Abstract
Treatment of filamentous fungal infections relies on a limited repertoire of antifungal agents. Compounds possessing novel modes of action are urgently required. N-myristoylation is a ubiquitous modification of eukaryotic proteins. The enzyme N-myristoyltransferase (NMT) has been considered a potential therapeutic target in protozoa and yeasts. Here, we show that the filamentous fungal pathogen Aspergillus fumigatus possesses an active NMT enzyme that is essential for survival. Surprisingly, partial repression of the gene revealed downstream effects of N-myristoylation on cell wall morphology. Screening a library of inhibitors led to the discovery of a pyrazole sulphonamide compound that inhibits the enzyme and is fungicidal under partially repressive nmt conditions. Together with a crystallographic complex showing the inhibitor binding in the peptide substrate pocket, we provide evidence of NMT being a potential drug target in A. fumigatus.
Aspergillus spp. are ubiquitous filamentous fungi and together with three other genera (Candida, Cryptococcus, and Pneumocystis) account for more than 90% of invasive fungal infections worldwide.1 While invasive aspergillosis due to A. fumigatus is estimated to affect over 200 000 people globally each year, a further 3 million are affected by chronic pulmonary aspergillosis.1 At present, treatment is limited to three antifungal classes: polyenes, azoles, and echinocandins. Voriconazole, introduced in the 1990s, is a third generation azole and current “gold standard” for invasive disease and has increased response and survival rates by 15–20%.2 Overall mortality for invasive disease, however, remains unacceptable at around 50%.1 Moreover, there are inherent problems with drug interactions, toxicities, and increasingly reported resistant strains3,4 necessitating the urgent identification and characterization of novel targets against A. fumigatus.
The myristoyl coenzyme A (MCoA)/protein N-myristoyltransferase (NMT, EC: 2.3.1.97) catalyzes an irreversible co- and post-translational covalent attachment of the saturated 14-carbon fatty acid, myristate, from MCoA to the N-terminal glycine residue of many eukaryotic and viral proteins.5N-myristoylation is abundant in eukaryotic organisms including protozoa, fungi, and mammals. Membrane targeting and the function of many proteins in a variety of signal transduction cascades and other critical cellular functions is NMT dependent.5−7 Structural studies advocate an ordered Bi–Bi reaction mechanism8 whereby binding of MCoA to NMT opens a “lid” permitting access to the peptide-binding site. While the MCoA binding site is highly conserved, NMTs are striking in their remarkable diversity of peptide substrates.9 Diversity in the peptide-binding groove has been suggested to offer the potential of developing species-specific inhibitors.9 As a result, NMT has been considered as a potential anticancer,10 antiviral,11 and antiparasitic12−14 therapeutic target. A substantial pool of new chemical starting points for inhibitors of NMTs from parasites has recently been described.15−21 So far, NMT in filamentous fungi such as A. fumigatus remains uncharacterized although orthologues in other fungal pathogens such as Candida albicans(22) and Cryptococcus neoformans(23) are essential for viability. An NMT knockout in C. albicans resulted in avirulence in a murine model,22 and several prototype inhibitors have been reported.24−26 Attempts to develop broad-spectrum antifungal NMT inhibitors were unsuccessful and are now potentially redundant given impending improved diagnostics and a developing preference for targeted narrow spectrum therapy.
At present, there is a dearth of new antifungals in the drug discovery pipeline. Conclusive validation of novel antifungal targets from both a chemical and genetic perspective provides a critical first step in reversing this trend. Here, we show that NMT is a potential drug target in A. fumigatus.
Results and Discussion
A. fumigatus Possesses an Active NMT Enzyme
In previous reports, BLAST searches using the Arabidopsis thaliana NMT127 or Aspergillus nidulans NMT28 sequences predicted the presence of an nmt gene (AFUA_4G08070) in the A. fumigatus genome. The gene is 1630 bp in length and contains two introns and three exons. The 1479 bp mRNA encodes a protein of 492 amino acids (UniProt: Q9UVX3) sharing 50%, 52%, 38%, and 44% sequence identity with the NMTs of C. albicans, S. cerevisiae, Leishmania major, and Homo sapiens (Supporting Information Figure 1), respectively.
Alignment of A. fumigatus NMT (AfNMT) with NMT structures deposited in the Protein Data Bank (PDB) from C. albicans (1IYL29) suggested truncation of the N-terminal end, leaving the core catalytic domain intact. Therefore, to increase the chance of crystallization, the first 85 amino acids were removed and a truncated AfNMT construct (Δ1–85) generated covering residues 86–492. AfNMT Δ1–85 was overexpressed as a GST fusion protein in Escherichia coli, purified to homogeneity by glutathione sepharose affinity chromatography followed by proteolysis of the fusion protein and size-exclusion chromatography, yielding 2 mg of pure AfNMT per liter of bacterial culture.
AfNMT steady-state kinetics were assessed by a scintillation proximity assay [SPA30,31] using biotin-tagged cytoskeleton-associated protein 5.5 (CAP5.5) as the peptide acceptor substrate. The Km of myristoyl-CoA (MCoA) for AfNMT was 0.8 ± 0.1 μM (Supporting Information Figure 2A) and comparable to the Km of MCoA previously reported for S. cerevisiae [1.4 μM32], L. major [1.4 ± 0.3 μM30], and Trypanosoma brucei [1.8 ± 0.4 μM30]. Likewise, the Km of peptide CAP5.5 for AfNMT was 22 ± 2 μM (Supporting Information Figure 2B), similar for T. brucei [11 ± 1 μM30].
nmt Is Essential for Viability of A. fumigatus
Attempts to construct a deletion mutant by replacing the A. fumigatus nmt gene with a pyrG selection marker33 and phenotypic rescue by supplementing the culture medium with myristic acid failed. Although myristic acid has fully or partially restored the phenotype in other species where nmt is essential,22,23,28 all our transformants failed initial screening with this approach. Instead, a conditional inactivation mutant was constructed by replacing the native promoter of the nmt gene with the A. nidulans alcohol dehydrogenase promoter (PalcA) that is inducible by ethanol, glycerol, or threonine but repressed by glucose.34 A plasmid containing a 3′ truncation of nmt fused to PalcA was transformed into A. fumigatus KU80ΔpyrG– to generate a conditional mutant (from hereon referred to as the NMT strain) by homologous recombination (Supporting Information Figure 3A). PCR and Southern blotting (Supporting Information Figure 3B–D) verified that in the NMT strain, the inducible PalcA promoter replaced the native promoter of the nmt gene.
