Abstract
The development and maintenance of the many different cell types in metazoan organisms requires robust and diverse intercellular communication mechanisms. Relatively few such signaling pathways have been identified, leading to the question of how such a broad diversity of output is generated from relatively simple signals. Recent studies have revealed complex mechanisms integrating temporal and spatial information to generate diversity in signaling pathway output. We review some general principles of signaling pathways, focusing on transcriptional outputs in Drosophila. We consider the role of spatial and temporal aspects of different transduction pathways and then discuss how recently developed tools and approaches are helping to dissect the complex mechanisms linking pathway stimulation to output.
Keywords: signaling pathways, signaling dynamics, crosstalk
Signaling pathways have complex effects on cellular output
The development and homeostasis of multicellular organisms requires the coordinated activity of many different cell types. Dependable mechanisms allowing communication between cells within and between tissues are required to ensure the correct assignment of cell types during development and repair. Strikingly, although hundreds of cell types exist, relatively few signaling pathways have been identified [1], raising the question of how such a limited number of pathways can provide sufficient information to produce and maintain the diversity of cell types present in metazoans.
One aspect of signaling pathway function is that stimulation of a given pathway does not have a predefined outcome. This is illustrated by countless examples throughout development where activation of one pathway can lead to proliferation, senescence, differentiation (into multiple cell types), morphological changes, or cell death, with little obvious difference between signaling inputs in each case. However, despite this repertoire of responses, the correct outcome is invariably achieved.
Elucidating the mechanisms responsible for this variability has proved to be far from simple, partly due to the complexity of the transduction mechanisms used by signaling pathways. Furthermore, signaling pathways do not act in isolation, with different pathways sharing components and an increasing number of interactions being identified between components of different pathways. Finally, although activation of a signaling pathway has historically been considered as a binary switch, it is now clear that the dynamics of signaling pathway activation and transduction play important roles in determining signaling outcome. After reviewing some general principles of signal transduction, we describe here the importance of understanding signaling dynamics and context to tease apart the complex mechanisms leading to the selection of appropriate signaling outputs.
Signaling context and cellular history generate diversity in outputs
One of the purposes of intercellular signaling systems is to produce different transcriptional profiles in different cell types. The simplest way to achieve this, in theory, would be to induce each cell type with a different signaling pathway, each regulating different target genes. However, the discrepancy between the number of cell types and the number of signaling pathways indicates that this mechanism cannot explain the diversity of cell types identified. Signaling through different pathways does however lead to diverse transcriptional responses, such that the differential use of these pathways goes some way towards generating different cell types. The basic mechanism by which this occurs is relatively simple, with different signaling pathways regulating different transcription factors [1] (Figure 1, Table 1). For example, the BMP (bone morphogenetic protein) pathway regulates Mad, and the JNK (Jun N-terminal kinase) pathway regulates Jun and Fos.
Figure 1.
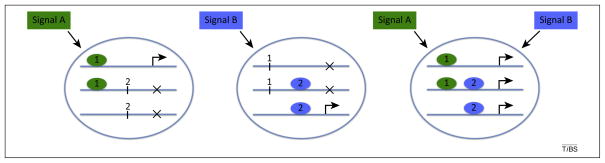
Signaling pathways produce distinct transcriptional outputs. Signaling pathways can produce distinct transcriptional outputs by binding to the regulatory sequences of different genes. Oval shapes labeled 1 and 2 represent transcription factors regulated downstream of signals A and B, respectively, and their effects on three classes of genes (blue lines) are illustrated. Arrows indicate gene activation and crosses represent the lack of activation. In some cases a single signal may be sufficient; in the first two examples, binding of transcription factor 1 or 2 is sufficient to activate the top or bottom target gene respectively. In other cases combinatorial inputs may be required for activation; in the third example, both transcription factors must be bound for activation of the middle gene. This leads to diversity of signaling outputs depending on the signaling contexts in which a pathway is activated.
Table 1.
