Abstract
Dissociated hippocampal neurons exposed to a variety of degenerative stimuli form neuritic cofilin-actin rods. Here we report on stimulus driven regional rod formation in organotypic hippocampal slices. Ultrastructural analysis of rods formed in slices demonstrates mitochondria and vesicles become entrapped within some rods. We developed a template for combining and mapping data from multiple slices, enabling statistical analysis for the identification of vulnerable sub-regions. Amyloid-β (Aβ) induces rods predominantly in the dentate gyrus region, and Aβ-induced rods are reversible following washout. Rods that persist 24 h following transient (30 min) ATP-depletion are broadly distributed, whereas rods formed in response to excitotoxic glutamate localize within and nearby the pyramidal neurons. Time-lapse imaging of cofilin-GFP-expressing neurons within slices shows neuronal rod formation begins rapidly and peaks by 10 min of anoxia. In ~50% of responding neurons, Aβ-induced rod formation acts via cdc42, an upstream regulator of cofilin. These new observations support a role for cofilin-actin rods in stress-induced disruption of cargo transport and synaptic function within hippocampal neurons and suggest both cdc42-dependent and independent pathways modulate cofilin activity downstream from Aβ.
Keywords: Actin, actin depolymerizing factor (ADF)/cofilin, actin inclusions, Alzheimer's disease, amyloid-β, ischemic brain injury
INTRODUCTION
Proteins of the actin depolymerizing factor (ADF)/cofilin family are major intracellular regulators of actin assembly dynamics [1]. In metazoans, these proteins are reversibly phosphoregulated on a conserved Ser-3 residue [2]. Inhibitory phosphorylation is catalyzed by LIM kinase (LIMK) and other kinases and activating dephosphorylation is catalyzed principally by slingshot (SSH) or chronophin phosphatases [3–7]. Because cofilin concentrations in mammalian hippocampal neurons are 5–12 fold higher than ADF [8,9], we will henceforth refer only to cofilin. Misregulation of neuronal actin and cofilin is associated with a range of cognitive impairment and degenerative conditions [10]. Exposure of cultured dissociated hippocampal neurons to peroxide (oxidative stress), excitotoxic levels of glutamate, ATP-depletion medium, or oligomers of amyloid-β (Aβ)1–42 induce the rapid activation of cofilin (dephosphorylation) leading to the formation of cofilin-actin rods [8,11,12]. Rods may provide transient neuroprotection from stress by sequestering most of the cofilin in non-dynamic structures that spare ATP utilized in actin turnover [13,14]. However, rods ultimately impair synaptic plasticity and induce synaptic loss [15].
Cofilin/actin containing inclusions (cofilin pathology) have been identified in brains of mice overexpressing a familial Alzheimer's disease (AD) mutation of amyloid-β protein precursor (AβPP) [11], where their formation is associated with Aβ-activated signaling pathways that lead to the loss of cognitive function [16]. Thus, stress-induced rods are a potential mechanism for cognitive and functional impairment associated with vascular diseases, anoxia, and AD. Although an interference in spine dynamics and synaptic plasticity during treatment with soluble forms of Aβ oligomers occurs in the excitatory pyramidal cells of the Cornu Ammonis (CA) region in the hippocampus [17,18], little is actually known concerning other major targets of Aβ oligomers within the hippocampus [19].
AD is characterized pathologically by the presence of amyloid plaques and neurofibrillary tangles in the brain. Here we characterize the properties and localization of an AD-related pathological feature, cofilin-actin rods, within subfields of ex vivo rodent hippocampus. Rod-like cofilin pathology was first described in the brains of human AD patients, in a rat hippocampal culture model [8], and in brains of AD transgenic mice [11,16]. In human AD brain, cofilin pathology was found associated with almost all amyloid plaques, however 45% of the rod-like pathology was not plaque-associated suggesting that rods may precede plaque deposition [8]. The feed forward hypothesis of rod-induced neurodegeneration proposes that neurodegenerative stimuli, including ischemic stress (stroke), glutamate excitotoxicity (seizure), oxidative stress (reactive oxygen species), and Aβ peptides (AD and Down syndrome), induce rod formation that blocks intraneu-rite transport [11]. It further proposes that Aβ production within endosomes [20], one of the subcellular sites of Aβ generation, may be either enhanced or altered within stalled vesicles to increase the release of Aβ species, such as dimers, which have a greater effect on disrupting communication within the hippocampus than do larger oligomers and fibrils [18]. Using organotypic slice cultures of rodent hippocampus, we sought to identify populations of neurons that may be responding to different neurodegenerative signals by forming rods and to examine the signaling pathway from Aβ1–42 in this process.
MATERIALS AND METHODS
Reagents
All chemicals are reagent grade and were obtained from Sigma-Aldrich Co. and all tissue culture reagents were from Life Technologies (Invitrogen, Carlsbad, CA). Aβ peptide (Aβ1–42 and a scrambled peptide with the same amino acid composition) were purchased from AnaSpec, Inc. (San Jose, CA).
Animals
Timed pregnant Sprague Dawley rats were obtained from Harlan (Indianapolis, IN).
Pups were sacrificed on postnatal days 6–10 and adults as needed as per NIH and AUCC approved protocols. Postnatal day 1–5 mouse pups were obtained from a breeding colony of Thy-1-YFP mice, line H, generously provided to us by Stuart Tobet but originally obtained from Jeff Lichtman [21]. Conditional neuronal cdc42 null mouse embryos were obtained from a Nestin-Cre+/–, Cdc42fl/wt × Cdc42fl/fl cross [9] and GFP-expressing wild type embryos were obtained from transgenic mice with GFP under the ubiquitous “CAG” promoter, composed of the CMV enhancer, a fragment of the chicken β-actin promoter and rabbit β-globin exons [22,23]. Animal studies were performed according to the National Research Council's guide for care and use of laboratory animals using protocols approved by the Institutional Animal Care and Use Committee.
Organotypic slice culture
Except as noted, transverse hippocampal slice cultures were prepared from P6-P10 Sprague Dawley rat pups essentially as described previously [24]. In one study, slices were prepared from adult female Sprague Dawley rats sacrificed during harvesting of E18 fetal pups, and in another study from P3-5 adult male/female Thy1-YFP mice (line H) in which Thy1-YFP is expressed in a limited number of hippocampal neurons allowing the identification of their neurites. Briefly, hippocampi were quickly dissected into filter sterilized ice-cold (4°C) Gey's Balanced Salt Solution plus 4% glucose, then sliced to a thickness of 400 μm on a McIlwain tissue chopper. We maintained some of the entorhinal cortex along with the hippocampus in these slices to minimize the degeneration of the perforant pathway (Supplemental Fig. S1). For many slice treatments, 3–6 slices were arranged onto 0.4 μm Transwell® Polyester membranes inserted into 6 well culture plates (Corning Costar® 3450, Lowell, MA). Beneath the membrane is added 1.7 mL of filter sterilized slice culture medium (50 mL horse serum, 50 mL Hank's Balanced Salt Solution (HBSS), 100 mL Minimal Essential Medium (MEM), 250 μL 200 mM GlutaMAX-1, 4 mL 25% glucose, 1 mL 100 U/mL Penicillin-Streptomycin). MEM is 4-(2-hydroxyethyl)-1-piperazineethane sulfonic acid (HEPES) and bicarbonate buffered. Slice culture medium was aspirated and replaced with 1.5 mL of fresh medium on day 3 and every 2–3 days thereafter or with treatment medium as required. For all experiments slices were cultured for about 10 days in a 95% air/5% CO2 incubator at 35.5°C.