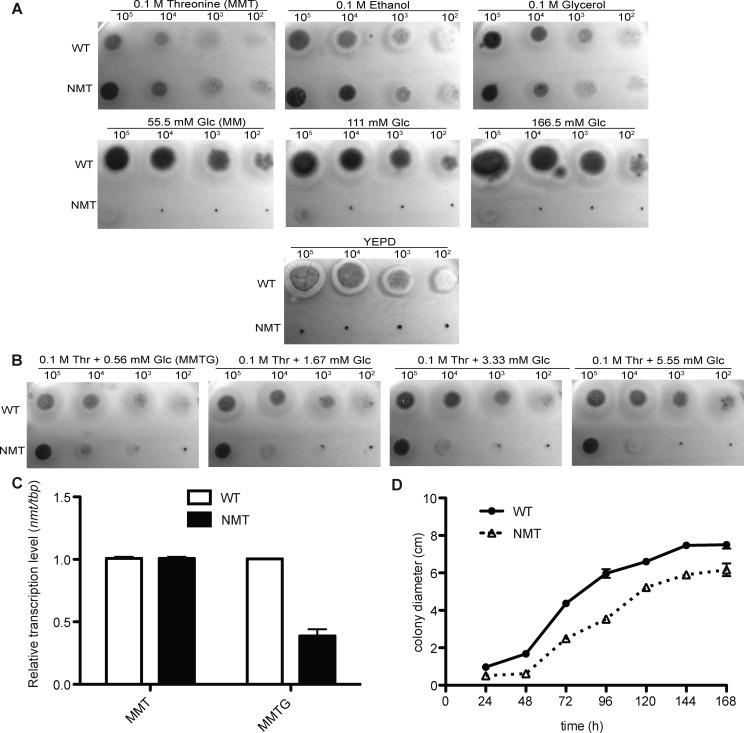
Growth of the NMT strain was comparable to the WT when grown under conditions of nmt induction on a solid minimal medium (MM) containing 0.1 M glycerol, 0.1 M ethanol, or 0.1 M threonine (MMT). However, growth of the NMT strain was completely inhibited under conditions of nmt repression on YEPD or MM containing 55.5–166.5 mM glucose after 48 h at 37 °C (Figure 1A). Thus, nmt expression is required for viability and is an essential gene in A. fumigatus.
Figure 1.
Growth phenotype of the NMT strain under varying repressive conditions. (A) Growth of A. fumigatus strains on solid MM supplemented with 0.1 M glycerol, 0.1 M ethanol, 0.1 M threonine or 55.5 mM, 111 mM, 166.5 mM glucose or YEPD. (B) Growth on solid MM supplemented with 0.1 M threonine and 0.56 mM, 1.67 mM 3.33 mM, or 5.55 mM glucose. (C) Real time PCR to amplify the tbp and nmt gene from MMT and MMTG conditions. (D) Growth curve of the WT and NMT strain on MMTG plates.
nmt Expression Affects A. fumigatus Cell Wall
In order to investigate the function of the essential nmt gene in A. fumigatus, various concentrations of threonine (inducer) and glucose (repressor) were assessed to identify a minimal level of nmt expression still producing sufficient mycelia for analysis (Figure 1B). MM with 0.1 M threonine and 0.56 mM glucose (MMTG) was selected for all subsequent phenotypic analysis. Under this condition, the transcription of nmt in the NMT strain was reduced to 39% of the WT (Figure 1C). Growth of the NMT strain was retarded compared to the WT (Figure 1D).
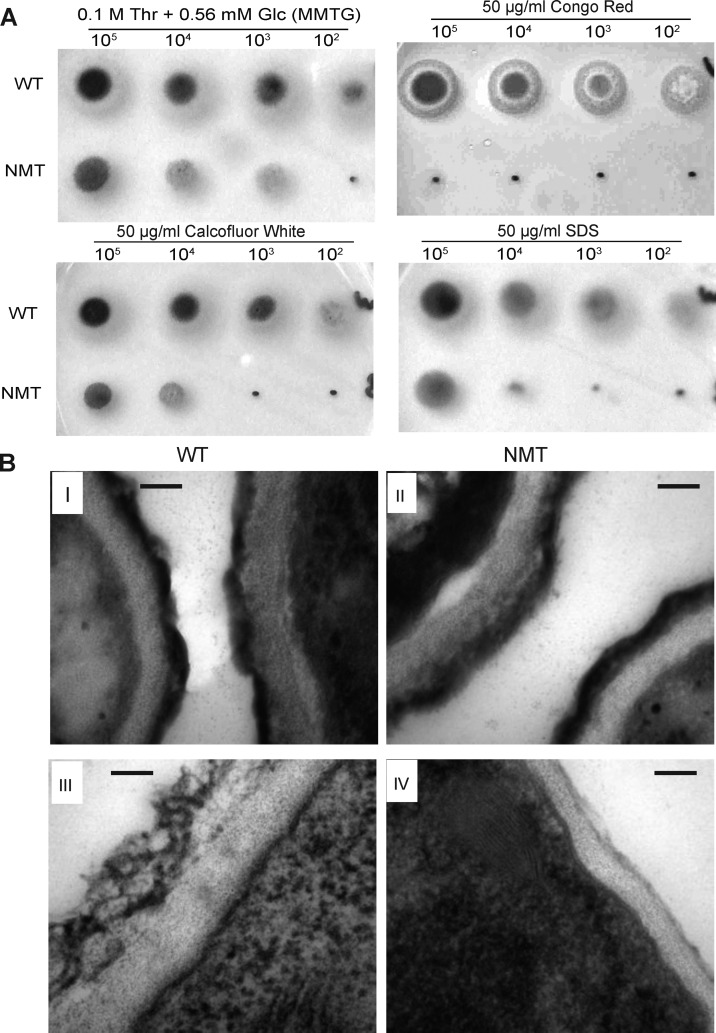
Surprisingly, increased sensitivity to agents compromising the cell wall (Congo red and Calcofluor white) or membrane (Sodium dodecyl sulfate) were observed under partial expression of the nmt gene indicative of cell wall defects and loss of membrane integrity (Figure 2A). Furthermore, examination of the cell wall ultrastructure by electron microscopy showed that the conidia of both NMT and WT strains were indistinguishable (Figure 2B, I and II), but the hyphal cell wall of the NMT strain was thinner than the WT (Figure 2B, III and IV).
Figure 2.
NMT strain sensitivity to chemical reagents and reduction in cell wall thickness. (A) Serial dilutions of conidia from 105 to 102 were spotted on solid MMTG containing 50 μg mL–1 Calcofluor white, Congo red, or SDS. (B) Cell wall architecture under repressive conditions (MMTG). I and II are conidia; III and IV are mycelium. Scale bar is 100 nm.
nmt Is Required for Cell Wall Morphogenesis
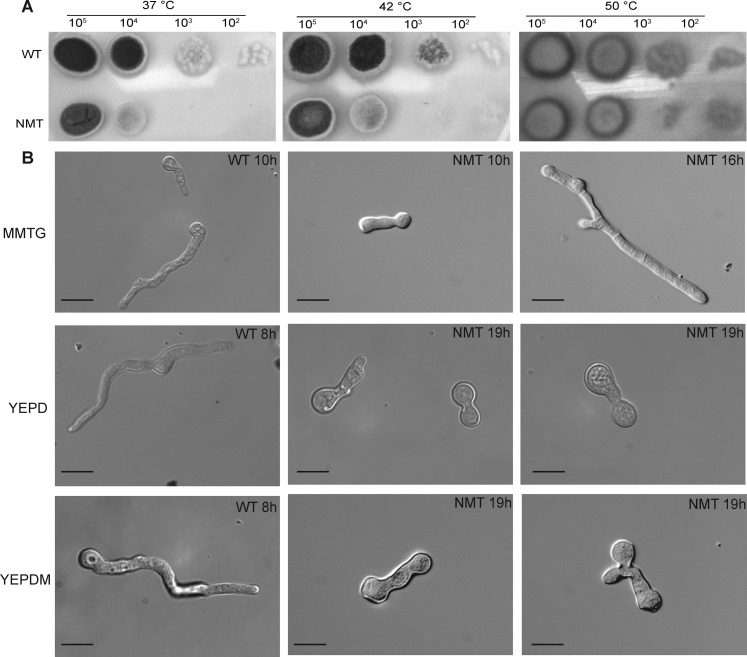
Unlike other fungi such as A. nidulans(28) where NMT mutants grew only at 37 °C, growth of the NMT strain was unaffected by temperature (37–50 °C) on MMTG plates (Figure 3A). This suggests that the target proteins of AfNMT appear to be neither temperature sensitive nor involved in temperature adaptation mechanisms.