Key components of Drosophila canonical signaling pathways
| Pathway | Receptor | Ligand | Transcription factor |
|---|---|---|---|
| Hormone receptors | e.g., EcR | e.g., Ecdysone | EcR |
| Notch | Notch | Delta, Serrate | Su(H) |
| JAK STAT | Domeless | Upd, Upd2, Upd3 | STAT92E |
| RTK | EGFR | Spitz, Kerren, Gurken, Vein | Pointed, Yan, Cic |
| FGFR (breathless) | Branchless, Heartless, Thisbe, Pyramus | Pointed,Yan | |
| InR | Dilp1-Dilp7 | Pointed, Yan, Foxo | |
| PVR | Pvf1-3 | Pointed,Yan | |
| Torso | Trunk, PTTH | Pointed,Yan | |
| ALK | Jelly belly | Pointed,Yan | |
| Sev | Boss | Pointed,Yan | |
| Hh | Patched | Hh | Ci |
| TGFβ (BMP) | Thickveins, Saxophone, Baboon, punt | Dpp, Gbb | Mad, Medea, Smox |
| Hippo | Fat | Dachsous | Yorkie |
| NF-κB | Toll | Spatzle | Dorsal |
| JNK | Eiger | Wengen | Jun, Fos |
| Wnt | Fz, Fz2, Fz3 | Wg, Wnt1-10 | Armadillo, Pan (TCF) |
Given that the differences in the transcriptional outputs between signaling pathways are insufficient to produce all of the diversity of responses, other mechanisms must exist that further diversify the outputs from signaling pathways. One way in which this occurs is via the combined effects of multiple signals, which generally produce non-additive effects compared to activation of the pathways in isolation [2–4]. Therefore, the signaling context in which pathway activation occurs can lead to regulation of a subset of genes distinct from activation of each pathway alone (Figure 1). For example, correct expression of the pax2 gene in the Drosophila eye requires direct inputs from Su(H) and Pnt, that are downstream of the Notch and EGFR (epidermal growth factor receptor) pathways, respectively, neither of which is sufficient alone [5].
Another way in which context can alter the output of signaling pathways is through the presence or absence of specific transcription factors. This is predetermined by the history of the cell, including previous signaling events, and provides a transcriptional background in which a signal is received. Therefore, integration of signaling pathway transcription factors with context-specific transcription factors can alter the response (Figure 2).
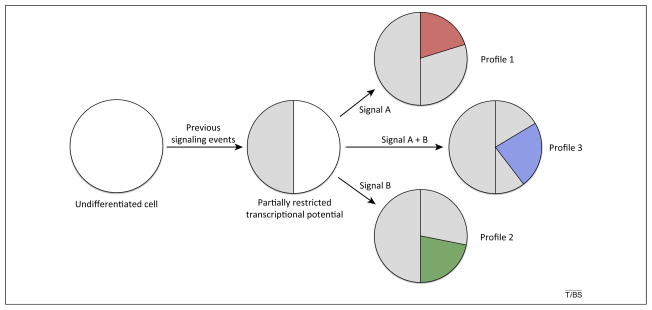
Figure 2.
Signaling pathway output depends on context. Circles represent the transcriptional potential of cells, which is initially unrestricted (white circle). As cells experience signaling events, the transcriptional potential becomes partially restricted (gray shading), providing a contextual background in which subsequent signaling is interpreted (i.e., the history of the cell). Signaling pathways then act on this limited set of accessible genes to further define the transcriptional profile (signals A, B, or A + B lead to profiles 1, 2, or 3 respectively). As illustrated in Figure 1, different signaling pathways or combinations of pathways will lead to distinct but possibly overlapping transcriptional profiles (red, blue, and green shading).
One example of this is the interaction between Notch and EGFR signaling pathways in different cell types within the developing wing disc. Notch and EGFR pathways have an antagonistic relationship in the wing pouch, with EGFR promoting wing vein formation and Notch inhibiting it [6–12]. EGFR activity in the vein tissue stimulates expression of Delta [6], which activates Notch signaling in the neighboring intervein tissues. Notch, in turn, inhibits the expression of EGFR components including argos and rhomboid in the intervein tissues [6,12]. This leads to exclusive activity of the two pathways in their respective tissues and is essential for correct patterning of the wing veins. However, a recent investigation into the role of Notch signaling in the adult muscle progenitor cells (AMPs) in the wing disc identified a different interaction between these pathways, with Notch signaling activating argos and the two pathways playing cooperative roles in maintaining the undifferentiated state of the cells [13]. How, then, could Notch inhibit argos in intervein cells and activate it in AMPs? Further investigation into the regulation of argos by Notch in these two contexts demonstrated that different enhancer sequences are used in each tissue. One mediates direct activation of argos by Su(H) in the AMPs, whereas the other mediates its indirect repression via HLHmβ [14], a direct target of Notch signaling in the wing pouch. These results suggest that enhancers are selected based on the expression of tissue-specific transcription factors that modulate enhancer accessibility. In cultured cells derived from the AMPs, expression of the transcription factor Twist is required for Su(H) binding to the relevant argos enhancer, and over-expression of Twist in the wing pouch can convert the regulation of argos downstream of Notch from repression to activation [14,15]. A similar factor (Vvl) was identified that may play a role analogous to that of Twist in regulating enhancer activity in the wing pouch [14].
