Abstract
Bacterial pathogens adapt to changing environments within their hosts, and the signaling molecule adenosine 3′, 5′-cyclic monophosphate (cAMP) facilitates this process. In this study, we characterized in vivo DNA binding and gene regulation by the cAMP-responsive protein CRP in M. bovis BCG as a model for tuberculosis (TB)-complex bacteria. Chromatin immunoprecipitation followed by deep-sequencing (ChIP-seq) showed that CRP associates with ∼900 DNA binding regions, most of which occur within genes. The most highly enriched binding region was upstream of a putative copper transporter gene (ctpB), and crp-deleted bacteria showed increased sensitivity to copper toxicity. Detailed mutational analysis of four CRP binding sites upstream of the virulence-associated Rv0249c-Rv0247c succinate dehydrogenase genes demonstrated that CRP directly regulates Rv0249c-Rv0247c expression from two promoters, one of which requires sequences intragenic to Rv0250c for maximum expression. The high percentage of intragenic CRP binding sites and our demonstration that these intragenic DNA sequences significantly contribute to biologically relevant gene expression greatly expand the genome space that must be considered for gene regulatory analyses in mycobacteria. These findings also have practical implications for an important bacterial pathogen in which identification of mutations that affect expression of drug target-related genes is widely used for rapid drug resistance screening.
INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is an ancient disease that continues to cause significant morbidity and mortality worldwide. Increasing levels of drug resistance and a complex synergy with human immunodeficiency virus (HIV) complicate efforts to control this deadly pathogen (1). There is an urgent need for new therapeutics against Mtb, and development of effective new drugs requires better understanding of Mtb physiology.
Bacterial pathogens must adapt to changing environments within the host during infection, and they often use cyclic nucleotides as ‘second messengers’ to sense and respond to their external environments (2,3). Adenosine 3′, 5′-cyclic monophosphate (cAMP) is one such signaling molecule that is widely used by both microbial pathogens and their mammalian hosts (4–8). Mtb is a particularly unusual microbe in that it has ∼15 biochemically distinct adenylyl cyclases (AC), which generate cAMP and allow Mtb to respond to multiple environmental cues (9,10). Mtb also encodes 10 putative cyclic nucleotide monophosphate (cNMP) binding proteins, of which three have been characterized. Rv0998 (Mt-PatA) is a cAMP-activated protein lysine acetylase that has several biological targets in mycobacteria, including Mtb acetyl-CoA synthase (11,12). Rv1675c (called Cmr for cAMP and macrophage regulator) and Rv3676 (named CRP for cAMP-responsive protein) both contain helix turn helix domains and belong to the CRP/FNR family of transcription factors (13–16).
CRP is important for Mtb pathogenesis, as Mtb mutants deleted for crp show impaired growth in murine macrophages (15,17) and in a mouse model of TB (15). The CRP ortholog in M. bovis BCG Pasteur differs from Mtb CRP by two amino acids (L47P and E178K), and has approximately 2-fold higher affinity for DNA in vitro compared to CRP from Mtb (13). However, virulence of Mtb H37Rv crp deletion mutants can be fully restored by expression of crp from either BCG or Mtb, confirming that both orthologs function similarly as virulence-associated transcriptional regulators in vivo (18). For simplicity, we use CRP throughout the text to refer to protein from either Mtb or BCG except when referring to a species-specific CRP behavior.
The molecular basis of CRP's role in virulence is not known, but it directly regulates expression of several biologically important genes. For example, CRP controls expression of rpfA, which encodes a resuscitation-promoting factor thought to play a role in the reactivation of dormant Mtb cultures (19). CRP also upregulates expression of serC, which encodes a phosphoserine aminotransferase (14,17,20), and growth of Mtb crp mutants is slowed by a resulting defect in serine biosynthesis (17). Correction of this deficiency by serine supplementation or by constitutive expression of serC restores normal growth levels of Mtb in culture media but not within macrophages (17). More recently, CRP was shown to directly regulate expression of whiB1 (21,22), which encodes an essential transcription factor that contains a nitric oxide (NO) sensing [4Fe-4S]2+ cluster (23,24). DNaseI-footprinting showed CRP's ability to bind cooperatively to each of two CRP sites within the whiB1 promoter region, with slightly enhanced binding in the presence of cAMP (21).
A combination of in silico and experimental studies has defined CRP as a global regulator within Mtb, but the full extent of this regulation is not known. Whole genome expression microarrays of Mtb H37Rv demonstrated potential regulation of 16 genes from 13 individual promoters, including those of lprQ, rpfA, ahpC, lipQ, fadD26 and whiB1(15). Another study (14) combined BCG DNA sequences recovered by affinity capture with previously characterized E. coli CRP binding sites to seed a computational analysis of the potential CRP regulon in TB complex bacteria. This affinity capture study predicted 114 CRPMt regulon members, based on conserved binding motifs, corresponding to 73 promoter regions in Mtb (14). A subsequent in silico study (25) used putative promoter sequences from the regulon of C. glutamicum GlxR, an ortholog of CRP, as seed sequences to predict 135 CRP binding sites with the potential to regulate expression of 207 genes within 121 transcriptional units.
Surprisingly little overlap was found among the regulons predicted from these prior studies, despite their identification of similar binding motifs. In this study, we characterize in vivo CRP-DNA binding and gene regulation in M. bovis BCG as a model system for TB complex bacteria. We found using Chromatin Immunoprecipitation followed by deep-sequencing (ChIP-seq) that CRP is associated with ∼900 DNA binding regions in M. bovis BCG, only ∼14% of which occur within linear or divergent intergenic DNA sequences. The remaining CRP binding regions were found within intragenic regions (83%) or between convergent intergenic sequences (3%). Blind deconvolution (26) of the CRP binding profile within these enriched regions revealed four CRP binding sites at the Rv0250c-Rv0249c succinate dehydrogenase locus, including one within the Rv0250c open reading frame (ORF). Deletion analyses demonstrated that CRP binding contributed to regulation of Rv0249c-Rv0247c expression from each of two promoters. An Rv0249c proximal promoter required upstream Rv0250c intragenic sequences for maximum expression, while the second promoter is located upstream of Rv0250c. These findings show that CRP regulates a critical metabolic step in central metabolism and that intragenic binding sites can significantly affect regulation of biologically important gene expression in TB complex bacteria. The extremely large number of binding regions further suggests that CRP may function as a nucleoid associated protein (NAP) in addition to its established role as a canonical transcription factor in TB complex mycobacteria.
MATERIALS AND METHODS
Bacterial strains and culture
M. tuberculosis H37Rv (ATCC 25618) and M. bovis BCG (Pasteur strain, Trudeau Institute) was grown in mycomedia: Middlebrook 7H9 medium supplemented with 0.2% glycerol, 10% oleic acid-albumin-dextrose-catalase (OADC), 0.05% Tween-80 or on Middlebrook 7H10 (Difco) supplemented with 0.5% glycerol, 10% OADC and 0.01% cycloheximide. Where indicated, heat-killed M. tuberculosis H37Rv (ATCC 25618) genomic DNA was used. Fresh cultures were inoculated from frozen seed stocks for every experiment and transferred to the desired culture condition, as previously described (27). Cultures for ChIP Sequencing were grown from seed stocks in mycomedia for seven days to late log phase in shallow (2 mM) cultures 1.3% O2 + 5% CO2 in 225 cm2 flasks with gentle rocking as described (28). On day 3, dibutyryl-cAMP (dbcAMP) was added to one culture flask for a final concentration of 10mM and returned to the incubator. For starvation experiments, cultures were grown in ambient air from seed stocks in mycomedia for 10 days, pelleted and washed with Dulbecco's phosphate buffered saline (PBS) or fresh mycomedia and returned to the incubator for 24 h. Transformations into M. bovis BCG were done by electroporation (29). E. coli was grown in Luria Broth or on Luria Broth agar plates. Kanamycin was used at 25 μg/ml as needed. All cultures were grown at 37°C.
Chromatin Immunoprecipitation and sequencing
50 ml culture volumes of M. bovis BCG Pasteur were grown to late-log phase under 1.3% O2 + 5% CO2 and cross-linked with 1% formaldehyde at room temperature for 30 min with gentle rocking. Crosslinking was quenched with glycine (250 mM final concentration) for 15 min at room temperature. The cells were harvested, washed twice with ice-cold PBS, and resuspended in 0.5–0.6 ml of Buffer I (20 mM HEPES pH 7.9, 50 mM KCl, 0.5 M DTT, 10% glycerol) with protease inhibitor cocktail (Sigma). Bacterial cells were lysed and DNA sheared by sonication for 25 min with a Covaris S2 (Covaris, Inc., Woburn, MA). The salt concentration of the cleared cell lysate was adjusted to a final concentration of 10mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% NP-40 (IPP150 buffer). Immunoprecipitation was carried out by incubation of lysate with 10 μl of CRP antiserum 4°C. 50 μl of Protein A agarose beads were rinsed with IPP150 buffer, and added to the lysate-antiserum mixture. Protein A agarose-lysate-antiserum mixture was incubated at 4°C for 30 min and room temperature for 1.5 h. The beads were washed at least five times with IPP150 buffer followed by two washes with TE buffer (10 mM Tris HCl pH 8.0, 1mM EDTA). DNA from agarose beads was eluted by incubation with 150 μl elution buffer (50 mM Tris HCl pH 8.0, 10 mM EDTA, 1% SDS) at 65°C for 15 min. A second elution was carried out by incubation of pellet with 100 μl of TE + 1% SDS at 65°C for 5 min. Both elutions were pooled and treated with 1mg/ml Proteinase K at 37°C for 2 h and 65°C overnight. DNA was purified using PCR purification kit (QIAGEN).