For live cell imaging, 1–2 hippocampi were placed onto 12 × 22 mm coverslips, and embedded in 20 μL of chicken plasma (Cocalico Biologicals, Inc., Reamstown, PA) containing 6 μL of freshly added thrombin (150 NIH units/mL in water; MP Biomedicals, Inc.). Slides were placed hippocampal side up on flat bottom test tubes (Nunclon Delta Tubes, Nalge Nunc, Rochester, NY) and 700 μL slice culture medium was added. Tubes were placed at a 5° angle in a roller incubator (10 revolutions per hour) at 35°C and medium was replaced every 2–3 days. Imaging was performed on day 10.
Slice culture treatments
Aβ oligomer was made by solubilizing the synthetic peptide in hexafluoroisopropanol and drying 10 μg aliquots. Each 10 μg of synthetic Aβ1–42 was solubilized in 10 μL of DMSO, diluted with 78.6 μl of sterile Ham's F-12 (25 μM stock) and incubated 24 h at 4°C [11,25,26]. Scrambled peptide was prepared identically. For treatments of slices on membrane (all on day 8), hydrogen peroxide was added to a final concentration of 10 μM, glutamate was added to a final concentration of 125 μM, and Aβ1–42 oligomer was added to final concentration of 1 μM. A small amount of the treated medium was layered over each slice then the cultures were placed back in the 95% air/5% CO2 incubator at 35.5°C for 48 h before the medium was removed and replaced with fresh medium, or slices were fixed. For ATP depletion experiments, the slice culture medium was aspirated and replaced with ATP depletion medium (6 mM 2-deoxyglucose, 10 mM NaN3 in Phosphate Buffered Saline (PBS)) and cultures were placed into a humidified air incubator at 37°C for 1 h before being immediately fixed or having the medium removed and replaced with fresh slice medium. For TUNEL staining, slices were treated as above, or for 24 h with 50 μM etoposide (an inducer of apoptosis as a positive control), rinsed in PBS, and fixed in 4% paraformaldehyde for 1 h. Samples were rinsed 3× with PBS and permeabilized with 0.1% Triton X-100 for 10 min at 4°C. TUNEL staining with labeled UTP was performed according to the manufacturer's directions for 60 min at 37°C (Roche Diagnostics, Indianapolis, IN). After TUNEL labeling, slices were immunostained for cofilin (Alexa 488 secondary) and nuclei were stained with Hoechst prior to mounting in ProLong Gold Antifade (Molecular Probes).
Dissociated hippocampal neuronal cultures
Primary hippocampal neurons from E16.5-17.5 mouse embryos [9] or E18 rat embryos [8] were cultured essentially as described. For the mixed wild-type (GFP-positive)/Cdc42 knockout (GFP-negative) cultures, the hippocampi of embryos from a Nestin-Cre+/–, Cdc42fl/wt × Cdc42fl/fl cross were dissected, trypsinized and dissociated individually for each embryo. In parallel, hippocampi from wild type, GFP positive embryos were dissected, trypsinized, and dissociated. The cells were then washed in HBSS containing 7 mM HEPES, pH 7.25, and 6–6.5 × 104 wild type cells were plated together with 8–9 × 104 Cdc42 knockout cells onto polylysine-coated glass coverslips in 6 cm tissue culture dishes containing MEM and 10% heat-inactivated horse serum. The cultures were grown in a humidified tissue culture incubator at 36.5°C, 5% CO2, and after 12–20 h the coverslips were inverted in 6 cm dishes containing astrocytes in N2 medium. E16.5 mouse hippocampal neurons used for the experiments with adenoviral-mediated expression were cultured as described [27].
Adenoviral-mediated gene expression
Adenoviruses for expressing the myc-tagged small GTPase cdc42 in constitutively active (V12cdc42) and dominant negative (N17cdc42) forms have been described [28] and were used at a multiplicity of infection (m.o.i.) of 100–300 for infecting dissociated neurons. Adenoviruses for expressing a human cofilin green fluorescent protein (hCof-GFP) chimera were made using the AdEasy system [29] as previously modified [30]. The human cofilin cDNA sequence was isolated from a pET vector (a gift from Alan Weeds, MRC Laboratory of Molecular Biology) and cloned into pEGFP-N1 (Clonetech), which was subsequently used to clone into pShuttle-CMV for virus production. About 107 adenoviral particles were added directly to the slice culture medium on day 7 and the cultures were returned to the incubator until viewed on day 10. Slices cultured on membranes were infected with adenovirus by placing a drop of the adenovirus directly on the slice and adding the excess to the culture medium below the slice. One to two hours later the liquid on top of the slice was mixed with the bath medium. Slices excised from the membrane were made anoxic by placing them face down onto glass-bottomed 35 mm culture dishes and covering them with a glass coverslip. Slice cultures grown on coverslips in roller tubes were rinsed in medium without virus, mounted slice down on a microscope slide and quickly sealed around the edges with paraffin.
Fixation and immunostaining
Slices were fixed for 4 h at room temperature in 4% paraformaldehyde in either cytoskeletal buffer (CBS; 10 mM MES pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA pH 7.0, 4% PEG, 0.32 M sucrose) or PBS adjusted to pH 7.0, with no apparent differences between buffers. Slices were methanol (–20°C) permeabilized for 10 min and blocked in 2% goat serum/1% bovine serum albumin in Tris-buffered saline before immunostaining. In experiments where fluorescent phalloidin was used to stain actin filaments, slices were permeabilized with 0.1% Triton X-100 in PBS for 10 min. Primary antibodies include: affinity purified rabbit IgG to chick ADF (1439; 2 ng/μL), which cross-reacts with mammalian ADF and cofilin [8,31], protein A purified monoclonal mouse anti-cofilin (MAb22; 10 ng/μL IgG) [32], affinity purified rabbit IgG to the phosphorylated peptide of chick ADF (rabbit 4321) [33], and mouse monoclonal antibody to actin (clone C4; ICN Biomedicals, Inc., Costa Mesa, CA). Secondary antibodies, all used at 1:450, include fluorescein goat anti-rabbit and goat anti-mouse and Texas-Red goat anti-rabbit and goat anti-mouse (Molecular Probes, Eugene, OR). Texas Red-X phalloidin (Molecular Probes) was used at 1:50 in PBS (final concentration 150 nM). DAPI (4’-6-Diamidino-2 phenylindole) or Hoechst 33342 were used to stain DNA. After blocking and staining, slices on membrane were cut out and mounted on 22 × 22 mm cover glasses with ProLong Gold Antifade.
Light microscopy
Phase-contrast and non-confocal fluorescence micrographs were obtained on a Nikon Diaphot using 4× (0.13 NA), 10× (0.25 NA), 20× (0.75 NA) air objectives or 40× (1.3 NA), 60× (1.4 NA), 100× (1.4 NA) oil objectives as previously described [8]. An Olympus IX81 microscope equipped with an ASI piezo stage (Applied Scientific Instrumentation, Eugene, OR), CSU22 spinning disk confocal head (Yokogawa Instruments, Japan), 440 nm, 473 nm and 561 nm diode lasers, and a Cascade II EMCCD Camera (Roper Scientific, Tucson, AZ), all integrated and operated by SlideBook software (Intelligent Imaging Innovations, Denver, CO), was used to obtain confocal sections through organotypic slices. The objectives used include a 4× Fluorite (0.13 NA), UAPO40X/340W-DIC (1.35 NA), or Plan-APO 60× (1.42 NA). For experiments in which we localized active cofilin across an entire slice, fixed slices were immunostained for phospho-cofilin (rabbit antibody 4321) and total cofilin (mouse MAb22), stained with different fluorescently tagged secondary antibodies, and imaged using the 4× objective, which allowed capture of most of the hippocampal area in one field. The two images were overlaid (Total cofilin/phospho-cofilin) and a hot scale applied to the ratio image such that the hottest colors correspond to the regions of most active (dephosphorylated) cofilin.