It is clear that differential regulation of transcription factors by signaling pathways can only explain a small subset of the observed range of outputs. Therefore, to improve our understanding of signaling output we must consider both the pathway used and the context in which the signal is received, including both the activity of other signaling pathways and the presence of other transcription factors.
Complexity of signaling mechanisms
Given that a large determinant of signaling output appears to be the context of signaling, rather than the properties of the signaling pathway itself, this raises the question of what the functional differences between pathways are and why different pathways are used in different situations. One possibility is that the different signaling mechanisms used by each pathway enable different modes of signaling rather than different signaling outputs per se (Figure 3).
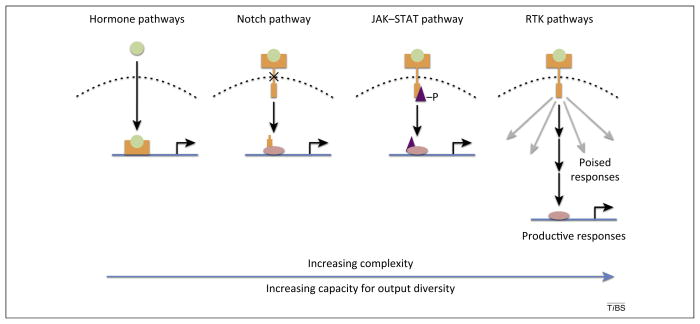
Figure 3.
Four classes of signaling pathways with different transduction mechanisms. Signaling pathways can be broadly classified based on the complexity of their signal transduction mechanisms. Hormone pathways are relatively simple, with few components. A ligand (green) interacts with a receptor (orange) that also functions as a transcription factor, thereby generating a direct link between the signal and a transcriptional effect. The Notch pathway illustrates a more complex class of signaling, with the receptor (orange) and transcription factor (pink) acting as separate components. The JAK/STAT (Janus kinase/signal transducer and activator of transcription) pathway has a further increase in complexity, with the recruitment of a signal transduction component (purple) between the receptor and transcription factor. Regulation of this transduction component by phosphorylation generates an additional level at which cross-regulation with other pathways can occur. Finally, RTKs (receptor tyrosine kinases) are the most complex class of signaling pathways, with multiple parallel transduction pathways. Many of these pathways are likely ineffective in a given context (gray arrows), and productive responses (black arrows) are defined by the components present (history of the cell) and inputs from other pathways (crosstalk). As signaling pathway complexity increases, opportunities for crosstalk with other pathways increase, as well leading to potentially new outputs.
The simplest pathways involve hormone receptors that function as transcription factors; they are activated upon binding of ligands that can enter the cell with no requirement for signal transduction machinery [16,17]. This mechanism provides direct regulation of gene expression but offers little control over signaling range or output diversity. In many cases the purpose of a signal is to alter gene expression in a small number of localized cells, requiring a limited signaling range and therefore more complex pathway organization. In addition, by limiting range of signaling, the same pathway can be used simultaneously to regulate multiple events in different locations, therefore reducing the number of pathways required.
Notch is one example of a pathway allowing a highly restricted range of signal transmission [18–21]. In this case, both the ligand (Delta or Serrate in Drosophila) and the receptor (Notch) are membrane-bound, limiting the range to immediately neighboring cells. Following ligand binding, the intracellular domain of Notch is cleaved and travels to the nucleus where it interacts with a transcription factor of the CSL family [Su(H) in Drosophila] to regulate gene expression. This simple signal transduction mechanism provides tight spatial control over pathway activation while maintaining a close link between pathway stimulation and transcriptional effects. This mode of signaling therefore allows a compromise between directly affecting transcription and providing more control over the signaling event by including potentially regulatable events such as ligand presentation, receptor cleavage, internalization, and trafficking.
Other pathways such as JAK/STAT (Janus kinase/ signal transducer and activator of transcription) have an additional step in signal transduction. Ligand-mediated receptor activation leads to recruitment of JAK. Activated JAK phosphorylates the transcription factor STAT, which regulates transcription [22–24]. This additional step allows the possibility of alternative outputs because JAK may phosphorylate targets other than STAT. Furthermore, additional regulation of the pathway can occur by altering the expression or activity of JAK. As pathway transduction mechanisms become more complex the opportunities for output diversification increase, as do the possible mechanisms of crosstalk between pathways. Other pathways, including Hh and BMP, are also reminiscent of this mode of signaling, with relatively simple transduction cascades [25,26].