Sequencing was performed as described previously (30). Briefly, sequencing was performed on the Illumina platform, using a GAIIx (Boston University, sequencing core). Coverage along the genome was calculated using Bowtie2 (31) and SamTools (32). Enriched regions were called using log-normal distributions as previously described (30). The minimum region length was at least 150 nt long and had a minimum 60 nt shift between its forward and reverse peaks. Region coverage was normalized using mean coverage of an experiment, correcting for the number of reads amongst experiments. Exact binding sites were determined as described by Gomes et al. (26), using the BRACIL blind deconvolution method. BRACIL integrates ChIP-seq coverage and genome sequence to jointly identify binding sites with single nucleotide resolution and a corresponding consensus binding site motif. Regions from the BCG genome were mapped to the Mtb H37Rv genome by sequence similarity using BLAST.
cAMP treatments
Levels of cAMP within bacteria were increased exogenously or endogenously for some experiments. For the exogenous method, dibutyryl-cAMP (dbcAMP) was added to the culture media for four days at a final concentration of 10mM. dbcAMP is a cell permeable molecule that is cleaved to produce cAMP upon internalization (16). Alternatively, excess cAMP was endogenously produced by expression of the adenylyl cyclase Rv1264 catalytic domain (Rv1264Cat) under the strong, constitutively active promoter of Rv0805 (16). This construct is cloned on the pcRII.oriM, a multicopy plasmid that contains the oriM origin of replication from pAL5000, which was shown to have a copy number of 3–10 (33). This approach was previously shown to elevate intracellular cAMP levels within bacteria by up to 40X (16).
Functional analysis of genes
Regulatory targets of binding sites were assigned based upon their locations relative to adjacent genes. Immediate downstream genes were assigned as targets for intragenic and intergenic binding sites. Therefore, linear intergenic sites were assigned only one target while divergent intergenic sites were assigned two. Intragenic binding sites were also assigned their overlapping gene as a target, yielding a possible total of up to three targets. Non-coding RNA genes currently listed in the Tuberculist annotations were treated similarly to protein coding genes with respect to their assignment as potential regulatory targets of proximal binding sites.
We next used Tuberculist functional annotations (http://tuberculist.epfl.ch/) (34) to assign the functional classification to each target gene within the CRP ChIP-seq data set. The percentage of potentially regulated target genes was calculated for each functional classification and segregated according to the location of the binding site to compare target distributions for intragenic binding sites, intergenic binding sites and all ChIP-seq sites combined. The expectation of functional classification for the Mtb H37Rv genome was also calculated. Hypergeometric calculations with a Bonferroni correction of 11 were performed using Excel to determine the significance of differences between the genome and the type of ChIP-seq site. P < 0.05 was considered significant.
Electrophoretic mobility shift assays (EMSA)
DNA probes were generated by PCR using DNA primers based on the Mtb H37Rv sequences denoted in Tuberculist (34) with the addition of BamHI restriction sites for downstream cloning as needed. The PCR forward primer was labeled with [γ- 33P]-ATP using T4 DNA polynucleotide kinase (New England Biolabs). DNA fragments were then labeled by PCR (30 cycles) using Mtb genomic DNA as a template, diluted 1:3 and 1 μl DNA probe was used in each 10 μl binding reaction. Samples were electrophoresed on an 8% (29:1) non-denaturing polyacrylamide gel for 2.5 h with a constant voltage of 150 v. Gels were vacuum dried, exposed on a phosphor screen, scanned with a Storm 860 PhosporImager (Molecular Dynamics), and analyzed with ImageQuant software (Molecular Dynamics).
Growth in copper
Wild-type BCG, BCG Δcrp and BCG Δcrp::pMBC1029, a strain with a single-copy, integrative vector containing the crp gene with its native promoter (17), were grown in 25 cm2 tissue culture flasks at 37°C in mycomedia with the indicated CuSO4 concentrations for 14 days under ambient air conditions or in hypoxia supplemented with 5% CO2. After testing a range of concentrations, based on results of prior studies (35,36), we chose 100 μM as our standard CuSO4 assay concentration for media that included OADC, and 50 μM for media that lacked OADC. At the time points indicated, aliquots were removed, cells were declumped by light sonication in a cup horn sonicator (Virtis Virsonic, setting 4, 10 s total time for 5 s on and 5 s off), and OD620 was determined using a Sunrise plate reader (Tecan).
Construction of lacZ fusions
Lists of plasmids and primers used in this study are in Supplementary Figures S1 and S2, respectively. Promoters of interest were amplified using primers that contained either BamHI or ScaI (due to an internal BamHI restriction site within Rv0250c) restriction sites flanking the ends. These PCR products were cloned into pCR2.1 or pCRII (Life Technologies) and confirmed by sequencing with either M13–20 or M13-R (Core Services, Wadsworth Center). DNA fragments were cut with the appropriate restriction enzyme, gel purified and recovered (Qiagen) and ligated into pLacInt, a transcriptional lacZ reporter fusion that contains the attachment and integration site (attP/int) of mycobacteriophage L5 and transformed into DH5α (27). Clones were sequenced verified with KM1674 or KM1029. Positive clones were transformed into M. bovis BCG Pasteur and checked for presence of insert using plasmid specific primer combinations.
DNA regions of interest were replaced using sequence overlap extension (SOE), a PCR-based overlap extension method. PCR primers complementary to the desired sequence were used to generate two partially overlapping PCR fragments, which were annealed for use as template for amplification with the outermost end primers containing the restriction sites. These products were then cloned into pCR2.1 or pCRII (Life Technologies) and sequence verified (Wadsworth Center Molecular Genetics Core). Digestion was used to subclone into either the BamHI or ScaI sites of pLacInt as described above.
β-galactosidase assays
Cells were grown as described within text and assays were done as previously described (37). Briefly, cells were declumped as described above, mixed with 20mM C2FDG (38) (Life Technologies) and incubated for 3 h at 37°C. The relative expression level for each sample was normalized to 106 cfu based on OD620 readings, with an OD620 equal to 1 previously determined to have 5×108 cfu by plate counts (37).
RNA and cDNA
RNA was harvested from M. bovis BCG Pasteur after being grown as described within text. Briefly, cells were treated with 5M guanidinium thiocyanate (GTC), harvested with a mini-beadbeater (BioSpec) in the presence of TriZol (Life Technologies) and processed according to manufacturer's instruction. RNA was determined to be DNA free by PCR amplification with 1 μg of RNA and gene specific primers for 23S and 16S determination of no bands present. cDNA was generated using Superscript III reverse transcriptase (Life Technologies) using random oligos as previously described (16).
RESULTS
Genome wide analysis of the CRP regulon
Hypoxia and starvation are thought to be biologically important environmental signals for Mtb during host infection, so we used ChIP-Seq to investigate the genome-wide binding of CRP during log phase growth of M. bovis BCG under low oxygen conditions (1.3% O2 supplemented with 5% CO2, which is referred to as baseline throughout the text) (28). Alternatively, cells were grown under ambient air conditions to stationary phase and exposed to PBS for 24 h to induce a starvation response. The union of all experiments generated over 900 enriched CRP binding regions in vivo, and the BRACIL blind deconvolution method (26) was used to predict ∼2000 individual binding sites within these binding regions. The binding sites were distributed evenly across the genome when normalized peak heights were plotted against their genomic coordinates (Figure 1A) and represented 1365 potential gene regulatory targets (see below and Methods).
Figure 1.
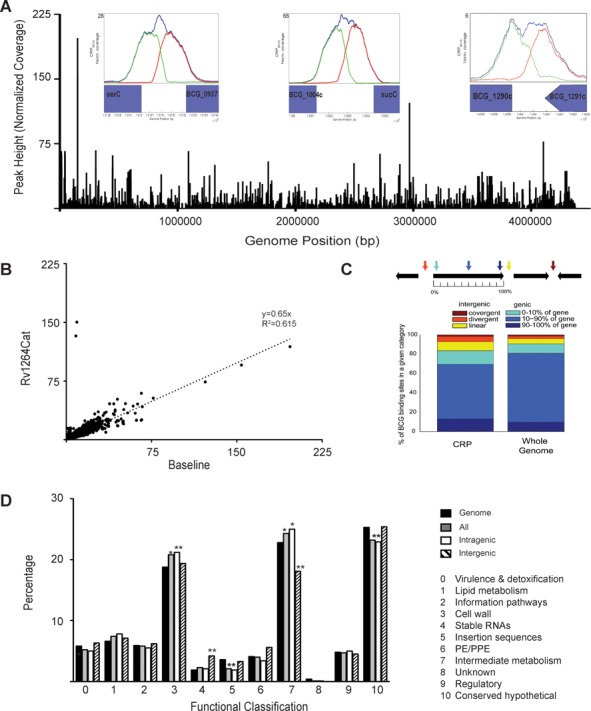
Binding of the CRPBCGin vivo. (A) Distribution of CRPBCG binding across the genome is evenly dispersed. Within insets are select, known binding sites and the correlating ChIP-Seq binding peaks. (B) Side-by-side plot of one repeat of untreated cells (baseline) against peaks for the Rv1264Cat. (C) Distribution of the binding sites within the various regions of genes. (D) Functional classification of the ChIP-Seq binding peaks within various categories including only intergenic sites, intragenic sites and all sites plotted together. *P at least <0.05 relative to the genome using a hypergeometic calculation with a correction factor of 11, **P < 0.005.
Expected peaks were observed for each of 10 regions previously shown to bind CRP by using electrophoretic mobility shift assays (EMSA) (13,15,17). These DNA sequences include the orthologous promoter regions of serC-Rv0885, frdA, Rv0950c-sucC, Rv0145, Rv1386, Rv1158-Rv1159, Rv3857c and Rv1230c, and showed a wide range of normalized peak heights in vivo. For example, the peak between BCG_1290c and BCG_1291c (corresponding to the Rv1230c - Rv1231c intergenic region in Mtb) had a height six times above the experimental mean coverage (also referred to as normalized coverage), while the region between BCG_1004c and sucC (orthologous to Mtb intergenic region of Rv0950c-sucC) had a normalized height of 68. By comparison, the highest normalized signal observed was 196.7, which corresponded to an intergenic site between BCG_0136c-BCG_0137 (orthologous to Rv0103c-Rv0104 in Mtb (Table 1)).