For live cell imaging, slices infected with adenoviruses for expressing hCof-GFP or hCof-RFP were made anoxic and images captured immediately and every 20 s for 10–15 min at 40× or 60× on the spinning disk confocal microscope at various stage positions for cells expressing fluorescent protein. Meta-Morph v7.03 software (MDS Analytical Technologies, Toronto, Canada) was used for all digital processing. Following time-lapse imaging the slice was scanned for rod formation. All experiments were repeated a minimum of three times. To ascertain the regional distribution of rods, the total number of rods per field was counted using a 60× oil objective. Slices were scanned in a grid-like pattern to sample a large area of the slice (Supplemental Fig. 2). By mapping the rod counts per 60× field onto a low power (4×) image (or montage) of the slice, the location of the high-power field within the slice was determined. This mapping was facilitated by the bleaching of background fluorescence in the region examined at high power. Data from multiple slices were combined onto a schematic of the hippocampus as discussed in the Results.
Electron Microscopy
Organotypic slices subjected to the same treatments as slices used for immunofluorescence analysis were fixed in 2% glutaraldehyde in MOPS buffer (20 mM MOPS, pH 7.0, 8 mM MgCl2, 5 mM NaN3, 5 mM EGTA) for actin cytoskeletal preservation. The tissue was processed as previously described [34]. Briefly, after primary fixation, the slices were rinsed 4× in MOPS buffer, post-fixed with 1% OsO4 at 4°C, rinsed 6× in cold water, incubated in 2% uranyl acetate for 25 min, and dehydrated in graded ethanol. The slices were transitioned into Epon812 and polymerized at 60 °C for 24 h. Thin (60–70 nm) and thick (200–300 nm) sections were cut and post stained with uranyl acetate and lead citrate. Thick sections were topically labeled with 12–15 nm gold particles to serve as fiducial markers in tomographic reconstruction. Images were captured on a Gatan 4 k × 4 k CCD camera with a FEI Technai TF20 200kV IVEM (Boulder Lab for 3D Electron Microscopy, University of Colorado, Boulder, CO). Tilt sections ranging from +/– 70° were obtained with post acquisition analysis done with IMOD software designed in this facility. Thin sections were analyzed with a JEOL TEM 1200EX operating at 100 kV. Images were capture on conventional film prior to digitization.
Statistics
Statistical analyses were done with either MATLAB or SPSS v13 with a set significance level described in the figure legends. Any post hoc tests are reported.
RESULTS
For investigation of regional rod distribution within the hippocampus, we utilized organotypicslice cultures because they largely retain the cytoarchitecuture and connectivity of the in vivo hippocampus. Slice cultures were prepared with care to preserve entorhinal cortex (EC) axons that comprise a substantial fraction of the perforant pathway. We verified maintenance in our cultures by DiI labeling within the EC and following the tracks into the hippocampus proper (Supplemental Fig. S1; Supplement available online at http:/www.jalz.com/issues/18/davis_supplement.pdf). All analysis for rod formation was performed only within the hippocampal formation.
Properties of cofilin-actin rods in organotypic hippocampal slice cultures
A rod is defined as a tapered cylindrical inclusion immunostained for cofilin and actin (Fig. 1A–C), but which does not label with fluorescent phalloidin, because rods contain a high ratio of cofilin/actin, blocking the phalloidin binding site [8,35]. By definition rods are readily distinguished from ovoid actin-containing aggresomes and Hirano Bodies [36]. Methanol permeabilization alters F-actin structure, preventing phalloidin binding, but preserves rods and provides better immunostaining of cofilin in the rods. Triton X-100 permeabilization reduces the intensity of cofilin immunostaining, yet permits the demonstration that cofilin-stained rods do not bind fluorescent phalloidin whereas other F-actin structures are clearly stained (Fig. 1D–F). Thus, for subsequent experiments, slices were permeabilized with methanol unless noted.
Fig. 1.
Rods in brain slices immunostain for cofilin and actin but do not stain with fluorescent phalloidin. Cold methanol permeabilization is optimal for immunostaining rods (arrowheads) for both cofilin (A) and actin (B) shown as a merged image in (C), but is not compatible with phalloidin staining. Permeablizing with Triton X-100 preserves cofilin immunostaining in rods (arrowheads in D-F) and permits Texas-red phalloidin staining of some F-actin structures, but not rods (E). The merged overlays (F) show that the cofilin rods (green) are distinct from other cofilin and phalloidin stained structures (F). Scale bar = 10 μm.
Owing in part to the low probability of locating a rod (0.1–0.2 μm × 2–4 μm) within thin sections of brain tissue, we have been unsuccessful in our attempts to identify rods using electron microscopy on human AD brain or brains of AD transgenic mice (Tg2576). The high density of rods present in Aβ-treated slices (see below) improved these odds, thus making it possible to conduct high resolution ultrastructural analysis. At the EM level of resolution, a rod is composed of many individual parallel filaments arranged into tightly bundled structures (Fig. 2), often with tapering ends. Of interest, and not previously reported, several rods contained entrapped mitochondria and vesicles (Fig. 2A, B). 3-D tilt series detailed some rods filling most of the neurite and disrupting microtubules (Fig. 2A, arrow), a finding previously reported in dissociated neuronal cultures both by immunostaining of microtubules and by ultrastructure of rods induced by ATP-depletion in dissociated neurons [8]. Furthermore, rod ultrastructure and their appearance in tandem arrays are strikingly similar to filaments in striated neuropil threads [37], an early pathological feature of human AD brain.
Fig. 2.
Transmission electron micrographs of thick (250 nm) sections of organotypic rat hippocampal slices that were treated with 1 μM Aβ for 48 h, fixed, stained and processed for TEM as described in Materials and Methods. Colloidal gold fiduciary labels were added to the tops of sections prior to accumulation of X and Y tilt series of images. (A) Linear filament array within a neurite in which two mitochondria (asterisks) appear to have become entrapped. Arrow points to a microtubule that can be followed in the tilt series as entering the rod structure and terminating there. (B) Vesicle (asterisk) surrounded by filament bundles. Bars = 100 nm.