RTK (receptor tyrosine kinase) and Wnt pathways have a further increase in complexity [27–29]. In the case of RTKs, the transmembrane tyrosine kinases regulate complex cascades of signal transduction components that include many other kinases. These pathways therefore have many opportunities for crosstalk and integration with other pathways. Furthermore, the branching cascade of transduction pathways can result in many different outputs, whether regulating different transcription factors or directly altering effector proteins via phosphorylation. It is therefore unlikely that each transduction pathway is used for every signaling event. Instead, only a subset of network components are likely to be expressed and activated in each context, resulting in defined outcomes depending on the history of the cell and the signaling status of other pathways. In addition, the presence of seemingly redundant components in complex signal transduction networks allows different modes of information processing and thus further response diversity. For example, a recent study demonstrated that the apparently redundant NFAT1 and NFAT4 transcription factors respond differently to changes in ligand dynamics, leading to different effects on pathway output [30]. RTK pathways likely provide not only the greatest range of possible outputs but also the greatest potential for integration of other signals to regulate those outputs. Given the likelihood that most transduction pathways downstream of RTKs are ineffective in each context, it is possible that crosstalk between transduction networks will profoundly affect the outputs of signaling by regulating the selection of these pathways.
For each of the different pathway structures described above, crosstalk is likely to occur at different levels (Figure 4). For example, because all pathways eventually regulate transcription factors, integration of these transcriptional regulatory signals can occur at the enhancer level for all pathway types. However, the more complex pathways such as RTKs have the additional potential to regulate directly components of other pathways via phosphorylation. In addition, recent studies have demonstrated that pathway components, including ligands, are often targets of other signaling pathways [2,4,13]. This raises the further possibility that sequential crosstalk may occur whereby activation of a primary pathway leads to production of a ligand, or a central component, for a secondary pathway and hence a second signaling event. When considering systems where sequential pathway regulation may occur, it is important to be aware of the limitations of current approaches used to investigate signaling systems. For example, the majority of in vivo assays used to study signaling pathways are performed on populations of cells and have low temporal resolution [31]. Therefore, care must be taken when assessing whether signals are acting on the same cells or different cells, and whether pathways are activated simultaneously or sequentially.
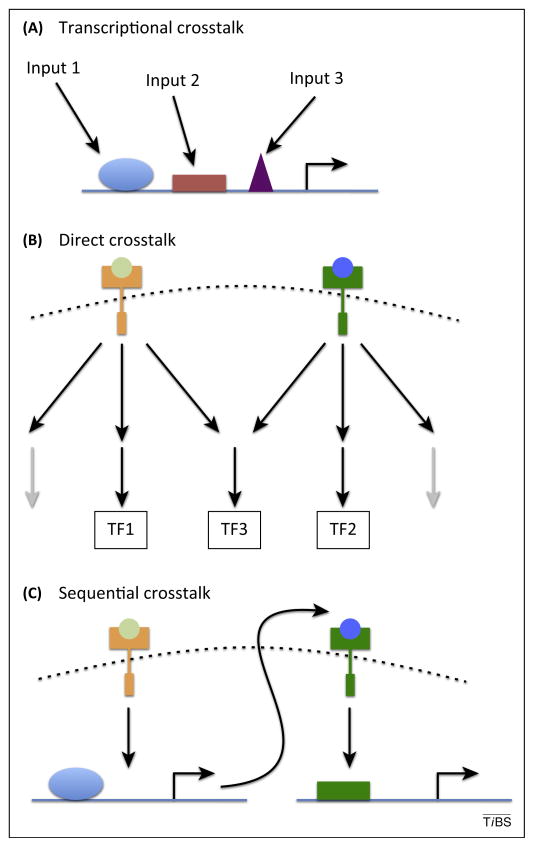
Figure 4.
Mechanisms of crosstalk. Depending on the category of pathways used, communication between pathways can occur at different levels. (A) Integration of transcription factors (blue, red, and purple shapes) at the enhancer level (all pathways). (B) Direct crosstalk between components of the transduction machinery can lead to activation of different transcription factors (TF) and/or the use of different transduction pathways (black arrows represent active transduction pathways, gray arrows represent inactive transduction pathways). This is more likely with increased pathway complexity. (C) Sequential activation of pathways (possible for all pathways). Note that sequential activation of pathways can be difficult to distinguish from simultaneous signaling, depending on the assays and tools used to monitor pathway activity.
