti-BrDU mouse monoclonal conjugated with Alexa Fluor 488; Invitrogen) and 7-AAD (Life Technologies). Samples were analyzed using flow cytometry on an LSRII (Becton Dickinson, San Jose, CA) and FlowJo software (TreeStar, Inc., Ashland, OR). At least 10,000 events were collected per sample.
Senescence Assays
Transfected, GDM-exposed ECFCs were plated at a density of 10,000 cells per well of a six-well plate. After 3 days of culture, staining for senescence-associated β-galactosidase was performed to assess senescence as previously described (10). At least 100 total cells per well were scored, and the percentage of senescence-associated β-galactosidase–positive cells was calculated. Senescence was quantified in five independent transfection experiments using two different GDM samples.
Apoptosis Assay
ECFCs were treated with or without 84 μmol/L etoposide (Cayman Chemical, Ann Arbor, MI) for 24 h to induce apoptosis. Adherent ECFCs were collected by trypsinization and combined with nonadherent cells. Apoptosis was assayed using FITC-Annexin-V/Propidium Iodide kit following the manufacturer’s instructions (Biolegend, San Diego, CA). Cells were assayed on an LSRII Flow Cytometer and analyzed using FlowJo software.
Statistical Analyses
Data illustrated in graphs are mean ± SEM. Statistical analyses used are described in the figure legends. Pearson and Spearman correlation analyses were performed on normal and nonnormal distributions, respectively. Prism 6 (GraphPad Software, La Jolla, CA) was used for all statistical analyses. Significance was noted when P < 0.05. For DNA methylation and RNA expression correlations, P values were corrected for multiple comparisons by the Benjamini-Hochberg method (16).
Results
PLAC8 Is Increased in ECFCs From GDM Pregnancies
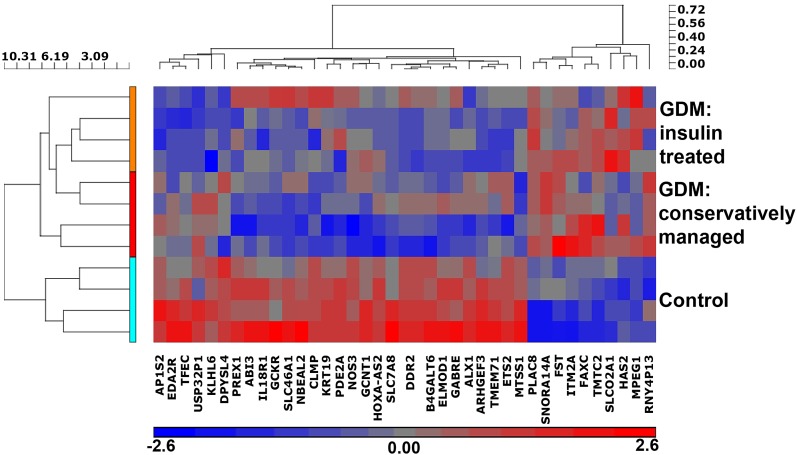
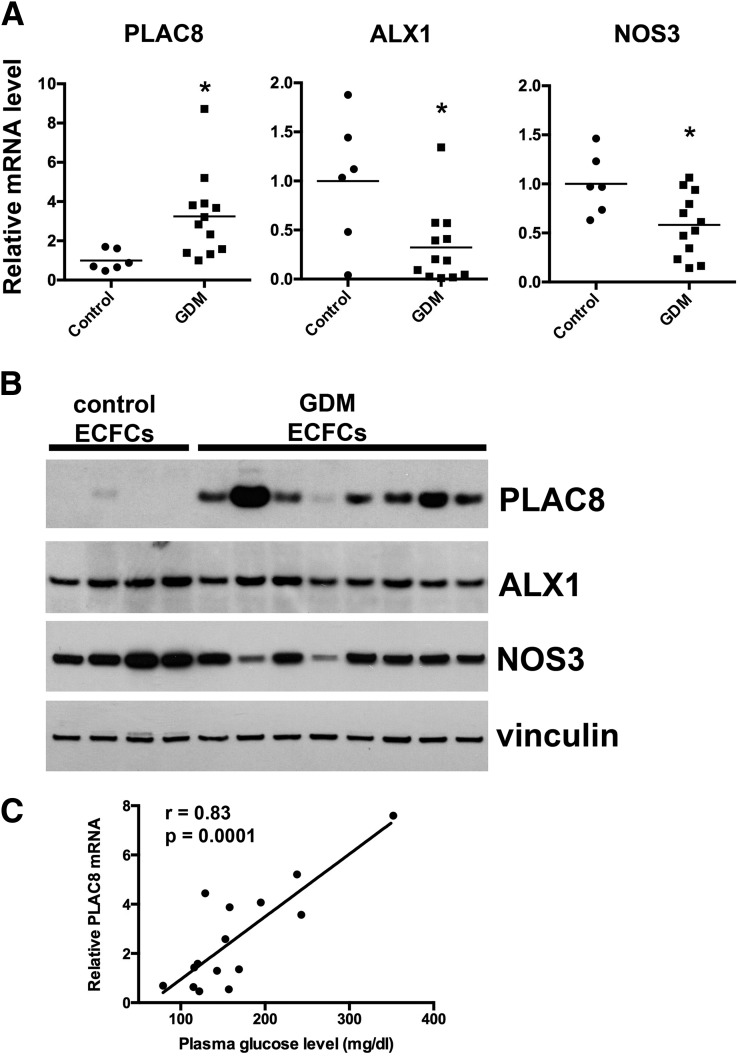
To identify genes in ECFCs that have altered expression after intrauterine exposure to GDM, we performed a microarray analysis. ECFC samples from both conservatively managed GDM patients (treated with diet and exercise) and insulin-treated GDM patients were included. Of the 28,000 genetic loci tested, 596 mRNAs were altered between control and GDM ECFCs (P < 0.01). More stringent criteria identified genes for further investigation by limiting analysis to genes that exhibited increased or decreased expression by at least 50%, with a P < 0.01. Figure 1 illustrates a hierarchical clustering analysis of the 38 genes that fulfilled the criteria, comparing control ECFCs with the two GDM groups independently. With use of this analysis strategy, 26 genes were differentially expressed between conservatively managed GDM and control ECFCs (7 increased and 19 decreased) (Table 1), and 15 genes were differentially expressed between GDM insulin-treated and control ECFCs (4 increased and 11 decreased) (Table 1). While there may be subtle differences in microarray data between the conservatively managed and insulin-treated GDM groups, no differences in ECFC function have been detected between these two groups (data not shown). Given this observation, we focused follow-up studies on gene products that were differentially expressed between combined GDM data and controls. qRT-PCR was used to validate the microarray results using gene-specific primers for 18 genes (Supplementary Table 4). Of the genes tested by qRT-PCR, 72% (13 of 18) had significantly altered mRNA expression (Supplementary Table 4). One highly upregulated gene was PLAC8, and one highly downregulated gene was ALX homeobox 1 (ALX1); both were confirmed by qRT-PCR (Fig. 2A). Endothelial nitric oxide synthase (NOS3) mRNA was also reduced in ECFCs from GDM pregnancies (Fig. 2A). Western blot analyses confirmed increased PLAC8 expression in GDM-exposed ECFCs with no detectable PLAC8 in control ECFCs (Fig. 2B). ALX1 and NOS3 were modestly decreased overall in GDM ECFC samples (Fig. 2B). Interestingly, PLAC8 mRNA levels positively correlated with maternal glucose levels during the screening glucose tolerance test (Fig. 2C) (r = 0.83, P = 0.0001).