Table 1. Top CRPBCG binding sites and their potential regulatory targets.
| Peak | Baseline Height | Genome coordinates (BCG) | Genome Coordinates (Mtb H37Rv) | Location | Potential Targets | Functional description |
|---|---|---|---|---|---|---|
| 1 | 196.7 | 152045–152088 | 122303–122346 | Intergenic | Rv0103c/ctpB | Cation-transporter P-type ATPase B |
| 2 | 154.1 | 152114–152154 | 122372–122412 | Intragenic | Rv0104* | cNMP binding protein |
| Rv0106 | CHP | |||||
| 3 | 122.3 | 2968736–2968778 | 3015051–3015093 | Intergenic | Rv2699c | CHP |
| Rv2700 | Secreted essential protein | |||||
| 4 | 76.5 | 410455–410597 | 4131442–4131484 | Intragenic | Rv3688c | CHP |
| Rv3689* | Conserved Membrane Protein | |||||
| Rv3690 | Conserved Membrane Protein | |||||
| 5 | 66.9 | 305864–305930 | 277834–277900 | Intergenic | Rv0231/fadE4 | Acyl-CoA dehydrogenase FadE4 |
| Rv0232 | TetR/AcR family of regulators | |||||
| Rv0233/nrdB | Ribonucleoside-diphosphate reductase | |||||
| 6 | 66.4 | 13448–13490 | 13449–13491 | Intragenic | Rv008 | Unknown |
| Rv0010c* | Conserved Membrane Protein | |||||
| Rv0111 | Conserved Membrane Protein | |||||
| 7 | 66.1 | 1092254–1092333 | 106177–1061856 | Intergenic | Rv0950c | Unknown |
| Rv0951 | Succinyl-CoA synthetase | |||||
| 8 | 65.9 | 43115–43157 | 13449–13491 | Intragenic | BCG_0038+ | Unknown |
| BCG_0040+* | Conserved Membrane Protein | |||||
| BCG_0041c+ | Conserved Membrane Protein | |||||
| 9 | 62.3 | 2717743–2717807 | 2752573–2752637 | Intragenic | Rv2451* | Hypothetical proline and serine rich protein |
| Rv2450c/rpfE | Resuscitation-promoting factor RpfE | |||||
| 10 | 61.8 | 44779–44823 | 15113–15157 | Intragenic | BCG_0041c+ | Conserved Membrane Protein |
| BCG_0043/trpG2+* | Glutamine Amidotransferase |
+binding site is within repeat region of BCG.
*gene to which binding site is intragenic.
Binding of CRP to DNA is enhanced by cAMP in vitro (13,21–22), so we also examined CRP's in vivo DNA binding profile in BCG exposed to elevated levels of cAMP. Excess cAMP was generated endogenously for these experiments by over-expression of the constitutively active Rv1264 adenylyl cyclase catalytic domain (Rv1264Cat) using the Mtb Rv0805 promoter on the multicopy plasmid pMBC621 (16). This approach was previously shown to elevate intracellular cAMP levels by up to 40X (16). No significant differences were observed for in vivo CRP binding in the presence of excess versus baseline cAMP levels with the exception of two strong peaks in the Rv1264Cat samples (Figure 1B). One of these two peaks mapped to a previously identified CRP binding site upstream of Rv0805 that is present within the pMBC621 expression plasmid (14), while the other peak represents a newly identified site. The observed enrichment of these two sites (normalized coverage of 15.7 and 16) correlates well with the previously determined estimate of 3–10 copies/cell for the plasmid's origin of replication (33). These binding peaks in the Rv1264Cat samples thus serve as a useful watermark for the binding of CRP to the additional plasmid-based copies of these two sites within the Rv0805 promoter region (Figure 1B, Supplementary Figure S1D, F). Dibutyryl-cAMP (dbcAMP) was also added exogenously for some experiments as an alternative means of elevating bacterial cAMP levels, as previously described (39). The cAMP supplementation results were similar to those of the Rv1264Cat experiments, as no CRP binding differences were observed in the cAMP-supplemented versus untreated baseline samples (Supplemental Figure S1). Additionally, no significant differences in CRP binding occurred in rich media compared to the starvation condition (Supplemental Figure S1F).
A high percentage of intragenic binding sites
CRP binding sites from all conditions were classified according to whether they were located within intragenic (within annotated open reading frames (ORFs)) or intergenic (between ORFs) sequences (Figure 1C), and compared with the distribution of total genomic nucelotides in intragenic (91%) versus intergenic (9%) regions. A majority (∼83%) of the binding sites we identified in these ChIP-seq analyses were intragenic and approximately two-thirds of these intragenic sites were located within the middle-coding region of the ORF (Figure 1C). The remaining third fell within the first or last 10% of an ORF. The high percentage of intragenic CRP binding sites was surprising, because transcription factors have been thought to function primarily through binding of intergenic sequences (40–42). While only 14% of the CRP binding sites occurred within canonical intergenic sequences (divergent or linear), an additional 3% occurred in convergent intergenic sequences. Combined, we observed ∼2-fold enrichment for CRP binding sites among intergenic sequences relative to percent of total genomic sequences. Approximately 55% of these intergenic sites were located upstream of ORFs in a linear orientation (lin, Figure 1C), while 32% were located between two divergent ORFs (div, Figure 1C). Similar distributions of intragenic versus intergenic binding sites were found in the baseline and Rv1264Cat samples (data not shown).
Broad functional roles of putative CRP regulated genes
We separately analyzed intragenic versus intergenic CRP binding sites in relation to their potential gene regulatory targets (as described in Methods), relative to all sites combined, using the orthologous Mtb DNA sequences for each binding site identified by ChIP-seq. Genes that contained or were immediately downstream of a CRP binding site were counted as potential regulatory targets of the CRP that bound at that locus. The distribution of all genes within the genome was calculated using the Tuberculist (43) assignment and the distribution for each functional classification for the genome was compared to the target gene calculations above (Figure 1D). CRP's potential regulatory targets were broadly distributed among functional classes, with fairly little bias. However, there was an overall enrichment among potential CRP regulatory targets for genes involved in cell wall processes (Category 3) and intermediary metabolism (Category 7), with intragenic binding sites driving these increases. Fewer than expected intergenic sites were associated with intermediary metabolism targets, but stable RNAs (Category 4), including tRNAs and ncRNAs, were enriched for intergenic binding sites (Figure 1D). Genes that encode insertion sequences (Category 5) were also underrepresented as potential CRP regulatory targets when compared to their total representation in the genome.
Characterization of in vivo binding sites
ChIP-seq may detect indirect as well as direct TF binding, so we used EMSA to further characterize binding of CRP to four regions that contain genes of biological interest. In contrast to E. coli Crp, CRP expression is not autoregulated in mycobacteria and no associated CRP binding sites were found at the crp locus in either BCG or Mtb. However, CRP binding was found upstream of BCG_1686 and BCG_3703, whose orthologs in Mtb encode for adenylyl cyclases (AC) (Rv1647 and Rv3645, respectively), and BCG_0137, which encodes a putative cNMP binding protein orthologous to Rv0104 (9) (Figure 2). An Rv0104 transposon insertion mutant is attenuated for virulence in SCID mice (9,44), and the Rv0103c/Rv0104 orthologous locus produced the most highly enriched ChIP-seq region found in this study. The divergently expressed Rv0103c (ctpB) encodes a putative P-type ATPase with a possible role in copper transport (34,43). The enriched intergenic region between the Rv0347 ortholog and ansP2 (Rv0346c), a possible L-asparagine transporter (34), was also chosen for analysis because previous mutagenesis studies suggest essentiality of Rv0347 for Mtb growth on 7H10 agar with 0.2% glucose (45), or in minimal media containing 0.01% cholesterol (46).
Figure 2.
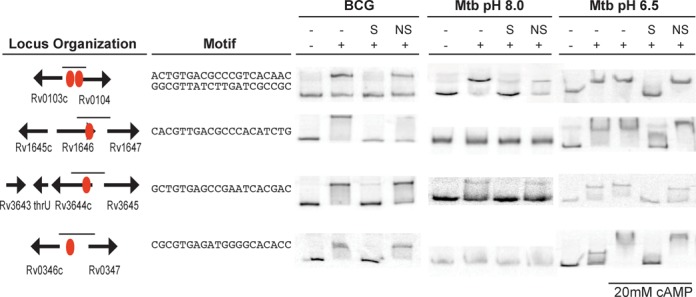
in vitro binding of CRPBCG and CRPMt by electrophoretic mobility shift assays. ChIP-seq sites are shown as red circles. The binding motif is shown for the identified binding sites. Probes used in each shift are labeled with small lines. EMSAs were performed with 100ng CRPBCG (first column), 100ng CRPMt (second column) or 300ng CRPMt (third column). Excess cold double stranded specific (S) and nonspecific (ns) probes used as competitors are labeled as such.
CRPBCG showed specific EMSA interactions with all four DNA regions (Figure 2), correlating well with the in vivo binding observed using ChIP-seq and confirming direct binding to these DNA sequences. CRPMt also bound all sequences, but CRPMt's DNA binding was more sensitive to environmental conditions. We previously showed robust binding of CRPMt to a variety of DNA sequences in the absence of added cAMP using EMSA at pH 8.0 (13). However, CRPMt bound only to the ctpB/Rv0104 and Rv3644/Rv3645 probes under these standard conditions in the present study. We reasoned that CRP encounters a range of pH environments and/or cAMP concentrations in vivo, so we re-tested CRP binding with all four probes at pH 9.5 and pH 6.5. We found that CRPMt bound specifically with all four DNA probes at pH 6.5, and that addition of cAMP enhanced binding of CRPMt to the Rv0346c/Rv0347 probe (Figure 2). CRPMt's DNA binding properties at pH 9.5 were similar to those at pH 8.0, while CRPBCG bound all four probes regardless of pH (data not shown).