Figure 3.
NMT strain temperature insensitivity and delayed germination. (A) Serial dilutions of 105 to 102 conidia were spotted on MMTG plates and incubated at 37 °C for 36 or 42 h and 50 °C for 60 h before imaging. (B) Comparison of germination times of the WT and NMT strains in MMTG, YEPD, or YEPD supplemented with myristic acid (YEPDM). Scale bar is 10 μm.
Polarized hyphal growth is essential in filamentous fungi and thought to be important for pathogenesis. Our NMT strain showed a marked polar growth defect under conditions of nmt repression including delayed germination, abnormal development, and retention of polarity (Figure 3B). When nmt was completely repressed in a YEPD medium, the conidia remained swollen or displayed very short germ tubes (Figure 3B), indicating NMT is required for germination and hyphal growth.
Distinct to A. fumigatus was the inability of myristic acid supplementation to fully restore the NMT strain to the WT phenotype as has been demonstrated for C. albicans, S. cerevisiae, and C. neoformans.22,23,35 Only short germ tubes with a ballooned tip or abnormal polarity were evident after 19 h incubation in supplementation medium (Figure 3B).
AfNMT Crystal Structure Reveals a Molecular Handle on Selectivity
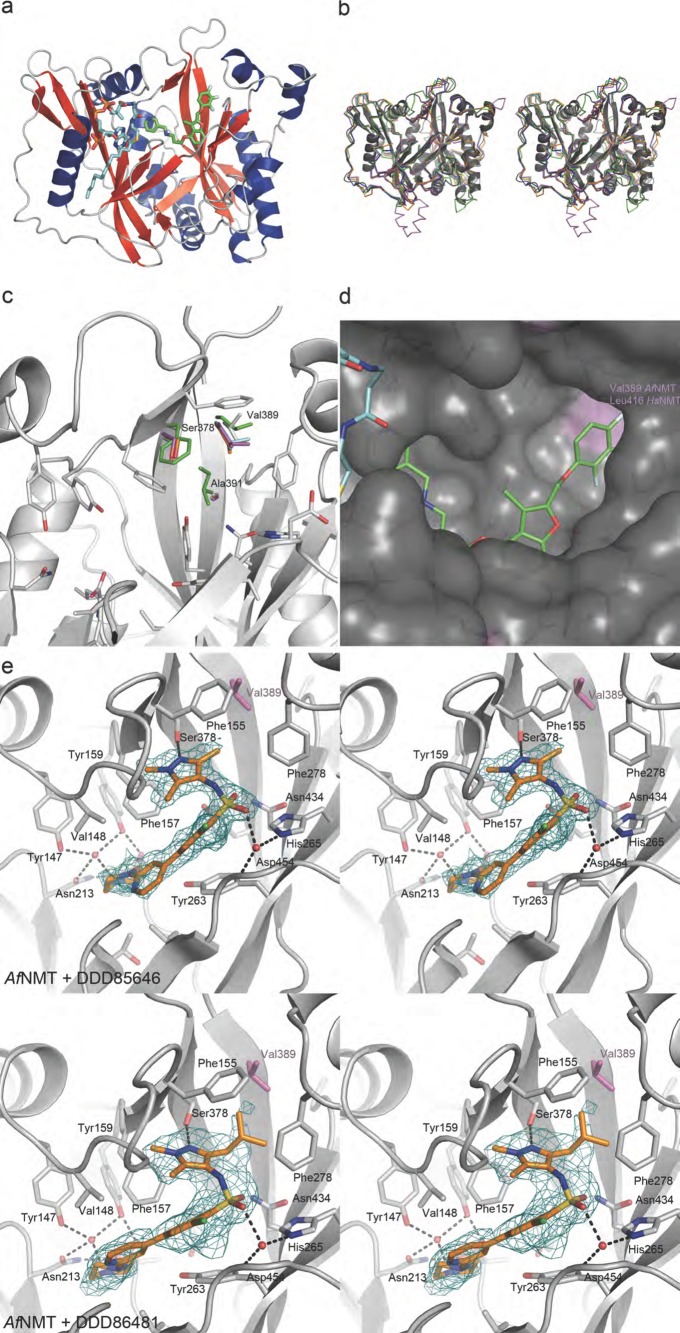
As a first step toward the design of selective inhibitors, we determined the crystal structure of AfNMT and compared it to the human enzyme. The purified AfNMT was cocrystallized with MCoA and the benzofuran compound Ro-09-4879 that is known to compete for peptide binding in other NMTs.29 The structure of the inhibitor complex was solved by molecular replacement using C. albicans NMT (PDB: 1IYK(29)) as a search model and refined against 1.9 Å synchrotron diffraction data (Supporting Information Table 1). No interpretable electron density was visible for the first 15 residues of the polypeptide chain.
The overall crystal structure of AfNMT consisted of a monomer of 391 residues (101–492) forming two GCN5 folds with MCoA bound to AfNMT in a horseshoe conformation (Figure 4A). To date, crystal structures of NMT from C. albicans, S. cerevisiae, H. sapiens, L. major, L. donovani, and P. vivax have been elucidated showing extensive conservation of the core fold and active site residues (Figure 4B). The structure of AfNMT reflects the high level of conservation between species at a primary, secondary, and tertiary sequence level, showing the highest level of conservation with C. albicans (Cα RMSD = 1.301 Å with 346 atoms matched) and S. cerevisiae NMT (Cα RMSD = 0.840 Å with 350 atoms matched). Although similar to NMT:MCoA structures previously determined, the CoA phosphate group was not coordinated by the polypeptide due to the truncated construct lacking the YKFW motif. Electron density for the benzofuran ligand Ro-09-4879 was observed in the peptide-binding groove of AfNMT where it competes with acceptor substrate binding (Figure 4A). Here, it adopts a similar conformation to a CaNMT:benzofuran complex [PDB 1IYL(29)]. The peptide binding site of NMT is highly conserved with minimal sequence differences between species. However, comparison of the known NMT crystal structures reveals three nonconserved residues; Ser378 and Ala391 are conserved apart from C. albicans and S. cerevisiae species where the corresponding residues are Phe and Ile. The residue Val389 is unique to AfNMT, and it forms part of a hydrophobic pocket with Phe155, Leu273, Phe278, Ser378, and Val432 that accepts the Ro-09-4879 ligand. In other species, notably human, this residue is a larger leucine (Leu416; Figure 4C,D). Such a difference may be exploitable in the design of inhibitors to selectively target AfNMT.
Figure 4.