Aside from differences in the opportunity for pathway crosstalk, it is also likely that the different mechanisms of stimulation make pathways suitable for different roles. Indeed, the lack of spatial restriction on hormone signaling pathways makes them well suited for regulating organism-wide responses such as the control of developmental timing. For example, the ecdysone pathway is used to signal particular stages of development such as larval molting and initiation of pupation [32]. Although they do not have spatial resolution in isolation, hormone pathways can integrate spatial information from other pathways to generate appropriate responses both in space and time [33–35]. By contrast, the structure of the Notch pathway results in an extremely restricted range of signaling that is limited to adjacent cells. This makes the Notch pathway ideal for generating differences between neighboring cells, such as those emerging during lateral inhibition, dorsal–ventral boundary formation in the wing disc, or asymmetric division of neuroblasts [18]. RTK, Hh, and Wnt pathways have an increased range of signaling that is determined by the ligand. This can result in signaling over large distances, exemplified by the Wg morphogen gradient in the wing disc, which extends many cell diameters from the signal-producing cells [36]. Finally, the JAK/STAT pathway offers multiple possibilities for signaling range. Two of the ligands in Drosophila (Upd and Upd3) interact with the extracellular matrix, limiting diffusion and resulting in short-range signals. Upd2, however, is able to signal over larger distances [37], suggesting that this pathway may be suitable for signaling over both short and long ranges. Interestingly, in adult flies, Upd2 has been proposed to act as a leptin-like hormone produced from the fat body and to regulate the activity of GABAergic neurons in the brain [38].
The profound differences in signaling pathway structure suggest that they may have different potential for integrating contextual information to generate output diversity, and may be best suited to very different roles in terms of spatial and temporal information transfer.
Dynamics of signaling
In addition to the context of signaling, the intensity, duration, and fluctuation of signaling pathway stimulation are important in determining the eventual output from the downstream transduction network. Improving our understanding of how these differences in input are converted into distinct outputs will be essential to unraveling the mechanisms of cell fate decisions.
One example of differential use of ligand concentration is with respect to morphogen gradients. In the classic view, a ligand is produced from a localized source and then diffuses away to form a gradient with high ligand levels near the source and lower levels further away. For example, the Spätzle ligand forms a gradient across the dorsal-ventral axis of the Drosophila embryo, resulting in graded translocation of the transcription factor Dorsal to the nuclei of responding cells. This gradient results in the induction of multiple different cell types depending on the level of pathway activation [36,39]. The mechanism of this patterning process relies at least partly on the presence of high- or low-affinity Dorsal binding sites at target genes. Thus, genes with low-affinity sites will be bound only when nuclear Dorsal is high, and genes with high-affinity sites will be regulated over a broader range [40–42]. Similar morphogen-based mechanisms are used in vertebrate systems such as dorsal–ventral patterning in zebrafish and patterning of the mammalian brain [43–45].
The topology of the signaling network is crucial for understanding how ligand dynamics affect signaling outputs. The presence of network motifs such as feed-forward and feedback loops enable a signaling pathway to respond selectively to particular aspects of a dynamic signal, and therefore affect the response. The properties of network motifs have been reviewed elsewhere [46–48], and we will therefore only describe a few general examples and discuss how they may affect the encoding of signaling dynamics.
A common network motif is a feedback loop where a downstream component of a pathway activates (positive feedback) or represses (negative feedback) an upstream component. Positive feedback can be used to control the response of a pathway to different stimulation durations by locking the system into either an on- or off-state following a transient signal [49,50]. Positive feedback is also involved in the interpretation of signal strength such as in morphogen gradients by converting graded stimulation into all-or-none responses [39,51,52]. Negative feedback, by contrast, can cause a change in the dynamics of signaling because activation of the pathway subsequently leads to repression of the same pathway. This may have several different effects depending on the kinetics of the feedback loop. For example, negative feedback can speed up responses, increase input dynamic range, or filter out noisy signals [46,53,54]. The presence of such motifs may mean that continuous stimulation causes a short pulse of pathway activity followed by a sustained attenuation of the signal or multiple pulses of activity [55–58].
A second type of network motif is the feed-forward loop. In this case, an upstream component regulates a downstream component by two parallel routes. These routes may act in the same manner (both activatory or both repressive), forming a coherent feed-forward loop, or instead activate and repress the downstream component respectively, forming an incoherent feed-forward loop. Coherent feed-forward loops may lead to a delay in activation or deactivation, allowing modification of the length of response to a transient signal. By contrast, incoherent feed-forward loops may cause a pulse of activation or repression, depending on which route is fastest, allowing a transient response to a sustained signal [46,59–61].
Although usually considered within transduction networks, motifs such as feed-forward loops have also been identified at the transcriptional level following signaling pathway activity. For example, as described earlier in this Review, the Notch and EGFR pathways have a cooperative role in the AMPs associated with the developing wing disc [13]. However, activation of Notch in cultured cells derived from the AMPs resulted in transcription of both egfr and the EGFR pathway inhibitor argos, forming an incoherent feed-forward loop [13]. Interestingly, these two targets were activated with different dynamics [62], with a transient increase in argos, and a more sustained activation of egfr. In this case, it is possible that the initial response to Notch signaling is a pulse of EGFR repression followed by a more sustained activation, therefore only leading to EGFR pathway stimulation if the Notch signal is sustained.