The robust direct binding of CRP to the DNA region upstream of the putative copper transporter gene ctpB (43) suggests a role for CRP in controlling the bacterium's response to copper, which is part of the antimicrobial response within macrophages (47). We tested the sensitivity of crp -deleted BCG mutants to copper over a two week period, and found that the presence of 100 μM copper significantly inhibited growth of crp mutants relative to that of wt bacteria (Figure 3). As albumin is known to sequester copper (48), we repeated these experiments in Sauton's media without OADC, and found that 50 μM copper was sufficient to slow the growth of BCG crp in the absence of albumin (data not shown). In both cases, normal growth was restored by complementation of the mutation with a wt crp allele, establishing a role for CRP in mycobacterial copper resistance.
Figure 3.
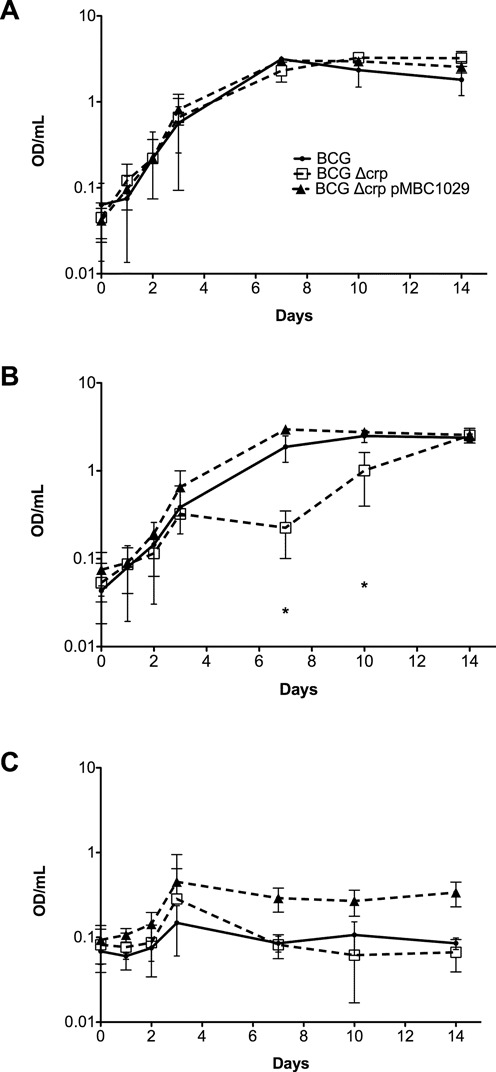
Effect of copper exposure on growth in BCG Δcrp strains. Ambient shaking cultures were monitored for growth in (A) untreated, (B) 100uM CuSO4 and (C) 250uM CuSO4 for 14 days. Black line, BCG wild-type, red dashed line, BCG Δcrp, blue line, BCG Δcrp single complement, encoded on pMBC1029. The standard deviations are shown from three independent experiments. (*, P < 0.05 as determined by a by student's t-test).
Comparison of CRP regulon studies
A CRP binding motif was generated using FIMO (Find Individual Motif Occurrences), which is part of the MEME suite (49–51) for all in vivo binding sites identified in the ChIP-seq analyses (Figure 4A). The M. bovis BCG Pasteur genome was then searched for potential binding sites using this motif. Only ∼30% of the strong CRP binding motifs instances (defined by FIMO as a P < 0.0001 in which p measures the similarity between the motif and motif instance) in the genome were enriched for binding in the ChIP-seq assays (Supplemental Figure S2), suggesting a high degree of selectivity for the sites that were bound and recovered in these experiments.
Figure 4.
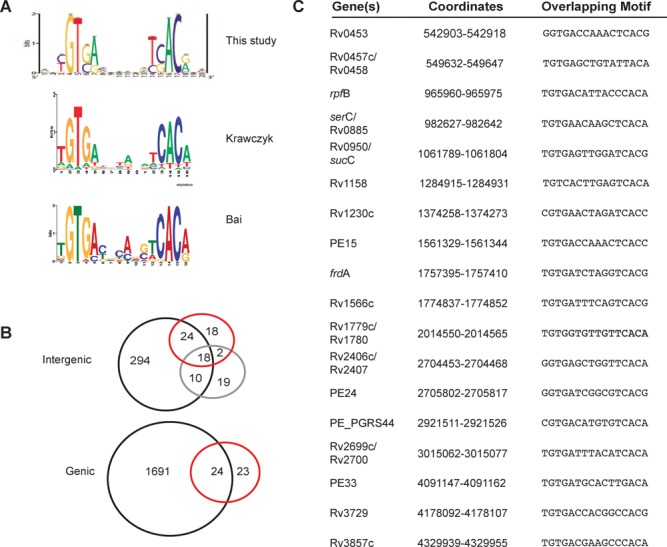
Comparison of in silico prediction and in vivo binding of CRP in Mycobacterium. (A) CRP binding motifs predicted from this study, Krawczyk et al. (25) [Figure copyright (c) 2009, Oxford University Press, Nucleic Acids Research, Vol 37, 2009, e97] and Bai et al. (14) [Figure copyright (c) 2005, American Society for Microbiology, Journal of Bacteriology, Vol. 187, 2005, pp 7795–7804. doi:10.1128/JB.187.22.7795-7804.2005]. (B) Top: comparison of the three studies, examining intergenic binding: this study, black, Krawczyk, red, Bai purple. Bottom: comparison of this study and Krawczyk et al. intragenic binding sequences. (C) Core intergenic binding sites from all three studies with overlapping Mtb H37Rv coordinates and corresponding sequences.
The in vivo binding model in this study closely resembles motifs from two previous studies, each of which used different approaches for prediction of CRPMt binding sites (14,25) (Figure 4A). Bai et al. (14) used a combination of E. coli CRP (CRPEc) binding sites and M. tuberculosis DNA sequences recovered by affinity capture using CRPMt as a seed their computational analysis. In contrast, Krawczyk et al. (25) approached their analysis from the perspective of the transcription factor, by using the regulon of the C. glutamicum CRP orthologue GlxR to predict orthologous CRPMt binding sequences. The strong agreement between binding models in all three studies suggests the presence of a robust core CRP binding site, which is supported by in vitro binding studies (13,14). All three binding models are comprised of a palindromic sequence with two 5 bp half sites separated by a 6 bp spacer with highly conserved GT in the 5′ half and an AC dinucleotide in the 3′ half-sequence. Nonetheless, there was limited overlap (40%) among the intergenic binding sites predicted in the Bai et al. (14) and Krawczyk et al. (25) studies despite their nearly identical binding motif models (Figure 4B).
We examined the concordance of 415 intergenic binding sites identified in this ChIP-seq analysis with predictions made in each of two previous studies to determine whether the CRP binding site discovery algorithm also affected binding site predictions (Figure 4B, top). This comparison was limited to intergenic sequences because intragenic sequences were excluded from the Bai et al. study (14). Eighteen of the total predicted binding sites from Bai et al. (37%) and Krawczyk et al. (29%) were common to all three studies (Figure 4C), but 57% of the Bai et al. sites and 68% of the Krawczyk et al. binding sites were also found in this ChIP-seq study (Figure 4B and C). Krawczyk et al. also identified 47 intragenic binding sites of which 24 (51%) were among our ChIP sites (Figure 4B, bottom). These results suggest that the discovery algorithms biased the specific regulon predictions in the prior studies, but the different approaches were similarly effective for predicting in vivo binding sites overall.
CRP binds upstream of the fumarate reductase and succinate dehydrogenase genes
We examined CRP's regulatory activity in more detail by focusing on the Rv0250c-Rv0249c locus, which contained four CRP binding sites in the ChIP-seq analysis. Rv0249c-Rv0247c is predicted to encode one of three fumarate reductase/succinate dehydrogenase enzyme complexes in Mtb; the remaining two complexes are encoded by sdhCDAB (Rv3316–3319) and frdABCD (Rv1552–1555) (34). An established CRP binding site upstream of frdA (Rv1552) (13) was confirmed in this ChIP-seq study, but a predicted CRP binding site upstream of sdhCD (Rv3316/Rv3317) (25) was not. CRPMt also failed to bind to the upstream region of sdhCD in vitro (Figure 5C), even at low pH, although CRPBCG was positive for this DNA fragment by EMSA (data not shown).
Figure 5.
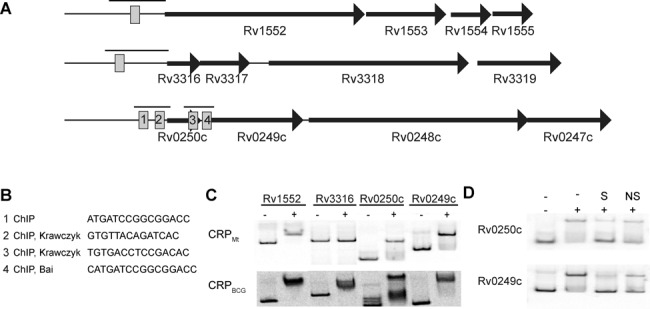
Binding of CRP to the predicted fumarate reductase and succinate dehydrogenase complexes in Mycobacteria. (A) Locus organization of frdABCD, sdhCDAB and Rv0250c-Rv0247c. (B) Source of CRP binding sites with sequences in relation to the upstream region of Rv0250c and Rv0249c. (C) Electrophoretic mobility shift assays of CRPBCG and CRPMt to the upstream promoter regions of indicated genes. Black bars above each of the suggested CRP sites in panel (A) indicate probe used for EMSA. (D) Competition assays with excess cold specific (S) or nonspecific (NS) double stranded DNA probes.
Only three of the four CRP sites identified by ChIP-seq in the Rv0250c-Rv0249c region were previously predicted (Figure 5A and B). Krawczyk et al. (25) predicted binding sites 2 and 3, while Bai et al. (14) identified site 4. Sites 2, 3 and 4 were also identified in another recent ChIP-seq study in Mtb (52). EMSA was used to investigate the in vitro binding of CRPBCG and CRPMt to the upstream regions of Rv0249c and Rv0250c, with the frdAbinding site serving as a positive control (Figure 5C). Both CRPMt and CRPBCG proteins bound to the upstream regions of Rv0250c and Rv0249c, and competition with excess cold specific or non-specific DNA fragments demonstrated binding specificity for both DNA probes (Figure 5D).