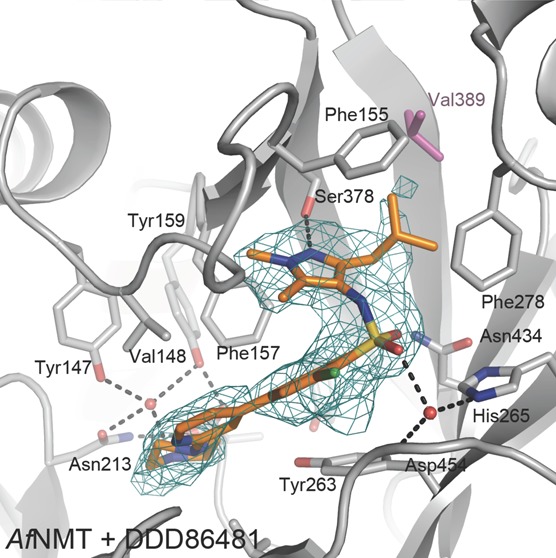
Crystal structure of AfNMT complexes with inhibitors. (A) Overall fold of AfNMT (residues 86–492) in complex with MCoA (stick representation, Cα cyan) and RO-09-4879 (stick representation, Cα green). (B) Representative crystal structures of NMTs. AfNMT shown in gray cartoon superimposed upon ScNMT (PDB 1IIC, green), LmNMT (PDB 3H5Z, magenta), HsNMT1 (PDB 3IU1, cyan), and PvNMT (PDB 4B10, orange). (C) Conservation of NMT active site residues. AfNMT backbone shown in gray cartoon with active site side chains shown in stick representation (C atoms gray). Strictly conserved residues between species are shown for AfNMT only. Where residues are not conserved, they are shown for all species in stick representation, ScNMT (green), LmNMT (magenta), HsNMT1 (cyan), and PvNMT (orange). (D) Close-up view of the AfNMT active site, showing the MCoA (Cα cyan) and RO-09-4879 (Cα green). A molecular surface is shown, colored by sequence conservation with HsNMT1 (gray, identical side chains; purple, nonidentical side chains). (E) Stereoscopic view of the active site of the AfNMT in complex with DDD85646 and DDD86481. Active site side chains are shown as sticks with gray Cα, whereas the compound is shown as sticks with gold Cα. The unbiased |Fo| – |Fc| map (2.5 σ) is shown as a green mesh.
Screening-Based Discovery of a Low Nanomolar AfNMT Inhibitor
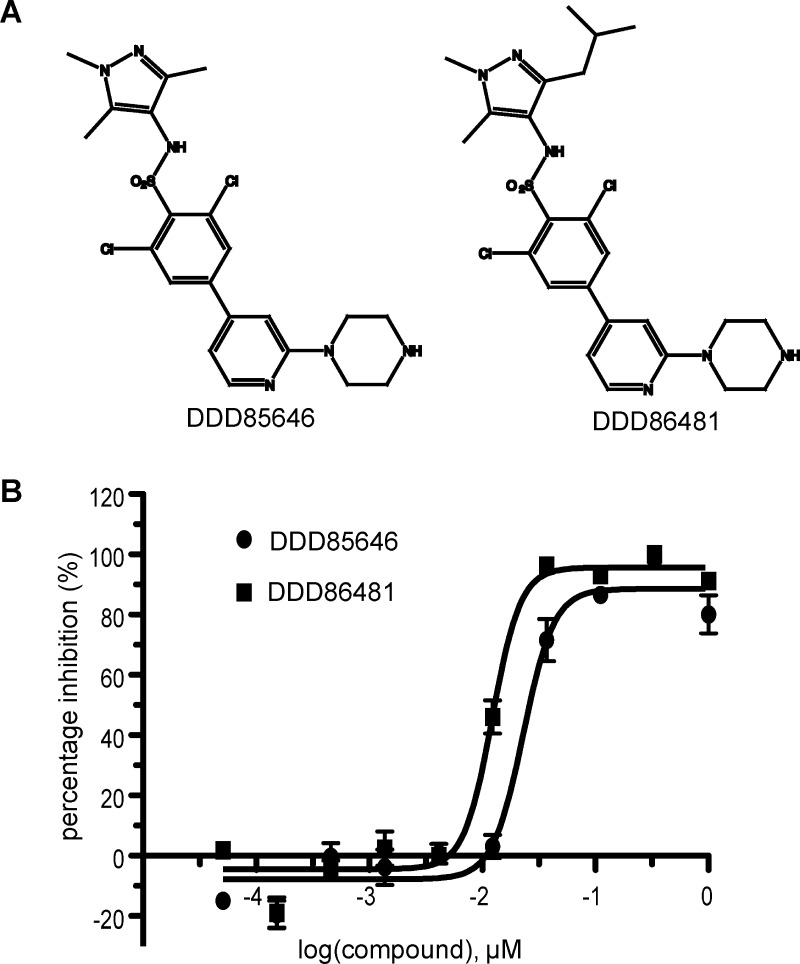
Pyrazole sulphonamide ligands act as highly potent inhibitors against T. brucei NMT (TbNMT).12 A panel of antitrypanosome NMT inhibitors was screened against recombinant AfNMT (Supporting Information Table 2), and the fungicidal effect on A. fumigatus was determined. As shown in Figure 5, two potent nanomolar pyrazole sulphonamide inhibitors were identified using the SPA assay with IC50 values of 23 nM (DDD85646) and 12 nM (DDD86481).
Figure 5.
Inhibition of AfNMT. (A) Chemical structures of compounds DDD85646 and DDD86481. (B) Dose response IC50 curves of DDD85646 and DDD86481 against AfNMT using an SPA assay.
To investigate the binding mode of these compounds, further crystal structures of AfNMT:MCoA in complex with the pyrazole sulphonamide inhibitors were solved (Supporting Information Table 1), demonstrating that they also occupied the peptide binding site. DDD85646 adopted a similar conformation to that previously observed in a L. major NMT complex (PDB 2WSA) used as a model for T. brucei.12 Specifically, the piperazine group interacts with the C-terminal carboxylate group via two bridging water molecules. The first water molecule is tightly coordinated by the C-terminal carboxylate and the phenolic hydroxyl groups of Tyr147 and Tyr159 while a second water molecule bridges the piperazine group and the carboxylate (Figure 4E). The biaryl system makes no specific interactions with the protein and sits on a hydrophobic surface formed by the side chains of Tyr263 and Leu457. A significant bend in the molecule introduced by the sulphonamide allows the pyrazole moiety to penetrate a hydrophobic pocket. Oxygen atoms of the sulphonamide interact with the protein through water-bridged interactions with the side chains of Asn434, His265, and the backbone amides of Asp454 and Gly455. The trimethyl pyrazole group sits in the same hydrophobic pocket occupied by the trifluoro-phenyl group of Ro-09-4897; however the pyrazole forms a hydrogen bond with the side chain of Ser378 in addition to strong hydrophobic packing interactions (Figure 4E).
The isobutyl pyrazole derivative DDD86481 has a very similar binding mode to DDD85646 (Figure 4E). The sulphonamide, however, sits lower in the active site losing interaction with Asn434, and the pyrazole group rotates slightly to accommodate the larger moiety at the 3- position (Figure 4E). As the isobutyl group packs against the side chains of Phe155, Phe278, Val389, Ala391, and Val432, this explains the increased potency over DDD85646 by increasing the hydrophobic complementarity in this pocket.