Although the temporal aspects of stimulation and information processing have profound effects on the outcome of signaling, these processes are poorly understood. This is partly due to the limited set of suitable tools to study signaling dynamics, and new approaches will be necessary to further our knowledge of the dynamic nature of signaling pathways.
Analysis of signaling dynamics in cultured cells
The majority of previous in vivo investigations into the function of signaling pathways have relied on tools and readouts with low temporal resolution [31]. For example, pathways have been activated using methods such as heat-shock induction or the Gal4/UAS system. Therefore, the exact time at which pathway activity is initiated is unknown, and could be several hours following the initial treatment. Furthermore, stimulation of one signaling pathway often results in the secondary activation of other pathways via crosstalk. Therefore, the outputs measured following these stimulations are likely to include both primary and secondary effects, and these cannot be easily distinguished.
To distinguish between primary and secondary effects, methods for signaling pathway activation with higher temporal resolution are required. Several methods have been developed for this purpose, including the use of chemicals or small molecules to regulate specific pathways, and genetically encoded optogenetic tools [31,63]. However, such systems are generally only applicable in cultured cells. For example, several studies have activated the Notch signaling pathway via chelation of calcium ions from the culture media using chemicals such as EDTA (ethylenediaminetetraacetic acid) [13,62,64–67]. This method provides extremely high temporal resolution of pathway activation. Similarly, purified ligands are available for some pathways (e.g., insulin, EGF, Hh), which can be used in a similar manner.
Optogenetic tools can also be used to provide high temporal resolution of signaling pathway stimulation [68,69]. One study used an optogenetic switch based on the Phy–PIF system to regulate signaling through the Ras pathway [70]. The system consists of two components that dimerize when stimulated with red light, but dissociate in the presence of far-red light. The system was designed such that one component (Phy) was localized to the cell membrane and the other (PIF) fused to a RasGEF, which is able to activate Ras when membrane-localized. Therefore stimulation with red or far-red light could be used to control activation of the Ras protein. In combination with a blue fluorescent reporter protein fused to the downstream kinase Erk, stringent temporal control of pathway activity and measurement of output were possible.
Early studies showed that different RTK ligands cause different signaling outputs from the Ras transduction cascade in PC-12 cells [71–73]. Stimulation with nerve growth factor (NGF) resulted in differentiation, whereas EGF caused proliferation and these differences were linked to sustained or transient activity of the downstream MAPK, respectively. Further studies suggested that this difference was caused by the use of different network motifs, with EGF activating a negative feedback loop involving MAPK, and NGF activating of a positive feedback loop [57]. Using optogenetic tools, Toettcher et al. demonstrated that the Ras/Erk module responds quantitatively to a wide range of stimulation intensities and dynamics [70]. In addition, proteomic analysis was used to identify downstream components that respond only to transient or sustained stimulation. Strikingly, the STAT3 transcription factor was induced non-autonomously only by sustained pathway activation, indicating that sustained MAPK activation leads to crosstalk with the JAK/STAT pathway, whereas transient stimulation does not.
Another study treated cultured cells with variable intensity, duration, and periodicity of ligand exposure to study dynamic multiplexing within the insulin signaling network [58]. By measuring four different outputs within the transduction network (phosphorylation states of Akt, S6k, and Gsk3β, and transcription of G6Pase), different downstream components were found to respond to different aspects of the initial stimulation. For example, transient or sustained stimulations with insulin resulted in transient or sustained phosphorylation-induced activation of Akt, but two of the downstream components selectively responded to transient (S6k) or sustained (G6Pase) stimulations. Furthermore, these selective responses could be explained by the kinetics of the molecules involved and the network structure including feed-forward loops.
The above examples illustrate some of the methods that have been developed to allow the study of signaling pathway dynamics. However, the generation of a broader toolkit will be necessary to allow deeper understanding of these systems in a wider range of contexts.
Future study of signaling pathway organization
Studies of signaling dynamics in cultured cells have demonstrated the importance of considering temporal variations in ligand levels. However, current tools are insufficient to perform similar assays in vivo. For example, the current tools for pathway activation (e.g., Gal4/UAS or heat-shock) generally result in a delay of several hours between initiation of treatment and onset of the signal. In addition, these tools allow little control over levels of induction or signal termination, and are therefore inappropriate for fine-scale temporal pathway control. Similar to the tools for pathway activation, the tools used as readouts of pathway activity in vivo also have low temporal resolution. For example, a GFP reporter protein is likely to persist for some time after transcription has ceased, and therefore is not an accurate representation of pathway activity. To overcome this problem, unstable fluorescent reporters have been developed [74,75], but these have the disadvantage that expression levels are low and therefore sensitivity is low. An alternative method to improve temporal readout is to analyze the rate of change of fluorescence from fast-folding, highly stable reporters, as has been done in bacterial systems [76,77].