Role of intragenic binding in CRP mediated regulation of Rv0250c and Rv0249c
Having established direct binding of CRP upstream of each coding region, we took advantage of binding site 3 within the Rv0250c ORF to test the role of intragenic DNA sequences in the regulation of gene expression in mycobacteria. The high density of CRP binding sites at this locus also provided an ideal opportunity to better understand CRP activity at a potentially complex virulence-associated regulatory region, as relatively few mycobacterial promoters have been carefully analyzed to date. Transcriptional reporter fusions were used to evaluate the role of CRP in regulation of Rv0250c or Rv0249c expression. PCR products A (Rv0250c) or D (large Rv0249c) (Figure 6A), which also corresponds to the EMSA probes shown in Figure 5, were cloned into the BamHI site of pLacInt, an integrative shuttle plasmid that contains a promoterless lacZ gene, to generate transcriptional promoter: reporter fusions. A smaller upstream region of Rv0249c (PCR product E Figure 6A) was also initially tested for promoter activity.
Figure 6.
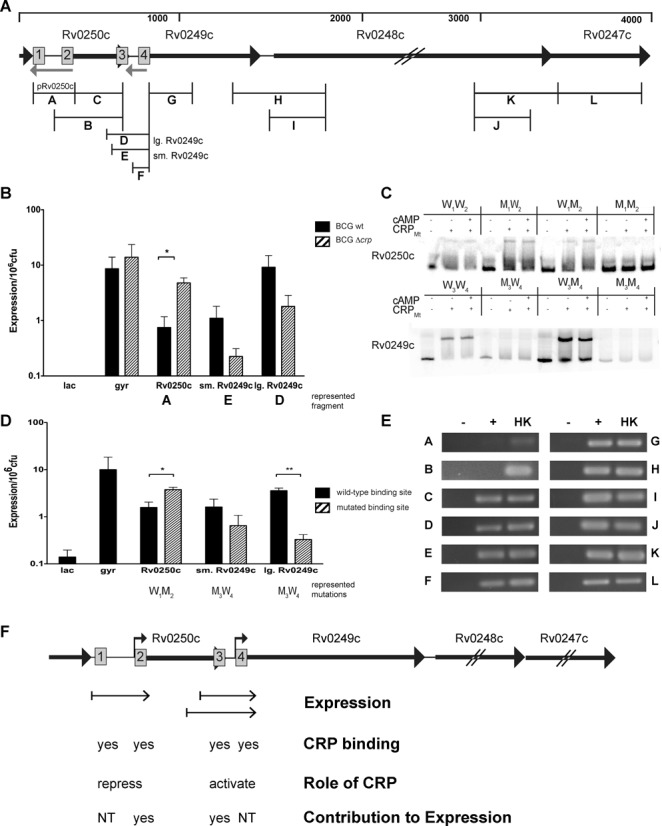
CRP binds and regulates at the Rv0250c locus. (A) Organization of the region. ∼60nt of the 3′ end of hsp are shown on the far left. The top bar shows approximate distance in nt. Gray arrows represent the anti-sense ncRNAs IG200 (ncRv10250) and IG199 (ncRv10249), identified by Arnvig et al. (72,75). (B) ß-galactosidase assay of the individual promoter fusion constructs in M. bovis BCG wild type (black bar) and crp background (hashed bars). (C) Purified CRPMt's ability to bind to either site 1 or 2 in the upstream region of Rv0250c or sites 3 or 4 in the upstream region of Rv0249c. (D) Contribution of sites 2 and 3 to expression. Black bars, wild type, hashed bars, replaced promoter fusion construct at site 2 or site 3 as indicated by x-axis. (E) RT-PCR using cDNA generated in the absence (−) or presence (+) of reverse transcriptase. Heat killed genomic DNA from M. tuberculosis H37Rv (H). Corresponding products are indicated in panel (A). (F) Summary of data. Small arrows above sites 2 and 4 indicate the positions of the binding sites relative to the transcriptional start sites (tss) as published (71). Binding site 1 is centered 64 nt from the +1 of Rv0250c, while the center of binding site 3 is 67.5 nt from the +1 of Rv0249c. Sites 2 and 4 overlap tss's for Rv0250c and Rv0249c promoters, respectively. Small arrows under each locus indicate promoter fragments used in these studies. NT not tested. Shown are the means with standard deviations from three independent experiments. (*, P < 0.05; **, P < 0.005 as determined by a student's t-test).
β-galactosidase expression levels were measured for each of these reporter constructs in wild-type or Δcrp M. bovis BCG grown shaking to late log phase in ambient air conditions (17) (Figure 6B). All three DNA fragments were able to drive expression of lacZ, demonstrating the presence of functional promoter elements upstream of each gene. However, expression of the Rv0249c promoters was decreased, while that of Rv0250c was increased, in Δcrp BCG compared to wild-type BCG. Therefore, CRP behaved as an activator of Rv0249c expression and a repressor of Rv0250c when the promoters were tested individually. The reporter with the extended Rv0249c promoter fragment (D) also had higher expression levels than the construct with the small Rv0249c fragment (E) (P < 0.05, Figure 6B), indicating that intragenic Rv0250c sequences upstream of CRP binding site 3 further contributed to Rv0249c expression.
We next examined the potential roles of individual CRP binding sites in the regulation of Rv0250c and Rv0249c. CRPMt binding was tested for each site by performing EMSAs with DNA probes in which one of the binding sites was replaced with an altered sequence. Results indicated that CRPMt bound at all four wild-type sites, with sites 2 and 3 showing the most robust interactions in vitro (Figure 6C). DNA fragments containing mutated sequences at sites 2 (W1M2) or 3 (M3W4) were then tested for promoter activity with the lacZ reporter system. The importance for promoter activity of each binding site was determined by comparing expression of the wild type versus the mutated promoter sequences in a wild type BCG background. Consistent with the crp knockout results (Figure 6B), mutation of site 2 increased β-galactosidase expression, while modification of site 3 decreased Rv0249c promoter activity (Figure 6D). Together, these results indicate that CRP regulates expression of both Rv0250c and Rv0249c through its binding of specific DNA sequences upstream of each gene.
We used RT-PCR to determine whether Rv0249c can be expressed from the Rv0250c promoter as part of an operon as well as from its own internal promoter. RT-PCR products D, E and F are consistent with co-transcription of Rv0249c and Rv0250c (Figure 6E). Together, these results indicate that Rv0249c can be expressed from the Rv0250c promoter as well as from the internal promoter that requires a portion of the Rv0250c coding sequences for maximal activity (Figure 6B–D). Amplification of junction fragments H and K (Figure 6E) also confirm earlier predictions that Rv0249c, Rv0248c and Rv0247c form an operon (34).
Expression of Rv0249c from the Rv0250c promoter
We next assessed the effects of individual binding sites on expression of lacZ when both the Rv0250c and Rv0249c promoters were present. We constructed a promoter: lacZ fusion using a DNA fragment that spanned both promoters and included all four CRP binding sites (designated ‘WWWW’ for the wild-type sequence of each of the four CRP binding sites). The expression of this construct was compared to that of each individual, wild type promoter (Figure 7A). The expression of the combined wild type construct was at least 10 times higher than either the Rv0250c or Rv0249c promoter fragment and ∼2–3-fold greater than that of the large Rv0249c promoter fragment.
Figure 7.
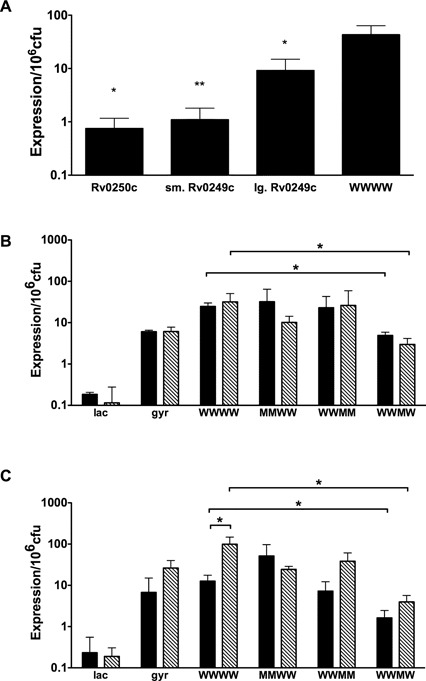
Contribution of binding sites to regulation of Rv0250c and Rv0249c. (A) The expression of the WWWW construct was compared to each of the individual promoters using ß-galactosidase assays. Significance is shown for individual promoters relative to the WWWW promoter. (B and C) Activity of the mutated dual promoter fragments was assessed in late-log (solid bars) or stationary (striped bars) cultures grown in (B), ambient oxygen or (C) hypoxic (1.3% oxygen + 5% CO2) conditions. The means with standard deviations are shown from three independent experiments. (*P < 0.05, **P < 0.005 as determined by a two-tailed students t-test).
Additional reporter constructs with altered CRP binding sequences were also generated and tested in wild-type BCG grown to late-log or stationary phase in ambient air (Figure 7B) or hypoxia (1.3% O2 + 5% CO2) (Figure 7C). Expression from the WWMW construct, which contained a mutated binding site 3, was significantly reduced compared to the WWWW in all conditions tested (Figure 7B and C). These observations correlate well with those obtained with the individual promoter fusions in which decreased expression was observed in the small and large Rv0249c promoter fragments that had CRP binding site 3 replacements (Figure 6D). However, when expression from the WWMM construct was measured, activity was similar to that of the WWWW construct. This observation suggests that site 4 has repressing activity that is suppressed by CRP binding to site 3.