Aβ1–42 oligomers induce rod formation within neurons in organotypic slice culture
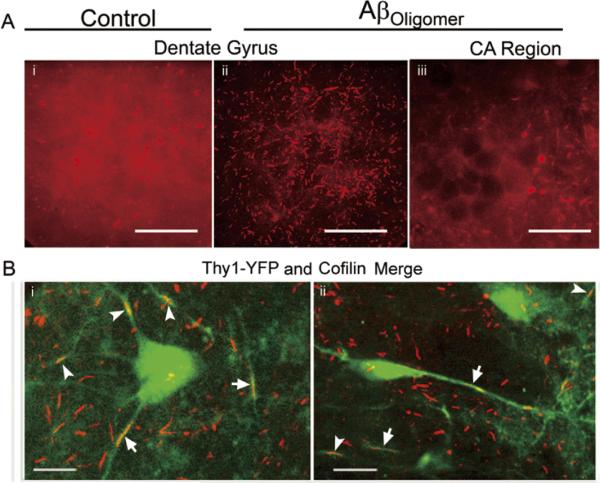
We first wanted to determine if Aβ-induced rods were being formed within subregions of the hippocampus and if they occurred in neurons. Rods formed in a maximum of 18–19% of the neurons in dissociated hippocampal cultures treated with synthetic Aβ1–42 oligomers at the highest sublethal concentration (1 μM) [11]. Organotypic slices treated 48 h with 1 μM synthetic Aβ1–42 display a marked regional distribution of rods within the hippocampus (Fig. 3A). Aβ1–42 oligomers induce rods primarily within the dentate gyrus (DG) while the CA1 and CA3 sub-regions remain largely unaffected. Neuronal rod formation was confirmed by confocal analysis in organotypic slices from Thy-1-YFP (line H) mice following 48 h treatment with 1 μM synthetic Aβ1–42 (Fig. 3B). In this particular line, Thy1-YFP is only expressed in a subset of hippocampal neurons making their neurites easier to follow. Identical results were obtained in slice cultures infected with an adenovirus for neuronal specific expression of RFP (data not shown). Uniform penetration of the slices by the oligomeric Aβ was assessed by immunostaining of Aβ-treated slices (data not shown).
Fig. 3.
Rods form in neurons in specific sub-regions of organotypic hippocampal slice cultures in response to Aβ-treatment. (A) Immunofluorescence staining of cofilin with a rabbit total cofilin antibody in the dentate gyrus (i, ii) or CA1 regions (iii) of rat hippocampal organotypic slices treated with 1 μM of synthetic Aβ1–42 oligomers (ii, iii) or left untreated (i). Projected images from a confocal Z-stack show abundant rod formation within the DG of Aβ-treated (ii) but not control slices (i). In CA and other regions (not shown) of the Aβ-treated slice, rod numbers are above background levels but are not abundant (iii). Less diffuse cofilin staining in (ii) suggests that most available cofilin in rod forming regions is sequestered into rods. In both Aβ-treated and untreated slices there are regions with globular accumulation of cofilin, which may represent aggresomes. (B) Merged confocal sections of Aβ1–42-induced rods (red; arrowheads) in Thy1-YFP-expressing neurons from a mouse organotypic hippocampal slice. (i, ii) YFP fluorescence (green) shows Thy1-YFP expressing neurons. Linear arrays of rods in the same neurite are designated with arrows. Scale bars = 10 μm.
Activation of cofilin occurs in stimulus sensitive sub-regions of organotypic slices
We next wanted to determine if Aβ-induced cofilin dephosphorylation was widespread throughout the hippocampus or was enhanced in the subregions where rods formed. Regardless of the precipitating insult, rod formation in dissociated hippocampal neurons follows activation (dephosphorylation) of the cofilin pool [8, 11]. As a result, rods are refractory to labeling with antibodies recognizing Ser-3 phosphorylated cofilin (Fig. 4A). Integrating the ratio of total cofilin immunostaining divided by phosphorylated cofilin immunostaining over the entire hippocampal slice reveals elevated cofilin activity especially in the DG, the region where Aβ-induced rod formation is most prominent (Fig. 4B,C).
Fig. 4.
Rods contain active (dephospho) cofilin but not phospho-cofilin. (A) Rat hippocampal organotypic slice treated with Aβ1–42 was fixed and co-stained with a mouse monoclonal antibody for total cofilin and with a rabbit antibody for phospho-cofilin; different fluorescent secondary antibodies were used. In the left panel total cofilin stained rods (arrowheads) which are not visible in the channel for the phospho-cofilin antibody (middle panel). Overlay of these images with a hot scale for the ratio of total cofilin/phospho-cofilin generated the panel on the right. Bar = 10 μm. (B) The same immunostaining and ratio imaging was applied to low magnification (4× objective) images of entire rat hippocampal slices, either untreated (left) or treated with Aβ-oligomers for 24 h (right). Only image ratios with an overlay of idealized CA and dentate gyrus regions are shown. Bar = 600 μm. (C) Higher magnification images collected from circled regions in (B) of control and Aβ treated slices reveal that a ratio image of a region of relative low rod counts (CA1 non-hot spot) in an Aβ-treated slice appears similar to the control dentate gyrus but the rod-forming regions have more active cofilin. Arrowheads point to rods. Bar = 10 μm.
Template mapping and matrix analysis of hippocampal slices
Morphological differences between slices obtained from the rostral, medial, and caudal regions of the hippocampus made a statistical analysis of rod numbers and location difficult to map over multiple slices. However, after counting rods within a region of an immunostained slice with a 60× objective, the background fluorescence was bleached leaving a dark circle corresponding to the region just viewed (Supplemental Fig. S2A, C). Rods remain bright as the background bleaches owing to the abundance of cofilin within the rods; background bleaching in the slice enhances the signal to noise ratio, thus facilitating rod counting and analysis. Low magnification (4× objective) pre- and post-analysis images stitched together into a montage identified the zones of analysis for rod counts and allowed us to compile a composite image of each slice in which the number of rods within each zone could be overlaid on a DAPI-stained montage showing the pyramidal cell and granule cell nuclear layers of the CA and DG, respectively (Supplemental Fig. S2B, D).
To facilitate a quantitative analysis, we generated a generic map of the hippocampus using conserved landmarks. Twenty fiduciary points were evenly spaced around the granule and pyramidal cell body regions (visualized with DAPI stain) for each slice. These points were then morphed to fit a template before being overlaid on a 20 × 20 matrix where the numbers of rods were placed within their respective boxes (Supplementary Figure S2E-G). Using MATLAB for matrix manipulations, we generated the mean and standard deviation for each matrix box across multiple slices exposed to identical conditions (Fig. 5A). A colorized hot scale was applied with maroon/red “warm” colors corresponding to regions with the highest average numbers of rods and the dark blue “cool” colors to the regions with lowest numbers of rods.
Fig. 5.
Positional information from rod quantification across identically treated rat hippocampal slices was combined as described in Materials and Methods section. (A) The means for rod quantification in each matrix box using the color scale are shown. Although rods were observed sporadically in different matrix boxes from control slices, averaging these across 13 slices eliminated the sporadic hot-spots. Rods formed in slices treated with 1 μM Aβ1–42 oligomers were predominantly in the DG and the mossy fiber regions compared to their distribution across the entire slice in response to 30 min transient ATP-depletion (Azide/2-deoxyglucose) and 24 h recovery. Rods in slices treated with glutamate (125 μM) were most plentiful in and near the pyramidal layer of the CA1 and CA3 regions. A statistical comparison of each matrix box from each treatment with its corresponding control matrix box is shown below each map. The boxes that show a significant difference (p < 0.01 using a two tailed table for p value assignments) for each of the treatments are colored in purple. The black matrix boxes lacked sufficient information to perform the comparison. A lack of significance (gray boxes) in regions where the colored boxes in the upper panel show a high average rod count usually arise from a strong rod response in one or two slices and background levels in others. Numbers of slices used for each composite: Control = 13, Aβ = 9, ATP-depletion = 12, Glutamate = 8. Graphical representation of average rod values per 60× field across the entire organotypic slice (B) or from the defined regions of the hippocampus (C). Application of 1 μM synthetic Aβ1–42 oligomer was the most potent rod-inducing treatment when averaged across the entire slice or just within the DG and mossy fiber regions. Peroxide treatment was above control levels but not significantly so because of a low sample number (n = 3). *Significantly different from scrambled Aβ-peptide-treatment (B) or control (B, C) at p < 0.05. Bars = SEM.