To further our knowledge of signaling dynamics, it will be important to develop tools for the manipulation and readout of pathway activity with high temporal resolution and high sensitivity. Furthermore, given the large repertoire of outputs from signaling pathways and crosstalk between them, a wide range of reporters, that can be multiplexed to simultaneously analyze multiple pathways while avoiding differences in context, are needed.
Concluding remarks and future directions
Signaling pathways can produce a wide range of different outputs from apparently similar signals, but the mechanisms regulating this are poorly understood. To understand this process, several factors must be considered–including the signaling pathway used, the status of other signaling pathways, the context in which a signal is interpreted, the temporal dynamics of the signal, and how those dynamics are processed by the transduction machinery.
Gaining a better understanding of how signaling pathways use context and dynamic information to determine output is necessary to determine the complex mechanisms underlying development and human diseases. Therefore, new tools and approaches must be developed that will allow multiplexed manipulation and monitoring of signaling pathways with high temporal and spatial resolution. The recent development of tools such as optogenetics has begun to enable such studies, but further innovation will be necessary to allow similar approaches to be performed in vivo.
Acknowledgments
We would like to thank David Doupé and Richelle Sopko for helpful comments. Work in the laboratory of N.P. is supported by the Howard Hughes Medical Institute and the National Institutes of Health (NIH).
References
- 1.Perrimon N, et al. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol. 2012;4:a005975. doi: 10.1101/cshperspect.a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurlbut GD, et al. Nodal points and complexity of Notch–Ras signal integration. Proc Natl Acad Sci USA. 2009;106:2218–2223. doi: 10.1073/pnas.0812024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsueh RC, et al. Deciphering signaling outcomes from a system of complex networks. Sci Signal. 2009;2:ra22. doi: 10.1126/scisignal.2000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natarajan M, et al. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8:571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 5.Flores GV, et al. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 6.de Celis JF, et al. Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development. 1997;124:1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- 7.de Celis JF, Garcia-Bellido A. Modifications of the notch function by Abruptex mutations in Drosophila melanogaster. Genetics. 1994;136:183–194. doi: 10.1093/genetics/136.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Benjumea FJ, Garcia-Bellido A. Behaviour of cells mutant for an EGF receptor homologue of Drosophila in genetic mosaics. Proc Biol Soc. 1990;242:36–44. doi: 10.1098/rspb.1990.0100. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Benjumea FJ, Hafen E. The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development. 1994;120:569–578. doi: 10.1242/dev.120.3.569. [DOI] [PubMed] [Google Scholar]
- 10.Huppert SS, et al. Feedback regulation is central to Delta–Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- 11.Sawamoto K, et al. The function of argos in regulating cell fate decisions during Drosophila eye and wing vein development. Dev Biol. 1994;164:267–276. doi: 10.1006/dbio.1994.1197. [DOI] [PubMed] [Google Scholar]
- 12.Sturtevant MA, Bier E. Analysis of the genetic hierarchy guiding wing vein development in Drosophila. Development. 1995;121:785–801. doi: 10.1242/dev.121.3.785. [DOI] [PubMed] [Google Scholar]
- 13.Krejci A, et al. Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci Signal. 2009;2:ra1. doi: 10.1126/scisignal.2000140. [DOI] [PubMed] [Google Scholar]
- 14.Housden BE, et al. Context-dependent enhancer selection confers alternate modes of notch regulation on argos. Mol Cell Biol. 2014;34:664–672. doi: 10.1128/MCB.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard F, et al. Specificity of Notch pathway activation: twist controls the transcriptional output in adult muscle progenitors. Development. 2010;137:2633–2642. doi: 10.1242/dev.053181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King-Jones K, Thummel CS. Nuclear receptors – a perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- 17.Pardee K, et al. Nuclear receptors: small molecule sensors that coordinate growth, metabolism and reproduction. Subcell Biochem. 2011;52:123–153. doi: 10.1007/978-90-481-9069-0_6. [DOI] [PubMed] [Google Scholar]
- 18.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 19.Hori K, et al. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- 22.Ihle JN, Kerr IM. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 23.Mohr A, et al. Dynamics and non-canonical aspects of JAK/ STAT signalling. Eur J Cell Biol. 2012;91:524–532. doi: 10.1016/j.ejcb.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Rawlings JS, et al. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 25.Ingham PW. Hedgehog signaling. Cold Spring Harb Perspect Biol. 2012;4:a011221. doi: 10.1101/cshperspect.a011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrana JL. Signaling by the TGFbeta superfamily. Cold Spring Harb Perspect Biol. 2013;5:a011197. doi: 10.1101/cshperspect.a011197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sopko R, Perrimon N. Receptor tyrosine kinases in Drosophila development. Cold Spring Harb Perspect Biol. 2013;5:a009050. doi: 10.1101/cshperspect.a009050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 29.Bray D. Protein molecules as computational elements in living cells. Nature. 1995;376:307–312. doi: 10.1038/376307a0. [DOI] [PubMed] [Google Scholar]