Expression from the WWWW fragment was significantly increased in stationary versus log phase bacteria subjected to hypoxia. This trend was also observed for WWMW and WWMM constructs, but not for the MMWW reporter (Figure 7C). This result suggests that the Rv0250c, but not the Rv0249c, promoter is responsive to growth phase during hypoxia, an observation that warrants further investigation. Together, these data indicate that DNA sequences within the Rv0250c coding region are required for maximal activity of Rv0249c promoter, and that CRP binding to intragenic site 3 within Rv0250c plays a significant role in the regulation of Rv0249c-Rv0247 expression in TB complex bacteria.
DISCUSSION
Regulation of succinate dehydrogenase expression by CRP is direct, but complex
ChIP-seq is a powerful technique for detecting global TF binding under biologically relevant conditions and it has been especially useful for identifying new biological roles for TFs within mycobacteria (30,53). In the present study, we combined ChIP-seq analysis with examination of the regulatory roles of individual CRP binding sites at the Rv0250c-Rv0247c locus. Rv0249c is essential for in vitro growth of H37Rv and an Rv0249c transposon mutant is attenuated in infected mice (45,46). Previous studies have also shown that crp is needed for Mtb virulence in a murine infection model (15), and it is likely that reduced expression of Rv0249c-Rv0247c expression in the crp mutant contributes to this attenuated virulence phenotype. Our results establish a significant role for CRP in regulating expression of this putative succinate dehydrogenase complex encoded by Rv0249c-Rv0247c, along with the adjacent core conserved mycobacterial gene Rv0250c (43). Importantly, an intragenic CRP binding site, along with additional Rv0250c sequences, was required for maximal expression of Rv0249c. The presence at this locus of multiple CRP binding sites with differing regulatory properties, along with the likelihood of additional regulatory sequences adjacent to some of these CRP binding sites, could provide opportunities for integration of multiple signal inputs. Similarly, the presence of two promoters allows modulation of Rv0249c-Rv0247c complex expression in the presence or absence of Rv0250c expression depending on the environment Mtb is experiencing at any given time.
Succinate dehydrogenases complexes are the only enzymes that are part of both the respiratory complex and the TCA cycle, serving as a bridge between oxidative phosphorylation and central metabolism. Succinate dehydrogenase oxidizes succinate to fumarate in a reaction coupled to the reduction of FAD, while fumarate reductase catalyzes the reverse reaction (54). Accumulation and secretion of succinate is essential for Mtb's adaptation to hypoxia (55), and it has been suggested that succinate secretion maintains the membrane potential for energy generation (56). The second half of the TCA cycle in Mtb operates in a reductive direction, which requires conversion of fumarate to succinate by fumarate reductase (56). Our previous observation that CRP also binds to the upstream region of frdA, a putative fumarate reductase complex (13) suggests that CRP is integrally involved in regulating succinate metabolism in Mtb. We find CRP's role in regulating the expression of multiple enzymes involved in the reductive conversion of fumarate to succinate intriguing, and think the importance of CRP for Mtb's adaptation to hypoxic stress during infection especially warrants further investigation.
It has been postulated that most mycobacteria harbor two succinate dehydrogenases because of their need to adapt to a range of environments (57). Recent studies by Pecsi et al. (58) examined the roles of the orthologous Rv0249c-Rv0247c (MSMEG_0416-MSMEG_0420 or sdh1) and sdhCDAB (MSMEG_1672-MSMEG_1669 or sdh2) complexes within M. smegmatis, which lacks a frdA ortholog (57). Deletion of the entire sdh1 operon had little effect on growth, while deletion of the sdh2 region severely inhibited growth of M. smegmatis. Generation of membrane potential in hypoxic M. smegmatis was also attributed to sdh2. Expression of sdh1 was high in energy limiting conditions and down regulated in response to hypoxia and fumarate, while the sdh2 operon showed increased expression in response to hypoxia and fumarate in M. smegmatis. In contrast, Rv0249c (orthologous to M. smegmatis sdh1) is required for Mtb growth and virulence (45,46), and we observed regulation of Rv0249c expression in hypoxic BCG cultures. The presence of a fumarate reductase in addition to two putative succinate dehydrogenases in TB complex mycobacteria may affect the specific roles of these enzyme complexes in response to changing environments. Future studies in TB complex mycobacteria should clarify how sdh complexes have evolved with frd complexes in Mtb's adaptation within the host.
The present study confirms that Rv0249c, Rv0248c and Rv0247c comprise an operon, but our finding that CRP can also regulate expression of Rv0249c as an extended operon from the Rv0250c promoter was unexpected. The presence of CRP-regulated promoters upstream of both Rv0249c and Rv0250c, either of which can drive expression of the succinate dehydrogenase complex genes in response to different signals (Figure 7B and C), allows for coordinated expression of succinate dehydrogenase with Rv0250c expression. The full significance of such coordinated expression is difficult to predict in the absence of more information on Rv0250c function. However, the presence of four CRP binding sites and the wide range of CRP's regulatory effects, including both activation and repression, on the isolated promoters provides a wide range of possible regulatory responses to changing environments. It is likely that other trans-acting elements also affect expression of these genes. DNA microarray analysis showed that expression of both Rv0250c and Rv0249c is down-regulated by HspR (Rv0353) (59), and the transcription factors RipA, DtxR and RamA affect expression of sdhCAB in C. glutamicum (60). Additional studies of the RipA, DtxR and RamA orthologs in Mtb may yield insights on additional layers of gene regulation.
Intragenic CRP binding contributes to gene expression
Our observation that intragenic coding sequences from Rv0250c that include CRP binding site 3 significantly increased expression of Rv0249c (Figures 6 and 7A) demonstrates the importance of intragenic DNA sequences for gene regulation in mycobacteria (Figures 6 and 7B–C). Expression of the combined Rv0250c-Rv0249c promoter regions was ∼2x higher than the cumulative expression from the individual promoters, suggesting a coordinated regulatory effect between the promoters (Figure 7A). However, it is possible that the inclusion of additional contiguous sequences from Rv0250c coding region, beyond what are included in the largest individual Rv0249c promoter reporter, rather than synergy between the promoters, provides this enhancing effect.
While TF binding within genes does not fit the canonical view of transcriptional regulation in prokaryotes (40–42), sigma factors have been shown to bind at non-canonical promoters located within intragenic regions (61,62). Specifically, SigF, one of 13 sigma factors identified in Mtb, was associated with promoters located within the intragenic regions of Rv1358 and Rv1870c (63). Intragenic binding of CRP orthologs in other organisms has also been reported, but its significance has not been addressed. For example, 43% of the total GlxR binding sites in C. glutamicum detected by ChIP-seq are intragenic, and 30% of these sites are upstream of a divergently transcribed gene (64). Similarly, 54% of the total CRPMt binding sites in a recent study, including several of the most enriched peaks (e.g. Rv0010c, Rv3689, Rv2245 and Rv2585c), are located within intragenic regions (52). Intragenic TF binding within prokaryotes is not limited to CRP orthologs, as E. coli RpoH is reported to bind many intragenic sites (65) and over half of the FliA and LeuO binding sites identified in recent ChIP-seq studies are intragenic in E. coli and Salmonella enterica serovar Typhimurium, respectively (66,67).
TF binding within intragenic regions also plays a critical role in eukaryotic transcription (68,69), and a recent study by Stergachis et al. (70) highlighted the co-dependence of intragenic TF binding sites and the protein coding sequences in which they reside. These sequences, termed duons, are intragenic coding sequences that must co-evolve with the TF binding sites to accommodate both codon usage and TF binding requirements. Thus, intragenic TF binding in eukaryotes not only plays a role in regulating downstream gene expression, but also puts evolutionary pressure on the codon usage of the genes within which the TF binds.
The position of CRP binding site 3 centered at −67.5 relative the internal Rv0249c tss (71) is most consistent with CRP functioning as a canonical transcription factor at this site, and its intragenic location may reflect an example of a bacterial duon. However, the possibility that CRP regulates Rv0249c expression by altering chromosome structure cannot be ruled out. Small antisense RNAs that overlap the intergenic regions between Rv0251c/Rv0250c and Rv0250c/Rv0249c have also been reported (72), but the position of CRP binding site 3 is not consistent with indirect regulation of Rv0249c expression through either of these sRNAs. Nonetheless, the presence of these sRNAs likely contributes to the overall regulatory complexity at this locus, and CRP-mediated regulation of sRNA expression from other intragenic, as well as intergenic, sites seems likely, given the abundance of sRNAs in mycobacteria (72–75).
The very high percentage of intragenic binding sites for CRP identified in this study has important implications for understanding gene expression and developing molecular diagnostics in Mtb. One area of particular relevance is the wide use of single-nucleotide polymorphisms (SNPs) to predict drug resistance in Mtb, because SNPs that do not cause amino acid changes in a drug target gene or its (upstream intergenic) promoter are not typically considered as strong candidates for drug resistance mutations. However, our results indicate that the relevant genome space considered for resistance-associated SNPs should be expanded to include coding regions upstream of known drug target genes. A recent study (76) investigating the mechanism of ethambutol resistance supports the use of this wider net, as the investigators identified a correlation between an SNP within the Rv3792 (aftA) gene and ethambutol drug resistance in Mtb. Rv3792 is immediately upstream of embC, an established target gene for ethambutol resistance. The SNP within Rv3792 is associated with elevated expression of embC, and it is hypothesized that this SNP is located within a intragenic promoter for embC. ChIP-seq data from the TB consortium show potential binding of four TFs within the Rv3792 ORF (30), consistent with the presence of gene regulatory sequences within the Rv3792 ORF.