Stimuli dependent patterning of rod formation
We analyzed rod formation across multiple organotypic slices from seven treatment conditions (untreated, vehicle treated, scrambled Aβ1–42, soluble Aβ1–42 oligomers, ATP depletion, hydrogen peroxide, and glutamate). We set the top limit of the hot scale at ≥ 15 rods per box over the selected color range. No significant differences in rod number or distribution were observed between untreated control slices or slices treated with 1 μM scrambled Aβ peptide or vehicle alone, although in each case there were sporadic hot spots, especially near slice edges. The data collected from these three treatments were subsequently combined to generate the control map (Fig. 5A). Following 48 h exposure to synthetic Aβ1–42 oligomers at 1 μM, we observed a robust increase in rod formation within the DG and mossy fiber region (Fig. 5A). By comparison, transient ATP-depletion and recovery produces rods distributed relatively uniformly across the slice. Large numbers of rods (too many to count) formed in slices fixed immediately after transient (30 min) ATP-depletion (data not shown), but after a 24 h recovery rod numbers (persistent rods) were reduced to quantifiable levels (Fig. 5A). Rods formed in response to excitotoxic levels of glutamate localized predominantly within CA1 and CA3 regions and less in the DG (Fig. 5A).
Regional significance of rod number data
We evaluated the statistical significance of the rod numbers in each matrix cell compared to the corresponding cell from controls (Fig. 5A, lower panels). Boxes in which the average rod number is significantly (p < 0.01) elevated from controls are colored purple, boxes for which p > 0.01 are colored gray and boxes that produced no T-test (too few values) are colored black. These analyses demonstrate that the uniquely localized formation of rods in response to each specific stress stimulus is significant.
Rod data averaging over entire slice
We also evaluated the average number of rods per slice in response to each experimental condition tested including hydrogen peroxide for which results were obtained from only 3 slices and were not mapped (Fig. 5B). All treatments, save peroxide, produced a significant (p < 0.05) increase in rod formation when averaged across the entire slice as compared to untreated, and 1 μM scrambled Aβ1–42 controls. We also selectively analyzed average rod formation in the DG, mossy fiber tract, entire CA, alveus, and other regions not included in the above (Fig. 5C). Of note, the DG and mossy fiber tract show heightened sensitivity to rod formation in response to Aβ treatment whereas the numbers of persistent rods following transient ATP-depletion was significantly (p < 0.05) above control in all regions except the DG and mossy fiber tract. Glutamate treatment was equal to or greater in rod-inducing ability to Aβ1–42 outside of the DG. Rod formation by glutamate treatment and ATP-depletion was similar in the CA, alveus, and other regions of the hippocampus and higher than rod numbers induced by Aβ1–42 in these regions.
Rod formation in postnatal versus adult slices
We next wanted to determine if organotypic slices from adult rodents formed rods similarly to the postnatal slices in response to Aβ treatment. Organotypic slices were prepared from both adult rat and mouse hippocampi and were treated with 1 μM amyloid peptide oligomers for 48 h. Four adult organotypic slices, two untreated and two treated with the Aβ1–42 oligomers, were analyzed for rod formation (Supplementary Fig. S3). Rods in adult slices were most prevalent in the DG and mossy fiber region but adult slices were larger and rod density per field was somewhat lower than that observed for the postnatal slices (compare color scales used between Figs 5A and S3) even when compressed to the same 20 × 20 matrix. However, there were no remarkable differences between rod formation and localization in adult mouse versus rat hippocampus in response to Aβ1–42 oligomers, but no statistical comparison could be made between the few slices analyzed.
Transient versus persistent rods
Next we investigated whether the Aβ-induced rods are transient in organotypic hippocampal slices by comparing slices treated 24 h with 1 μM Aβ1–42 oligomer followed by washout and a 24 h recovery to slices treated chronically for 48 h (Fig. 6). The numbers of rods declined to control levels by 24 h after Aβ1–42 washout. Because rod formation in response to Aβ1–42 is maximal by 12–24 h after treatment [11], and during this period of treatment there is neither a decline in the number of cells per field nor an increase in TUNEL positive cells in Aβ-treated versus untreated samples (supplemental Fig. S4), these results suggest that Aβ-induced rods are reversible and do not return soon after recovery. This is unlike the rods induced by transient ATP-depletion, which reform and become persistent by 24 h after washout of the ATP-depletion medium (Fig. 5A).
Fig. 6.
Aβ induced rods are reversible upon washout of the Aβ peptide and a 24 h recovery. Multiple postnatal rat hippocampal slices were used for each treatment and were fixed, immunostained and analyzed for rod numbers and distribution as described for Figure 5A. (A) Composite map from 6 control slices. (B) Composite map from 6 slices treated with 1 μM Aβ1–42 for 48 h. (C) Composite map from 4 slices treated with 1 μM Aβ1–42 for 24 h and then incubated another 24 h after washout of the Aβ. (D) Average number of rod counts per field for the three composite maps shown. Error bars = standard deviation. *Different from control at p < 0.01. #Not different from control but different from the no washout at p < 0.01.
Live imaging of rod formation during ischemic stress
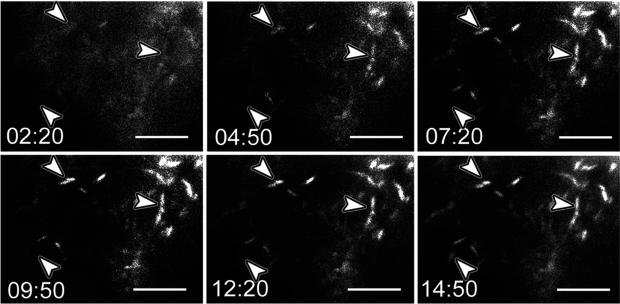
Rods form rapidly, within 5–10 min of ATP-depletion in dissociated hippocampal neurons [8], but we wanted to know if the more nurturing environment of an organotypic slice might provide some protection to rod formation. About 24–48 h before observation, slices were infected with adenovirus mediating human cofilin-GFP expression. Slices on coverslips were rinsed once in fresh 37°C medium immediately before microscopy and a cover slip was overlaid on the slice and sealed with paraffin to initiate anoxia. Slices on membrane were cut out with a scalpel, placed membrane side down on a microscope slide, and overlaid with a coverslip to initiate anoxia. In some areas of a hippocampal slice we observed regions where spontaneous rods were present, perhaps in response to over expression of cofilin-GFP [14], whereas in most other regions rods began to form within a few minutes of anoxia (Fig. 7). To test if hCof-GFP expression alone was inducing rod formation during time lapse imaging, we collected infrequent (30 min) time-lapse images from hippocampal organotypic slices expressing hCof-GFP in which slices were not made anoxic and over 12 h rods were not induced. From these experiments we conclude that anoxia is a potent inducing stimulus for rapid rod formation. As with rods formed in dissociated neurons during ATP depletion, rod length and width reached maximum proportions by 10 min of anoxia.
Fig. 7.
Live-cell imaging of rod formation in an organotypic postnatal rat hippocampal slice expressing cofilin-GFP. The slice shown here was cultured on a membrane. About 44 h before observation, the slice was infected with adenovirus for expressing human cofilin-GFP. Immediately before imaging, the slice was cut from the membrane, laid flat (membrane side down) on a microscope slide and a coverslip was overlaid (t = 0). Time-lapse video microscopy was begun as rapidly as possible. Fields containing cofilin-GFP were located by quickly scanning the sample and one field was selected for observation. New rods (arrowheads) form between 4–5 min of anoxia and both new rods and those that may have started to form before anoxia began reach their full intensity and size by 7–10 min after anoxia. Scale bars = 10 μm.