DDD86481 Is Fungicidal under Repressed nmt Expression
Initially, all compounds from Supporting Information Table 2 were taken for MIC determination in RPMI medium containing 0.1 M threonine and 0.56 mM glucose. Only three compounds (DDD86481, DDD90086, and Ro-09-4879) showed potent inhibition for the NMT mutant. The fungicidal activity of DDD85646 was >2.5 mM irrespective of the A. fumigatus strain tested (WT or NMT). Although the MIC of DDD86481 was 925 μM against the WT (at 0.625 mM, WT showed a mutant morphogenesis phenotype), it decreased to 116 μM for the NMT strain (Table 1). As this molecule is more lipophilic than DDD85646, this might account for marginally increased cell penetration and a lower MIC. In order to demonstrate target engagement inside the cell, the chemical labeling technology employing click chemistry with a myristate alkyne derivative13,16,36 was explored. However, this approach did not work in A. fumigatus cells, probably because the myristate derivative could not penetrate the fungal cell wall.
Table 1. MIC of Inhibitor Compounds against A. fumigatusa.
| strains | WT | NMT | ||||
|---|---|---|---|---|---|---|
| growth conditions | BMT + | 0.56 mM Glc | 0.56 mM Glc | 3.33 mM Glc | 55.5 mM Glc | 111 mM Glc |
| level of NMT repression | nmt transcription (%) | 100 ± 4 | 81 ± 10 | 78 ± 15 | 29 ± 7 | 24 ± 1 |
| MIC (mM) | DDD86481 | 0.925 | 0.116 | 0.058 | 0.014 | 0.007 |
| DDD85646 | >2.5 | >2.5 | ND | >2.5 | >2.5 | |
| DDD90086 | >2.5 | 1.25 | ND | 0.625 | 0.625 | |
| Ro-09-4609 | >2.5 | >2.5 | ND | >2.5 | >2.5 | |
| Ro-09-4879 | 0.625 | 0.625 | ND | 0.625 | 0.625 | |
| other compounds* | >2.5 | >2.5 | ND | ND | ND | |
| amphotericin B | 0.0017 | 0.0017 | 0.0017 | 0.0017 | 0.0017 | |
The CLSI broth microdilution method was used to determine the MIC (lowest concentration that completely inhibited growth) after 48 h of incubation at 37 °C. BMT, basic medium plus 0.1 M threonine; Glc, glucose; ND, not determined; *All other compounds listed in the Supporting Information, Table 2.
To investigate whether DDD86481 was acting through inhibition of AfNMT in the NMT strain, MIC values were determined under different repressive conditions and nmt transcript levels assessed by quantitative real time PCR. In 0.56 mM glucose medium, nmt transcription was 81% of the WT. As nmt transcription reduced, DDD86481 efficacy increased with correlating reductions in MIC (Table 1) to 7 μM with 24% nmt expression. As a control, the MICs of amphotericin B, DDD85646, Ro-09-4609, and Ro-09-4879 were unaffected by nmt transcript levels (Table 1), although the MIC of DDD90086 was decreased slightly. Nevertheless, there is still a significant drop in activity of DDD86481 on going from enzyme to cell. When NMT is normally expressed, there is an 80 000-fold drop in activity; when NMT is expressed at 24%, there is a 600-fold drop in activity. This could be due to poor penetration of DDD86481 through the fungal cell wall but alternatively could be due to the catalytic role of NMT not being essential or off-target effects of DDD86481.
Concluding Remarks
Early stage drug discovery suffers high attrition rates, and there are limited immediate offerings in the antifungal pipeline besides modifications of existing classes.37 Increased recognition of the clinical burden posed by chronic pulmonary aspergillosis threatens to place huge selective pressures on oral azoles (e.g., itraconazole) as they have been shown to partially alleviate symptoms.38 Consequently, there is profound medical need for alternative classes ideally with oral formulations. As a step toward this, we presented evidence that validates AfNMT as a potential drug target in A. fumigatus showing that protein N-myristoylation affects cell morphology and integrity of the cell wall, a structure essential to all pathogenic fungi and absent from the human cell.
A conditional NMT mutant strain showed that this gene was essential and also provided insights into the downstream effects of NMT inhibition on the cell wall of A. fumigatus. Reduced expression affected cell morphogenesis and cell wall integrity—effects that have not previously been observed in NMT genetic disruption studies in other organisms. The fungal cell wall is a dynamic three-dimensional structure composed predominantly of polysaccharides. The cell wall involves numerous biosynthetic pathways and hundreds of proteins for its synthesis and remodeling. A significant fraction (typically 0.5%) of eukaryotic proteins are thought to be myristoylated,27 assisting switching between cytoplasmic and membrane bound states. In fact, using the MYR Predictor (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm), 29 proteins from the A. fumigatus proteome are predicted to be myristoylated (Supporting Information Table 3). While most proteins are of unknown function, two classes, namely the G-protein alpha subunits and the ADP-ribosylation factors, are known myristoylation substrates.39,40 Myristoylation of G protein alpha subunits is required for localization to the plasma membrane and also facilitating formation of a heterotrimer for G protein signaling.41 As signal transducers, G proteins influence numerous signaling cascades especially activate the cyclic-AMP-dependent PKA pathway42 that regulates growth, development, stress response, morphogenesis, and virulence in A. fumigatus.43−45 There is also direct evidence that G proteins regulate cell wall carbohydrate composition in both plants and other fungi.46−49 One of the ADP-ribosylation factors in A. nidulans, ArfB, is involved in polarity establishment during isotropic growth and polarity maintenance during hyphal extension.50 Three ArfB orthologues (Afu3g12080, Afu1g11730, and Afu2g10980) in A. fumigatus are predicted to be myristoylated proteins (Supporting Information Table 3). Our NMT strain displayed the same polarized growth defect as the ArfB mutant. In summary, we propose that the G alpha subunits and these ArfB orthologues account for the changes in cell wall and defect in morphogenesis in the NMT strain.
Screening a library of compounds identified a 12 nM AfNMT inhibitor (DDD86481) that targeted the peptide binding site and interacted with an amino acid side chain that is different between the fungal and human enzyme. However, this failed to directly correlate with fungicidal activity against A. fumigatus (MIC 925 μM). Reducing nmt expression sensitized the organism to the inhibitor, reducing the MIC to 7 μM, yet retaining a 600-fold difference between activity on the target in vitro and the fungal cell, suggesting the need for more potent and cell-penetrant compounds to pursue full chemical validation.
Essential gene targets like NMT produce a lethal phenotype but a significant challenge will be discovering inhibitors that are highly selective for A. fumigatus. The peptide-binding sites of the human and A. fumigatus NMTs are exquisitely similar to only a single amino acid difference (Val389 in AfNMT and Leu416 in HsNMT1). However, there is evidence from TbNMT inhibitors that a substantial window of selectivity can be derived between HsNMT1 and TbNMT despite highly similar active site structures.12 Recently, further optimized TbNMT inhibitors not only possessed improved CNS penetrance but also improved selectivity that could be used to treat stage 2 human African trypanosomiasis.21 Highly potent Leishmania-selective NMT inhibitors have been identified either from compound screening15 or with a structure-based approach.20 More encouragingly, although structure–activity relationships (SARs) between Plasmodium NMT and HsNMT1 overlap,15 potent and selective Plasmodium NMT inhibitors have been discovered15 and optimized,18 demonstrating that even the small conformational differences of single equivalent tyrosine could be exploited to engineer selectivity over the human enzyme.17 Therefore, it may be possible to discover selective A. fumigatus inhibitors based on the structure differences between AfNMT and HsNMT1 described here.