Figure 1.
Intrauterine exposure to GDM induces altered mRNA expression in neonatal ECFCs. Thirty-eight genes in GDM-exposed ECFCs exhibited either increased or decreased expression by at least 50% compared with controls (P < 0.01). A hierarchical clustering analysis of the 38 genes is illustrated.
Table 1.
Genes with altered expression based on RNA microarray
| Gene symbol | GDM conservatively managed compared with control ECFCs |
|||
|---|---|---|---|---|
| Fold change | P | FDR | Gene name | |
| PLAC8 | 5.90 | 0.002 | 0.33 | Placenta-specific 8 |
| FST | 4.68 | 0.004 | 0.33 | Follistatin |
| SNORA14A | 3.05 | 0.003 | 0.33 | Small nucleolar RNA, H/ACA box 14A |
| ITM2A | 2.71 | 0.008 | 0.34 | Integral membrane protein 2A |
| FAXC | 2.37 | 0.010 | 0.34 | Failed axon connections homolog (Drosophila) |
| TMTC2 | 1.73 | 0.009 | 0.34 | Transmembrane and tetratricopeptide repeat containing 2 |
| RNY4P13 | 1.59 | 0.007 | 0.34 | RNA, Ro-associated Y4 pseudogene 13 |
| PREX1 | −1.50 | 0.010 | 0.34 | Phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| SLC46A1 | −1.54 | 0.001 | 0.33 | Solute carrier family 46 (folate transporter), member 1 |
| NBEAL2 | −1.57 | 0.003 | 0.33 | Neurobeachin-like 2 |
| GCNT1 | −1.61 | 0.005 | 0.33 | Glucosaminyl (N-acetyl) transferase 1, core 2 |
| ABI3 | −1.65 | 0.004 | 0.33 | ABI family, member 3 |
| GABRE | −1.68 | 0.008 | 0.34 | γ-Aminobutyric acid (GABA) A receptor, ε |
| HOXA-AS2 | −1.69 | 0.010 | 0.34 | HOXA cluster antisense RNA 2 |
| PDE2A | −1.71 | 0.009 | 0.34 | Phosphodiesterase 2A, cGMP-stimulated |
| IL18R1 | −1.74 | 0.009 | 0.34 | Interleukin 18 receptor 1 |
| B4GALT6 | −1.81 | 0.003 | 0.33 | β-1,4-galactosyltransferase 6 |
| MTSS1 | −1.83 | 0.001 | 0.26 | Metastasis suppressor 1 |
| TFEC | −1.86 | 0.006 | 0.33 | Transcription factor EC |
| SLC7A8 | −2.04 | 0.008 | 0.34 | Solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| GCKR | −2.28 | 0.004 | 0.33 | Glucokinase (hexokinase 4) regulator |
| NOS3 | −2.59 | 0.008 | 0.34 | Nitric oxide synthase 3 (endothelial cell) |
| DDR2* | −2.66 | 0.002 | 0.33 | Discoidin domain-containing receptor 2 |
| CLMP | −2.66 | 0.001 | 0.32 | CXADR-like membrane protein |
| KRT19 | −2.98 | 0.001 | 0.33 | Keratin 19 |
| ELMOD1 | −3.23 | 0.002 | 0.34 | ELMO/CED-12 domain containing 1 |
| Gene symbol | GDM insulin-treated compared with control ECFCs | |||
| Fold change | P | FDR | Gene name | |
| PLAC8 | 7.05 | <0.001 | 0.84 | Placenta-specific 8 |
| SLCO2A1 | 2.85 | 0.001 | 0.86 | Solute carrier organic anion transporter family, member 2A1 |
| MPEG1 | 1.86 | 0.004 | 0.87 | Macrophage expressed 1 |
| HAS2 | 1.84 | 0.006 | 0.87 | Hyaluronan synthase 2 |
| AP1S2 | −1.56 | 0.001 | 0.84 | Adaptor-related protein complex 1, σ 2 subunit |
| ARHGEF3 | −1.62 | 0.006 | 0.87 | Rho guanine nucleotide exchange factor (GEF) 3 |
| KLHL6 | −1.64 | 0.005 | 0.87 | Kelch-like family member 6 |
| DPYSL4 | −1.88 | 0.004 | 0.87 | Dihydropyrimidinase-like 4 |
| ETS2 | −2.05 | 0.009 | 0.87 | V-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| DDR2* | −2.46 | 0.002 | 0.87 | Discoidin domain-containing receptor 2 |
| TFEC | −2.69 | <0.001 | 0.84 | Transcription factor EC |
| TMEM71 | −2.89 | 0.006 | 0.87 | Transmembrane protein 71 |
| EDA2R | −2.94 | 0.004 | 0.87 | Ectodysplasin A2 receptor |
| USP32P1 | −3.01 | 0.006 | 0.87 | Ubiquitin-specific peptidase 32 pseudogene 1 |
| ALX1 | −6.72 | 0.009 | 0.87 | ALX homeobox 1 |
DDR2 was detected by two probe sets. Fold change is based on the average fold change. FDR, false discovery rate.