Signaling pathways to rod formation
There are a multitude of signaling pathways in neurons reported to be altered by Aβ, several of which could potentially modulate cofilin activity. One major upstream signaling molecule that modulates levels of phospho-cofilin in neurons is cdc42, a small Rho family GTPase. Neuronal expression of constitutively active (CA) cdc42 (either V12 or L61), enhances ADF/cofilin dephosphorylation whereas expression of dominant negative (DN) N17cdc42 increases the phospho-ADF/cofilin pool [38]. Adenoviruses that were used to deliver either CA or DN cdc42, also expressed GFP under a separate promoter so that infected cells could be identified. Rod formation was scored in GFP positive (infected) and negative (uninfected) cells within the same cultures (Fig. 8A). Control-infected cells (GFP or tubulin-GFP) behaved similarly to uninfected cells in that 1–3% had rods without Aβ treatment and 18–19% had rods after Aβ treatment. Expression of wt or CA V12cdc42 increased rod formation in non-Aβ-treated cultures to 10–13%, whereas rod formation in cells expressing the DN N17cdc42 was identical to controls. Following Aβ-treatment, rod formation showed a non-significant increase in cells expressing the wt cdc42 (23%), and a significant increase (30%) in cells expressing the CA cdc42 as compared with uninfected control cells (19%). This finding suggests that a larger population of hippocampal neurons have the capacity to respond to externally applied Aβ but do not reach the threshold required for rod formation unless more active cdc42 is present. Following Aβ treatment, only 7.5% of neurons expressing DN N17cdc42 formed rods compared with 19% of uninfected or control-infected neurons within the same culture, a 60% reduction. Similarly, in organotypic slices, we quantified rods per field and found that CA cdc42 expression significantly increased rod numbers over uninfected and control infected cells both in untreated and Aβ-treated slices. Expression of DN cdc42 decreased the rod numbers in slices treated with Aβ1–42 oligomers by about 65% (Fig. 8B).
Fig. 8.
Rod formation downstream of Aβ1–42 is mediated by cdc42 in a subset of rod forming neurons. (A) Dissociated E18 rat hippocampal neurons were treated with adenovirus infection medium (uninfected) or infected at 4 DIV with adenoviruses for expression of GFP or cdc42 wild type (wt), constitutively active (CA) V12cdc4 or dominant negative (DN) N17cdc42. At 6 DIV half of the cultures for each treatment were exposed to 1 μM Aβ1–42 oligomers. At 7 DIV, cultures were fixed and immunostained for cofilin and the percent of neurons with rods was quantified. Included also are the rod counts in response to Aβ1–42 for control and cdc42 null (KO line) mouse hippocampal neurons in the same culture (see panels C and D). (B) Similar treatments to (A) performed on organotypic postnatal rat hippocampal slices. Slices were infected on day 10 with CA and DN cdc42 adenoviruses, challenged with Aβ1–42 on day 12 and fixed and stained for rods on day 13. Rods per field were counted across the slices. (C, D) Co-cultures of neurons from cdc42 conditional knockout mouse with wild type neurons from GFP expressing mouse. Phase pictures show wild type (GFP) and knockout (unlabeled) mouse hippocampal neurons in same field, 24 h after exposure to Aβ1–42 followed by fixation, permeabilization, and immunostaining for cofilin. Tandem arrays of rods (red arrows) are typically seen in 18–19% of wild type neurons but in about half this number of cdc42 knockout neurons within the same cultures, as is quantified by the green bars in (A). Experiments in A and B were repeated three or more times with multiple replicates for each condition in every experiment.
To more specifically examine the role of cdc42 in Aβ signaling to rod formation, we utilized hippocampal neurons from the brains of conditional cdc42 null mice [9]. We co-cultured cdc42 null neurons along with wild type neurons from a GFP-mouse, treated these cultures with Aβ1–42 oligomers for 24 h, and, after fixing and staining, scored rod formation in both GFP-expressing (wt) and non-expressing (cdc42 null) neurons (Fig. 8C, D). About 19% of the wt mouse neurons formed rods in response to Aβ (Fig. 8A), similar to what we found above and previously reported for rat neurons [11]. The percentage of cdc42 null neurons with rods in response to Aβ is 8% (Fig. 8A), a 60% decline that is in agreement with that observed above in neurons expressing DN cdc42. Taken together these results demonstrate that neurons have both cdc42-dependent and -independent pathways downstream of Aβ1–42 for cofilin activation and rod formation.
DISCUSSION
Here we report that stress-induced cofilin-actin rod formation occurs within specific hippocampal sub-regions in a stimulus dependent manner. These results extend our work in dissociated neurons into a more physiologically relevant context. Through the maintenance of vital neuronal connectivity, the ex vivo organotypic hippocampal slice provides a much more “in vivo” like setting for studying neuronal behavior compared to cultures of dissociated cells [39]. Rods in slice culture possess the same characteristics as those in dissociated neurons; they are refractory to phalloidin staining yet readily label with antibodies to actin and dephosphorylated but not phosphosphorylated cofilin, and their filamentous ultrastructural organization is identical. Rod formation in neurons was verified by performing identical experiments in organotypic slices from the Thy1-YFP Line H mice in which a subset of neurons express YFP. We additionally confirmed neuronal rod formation by observing many RFP positive neurons containing rods in slices infected with adenovirus expressing RFP behind a neuronal specific promoter.
We developed a method for mapping rod formation onto a template of the hippocampus so that we could directly compare different rod inducing stimuli across multiple hippocampal slices.
Acute stress, such as ATP-depletion, resulted in a global response. This was expected, as our studies on dissociated neurons showed > 80% initially form rods after 30 min of ATP-depletion, and 35% of neurons contained rods following 24 h recovery [8]. Cofilin-GFP expressing cells within organotypic slices formed rods about 5 min after being rendered anoxic (Fig. 7) or following addition of ATP-depletion medium (data not shown). This temporal feature is also strikingly similar to the speed with which dissociated neurons form rods from endogenous proteins after ATP-depletion [8]. With live-cell imaging we observed that rods start forming between 3–5 min after onset of anoxia and reach their maximum length and width by 10 min, which is the late time frame for the onset of irreversible brain damage in patients who suffer brain oxygen deprivation [40]. Given that rods can rapidly block distal synaptic function in processes where they form [15], it is reasonable to speculate that rod formation may also contribute to regional loss of synaptic connections and localized brain function during stroke.
Sub-regions of the hippocampus are known to display heightened sensitivity to various forms of Aβ [41–43]. Here we show that rod formation in and around the DG is very robust following treatment of slices with Aβ oligomer (1 μM). The DG, a termination zone of the perforant pathway connecting the hippocampal formation with the associative and limbic cortices [44], plays a crucial role in associative memory, regulation of stress responses, and in cognitive aspects of depression [45]. Pathology within the perforant pathway is the key determinant of age-related dementia [46]. Furthermore, the “normal” age related decline in human cognitive function also is manifested in a disruption of dentate circuitry [47] for which stress-induced rod formation could provide a mechanism. However, other cofilin-dependent processes need also to be considered.