In summary, we have provided the first comprehensive characterization of NMT in a filamentous fungal pathogen and combined genetic, structural, and chemical approaches to validate AfNMT as a target illustrating NMT dependent inhibition of A. fumigatus growth. While the genetic work confirmed NMT is essential for viability in A. fumigatus, the mutant NMT strain allows controlled gene expression and provides a research tool for further probing the effects of (i) N-myristoylation and (ii) host-fungal interactions.
Methods
Construction of the Conditional Inactivation Mutant
Plasmid pAL3 containing the A. nidulans alcA promoter (PalcA) and the Neurospora crassa pyr-4 gene as a fungal selectable marker51 was used to construct a vector replacing the native nmt promoter with PalcA. An 1120 bp fragment from −50 to +1070 of the nmt genomic DNA sequence was amplified with primers P3 and P4 (see Supporting Information Table 4 for all primer sequences) containing a KpnI and XbaI restriction site, respectively. The PCR-amplified fragment was cloned into expression vector pAL3 to yield pALnmtN and confirmed by sequencing. The pALnmtN was transformed into A. fumigatus strain KU80ΔpyrG– by PEG-mediated fusion of protoplasts52 and screened for colonies with uridine/uracil autotrophy. The transformants were confirmed by PCR and Southern blot analysis.
For PCR analysis, three sets of primers were designed: primers P5 and P6 for the 1.6 kb nmt gene, P7 and P8 for the 1.2 kb pyr-4 marker, and P9 and P10 for a 2.5 kb fragment from PalcA to a downstream region of the nmt gene. For Southern blotting, genomic DNA was digested with BamHI, separated by electrophoresis and transferred to a nylon membrane (Zeta-probe+, Bio-Rad). The 1120 bp fragment of the nmt genomic sequence and a 1.2 kb HindIII fragment of the N. crassa pyr-4 gene from pAL3 were used as probes. Labeling and visualization were performed using the DIG DNA labeling and detection kit (Roche) according to the manufacturer’s instructions.
Crystallization, Data Collection, and Structure Determination
AfNMT Δ1–85 (20 mg mL–1) was crystallized using the sitting drop method. Prior to crystallization, AfNMT was incubated with 10 mM myristoylCoA (MCoA). To obtain ligand complexes, AfNMT was incubated with 10 mM of compound. Crystals grew after 2 days in a mother liquor of 0.2 M NaCl and 25% (v/v) PEG3350 and were cryoprotected for flash freezing in liquid nitrogen. Diffraction data were collected at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) and all data processed and scaled with HKL software.53
The AfNMT-MCoA-RO-09-4879 complex structure was solved by molecular replacement using MOLREP54 from the CCP4 suite programs55 with the C. albicans NMT structure (PDB: 1IYK(29)) as a search model giving a solution with one molecule in the asymmetric unit. The resulting electron density map was of good quality, allowing WarpNtrace56 to build virtually all residues except for a few disordered regions. REFMAC557 was used for further refinement, iterated with model building using COOT,58 producing a final model with the statistics shown in Supporting Information Table 1. Other compound complexes were solved by molecular replacement using the AfNMT-MCoA-RO-09-4879 structure as a search model. The model for ligands was not included until their conformations were fully defined by unbiased |Fo| – |Fc| ϕcalc electron density maps. Ligand structures and topologies were generated by PRODRG.59 PyMol was used to generate figures.60
Minimal Inhibitory Concentration (MIC) Assay
Selected compounds were tested against A. fumigatus (WT [KU80] and NMT strains) according to the Clinical and Laboratory Standards Institute (CLSI) M38-A broth microdilution methodology.61 All compounds were dissolved in DMSO, and the final DMSO concentration was 2.5% (v/v) in all wells. The MIC end point was defined as the lowest concentration producing complete inhibition of growth (MIC100) for all compounds studied. The MIC values for each compound were determined in triplicate. Amphotericin B (Sigma) and A. fumigatus ATCC 204305 served as a positive control and the MIC within the recommended limits of CLSI M38-A.
Acknowledgments
The authors would like to thank the European Synchrotron Radiation facility (ESRF), Grenoble, for beamline time. We thank J. James of the College of Life Sciences Microscopy Facility for his help with electron microscopy. We also thank F. Eisenhaber, Bioinformatics Institute Singapore, for predicting myristoylated proteins. This work was funded by an MRC Programme Grant (M004139) and a Wellcome Trust Senior Research Fellowship (WT087590MA) to D.M.F.v.A. D.E.A.L was supported by an MRC Clinical Research Training Fellowship (G1100430). We also would like to acknowledge the Wellcome Trust (WT077705, WT083481) for support.
Supporting Information Available
Supporting Figures S1–S3, Supporting Tables S1–S4, and further details of experimental procedures. This material is available free of charge via the Internet from http://pubs.acs.org.
Accession Codes
The atomic coordinates and structure factors of AfNMT:MCoA with RO-09-4879, DDD85646, and DDD86481 have been deposited in the Protein Data Bank with accession codes 4CAV, 4CAX, and 4CAW.
Other procedures are reported in the Supporting Information.
The authors declare no competing financial interest.