Figure 2.
PLAC8 is increased in GDM ECFCs, while NOS3 and ALX1 are decreased. A: qRT-PCR was performed to validate the results of the microarray analysis. Results were normalized to hypoxanthine phosphoribosyltransferase and to the mean control expression for each gene (n = 6 control and 12 GDM, *P < 0.05 by unpaired t test with Welch correction). B: Western blot analysis showed that PLAC8 was increased in most GDM ECFCs compared with controls. ALX1 and NOS3 were decreased in several GDM samples compared with controls. Vinculin is the loading control. C: Maternal plasma glucose levels in the glucose tolerance screen correlate with PLAC8 mRNA levels in neonatal ECFCs (r = 0.83 and P = 0.0001 by Pearson correlation).
PLAC8 Expression Correlates With CpG Methylation
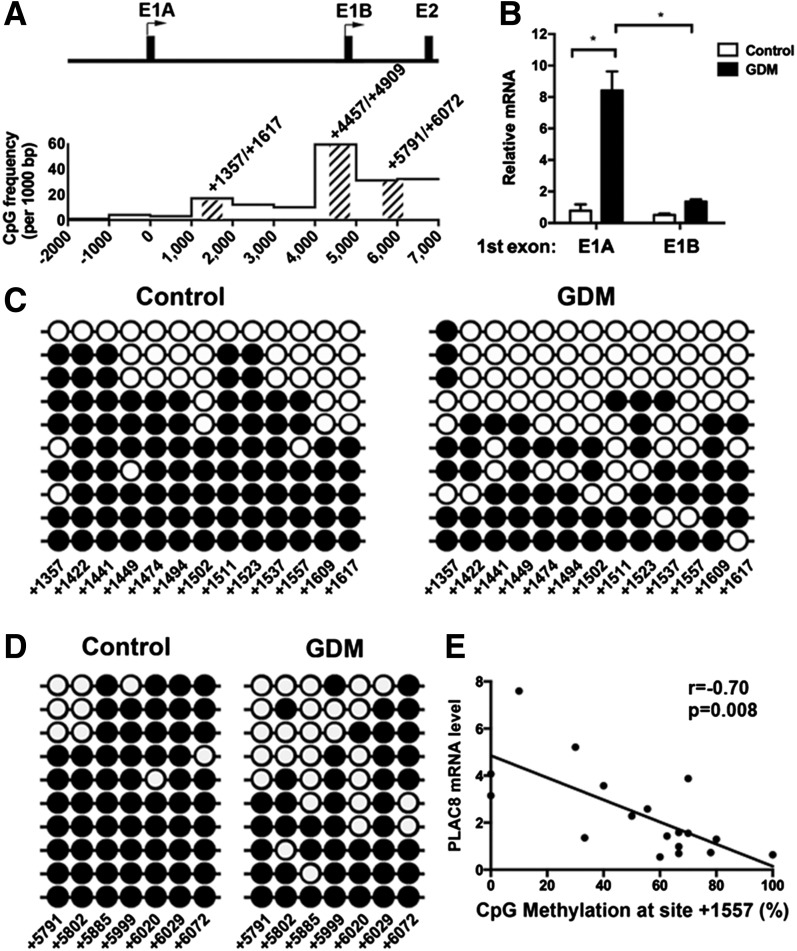
After confirming that PLAC8 was significantly upregulated in GDM-exposed ECFCs, we next evaluated the mechanism by which PLAC8 expression is regulated. We speculated that an epigenetic mechanism was mediating the long-term changes in gene expression of ECFCs from GDM pregnancies. Therefore, our next studies focused on identifying alterations in DNA methylation. Examination of the PLAC8 gene revealed two putative transcriptional start sites, denoted as exon 1A and exon 1B (E1A and E1B [Fig. 3A]). For determination of PLAC8 isoforms present in ECFCs, 5′-rapid amplification of cDNA ends (RACE) analysis was conducted. Amplification of cDNA using a reverse primer complementary to the sequence in the shared exon 3 of PLAC8 revealed two transcript types: one that contained E1A and another with E1B (data not shown). RT-PCR analysis was conducted to determine whether the levels of the two types of transcripts differed in control and GDM ECFCs. In control cells, there were approximately equal quantities of transcripts containing E1A and E1B (Fig. 3B). However, in GDM ECFCs, E1A transcripts were elevated compared with E1B (Fig. 3B), suggesting an increase in promoter activity for E1A. Given our hypothesis that alterations in DNA methylation were responsible for increased PLAC8 expression in ECFCs from GDM pregnancies, the CpG density of a 9-kb region beginning 2,000 nucleotides upstream of E1A (−2,000) and ending in intron 1 (7,000) was evaluated for CpG-rich areas that could serve a regulatory function (Fig. 3A). Based on the CpG densities, a targeted approach was used to examine the regions surrounding PLAC8 E1A and E1B with higher-than-average CpG density (>10 sites per 1,000 bp or 1.0%) (17). With use of this strategy, three areas were interrogated. (See shaded areas in Fig. 3A.) First, a CpG island was identified in a region encompassing E1B (4,457–4,909). Bisulfite sequencing demonstrated that the CpG island was unmethylated at 35 CpG sites in both control and GDM ECFCs (data not shown). As a control, genomic DNA from 293/HEK cells, which have minimal detectable PLAC8 by Western blotting, was assessed. In these cells, the PLAC8 CpG island was 90–100% methylated at all 35 CpG sites tested (data not shown). Together, these data suggest that methylation of the CpG island is not involved in upregulated PLAC8 expression in ECFCs, though it may be important in other cell types. Next, the methylation status of two CpG-dense regions surrounding the E1A and E1B start sites was evaluated by bisulfite sequencing (Fig. 3A) (1,357–1,617 and 5,791–6,072). Control ECFCs had consistently higher CpG methylation frequencies across the region spanning from 1,357 to 1,617, while GDM ECFCs were hypomethylated (Fig. 3C). Similarly, the CpG-rich region in intron 1 (5,791–6,072) was hypomethylated in GDM-exposed ECFCs compared with control cells (Fig. 3D). Together, these data are consistent with the hypothesis that decreased DNA methylation in putative regulatory regions of PLAC8 