An expanded view of the CRP regulon
Our 10 most enriched CRP binding sites include both intragenic and intergenic binding sites, only two of which were previously predicted (site 1 within Rv0104, and site 7 between Rv0950c and Rv0951 (sucC), Table 1) (14,25). Site 1 within Rv0104 was also one of the most enriched binding sites in Mtb identified in another ChIP-seq study (52). Functions are not known for more than half the potential regulatory targets of these top 10 binding sites, suggesting that the majority of CRP's regulatory impact in Mtb remains to be elucidated. We previously demonstrated binding of CRP binding upstream of sucCD (14), whose expression is regulated by CRP under starvation conditions (13). The in vivo binding of CRP at this locus in the present study provides additional support for direct sucCD regulation by CRP and another link between CRP and succinate metabolism. Similarly, the increased sensitivity of the crp mutant to copper provides the first evidence for CRP-mediated regulation of copper homeostasis in mycobacteria, which could contribute to Mtb survival within macrophages. As noted, two of the most enriched binding sites from this study are located upstream of the ctpB gene, which encodes a putative copper transporter. Copper is toxic to Mtb, so its levels within the cell must be tightly controlled (35,36). The possibility that CRP controls copper resistance through regulation of ctpB expression in Mtb provides another potential role for CRP during host infection that should be explored.
CRP binding in the DNA region upstream of serC is consistent with our previous studies demonstrating CRP's role in the regulation of serC expression (17), and several other highly enriched binding sites have the potential to regulate cAMP-associated genes. In addition to binding the putative cNMP encoded by the Mtb ortholog Rv0104, CRP directly bound upstream of AC genes Rv1647 (77) and Rv3645, suggesting multiple levels of regulation within the cAMP signaling network in mycobacteria (9). We were interested to note that elevated levels of cAMP did not affect the CRP's in vivo binding profile (Figure 1B and Supplementary Figure S1), despite evidence that cAMP enhances interactions in vitro (Figure 2, (13–14,22,78)). It is possible that the baseline levels of cAMP under the conditions examined are already sufficient to fully bind CRP (16,79).
There was general agreement between our ChIP results and those of another recent ChIP study in Mtb (52), although we found nearly 10x as many binding sites in our study. The percentage (∼83%) of intragenic sites in our study was also greater than the proportion (∼50%) in the Kahramanoglu et al. (52) study. Overall, we identified orthologous sequences for ∼90% of the Mtb CRP sites recovered by Kahramanoglu et al. in Mtb (52), supporting our use of BCG as a model for identification of in vivo CRP binding in TB complex bacteria. Both studies identified the intragenic Rv0104 binding site as the most enriched site, and our third, fourth and sixth most enriched peaks were also among the top five in the Kahramanoglu et al. study (52). Binding sites in our study with peak heights as low as 4.5 were also found in the Mtb ChIP experiments, further validating CRP binding of these sites in Mtb.
The reason for the much larger number of binding sites in our study is not clear, but we think a higher level of sensitivity in our ChIP analysis is a major contributor to this difference. We used BCG as a model for these studies in part because the more stringent fixation conditions required for Mtb to ensure its biological inactivation led to inefficient chromosomal shearing and reduced the robustness of our ChIP assays. Biological differences could also contribute to the disparity in number of binding sites between the different studies. CRPBCG has a ∼2x higher affinity than CRPMt in vitro (13). In vitro binding affinity does not directly translate to in vivo binding, and we previously noted only subtle differences between BCG and Mtb in CRP binding by ChIP-PCR (13). Nonetheless, higher affinity could increase in vivo CRP binding and/or contribute to more efficient recovery of bound DNA fragments by immunoprecipation. While growth conditions also varied between studies, the relative lack of variation in CRP binding profiles across multiple conditions in our study (Supplemental Figure S1) argues against this being a significant variable.
Significance of low affinity binding sites
We observed a wide range of binding site enrichment, with roughly half of the binding sites having a normalized peak height less than 5. We retained all observed binding sites following background normalization because we think that the many of the less enriched sites have potential to be biologically relevant despite the likelihood of some false positives. Independent experiments across a range of environmental conditions, including the biological repeats of baseline versus Rv1264Cat, generated highly reproducible CRP binding profiles (Figure 1B and Supplemental Figure S1). In addition, identification of binding sites within the duplicated region of BCG_001-BCG_0040, served as an internal control of sorts. For example, an enriched binding region was found between 9534–9574 (BCG coordinates) and at the equivalent position in the duplicated region, between 39201–39242 (BCG coordinates). Both regions were similarly enriched, with peak heights of 2.64 and 2.27, respectively. The high number of strong binding motifs that were not bound by CRP further suggests that even low level binding is specific and should be considered in the analysis (Supplemental Figure S2). Moreover, highly enriched binding sites were often associated with closely linked sites that were less enriched, including three sites downstream of the two highest normalized peaks in this study that are intragenic to Rv0104c. These observations fit well with previous ChIP-seq studies in which the binding of eight different Mtb transcription factors showed highly reproducible weak binding profiles (30). In addition, thousands of weak CRP binding sites were identified within the E. coli genome by using ChIP-chip analysis (80), and over 2000 binding sites were found using ChIP-seq with NtcA, a CRP ortholog in cyanobacteria (81).
This theme of extensive weak binding by some transcription factors emerging from global ChIP analyses suggests that cooperative binding of transcription factors at multiple weak sites is a more common mechanism for modulation of promoter activity than has been previously appreciated. Nucleoid associated proteins (NAPs) can also contribute to gene regulation by modifying chromatin structure rather than through the types of direct interaction with RNA polymerase associated with canonical transcription factors. In Mtb, Lsr2 is a NAP that binds DNA at AT rich regions (82–84), while HupB is an HU ortholog that interacts with CRP binding regions, such as one associated with Rv1230c (85,86). EspR, was first described as a transcription factor that modulates expression of the virulence-associated espACD operon in Mtb (87). However, subsequent biophysical studies demonstrated that EspR regulates transcription over long distances by physically looping the DNA (53,88).
CRP in TB-complex mycobacteria may represents an example of a ‘dual acting’ transcription factors with NAP activity, as has been suggested for CRP orthologs in E. coli and cyanobacteria. The localization of binding sites at positions associated with direct polymerase interaction reported previously (17,21,89) and in this study, including the four binding sites at the Rv0250c-Rv0249c locus, is consistent with canonical transcription factor function. However, the large number of low level binding sites along with the long distances between many binding sites and their most proximal transcription start sites are NAP-like properties. In addition, CRP's ability to bend DNA (14) could contribute to chromatin remodeling. Therefore, CRP may have a spectrum of activities in TB complex bacteria, ranging from canonical transcription factor to those of a NAP, and are currently addressing this possibility.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We gratefully acknowledge Damen Schaak, Miralem Prijic and Dr. Sang Tae Park for technical assistance. We also thank Dr. Guanghcun Bai and members of the McDonough lab for their helpful discussions. DNA sequencing was performed in the Wadsworth Center Molecular Genetics Core.
Footnotes
Present addresses:
Zhuo Ma, Department of Basic and Social Sciences, Albany College of Pharmacy and Health Sciences, Albany, NY 12208, USA.
Antonio L.C. Gomes, Department of Systems Biology, Columbia University, New York, NY 10032, USA.
FUNDING
National Institutes of Health [F32GM095038 to G.S.K., HHSN272200800059C to J.E.G and 5RO1A1063499 to K.A.M]. Funding for open access charge: NIH NIAID.
Conflict of interest statement. None declared.
REFERENCES
- 1.World Health Organization. Global tuberculosis report 2013. 2013 [Google Scholar]
- 2.Corrigan R.M., Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat. Rev. Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 3.Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., Guo M., Roembke B.T., Sintim H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013;42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 4.Botsford J.L., Harman J.G. Cyclic AMP in prokaryotes. Microbiol. Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen S., Young G.M. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 2002;70:3665–3672. doi: 10.1128/IAI.70.7.3665-3672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith R.S., Wolfgang M.C., Lory S. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 2004;72:1677–1684. doi: 10.1128/IAI.72.3.1677-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtiss R. 3rd, Kelly S.M. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect. Immun. 1987;55:3035–3043. doi: 10.1128/iai.55.12.3035-3043.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonough K.A., Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat. Rev. Microbiol. 2012;10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCue L.A., McDonough K.A., Lawrence C.E. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 2000;10:204–219. doi: 10.1101/gr.10.2.204. [DOI] [PubMed] [Google Scholar]