Supplementary Material
References
- Brown G. D.; Denning D. W.; Gow N. A.; Levitz S. M.; Netea M. G.; White T. C. (2012) Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113. [DOI] [PubMed] [Google Scholar]
- Nivoix Y.; Velten M.; Letscher-Bru V.; Moghaddam A.; Natarajan-Ame S.; Fohrer C.; Lioure B.; Bilger K.; Lutun P.; Marcellin L.; Launoy A.; Freys G.; Bergerat J. P.; Herbrecht R. (2008) Factors associated with overall and attributable mortality in invasive aspergillosis. Clin. Infect. Dis. 47, 1176–1184. [DOI] [PubMed] [Google Scholar]
- Bowyer P.; Moore C. B.; Rautemaa R.; Denning D. W.; Richardson M. D. (2011) Azole antifungal resistance today: focus on Aspergillus. Curr. Infect Dis. Rep. 13, 485–491. [DOI] [PubMed] [Google Scholar]
- van der Linden J. W.; Camps S. M.; Kampinga G. A.; Arends J. P.; Debets-Ossenkopp Y. J.; Haas P. J.; Rijnders B. J.; Kuijper E. J.; van Tiel F. H.; Varga J.; Karawajczyk A.; Zoll J.; Melchers W. J.; Verweij P. E. (2013) Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin. Infect. Dis. 57, 513–520. [DOI] [PubMed] [Google Scholar]
- Farazi T. A.; Waksman G.; Gordon J. I. (2001) The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276, 39501–39504. [DOI] [PubMed] [Google Scholar]
- Knoll L. J.; Johnson D. R.; Bryant M. L.; Gordon J. I. (1995) Functional significance of myristoyl moiety in N-myristoyl proteins. Methods Enzymol. 250, 405–435. [DOI] [PubMed] [Google Scholar]
- Boutin J. A. (1997) Myristoylation. Cell. Signal. 9, 15–35. [DOI] [PubMed] [Google Scholar]
- Rudnick D. A.; McWherter C. A.; Rocque W. J.; Lennon P. J.; Getman D. P.; Gordon J. I. (1991) Kinetic and structural evidence for a sequential ordered Bi Bi mechanism of catalysis by Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J. Biol. Chem. 266, 9732–9739. [PubMed] [Google Scholar]
- Kishore N. S.; Wood D. C.; Mehta P. P.; Wade A. C.; Lu T.; Gokel G. W.; Gordon J. I. (1993) Comparison of the acyl chain specificities of human myristoyl-CoA synthetase and human myristoyl-CoA:protein N-myristoyltransferase. J. Biol. Chem. 268, 4889–4902. [PubMed] [Google Scholar]
- Selvakumar P.; Lakshmikuttyamma A.; Shrivastav A.; Das S. B.; Dimmock J. R.; Sharma R. K. (2007) Potential role of N-myristoyltransferase in cancer. Prog. Lipid Res. 46, 1–36. [DOI] [PubMed] [Google Scholar]
- Hill B. T.; Skowronski J. (2005) Human N-myristoyltransferases form stable complexes with lentiviral nef and other viral and cellular substrate proteins. J. Virol. 79, 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frearson J. A.; Brand S.; McElroy S. P.; Cleghorn L. A.; Smid O.; Stojanovski L.; Price H. P.; Guther M. L.; Torrie L. S.; Robinson D. A.; Hallyburton I.; Mpamhanga C. P.; Brannigan J. A.; Wilkinson A. J.; Hodgkinson M.; Hui R.; Qiu W.; Raimi O. G.; van Aalten D. M.; Brenk R.; Gilbert I. H.; Read K. D.; Fairlamb A. H.; Ferguson M. A.; Smith D. F.; Wyatt P. G. (2010) N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature 464, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. H.; Clough B.; Rackham M. D.; Rangachari K.; Brannigan J. A.; Grainger M.; Moss D. K.; Bottrill A. R.; Heal W. P.; Broncel M.; Serwa R. A.; Brady D.; Mann D. J.; Leatherbarrow R. J.; Tewari R.; Wilkinson A. J.; Holder A. A.; Tate E. W. (2014) Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat. Chem. 6, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate E. W.; Bell A. S.; Rackham M. D.; Wright M. H. (2014) N-Myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology 141, 37–49. [DOI] [PubMed] [Google Scholar]
- Bell A. S.; Mills J. E.; Williams G. P.; Brannigan J. A.; Wilkinson A. J.; Parkinson T.; Leatherbarrow R. J.; Tate E. W.; Holder A. A.; Smith D. F. (2012) Selective inhibitors of protozoan protein N-myristoyltransferases as starting points for tropical disease medicinal chemistry programs. PLoS Negl. Trop. Dis. 6, e1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinon E.; Serwa R. A.; Broncel M.; Brannigan J. A.; Brassat U.; Wright M. H.; Heal W. P.; Wilkinson A. J.; Mann D. J.; Tate E. W. (2014) Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 5, 4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Brannigan J. A.; Moss D. K.; Brzozowski A. M.; Wilkinson A. J.; Holder A. A.; Tate E. W.; Leatherbarrow R. J. (2012) Design and synthesis of inhibitors of Plasmodium falciparum N-myristoyltransferase, a promising target for antimalarial drug discovery. J. Med. Chem. 55, 8879–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham M. D.; Brannigan J. A.; Rangachari K.; Meister S.; Wilkinson A. J.; Holder A. A.; Leatherbarrow R. J.; Tate E. W. (2014) Design and synthesis of high affinity inhibitors of Plasmodium falciparum and Plasmodium vivax N-myristoyltransferases directed by ligand efficiency dependent lipophilicity (LELP). J. Med. Chem. 57, 2773–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaleye T. O.; Brannigan J. A.; Roberts S. M.; Leatherbarrow R. J.; Wilkinson A. J.; Tate E. W. (2014) Peptidomimetic inhibitors of N-myristoyltransferase from human malaria and leishmaniasis parasites. Org. Biomol. Chem. 12, 8132–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton J. A.; Goncalves V.; Brannigan J. A.; Paape D.; Wright M. H.; Waugh T. M.; Roberts S. M.; Bell A. S.; Wilkinson A. J.; Smith D. F.; Leatherbarrow R. J.; Tate E. W. (2014) Structure-based design of potent and selective Leishmania N-myristoyltransferase inhibitors. J. Med. Chem. 57, 8664–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S.; Norcross N. R.; Thompson S.; Harrison J. R.; Smith V. C.; Robinson D. A.; Torrie L. S.; McElroy S. P.; Hallyburton I.; Norval S.; Scullion P.; Stojanovski L.; Simeons F. R.; van Aalten D.; Frearson J. A.; Brenk R.; Fairlamb A. H.; Ferguson M. A.; Wyatt P. G.; Gilbert I. H.; Read K. D. (2014) Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis. J. Med. Chem. 57, 9855–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A.; McWherter C. A.; Freeman S. K.; Wood D. C.; Gordon J. I.; Lee S. C. (1995) Genetic studies reveal that myristoylCoA:protein N-myristoyltransferase is an essential enzyme in Candida albicans. Mol. Microbiol. 16, 241–250. [DOI] [PubMed] [Google Scholar]
- Lodge J. K.; Jackson-Machelski E.; Toffaletti D. L.; Perfect J. R.; Gordon J. I. (1994) Targeted gene replacement demonstrates that myristoyl-CoA: protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc. Natl. Acad. Sci. U. S. A. 91, 12008–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas B.; Freeman S. K.; Zupec M. E.; Lu H. F.; Nagarajan S. R.; Kishore N. S.; Lodge J. K.; Kuneman D. W.; McWherter C. A.; Vinjamoori D. V.; Getman D. P.; Gordon J. I.; Sikorski J. A. (1997) Design and synthesis of novel imidazole-substituted dipeptide amides as potent and selective inhibitors of Candida albicans myristoylCoA:protein N-myristoyltransferase and identification of related tripeptide inhibitors with mechanism-based antifungal activity. J. Med. Chem. 40, 2609–2625. [DOI] [PubMed] [Google Scholar]
- Masubuchi M.; Kawasaki K.; Ebiike H.; Ikeda Y.; Tsujii S.; Sogabe S.; Fujii T.; Sakata K.; Shiratori Y.; Aoki Y.; Ohtsuka T.; Shimma N. (2001) Design and synthesis of novel benzofurans as a new class of antifungal agents targeting fungal N-myristoyltransferase. Bioorg. Med. Chem. Lett. 11(Part 1), 1833–1837. [DOI] [PubMed] [Google Scholar]