facilitates increased expression in ECFCs from GDM pregnancies. To more directly assess this hypothesis, we examined whether DNA methylation frequency at specific CpG sites inversely correlated with PLAC8 mRNA expression. These analyses demonstrated a negative correlation between DNA methylation frequency and PLAC8 mRNA expression at 17 CpG sites (Fig. 3E and Supplementary Table 5). For validation of our findings, a second cohort of control and GDM ECFCs was interrogated for PLAC8 mRNA expression and CpG methylation. Consistent with our original cohort, PLAC8 mRNA was upregulated in GDM-exposed ECFCs (control 1.0 ± 0.6; GDM 4.9 ± 1.2; P = 0.009). To test the correlation between PLAC8 mRNA levels and DNA methylation, we performed bisulfite sequencing of the region from 1,357 to 1,617. This region contained 12 of the 17 CpG sites whose methylation inversely correlated with PLAC8 mRNA levels in cohort 1. The results of this experiment verify a significant negative correlation between mRNA expression and methylation of 11 of 12 CpG sites (Supplementary Table 6). Thus, DNA methylation of a region in intron 1 of PLAC8 negatively correlates with mRNA expression, suggesting a possible regulatory function.
Figure 3.
Several CpG sites in the PLAC8 promoter and 1st intron are differentially hypomethylated in GDM-exposed ECFCs. A: The schematic shows the promoter and intron 1 of PLAC8, where there are two transcriptional start sites, E1A and E1B. Below the schematic is a graph illustrating the CpG frequency over each 1,000 bp region. The first start site E1A is denoted as “0” on the graph. PCR primers for bisulfite sequencing were generated to amplify CpG-rich regions at 1,357–1,617, 4,457–4,909, and 5,791–6,072, as shown by hash marks on the schematic. B: qRT-PCR identified PLAC8 mRNA variants present in control and GDM-exposed ECFCs. Two primer sets differentiating E1A or E1B were used to quantitate PLAC8 isoforms. Data were normalized to hypoxanthine phosphoribosyltransferase. n = 4 control, n = 7 GDM ECFC samples; *P < 0.001 by two-way ANOVA, followed by Šidák multiple comparisons. C and D: Bisulfite sequencing was performed on regions amplified by the primer sets shown in A and in Supplementary Table 1. C (1,357–1,617 region) and D (5,791–6,072 region) illustrate bisulfite sequencing data from representative control and GDM-exposed ECFC samples. ●, methylated CpGs; ○, unmethylated CpGs. Individual rows denote data from a single clone. The CpG site numbers are listed along the bottom. E: A correlation between CpG methylation frequency and PLAC8 mRNA expression is shown for 18 ECFC samples (n = 6 control and n = 12 GDM) by Pearson analysis. CpG methylation at site 1,557 was measured in 8–12 clones for each ECFC sample and is expressed as percent methylation. RNA expression was measured by qRT-PCR on parallel samples.
Previous studies identified several methylated CpG sites in the NOS3 promoter that regulate NOS3 expression (18). Thus, after observing differential methylation of PLAC8, we questioned whether altered DNA methylation was responsible for decreased NOS3 expression in ECFCs exposed to GDM in utero. Bisulfite sequencing of −209 to −51 bp upstream of the NOS3 transcription start site revealed that this region contained very low methylation frequencies in both control and GDM-exposed ECFCs (0–20%). Moreover, there was no correlation between CpG methylation and mRNA expression. In contrast, Jurkat cells, which have undetectable NOS3 mRNA, were 100% methylated at six CpG sites and 40–80% methylated at the other two sites in this region. These data indicate that while this region is important in regulating NOS3 expression between endothelial cells and other cell types, it is not responsible for the change in mRNA levels between control and GDM-exposed ECFCs.