Normal synaptic consolidation at excitatory medial perforant path granule cell synapses in the DG requires the synthesis of the immediate early gene activity-regulated cytoskeletal-associated protein (Arc) [48]. Arc expression is induced by brain-derived neurotrophic factor or high frequency stimulation-driven long-term potentiation (LTP) in the rat DG in vivo [48]. The Arc mRNA is transported into dendrites where it is translated. Prolonged synthesis of Arc leads to phosphorylation (inactivation) of cofilin and decreased cofilin activity results in subsequent local expansion of actin filament structures and synaptic stabilization [48].
In an opposing pathway, Aβ has been shown to down regulate the expression of Pak1 [16]. Pak1, an effector of the GTP-bound form of cdc42, is an activator of LIM kinase [49], which inactivates cofilin; thus depletion of Pak1 may result in an over activation of cofilin [16]. Here we show that rod formation in response to Aβ is regulated in about half of the responsive neurons through a cdc42-dependent pathway, implying that extracellular Aβ1–42 activates cofilin in a subpopulation of hippocampal neurons through multiple pathways, only one of which requires cdc42. Expression of a constitutively active form of cdc42 would be expected to enhance the activity of Pak1 and through LIMK1 cause cofilin phosphorylation. In neurons, however, activation of cdc42 leads to cofilin dephosphorylation through inhibition of Rho A-dependent signaling [38], whereas knockout of cdc42 expression leads to increased cofilin phosphorylation even though the activation of Pak1 (a class I Pak) has been reduced [9]. Thus, it is not surprising that inhibiting cdc42 activity, either by knockout or expression of dominant negative cdc42, significantly decreases the percentage of rod-forming neurons downstream of Aβ. In the conditional cdc42 knockout mouse, a decline in cofilin-specific phosphatase activity was demonstrated [9], suggesting that other cdc42 mediators (e.g., class II Paks) may modulate cofilin activity through a phosphatase pathway. It would not be surprising to find upstream modulators of cofilin phosphoregulation that are involved in bifurcating pathways to spatially regulate cofilin activity through a cycle of phosphorylation and dephosphorylation in response to a single extracellular ligand. Evidence for a bifurcating pathway to cofilin phosphocycling was first reported in serum stimulated fibroblasts [33] but also occurs in neuronal growth cone pathfinding [50] and in postsynaptic N-methyl-D-aspartate regulation of dendritic spine morphology [51].
Dendritic spine architecture is controlled by the Ras/Rap GTPase activating protein SynGAP, which has recently been shown to regulate both steady-state and activity-dependent cofilin phosphorylation [51]. Excess active cofilin will displace the actin filament stabilizing protein drebrin from spines, which alters spine dynamics [52]. Drebrin levels are reduced in brains of patients with AD and Down syndrome [53], especially in regions where cofilin is activated [16]. Thus the activation of cofilin that occurs within the DG in response to treatment with Aβ1–42 oligomers may cause rapid alterations in synaptic function through its direct effects on the dynamics of the actin core of spines, perhaps mediated by SynGAP [51], or through formation of rods within the neurite. Rod formation has the dual property of blocking delivery of material required for normal spine function and sequestering cofilin so that it is less able to participate in spine dynamics.
To further examine the potential impact of rods on neuronal physiology, we examined if rods induced by Aβ could be reversed following washout of the Aβ. Surprisingly, the large increase in rod number returned to near baseline levels within 24 h of washout. It is of interest in this regard that the learning and memory deficits of adult rats induced by a single brain infusion of an Aβ peptide fraction containing dimers and trimers [54] or dimers extracted from human AD brain [18] returned to normal 24 h after the single treatment.
Rods observed within the organotypic slices have similar ultrastructure to rods in dissociated neurons [8]. Furthermore, within some rods we found mitochondria and vesicles that appear to have become entrapped, supporting the fluorescence microscopy observations in dissociated cells of AβPP-containing vesicles accumulating at rods [11], and that rapid axonal transport is inhibited in neurons treated with glutamate or Aβ-peptide [55]. Additionally, the normal microtubule network is disrupted, which contributes to the observed transport deficits in rod-containing neurons. Thus, rods, which form rapidly and in abundance in specific regions of the hippocampus in response to different stress inducing agents, have a dramatic effect on neuronal behavior. Taken together the physiological effect of rods appears to be important in the etiology of AD, but rods may play much broader roles in other cognitive disorders such as cerebrovascular dementia including cerebral amyloid angiopathy, developmental disorders such as Down syndrome, or even the decline in cognitive function associated with “normal” aging.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge funding from the National Institutes of Health Neurological Disorders and Stroke (NS40371 to J.R.B., NS43115 to RCD, MTM, KCF and JRB, and NS48660 to KCF). We are grateful for technical assistance of Chi Pak and Alisa Shaw and valuable discussions from these same individuals and from Drs. Barbara Bernstein, O'Neil Wiggan and Boyan Garvalov. We are especially grateful to Dr. Frank Bradke, Max Planck Institute for Neurobiology, Martinsreid, Germany for providing the cdc42 KO neurons and the outstanding staff of the Boulder Lab for 3D Electron Microscopy of Cells, University of Colorado, Boulder, CO for technical assistance.
REFERENCES
- 1.Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 2.Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- 3.Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 4.Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 5.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 6.Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- 7.Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol. 2000;2:628–636. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- 9.Garvalov BK, Flynn KC, Neukirchen D, Meyn L, Teusch N, Wu X, Brakebusch C, Bamburg JR, Bradke F. Cdc42 regulates cofilin during the establishment of neuronal polarity. J Neurosci. 2007;27:13117–13129. doi: 10.1523/JNEUROSCI.3322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maloney MT, Kinley AK, Pak CW, Bamburg JR. ADF/cofilin, actin dynamics and disease. Protein Reviews. 2008;8:83–187. [Google Scholar]
- 11.Maloney MT, Minamide LS, Kinley AW, Boyle JA, Bamburg JR. Beta-secretase-cleaved amyloid precursor protein accumulates at actin inclusions induced in neurons by stress or amyloid beta: a feedforward mechanism for Alzheimer's disease. J Neurosci. 2005;25:11313–11321. doi: 10.1523/JNEUROSCI.3711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang TY, Minamide LS, Bamburg JR, Bokoch GM. Chronophin mediates an ATP-sensing mechanism of cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev Cell. 2008;15:691–703. doi: 10.1016/j.devcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein BW, Chen H, Boyle JA, Bamburg JR. Formation of actin-ADF/cofilin rods transiently retards decline of mitochondrial potential and ATP in stressed neurons. Am J Physiol Cell Physiol. 2006;291:C828–C839. doi: 10.1152/ajpcell.00066.2006. [DOI] [PubMed] [Google Scholar]