- Ebiike H.; Masubuchi M.; Liu P.; Kawasaki K.; Morikami K.; Sogabe S.; Hayase M.; Fujii T.; Sakata K.; Shindoh H.; Shiratori Y.; Aoki Y.; Ohtsuka T.; Shimma N. (2002) Design and synthesis of novel benzofurans as a new class of antifungal agents targeting fungal N-myristoyltransferase. Bioorg. Med. Chem. Lett. 12(Part 2), 607–610. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S.; Eisenhaber B.; Eisenhaber F. (2002) N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 317, 541–557. [DOI] [PubMed] [Google Scholar]
- Shaw B. D.; Momany C.; Momany M. (2002) Aspergillus nidulans swoF encodes an N-myristoyl transferase. Eukaryot Cell 1, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogabe S.; Masubuchi M.; Sakata K.; Fukami T. A.; Morikami K.; Shiratori Y.; Ebiike H.; Kawasaki K.; Aoki Y.; Shimma N.; D’Arcy A.; Winkler F. K.; Banner D. W.; Ohtsuka T. (2002) Crystal structures of Candida albicans N-myristoyltransferase with two distinct inhibitors. Chem. Biol. 9, 1119–1128. [DOI] [PubMed] [Google Scholar]
- Panethymitaki C.; Bowyer P. W.; Price H. P.; Leatherbarrow R. J.; Brown K. A.; Smith D. F. (2006) Characterization and selective inhibition of myristoyl-CoA:protein N-myristoyltransferase from Trypanosoma brucei and Leishmania major. Biochem. J. 396, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer P. W.; Gunaratne R. S.; Grainger M.; Withers-Martinez C.; Wickramsinghe S. R.; Tate E. W.; Leatherbarrow R. J.; Brown K. A.; Holder A. A.; Smith D. F. (2007) Molecules incorporating a benzothiazole core scaffold inhibit the N-myristoyltransferase of Plasmodium falciparum. Biochem. J. 408, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocque W. J.; McWherter C. A.; Wood D. C.; Gordon J. I. (1993) A comparative analysis of the kinetic mechanism and peptide substrate specificity of human and Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J. Biol. Chem. 268, 9964–9971. [PubMed] [Google Scholar]
- d’Enfert C. (1996) Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30, 76–82. [DOI] [PubMed] [Google Scholar]
- Romero B.; Turner G.; Olivas I.; Laborda F.; De Lucas J. R. (2003) The Aspergillus nidulans alcA promoter drives tightly regulated conditional gene expression in Aspergillus fumigatus permitting validation of essential genes in this human pathogen. Fungal Genet. Biol. 40, 103–114. [DOI] [PubMed] [Google Scholar]
- Duronio R. J.; Rudnick D. A.; Johnson R. L.; Johnson D. R.; Gordon J. I. (1991) Myristic acid auxotrophy caused by mutation of S. cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J. Cell Biol. 113, 1313–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heal W. P.; Wright M. H.; Thinon E.; Tate E. W. (2012) Multifunctional protein labeling via enzymatic N-terminal tagging and elaboration by click chemistry. Nat. Protoc. 7, 105–117. [DOI] [PubMed] [Google Scholar]
- Howard S. J.; Lass-Florl C.; Cuenca-Estrella M.; Gomez-Lopez A.; Arendrup M. C. (2013) Isavuconazole susceptibility of Aspergillus and Candida species by EUCAST method. Antimicrob. Agents Chemother. 57, 5426–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. W.; Bowyer P. (2013) Voriconazole resistance in Aspergillus fumigatus: should we be concerned?. Clin. Infect. Dis. 57, 521–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby S. M.; Heukeroth R. O.; Gordon J. I.; Gilman A. G. (1990) G-protein alpha-subunit expression, myristoylation, and membrane association in COS cells. Proc. Natl. Acad. Sci. U. S. A. 87, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. C.; Shaw B. D. (2008) ArfB links protein lipidation and endocytosis to polarized growth of Aspergillus nidulans. Commun. Integr. Biol. 1, 51–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M. E.; Pang I. H.; Duronio R. J.; Gordon J. I.; Sternweis P. C.; Gilman A. G. (1991) Lipid modifications of G protein subunits. Myristoylation of Go alpha increases its affinity for beta gamma. J. Biol. Chem. 266, 4654–4659. [PubMed] [Google Scholar]
- Lafon A.; Seo J. A.; Han K. H.; Yu J. H.; d’Enfert C. (2005) The heterotrimeric G-protein GanB(alpha)-SfaD(beta)-GpgA(gamma) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebmann B.; Gattung S.; Jahn B.; Brakhage A. A. (2003) cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269, 420–435. [DOI] [PubMed] [Google Scholar]
- Zhao W.; Panepinto J. C.; Fortwendel J. R.; Fox L.; Oliver B. G.; Askew D. S.; Rhodes J. C. (2006) Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect. Immun. 74, 4865–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebmann B.; Muller M.; Braun A.; Brakhage A. A. (2004) The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect. Immun. 72, 5193–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Y.; Seo J. A.; Kim J. E.; Han K. H.; Shim W. B.; Yun S. H.; Lee Y. W. (2008) Functional analyses of heterotrimeric G protein G alpha and G beta subunits in Gibberella zeae. Microbiology 154, 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopffleisch K.; Phan N.; Augustin K.; Bayne R. S.; Booker K. S.; Botella J. R.; Carpita N. C.; Carr T.; Chen J. G.; Cooke T. R.; Frick-Cheng A.; Friedman E. J.; Fulk B.; Hahn M. G.; Jiang K.; Jorda L.; Kruppe L.; Liu C.; Lorek J.; McCann M. C.; Molina A.; Moriyama E. N.; Mukhtar M. S.; Mudgil Y.; Pattathil S.; Schwarz J.; Seta S.; Tan M.; Temp U.; Trusov Y.; Urano D.; Welter B.; Yang J.; Panstruga R.; Uhrig J. F.; Jones A. M. (2011) Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Cerezo M.; Sanchez-Rodriguez C.; Escudero V.; Miedes E.; Fernandez P. V.; Jorda L.; Hernandez-Blanco C.; Sanchez-Vallet A.; Bednarek P.; Schulze-Lefert P.; Somerville S.; Estevez J. M.; Persson S.; Molina A. (2012) Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol. Plant 5, 98–114. [DOI] [PubMed] [Google Scholar]
- Coca M. A.; Damsz B.; Yun D. J.; Hasegawa P. M.; Bressan R. A.; Narasimhan M. L. (2000) Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J. 22, 61–69. [DOI] [PubMed] [Google Scholar]
- Lee S. C.; Schmidtke S. N.; Dangott L. J.; Shaw B. D. (2008) Aspergillus nidulans ArfB plays a role in endocytosis and polarized growth. Eukaryot Cell 7, 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B.; May G. S.; Morris N. R. (1989) Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79, 119–130. [DOI] [PubMed] [Google Scholar]
- Langfelder K.; Gattung S.; Brakhage A. A. (2002) A novel method used to delete a new Aspergillus fumigatus ABC transporter-encoding gene. Curr. Genet. 41, 268–274. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.; Minor A. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 307–326. [DOI] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- (1994) The CCP4 suite: programs for protein crystallography, Acta Crystallogr., Sect. D: Biol. Crystallogr. 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Perrakis A.; Morris R.; Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Schuettelkopf A. W.; van Aalten D. M. F. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 1355–1363. [DOI] [PubMed] [Google Scholar]
- DeLano W. L. (2004) Use of PYMOL as a communication tool for molecular science. Abstr. Pap. Am. Chem. Soc. 228, 030-CHED. [Google Scholar]
- Espinel-Ingroff A.; Fothergill A.; Ghannoum M.; Manavathu E.; Ostrosky-Zeichner L.; Pfaller M. A.; Rinaldi M. G.; Schell W.; Walsh T. J. (2007) Quality control and reference guidelines for CLSI broth microdilution method (M38-A document) for susceptibility testing of anidulafungin against molds. J. Clin. Microbiol. 45, 2180–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.