Depletion of PLAC8 in GDM ECFCs Results in Decreased Proliferation and Increased Senescence
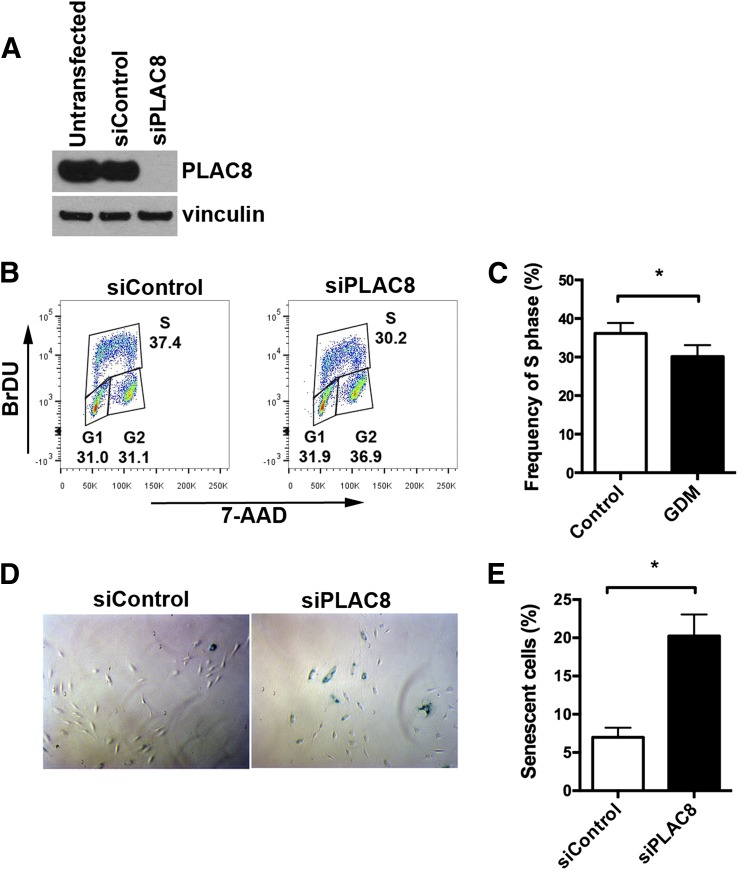
Previous studies showed that PLAC8 overexpression induces loss of cell cycle control, increased proliferation, and resistance to apoptosis (19). GDM-exposed ECFCs exhibit increased proliferation and resistance to senescence compared with control ECFCs (10). Therefore, we hypothesized that increased PLAC8 expression in GDM-exposed ECFCs may contribute to the aberrant proliferation and senescence observed in these cells. To test this hypothesis, we depleted PLAC8 from GDM-exposed ECFCs and evaluated the effect on proliferation, apoptosis, and senescence. GDM-exposed ECFCs were transfected with siControl or siPLAC8. Western blot analysis confirmed that the siControl-transfected ECFCs had no change in PLAC8 expression compared with untransfected controls and that the siPLAC8-transfected cells had effective depletion of PLAC8 (Fig. 4A). A 1-h, BrDU pulse of actively proliferating GDM-exposed ECFCs revealed a significant decrease in S-phase ECFCs that were transfected with siPLAC8 compared with siControl (Fig. 4B and C). These data suggest that PLAC8 overexpression may contribute to the hyperproliferative phenotype detected in ECFCs from GDM pregnancies. Since PLAC8 overexpression inhibits apoptosis (19), we next examined whether apoptosis was affected by reducing PLAC8 expression. These studies detected no differences in baseline or induced apoptosis in GDM-exposed ECFCs transfected with siPLAC8 or siControl (data not shown). However, ECFCs tend to undergo senescence rather than apoptosis in response to stress stimuli (20). Therefore, we speculated that increased PLAC8 expression in GDM-exposed ECFCs may participate in the resistance to senescence observed previously (10). GDM-exposed ECFCs transfected with siPLAC8 exhibited a dramatic increase in basal senescence compared with siControl-transfected cells (Fig. 4D and E). Together, these data suggest that PLAC8 overexpression in GDM-exposed ECFCs contributes to their abnormal phenotype by increasing proliferation and protecting from senescence.
Figure 4.
Depletion of PLAC8 reduces proliferation and increases senescence. PLAC8-specific siRNA was used to deplete PLAC8 from GDM-exposed ECFCs, and functional assays were performed. Transfection of an siControl was used as the control. A: Western blotting confirms efficient PLAC8 protein knockdown. The Western blot shows that the control siRNA did not affect protein levels. Vinculin is the loading control. B and C: Cell cycle analysis was conducted using flow cytometry. B: Dot plots from a representative experiment are shown. C: Quantitation of cells in S phase shows increased proliferation with PLAC8 depletion (n = 12 using five different GDM-exposed ECFC samples; *P < 0.05 by paired t test). D: Representative image of siControl- and siPLAC8-transfected GDM ECFCs stained for senescence-associated β-galactosidase (blue cells). E: Quantitation of senescent cells demonstrates increased senescence with PLAC8 depletion (n = 5 experiments using two different GDM-exposed ECFC samples; *P < 0.05 by paired t test).
Discussion
Developmental origins of cardiovascular disease are well established in humans and animal models (3–9,13,14,21). Elucidation of the mechanisms underlying disease predisposition is the current challenge for scientists and clinicians to develop innovative prevention and treatment strategies. This study provides strong evidence in neonatal endothelial progenitor cells that GDM exposure in utero leads to altered gene expression and that disrupted epigenetic regulation may contribute to aberrant expression of PLAC8.
Previous studies have identified global changes in DNA methylation of placenta and unfractionated cord blood cells after GDM exposure (22,23). Other studies examined these same tissues from GDM pregnancies for alterations in CpG methylation using a targeted gene approach (23–25) or unbiased screening (26–29). El Hajj et al. (23) examined the methylation status of 14 candidate genes and found reduced CpG methylation in GDM samples in regulatory regions of MEST (mesoderm specific transcript) and NR3C1, which encodes a glucocorticoid receptor. However, minimal differences were detected between GDM and control samples (4–7% in MEST and 2% in NR3C1), making it difficult to extrapolate biologic significance in the absence of functional data in placental or cord blood cells. A potential reason for small alterations in DNA methylation in GDM samples may be that heterogeneous cell populations from cord blood and placenta were used (23). Specific cell types have unique methylation patterns, in part to determine cell fate (30). Therefore, analyses of heterogeneous cell populations dilute the ability to detect meaningful changes in DNA methylation (23–28). Furthermore, if a disease state changes the proportion of cell types in an input population, then observed differences in methylation may be indirect. For example, neonates from GDM pregnancies display increases in nucleated red blood cells in their circulation (31). To circumvent this limitation, Cheng et al. (29) used a more homogeneous cell population, human umbilical vein endothelial cells, to determine whether GDM exposure impacts the proteome. In these studies, expression changes were identified in several proteins involved in redox signaling; however, epigenetic alterations were not detected in the two gene promoters examined (29). While this important study demonstrates a detrimental effect of GDM exposure on neonatal endothelial cells, it does not provide evidence that an epigenetic mechanism is involved.