- 15.Jang DH, Han JH, Lee SH, Lee YS, Park H, Lee SH, Kim H, Kaang BK. Cofilin expression induces cofilin-actin rod formation and disrupts synaptic structure and function in Aplysia synapses. Proc Natl Acad Sci USA. 2005;102:16072–16077. doi: 10.1073/pnas.0507675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, Ma QL, Calon F, Harris-White ME, Yang F, Lim GP, Morihara T, Ubeda OJ, Ambegaokar S, Hansen JE, Weisbart RH, Teter B, Frautschy SA, Cole GM. Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease. Nat Neurosci. 2006;9:234–242. doi: 10.1038/nn1630. [DOI] [PubMed] [Google Scholar]
- 17.Lacor PN, Buniel MC, Furlow PW, Clemente AS, Velasco PT, Wood M, Viola KL, Klein WL. Abeta oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer's disease. J Neurosci. 2007;27:796–807. doi: 10.1523/JNEUROSCI.3501-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viola KL, Velasco PT, Klein WL. Why Alzheimer's is a disease of memory: the attack on synapses by Abeta oligomers (ADDLs). J Nutr Health Aging. 2008;12:51S–57S. doi: 10.1007/BF02982587. [DOI] [PubMed] [Google Scholar]
- 20.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 22.Ikawa M, Yamada S, Nakanishi T, Okabe M. ’Green mice’ and their potential usage in biological research. FEBS Lett. 1998;430:83–87. doi: 10.1016/s0014-5793(98)00593-6. [DOI] [PubMed] [Google Scholar]
- 23.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ’Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 24.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 25.Dahlgren KN, Manelli AM, Stine WB, Jr., Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 26.Stine WB, Jr., Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 27.Meberg PJ, Miller MW. Culturing hippocampal and cortical neurons. Methods Cell Biol. 2003;71:111–127. doi: 10.1016/s0091-679x(03)01007-0. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn TB, Brown MD, Wilcox CL, Raper JA, Bamburg JR. Myelin and collapsing-1 induce motor neuron growth cone collapse through different pathways: inhibition of collapse by opposing mutants of rac1. J Neurosci. 1999;19:1965–1975. doi: 10.1523/JNEUROSCI.19-06-01965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant aden-oviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamide LS, Shaw AE, Sarmiere PD, Wiggan O, Maloney MT, Bernstein BW, Sneider JM, Gonzalez JA, Bamburg JR. Production and use of replication-deficient adenovirus for transgene expression in neurons. Methods Cell Biol. 2003;71:387–416. doi: 10.1016/s0091-679x(03)01019-7. [DOI] [PubMed] [Google Scholar]
- 31.Shaw AE, Minamide LS, Bill CL, Funk JD, Maiti S, Bamburg JR. Cross-reactivity of antibodies to actin-depolymerizing factor/cofilin family proteins and identification of the major epitope recognized by a mammalian actin-depolymerizing factor/cofilin antibody. Electrophoresis. 2004;25:2611–2620. doi: 10.1002/elps.200406017. [DOI] [PubMed] [Google Scholar]
- 32.Abe H, Ohshima S, Obinata T. A cofilin-like protein is involved in the regulation of actin assembly in developing skeletal muscle. J Biochem. 1989;106:696–702. doi: 10.1093/oxfordjournals.jbchem.a122919. [DOI] [PubMed] [Google Scholar]
- 33.Meberg PJ, Ono S, Minamide LS, Takahashi M, Bamburg JR. Actin depolymerizing factor and cofilin phosphorylation dynamics: Response to signals that regulate neurite extension. Cell Motil Cytoskel. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Davis RC, Furukawa R, Fechheimer M. A cell culture model for investigation of Hirano bodies. Acta Neuropathol. 2008;115:205–217. doi: 10.1007/s00401-007-0275-9. [DOI] [PubMed] [Google Scholar]
- 35.Nishida E, Iida K, Yonezawa N, Koyasu S, Yahara I, Sakai H. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc Natl Acad Sci U S A. 1987;94:5262–5266. doi: 10.1073/pnas.84.15.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano A. Hirano bodies and related neuronal inclusions. Neuropathol Appl Neurobiol. 1994;20:3–11. doi: 10.1111/j.1365-2990.1994.tb00951.x. [DOI] [PubMed] [Google Scholar]
- 37.Velasco ME, Smith MA, Siedlak SL, Nunomura A, Perry G. Striation is the characteristic abnormality in Alzheimer disease. Brain Res. 1998;813:329–333. doi: 10.1016/s0006-8993(98)01034-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen T-J, Gehler S, Shaw AE, Bamburg JR, Letourneau PC. Cdc42 participates in regulation of ADF/cofilin and retinal growth cone filopodia by brain derived neurotrophic factor. J Neurobiol. 2006;66:103–114. doi: 10.1002/neu.20204. [DOI] [PubMed] [Google Scholar]
- 39.Holopainen IE. Organotypic hippocampal slice cultures: a model system to study basic cellular and molecular mechanisms of neuronal cell death, neuroprotection, and synaptic plasticity. Neurochem Res. 2005;30:1521–1528. doi: 10.1007/s11064-005-8829-5. [DOI] [PubMed] [Google Scholar]
- 40.Safar P. Cerebral resuscitation after cardiac arrest: a review. Circulation. 1986;74:IV138–IV153. [PubMed] [Google Scholar]
- 41.Bahr BA, Hoffman KB, Yang AJ, Hess US, Glabe CG, Lynch G. Amyloid beta protein is internalized selectively by hippocampal field CA1 and causes neurons to accumulate amyloidogenic carboxyterminal fragments of the amyloid precursor protein. J Comp Neurol. 1998;397:139–147. [PubMed] [Google Scholar]
- 42.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HJ, Chae SC, Lee DK, Chromy B, Lee SC, Park YC, Klein WL, Krafft GA, Hong ST. Selective neuronal degeneration induced by soluble oligomeric amyloid beta protein. FASEB J. 2003;17:118–120. doi: 10.1096/fj.01-0987fje. [DOI] [PubMed] [Google Scholar]
- 44.Hyman BT, Van Hoesen GW, Kromer LJ, Damasio AR. Perforant pathway changes and the memory impairment of Alzheimer’s disease. Ann Neurol. 1986;20:472–481. doi: 10.1002/ana.410200406. [DOI] [PubMed] [Google Scholar]
- 45.Ohm TG. The dentate gyrus in Alzheimer’s disease. Prog Brain Res. 2007;163:723–740. doi: 10.1016/S0079-6123(07)63039-8. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Sierra F, Hauw JJ, Duyckaerts C, Wischik CM, Luna-Munoz J, Mena R. The extent of neurofibrillary pathology in perforant pathway neurons is the key determinant of dementia in the very old. Acta Neuropathol. 2000;100:29–35. doi: 10.1007/s004010051189. [DOI] [PubMed] [Google Scholar]
- 47.Patrylo PR, Williamson A. The effects of aging on dentate circuitry and function. Prog Brain Res. 2007;163:679–696. doi: 10.1016/S0079-6123(07)63037-4. [DOI] [PubMed] [Google Scholar]
- 48.Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signaling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 50.Wen Z, Han L, Bamburg JR, Shim S, Ming GL, Zheng JQ. BMP gradients steer nerve growth cones by a balancing act of LIM kinase and Slingshot phosphatase on ADF/cofilin. J Cell Biol. 2007;178:107–119. doi: 10.1083/jcb.200703055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlisle HJ, Manzerra P, Marcora E, Kennedy MB. Syn-GAP regulates steady-state and activity-dependent phosphorylation of cofilin. J Neurosci. 2008;28:13673–13683. doi: 10.1523/JNEUROSCI.4695-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kojima N, Shirao T. Synaptic dysfunction and disrup tion of postsynaptic drebrin-actin complex: a study of neurological disorders accompanied by cognitive deficits. Neurosci Res. 2007;58:1–5. doi: 10.1016/j.neures.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Shim KS, Lubec G. Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer's disease and Down syndrome. Neurosci Lett. 2002;324:209–212. doi: 10.1016/s0304-3940(02)00210-0. [DOI] [PubMed] [Google Scholar]
- 54.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 55.Hiruma H, Katakura T, Takahashi S, Ichikawa T, Kawakami T. Glutamate and amyloid beta-protein rapidly inhibit fast axonal transport in cultured rat hippocampal neurons by different mechanisms. J Neurosci. 2003;23:8967–8977. doi: 10.1523/JNEUROSCI.23-26-08967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.