- 30.Yissachar N, et al. Dynamic response diversity of NFAT isoforms in individual living cells. Mol Cell. 2013;49:322–330. doi: 10.1016/j.molcel.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Doupe DP, Perrimon N. Visualizing and manipulating temporal signaling dynamics with fluorescence-based tools. Sci Signal. 2014;7:re1. doi: 10.1126/scisignal.2005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamanaka N, et al. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol. 2013;58:497–516. doi: 10.1146/annurev-ento-120811-153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li TR, White KP. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 34.Jang AC, et al. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol. 2009;11:569–579. doi: 10.1038/ncb1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orme MH, Leevers SJ. Flies on steroids: the interplay between ecdysone and insulin signaling. Cell Metab. 2005;2:277–278. doi: 10.1016/j.cmet.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Rogers KW, Schier AF. Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol. 2011;27:377–407. doi: 10.1146/annurev-cellbio-092910-154148. [DOI] [PubMed] [Google Scholar]
- 37.Wright VM, et al. Differential activities of the Drosophila JAK/ STAT pathway ligands Upd, Upd2 and Upd3. Cell Signal. 2011;23:920–927. doi: 10.1016/j.cellsig.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–137. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–394. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- 40.Papatsenko D, Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc Natl Acad Sci USA. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 42.Ip YT, et al. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 43.Sansom SN, Livesey FJ. Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harb Perspect Biol. 2009;1:a002519. doi: 10.1101/cshperspect.a002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furthauer M, et al. A role for FGF-8 in the dorsoventral patterning of the zebrafish gastrula. Development. 1997;124:4253–4264. doi: 10.1242/dev.124.21.4253. [DOI] [PubMed] [Google Scholar]
- 45.Bokel C, Brand M. Generation and interpretation of FGF morphogen gradients in vertebrates. Curr Opin Genet Dev. 2013;23:415–422. doi: 10.1016/j.gde.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 47.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yosef N, Regev A. Impulse control: temporal dynamics in gene transcription. Cell. 2011;144:886–896. doi: 10.1016/j.cell.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandman O, et al. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 51.Becskei A, et al. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gould A, et al. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- 53.Madar D, et al. Negative auto-regulation increases the input dynamic-range of the arabinose system of Escherichia coli. BMC Syst Biol. 2011;5:111. doi: 10.1186/1752-0509-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 55.Lahav G, et al. Dynamics of the p53–Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 56.Lev Bar-Or R, et al. Generation of oscillations by the p53–Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci USA. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos SD, et al. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 58.Kubota H, et al. Temporal coding of insulin action through multiplexing of the AKT pathway. Mol Cell. 2012;46:820–832. doi: 10.1016/j.molcel.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 59.Basu S, et al. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci USA. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaplan S, et al. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol Syst Biol. 2008;4:203. doi: 10.1038/msb.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalir S, et al. A coherent feed-forward loop with a SUM input function prolongs flagella expression in Escherichia coli. Mol Syst Biol. 2005;1:0006. doi: 10.1038/msb4100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Housden BE, et al. Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/ Hes genes. PLoS Genet. 2013;9:e1003162. doi: 10.1371/journal.pgen.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sample V, et al. Genetically encoded molecular probes to visualize and perturb signaling dynamics in living biological systems. J Cell Sci. 2014;127:1151–1160. doi: 10.1242/jcs.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta-Rossi N, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 65.Ilagan MX, et al. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci Signal. 2011;4:rs7. doi: 10.1126/scisignal.2001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rand MD, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toettcher JE, et al. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat Methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aoki K, et al. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol Cell. 2013;52:529–540. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 70.Toettcher JE, et al. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 72.Sasagawa S, et al. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol. 2005;7:365–373. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- 73.Albeck JG, et al. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol Cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masamizu Y, et al. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci USA. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soroldoni D, Oates AC. Live transgenic reporters of the vertebrate embryo’s Segmentation Clock. Curr Opin Genet Dev. 2011;21:600–605. doi: 10.1016/j.gde.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 76.Bren A, et al. The last generation of bacterial growth in limiting nutrient. BMC Syst Biol. 2013;7:27. doi: 10.1186/1752-0509-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ronen M, et al. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc Natl Acad Sci USA. 2002;99:10555–10560. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]