The identification of ECFCs as endothelial progenitor cells that circulate in human peripheral blood and reside in the endothelium of vessel walls has expanded insight into vascular repair processes as well as postnatal angiogenesis and vasculogenesis (32). Under a variety of disease states, ECFC function is disrupted, which further contributes to the pathogenesis of vascular disease (33–37). The importance of ECFCs in maintenance of vascular health is highlighted by numerous clinical trials assessing the therapeutic potential of infusing ECFCs for cardiovascular diseases (38). Therefore, it is alarming that neonatal ECFCs exposed to GDM or pre-GDM in utero have significant impairments in function (10,39). This is not unique to diabetes exposure, as there is increasing evidence that intrauterine exposure to preeclampsia, obesity, growth restriction, and preterm delivery impair neonatal ECFC function and numbers as well (40–44). Therefore, it is paramount to elucidate underlying molecular mechanisms that may be exploited for future therapeutic benefit for these infants.
Our approach to understand the functional differences between control and GDM-exposed ECFCs was to conduct an unbiased microarray screen followed by validation of selected gene products and final functional assessment of PLAC8 in ECFCs. Control and GDM-exposed ECFCs exhibited modest differences in gene expression by microarray, which were subsequently verified by independent methods. These data are intriguing and suggest the possibility of defining a “molecular signature” that correlates with ECFC dysfunction, an approach that has been successful in driving the discovery of the molecular underpinnings of acute leukemias and exploited for prognostication and treatment decisions (45). Our finding that maternal hyperglycemia at GDM diagnosis directly correlates with PLAC8 expression in neonatal ECFCs provides rationale to pursue this ultimate goal. With this objective in mind, an important limitation of the current study is the likelihood of underestimating the number of gene products aberrantly expressed in GDM ECFCs owing to a relatively low sample size. In addition, our study populations in two independent cohorts had a high mean BMI in both control and GDM groups, which does not allow for the evaluation of a potential effect of maternal obesity on the gene expression profile of ECFCs. Therefore, it will be important to expand upon this data set using an increased number of samples from healthy–normal weight, healthy-obese, and GDM pregnancies to develop a robust molecular phenotype in ECFCs. However, the aberrantly expressed genes that were validated in ECFCs from GDM pregnancies lend themselves to exploration of the mechanisms responsible for altered expression and functional significance, similar to studies conducted for PLAC8.
A novel discovery from our work was that PLAC8 expression is highly dysregulated in GDM ECFCs, which was initially surprising, since PLAC8 has not been reported to be expressed in endothelial cells. PLAC8, which is also known as onzin, was originally identified as a placental-enriched protein (46). Subsequent studies showed that PLAC8 is expressed in epithelial cells, adipocytes, and hematopoietic cells (19,47–50). Although the precise endogenous biochemical function of PLAC8 is unclear, data suggest a role in regulating adipocyte differentiation, innate immune response, cell proliferation, and survival (19,49–51). Furthermore, recent studies demonstrate an important role of PLAC8 in promoting tumorigenesis through mechanisms involving proliferation, survival, autophagy, and epithelial-to-mesenchymal transition (19,47,48,52). However, no studies report expression or function of PLAC8 in endothelial cells, which may be because basal PLAC8 expression is negligible. Our data suggest that upregulated PLAC8 expression in GDM-exposed ECFCs contributes to the hyperproliferative phenotype previously reported (10), which is consistent with studies in other cell types (19). In addition, our data support PLAC8 overexpression as a protective mechanism for GDM-exposed ECFCs to avoid senescence. Given that hyperglycemia enhances ECFC senescence and impairs vasculogenesis, these findings suggest an adaptive response of fetal ECFCs to circumvent the untoward effects of a diabetic milieu.
To evaluate whether an epigenetic mechanism may be involved in the overexpression of PLAC8, the methylation status of the PLAC8 gene was interrogated. In GenBank, three transcript variants of PLAC8 are reported (variant 1: NM_001130716, variant 2: NM_016619, and variant 3: NM_001130715), though no information regarding transcriptional regulation of PLAC8 is available. The three PLAC8 isoforms differ only in the untranslated regions; thus, the gene products have identical amino acid sequences. Isoform 3 has a unique 3′-untranslated region and was minimally expressed in ECFCs (∼0.4 ± 0.3% of total PLAC8 mRNA). Isoform 2, which contains E1A, is highly upregulated in GDM ECFCs, while isoform 1, which contains E1B, is not changed in GDM ECFCs. Interestingly, differential methylation was detected in GDM-exposed ECFCs in the first intron of isoform 2. Moreover, the methylation status of several individual CpG sites in these regions negatively correlated with PLAC8 mRNA expression, suggesting a mechanistic link, possibly via altered transcription factor binding. Interrogation of ChIP-seq data from the ENCODE project suggests that the 5- to 6-kB region surrounding E1A and E1B of PLAC8 may have a role in regulating PLAC8 transcription, since an enrichment of transcription factor binding was observed in this region (53). Using these publicly accessible data, we found that several transcription factors bind near the PLAC8 transcription start sites including RUNX3, GATA3, EP300, TBP, RelA, MAX, PAX5, and IKZF1. Future studies will investigate whether the altered methylation observed in GDM-exposed ECFCs directly impacts PLAC8 transcriptional regulation. Our data suggest that intrauterine exposure to GDM may have induced epigenetic alterations in neonatal ECFCs that modified PLAC8 expression and ultimately ECFC function. Collectively, these findings provide the foundation for developing a molecular signature or a targeted biomarker to assess the impact of intrauterine GDM exposure on ECFCs so that novel interventions may be tested to prevent future endothelial dysfunction in offspring of mothers with GDM.
Article Information
Acknowledgments.The authors thank Dr. Jamie Case, Julie Mund, Matt Repass, Emily Sims (Indiana University Melvin and Bren Simon Cancer Center Angio BioCore), Dr. Paul Herring, Sarah Rust, and Cavya Chandra (Indiana University School of Medicine) for excellent technical assistance. The authors also thank Dr. Debbie Thurmond (Indiana University School of Medicine) and Dr. David Skalnik (Purdue University School of Science, Indianapolis, IN) for review and discussion of the manuscript and Elizabeth Rybak (Indiana University School of Medicine) for administrative support.
Funding. Funding for this study came from the National Institutes of Health (Bethesda, MD) R01 HL094725, U10 HD063094, and P30 DK090948 (to L.S.H.) and the Riley Children’s Foundation (Indianapolis, IN) (to L.S.H.). The microarray experiments were carried out using the facilities of the Center for Medical Genomics at Indiana University School of Medicine (Indianapolis, IN), which was initially funded in part by a grant from the Indiana 21st Century Research and Technology Fund and by the Indiana Genomics Initiative (INGEN). INGEN is supported in part by the Lilly Endowment (Indianapolis, IN).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. E.K.B., B.M.S., Z.V.N., F.A.B., C.M.H., C.R.G., K.M.V., and J.N.M. conducted the experiments and analyzed the data. E.K.B., B.M.S., and L.S.H. designed the studies and wrote the manuscript. L.S.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Part of this study was presented at the Pediatric Academic Societies and Asian Society for Pediatric Research Joint Meeting, Vancouver, Canada, 3–6 May 2014.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-1709/-/DC1.
References
- 1.Calkins K, Devaskar SU. Fetal origins of adult disease. Curr Probl Pediatr Adolesc Health Care 2011;41:158–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet 1989;2:577–580 [DOI] [PubMed] [Google Scholar]
- 3.Cho NH, Silverman BL, Rizzo TA, Metzger BE. Correlations between the intrauterine metabolic environment and blood pressure in adolescent offspring of diabetic mothers. J Pediatr 2000;136:587–592 [DOI] [PubMed] [Google Scholar]
- 4.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics 2005;115:e290–e296 [DOI] [PubMed] [Google Scholar]
- 5.Bunt JC, Tataranni PA, Salbe AD. Intrauterine exposure to diabetes is a determinant of hemoglobin A(1)c and systolic blood pressure in pima Indian children. J Clin Endocrinol Metab 2005;90:3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawlor DA, Lichtenstein P, Långström N. Association of maternal diabetes mellitus in pregnancy with offspring adiposity into early adulthood: sibling study in a prospective cohort of 280,866 men from 248,293 families. Circulation 2011;123:258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crume TL, Ogden L, Daniels S, Hamman RF, Norris JM, Dabelea D. The impact of in utero exposure to diabetes on childhood body mass index growth trajectories: the EPOCH study. J Pediatr 2011;158:941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Sloboda DM, Vickers MH. Maternal obesity and developmental programming of metabolic disorders in offspring: evidence from animal models. Exp Diabetes Res 2011;2011:592408 [DOI] [PMC free article] [PubMed]
- 9.Jawerbaum A, White V. Animal models in diabetes and pregnancy. Endocr Rev 2010;31:680–701 [DOI] [PubMed] [Google Scholar]
- 10.Blue EK, DiGiuseppe R, Derr-Yellin E, et al. Gestational diabetes induces alterations in the function of neonatal endothelial colony-forming cells. Pediatr Res 2014;75:266–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osborne-Majnik A, Fu Q, Lane RH. Epigenetic mechanisms in fetal origins of health and disease. Clin Obstet Gynecol 2013;56:622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinney SE, Simmons RA. Metabolic programming, epigenetics, and gestational diabetes mellitus. Curr Diab Rep 2012;12:67–74 [DOI] [PubMed] [Google Scholar]
- 13.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest 2008;118:2316–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinkhan EK, Fu Q, Wang Y, et al. Maternal hyperglycemia disrupts histone 3 lysine 36 trimethylation of the IGF-1 gene. J Nutr Metab 2012;2012:930364 [DOI] [PMC free article] [PubMed]
- 15.American College of Obstetricians and Gynecologists Committee on Practice Bulletins--Obstetrics. ACOG Practice Bulletin. Clinical management guidelines for obstetrician-gynecologists. Number 30, September 2001 (replaces Technical Bulletin Number 200, December 1994). Gestational diabetes. Obstet Gynecol 2001;98:525–538 [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B 1995;57:289–300 [Google Scholar]
- 17.Jabbari K, Cacciò S, Païs de Barros JP, Desgrès J, Bernardi G. Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene 1997;205:109–118 [DOI] [PubMed] [Google Scholar]
- 18.Chan Y, Fish JE, D’Abreo C, et al. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem 2004;279:35087–35100 [DOI] [PubMed] [Google Scholar]
- 19.Rogulski K, Li Y, Rothermund K, et al. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt-Mdm2-p53 pathway. Oncogene 2005;24:7524–7541 [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Herbert BS, Rajashekhar G, et al. Premature senescence of highly proliferative endothelial progenitor cells is induced by tumor necrosis factor-alpha via the p38 mitogen-activated protein kinase pathway. FASEB J 2009;23:1358–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonnold M, Tamayo E, Kechichian T, et al. The effect of prenatal pravastatin treatment on altered fetal programming of postnatal growth and metabolic function in a preeclampsia-like murine model. Am J Obstet Gynecol 2014;210:542.e1–e7 [DOI] [PubMed]
- 22.Nomura Y, Lambertini L, Rialdi A, et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci 2014;21:131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Hajj N, Pliushch G, Schneider E, et al. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes 2013;62:1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouchard L, Hivert MF, Guay SP, St-Pierre J, Perron P, Brisson D. Placental adiponectin gene DNA methylation levels are associated with mothers’ blood glucose concentration. Diabetes 2012;61:1272–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houde AA, St-Pierre J, Hivert MF, et al. Placental lipoprotein lipase DNA methylation levels are associated with gestational diabetes mellitus and maternal and cord blood lipid profiles. J Dev Orig Health Dis 2014;5:132–141 [DOI] [PubMed] [Google Scholar]
- 26.West NA, Kechris K, Dabelea D. Exposure to Maternal Diabetes in Utero and DNA Methylation Patterns in the Offspring. Immunometabolism 2013;1:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quilter CR, Cooper WN, Cliffe KM, et al. Impact on offspring methylation patterns of maternal gestational diabetes mellitus and intrauterine growth restraint suggest common genes and pathways linked to subsequent type 2 diabetes risk. FASEB J 2014;28:4868–4879 [DOI] [PubMed] [Google Scholar]
- 28.Ruchat SM, Houde AA, Voisin G, et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 2013;8:935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng X, Chapple SJ, Patel B, et al. Gestational diabetes mellitus impairs Nrf2-mediated adaptive antioxidant defenses and redox signaling in fetal endothelial cells in utero. Diabetes 2013;62:4088–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zilbauer M, Rayner TF, Clark C, et al. Genome-wide methylation analyses of primary human leukocyte subsets identifies functionally important cell-type-specific hypomethylated regions. Blood 2013;122:e52–e60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeruchimovich M, Mimouni FB, Green DW, Dollberg S. Nucleated red blood cells in healthy infants of women with gestational diabetes. Obstet Gynecol 2000;95:84–86 [DOI] [PubMed] [Google Scholar]
- 32.Ingram DA, Mead LE, Moore DB, Woodard W, Fenoglio A, Yoder MC. Vessel wall-derived endothelial cells rapidly proliferate because they contain a complete hierarchy of endothelial progenitor cells. Blood 2005;105:2783–2786 [DOI] [PubMed] [Google Scholar]
- 33.Güven H, Shepherd RM, Bach RG, Capoccia BJ, Link DC. The number of endothelial progenitor cell colonies in the blood is increased in patients with angiographically significant coronary artery disease. J Am Coll Cardiol 2006;48:1579–1587 [DOI] [PubMed] [Google Scholar]
- 34.Tan K, Lessieur E, Cutler A, et al. Impaired function of circulating CD34(+) CD45(-) cells in patients with proliferative diabetic retinopathy. Exp Eye Res 2010;91:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meneveau N, Deschaseaux F, Séronde MF, et al. Presence of endothelial colony-forming cells is associated with reduced microvascular obstruction limiting infarct size and left ventricular remodelling in patients with acute myocardial infarction. Basic Res Cardiol 2011;106:1397–1410 [DOI] [PubMed] [Google Scholar]
- 36.DiMeglio LA, Tosh A, Saha C, et al. Endothelial abnormalities in adolescents with type 1 diabetes: a biomarker for vascular sequelae? J Pediatr 2010;157:540–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duong HT, Comhair SA, Aldred MA, et al. Pulmonary artery endothelium resident endothelial colony-forming cells in pulmonary arterial hypertension. Pulm Circ 2011;1:475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sen S, McDonald SP, Coates PT, Bonder CS. Endothelial progenitor cells: novel biomarker and promising cell therapy for cardiovascular disease. Clin Sci (Lond) 2011;120:263–283 [DOI] [PubMed] [Google Scholar]
- 39.Ingram DA, Lien IZ, Mead LE, et al. In vitro hyperglycemia or a diabetic intrauterine environment reduces neonatal endothelial colony-forming cell numbers and function. Diabetes 2008;57:724–731 [DOI] [PubMed] [Google Scholar]
- 40.Sipos PI, Bourque SL, Hubel CA, et al. Endothelial colony-forming cells derived from pregnancies complicated by intrauterine growth restriction are fewer and have reduced vasculogenic capacity. J Clin Endocrinol Metab 2013;98:4953–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muñoz-Hernandez R, Miranda ML, Stiefel P, et al. Decreased level of cord blood circulating endothelial colony-forming cells in preeclampsia. Hypertension 2014;64:165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Luna R, Muñoz-Hernandez R, Lin RZ, et al. Maternal body-mass index and cord blood circulating endothelial colony-forming cells. J Pediatr 2014;164:566–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ligi I, Simoncini S, Tellier E, et al. A switch toward angiostatic gene expression impairs the angiogenic properties of endothelial progenitor cells in low birth weight preterm infants. Blood 2011;118:1699–1709 [DOI] [PubMed] [Google Scholar]
- 44.von Versen-Höynck F, Brodowski L, Dechend R, Myerski AC, Hubel CA. Vitamin D antagonizes negative effects of preeclampsia on fetal endothelial colony forming cell number and function. PLoS ONE 2014;9:e98990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shivarov V, Bullinger L. Expression profiling of leukemia patients: key lessons and future directions. Exp Hematol 2014;42:651–660 [DOI] [PubMed] [Google Scholar]
- 46.Galaviz-Hernandez C, Stagg C, de Ridder G, et al. Plac8 and Plac9, novel placental-enriched genes identified through microarray analysis. Gene 2003;309:81–89 [DOI] [PubMed] [Google Scholar]
- 47.Kinsey C, Balakrishnan V, O’Dell MR, et al. Plac8 links oncogenic mutations to regulation of autophagy and is critical to pancreatic cancer progression. Cell Reports 2014;7:1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C, Ma H, Wang Y, et al. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J Clin Invest 2014;124:2172–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimenez-Preitner M, Berney X, Uldry M, et al. Plac8 is an inducer of C/EBPβ required for brown fat differentiation, thermoregulation, and control of body weight. Cell Metab 2011;14:658–670 [DOI] [PubMed] [Google Scholar]
- 50.Ledford JG, Kovarova M, Koller BH. Impaired host defense in mice lacking ONZIN. J Immunol 2007;178:5132–5143 [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Rogulski K, Zhou Q, Sims PJ, Prochownik EV. The negative c-Myc target onzin affects proliferation and apoptosis via its obligate interaction with phospholipid scramblase 1. Mol Cell Biol 2006;26:3401–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMurray HR, Sampson ER, Compitello G, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature 2008;453:1112–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raney BJ, Cline MS, Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update). Nucleic Acids Res 2011;39:D871–D875 [DOI] [PMC free article] [PubMed] [Google Scholar]