- 10.Shenoy A.R., Sivakumar K., Krupa A., Srinivasan N., Visweswariah S.S. A survey of nucleotide cyclases in actinobacteria: unique domain organization and expansion of the class III cyclase family in Mycobacterium tuberculosis. Comp. Funct. Genomics. 2004;5:17–38. doi: 10.1002/cfg.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H., Hegde S.S., Blanchard J.S. Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry. 2011;50:5883–5892. doi: 10.1021/bi200156t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nambi S., Basu N., Visweswariah S.S. cAMP-regulated protein lysine acetylases in mycobacteria. J. Biol. Chem. 2010;285:24313–24323. doi: 10.1074/jbc.M110.118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai G., Gazdik M.A., Schaak D.D., McDonough K.A. The Mycobacterium bovis BCG cyclic AMP receptor-like protein is a functional DNA binding protein in vitro and in vivo, but its activity differs from that of its M. tuberculosis ortholog, Rv3676. Infect. Immun. 2007;75:5509–5517. doi: 10.1128/IAI.00658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai G., McCue L.A., McDonough K.A. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 2005;187:7795–7804. doi: 10.1128/JB.187.22.7795-7804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickman L., Scott C., Hunt D.M., Hutchinson T., Menéndez M.C., Walan R., Hinds J., Colston M.J., Green J., Buxton R.S. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 2005;56:1274–1286. doi: 10.1111/j.1365-2958.2005.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazdik M.A., Bai G., Wu Y., McDonough K.A. Rv1675c (cmr) regulates intramacrophage and cyclic AMP-induced gene expression in Mycobacterium tuberculosis-complex mycobacteria. Mol. Microbiol. 2009;71:434–448. doi: 10.1111/j.1365-2958.2008.06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai G., Schaak D.D., Smith E.A., McDonough K.A. Dysregulation of serine biosynthesis contributes to the growth defect of a Mycobacterium tuberculosis crp mutant. Mol. Microbiol. 2011;82:180–198. doi: 10.1111/j.1365-2958.2011.07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunt D.M., Saldanha J.W., Brennan J.F., Benjamin P., Strom M., Cole J.A., Spreadbury C.L., Buxton R.S. Single nucleotide polymorphisms that cause structural changes in the cyclic AMP receptor protein transcriptional regulator of the tuberculosis vaccine strain Mycobacterium bovis BCG alter global gene expression without attenuating growth. Infect. Immun. 2008;76:2227–2234. doi: 10.1128/IAI.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaprelyants A.S., Mukamolova G.V., Ruggiero A., Makarov V.A., Demina G.R., Shleeva M.O., Potapov V.D., Shramko P.A. Resuscitation-promoting factors (Rpf): in search of inhibitors. Protein Pept. Lett. 2012;19:1026–1034. doi: 10.2174/092986612802762723. [DOI] [PubMed] [Google Scholar]
- 20.Clarke S.J., Low B., Konigsberg W.H. Close linkage of the genes serC (for phosphohydroxy pyruvate transaminase) and serS (for seryl-transfer ribonucleic acid synthetase) in Escherichia coli K-12. J. Bacteriol. 1973;113:1091–1095. doi: 10.1128/jb.113.3.1091-1095.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stapleton M., Haq I., Hunt D.M., Arnvig K.B., Artymiuk P.J., Buxton R.S., Green J. Mycobacterium tuberculosis cAMP receptor protein (Rv3676) differs from the Escherichia coli paradigm in its cAMP binding and DNA binding properties and transcription activation properties. J. Biol. Chem. 2010;285:7016–7027. doi: 10.1074/jbc.M109.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal N., Raghunand T.R., Bishai W.R. Regulation of the expression of whiB1 in Mycobacterium tuberculosis: role of cAMP receptor protein. Microbiology. 2006;152:2749–2756. doi: 10.1099/mic.0.28924-0. [DOI] [PubMed] [Google Scholar]
- 23.Smith L.J., Stapleton M.R., Fullstone G.J., Crack J.C., Thomson A.J., Le Brun N.E., Hunt D.M., Harvey E., Adinolfi S., Buxton R.S., et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem. J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crack J.C., Smith L.J., Stapleton M.R., Peck J., Watmough N.J., Buttner M.J., Buxton R.S., Green J., Oganesyan V.S., Thomson A.J., et al. Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J. Am. Chem. Soc. 2011;133:1112–1121. doi: 10.1021/ja109581t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krawczyk J., Kohl T.A., Goesmann A., Kalinowski J., Baumbach J. From Corynebacterium glutamicum to Mycobacterium tuberculosis–towards transfers of gene regulatory networks and integrated data analyses with MycoRegNet. Nucleic Acids Res. 2009;37:e97. doi: 10.1093/nar/gkp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes A.L., Abeel T., Peterson M., Azizi E., Lyubetskaya A., Carvalho L., Galagan J. Decoding ChIP-seq with a double-binding signal refines binding peaks to single-nucleotides and predicts cooperative interaction. Genome Res. 2014;24:1686–1697. doi: 10.1101/gr.161711.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purkayastha A., McCue L.A., McDonough K.A. Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr aene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 2002;70:1518–1529. doi: 10.1128/IAI.70.3.1518-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florczyk M.A., McCue L.A., Purkayastha A., Currenti E., Wolin M.J., McDonough K.A. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 2003;71:5332–5343. doi: 10.1128/IAI.71.9.5332-5343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parish T., Stoker N.G. Electroporation of mycobacteria. Methods Mol. Biol. 1998;101:129–144. doi: 10.1385/0-89603-471-2:129. [DOI] [PubMed] [Google Scholar]
- 30.Galagan J.E., Minch K., Peterson M., Lyubetskaya A., Azizi E., Sweet L., Gomes A., Rustad T., Dolganov G., Glotova I., et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatfull G.F. Genetic transformation of mycobacteria. Trends Microbiol. 1993;1:310–314. doi: 10.1016/0966-842x(93)90008-f. [DOI] [PubMed] [Google Scholar]
- 34.Lew J.M., Kapopoulou A., Jones L.M., Cole S.T. TubercuList–10 years after. Tuberculosis (Edinb.) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Wolschendorf F., Ackart D., Shrestha T.B., Hascall-Dove L., Nolan S., Lamichhane G., Wang Y., Bossmann S.H., Basaraba R.J., Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowland J.L., Niederweis M. A multicopper oxidase is required for copper resistance in Mycobacterium tuberculosis. J. Bacteriol. 2013;195:3724–3733. doi: 10.1128/JB.00546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasudeva-Rao H.M., McDonough K.A. Expression of the Mycobacterium tuberculosis acr-coregulated genes from the DevR (DosR) regulon is controlled by multiple levels of regulation. Infect. Immun. 2008;76:2478–2489. doi: 10.1128/IAI.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rowland B., Purkayastha A., Monserrat C., Casart Y., Takiff H., McDonough K.A. Fluorescence-based detection of lacZ reporter gene expression in intact and viable bacteria including Mycobacterium species. FEMS Microbiol. Lett. 1999;179:317–325. doi: 10.1111/j.1574-6968.1999.tb08744.x. [DOI] [PubMed] [Google Scholar]
- 39.Gazdik M.A., McDonough K.A. Identification of cyclic AMP-regulated genes in Mycobacterium tuberculosis complex bacteria under low-oxygen conditions. J. Bacteriol. 2005;187:2681–2692. doi: 10.1128/JB.187.8.2681-2692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young B.A., Gruber T.M., Gross C.A. Views of transcription initiation. Cell. 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 41.Lee D.J., Minchin S.D., Busby S.J. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 42.Browning D.F., Busby S.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 43.Lew J.M., Mao C., Shukla M., Warren A., Will R., Kuznetsov D., Xenarios I., Robertson B.D., Gordon S.V., Schnappinger D., et al. Database resources for the tuberculosis community. Tuberculosis (Edinb.) 2013;93:12–17. doi: 10.1016/j.tube.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McAdam R.A., Quan S., Smith D.A., Bardarov S., Betts J.C., Cook F.C., Hooker E.U., Lewis A.P., Woollard P., Everett M.J., et al. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology. 2002;148:2975–2986. doi: 10.1099/00221287-148-10-2975. [DOI] [PubMed] [Google Scholar]
- 45.Sassetti C.M., Rubin E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffin J.E., Gawronski J.D., Dejesus M.A., Ioerger T.R., Akerley B.J., Sassetti C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White C., Lee J., Kambe T., Fritsche K., Petris M.J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halliwell B., Gutteridge J.M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 50.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kahramanoglou C., Cortes T., Matange N., Hunt D.M., Visweswariah S.S., Young D.B., Buxton R.S. Genomic mapping of cAMP receptor protein (CRPMt) in Mycobacterium tuberculosis: relation to transcriptional start sites and the role of CRPMt as a transcription factor. Nucleic Acids Res. 2014;42:8320–8329. doi: 10.1093/nar/gku548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blasco B., Chen J.M., Hartkoorn R., Sala C., Uplekar S., Rougemont J., Pojer F., Cole S.T. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 2012;8:e1002621. doi: 10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cecchini G., Schroder I., Gunsalus R.P., Maklashina E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta. 2002;1553:140–157. doi: 10.1016/s0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 55.Eoh H., Rhee K.Y. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe S., Zimmermann M., Goodwin M.B., Sauer U., Barry C.E. 3rd, Boshoff H.I. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berney M., Cook G.M. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One. 2010;5:e8614. doi: 10.1371/journal.pone.0008614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pecsi I., Hards K., Ekanayaka N., Berney M., Hartman T., Jacobs W.R. Jr, Cook G.M. Essentiality of succinate dehydrogenase in Mycobacterium smegmatis and its role in the generation of the membrane potential under hypoxia. MBio. 2014;5:e01093–e01114. doi: 10.1128/mBio.01093-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart G.R., Wernisch L., Stabler R., Mangan J.A., Hinds J., Laing K.G., Young D.B., Butcher P.D. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 60.Bussmann M., Emer D., Hasenbein S., Degraf S., Eikmanns B.J., Bott M. Transcriptional control of the succinate dehydrogenase operon sdhCAB of Corynebacterium glutamicum by the cAMP-dependent regulator GlxR and the LuxR-type regulator RamA. J. Biotechnol. 2009;143:173–182. doi: 10.1016/j.jbiotec.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 61.Stringer A.M., Currenti S., Bonocora R.P., Baranowski C., Petrone B.L., Palumbo M.J., Reilly A.A., Zhang Z., Erill I., Wade J.T. Genome-scale analyses of Escherichia coli and Salmonella enterica AraC reveal noncanonical targets and an expanded core regulon. J. Bacteriol. 2014;196:660–671. doi: 10.1128/JB.01007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonocora R.P., Fitzgerald D.M., Stringer A.M., Wade J.T. Non-canonical protein-DNA interactions identified by ChIP are not artifacts. BMC Genomics. 2013;14:254. doi: 10.1186/1471-2164-14-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hartkoorn R.C., Sala C., Magnet S.J., Chen J.M., Pojer F., Cole S.T. Sigma factor F does not prevent rifampin inhibition of RNA polymerase or cause rifampin tolerance in Mycobacterium tuberculosis. J. Bacteriol. 2010;192:5472–5479. doi: 10.1128/JB.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jungwirth B., Sala C., Kohl T.A., Uplekar S., Baumbach J., Cole S.T., Puhler A., Tauch A. High-resolution detection of DNA binding sites of the global transcriptional regulator GlxR in Corynebacterium glutamicum. Microbiology. 2013;159:12–22. doi: 10.1099/mic.0.062059-0. [DOI] [PubMed] [Google Scholar]
- 65.Wade J.T., Castro Roa D., Grainger D.C., Hurd D., Busby S.J., Struhl K., Nudler E. Extensive functional overlap between sigma factors in Escherichia coli. Nat. Struct. Mol. Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 66.Fitzgerald D.M., Bonocora R.P., Wade J.T. Comprehensive Mapping of the Escherichia coli Flagellar Regulatory Network. PLoS Genet. 2014;10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dillon S.C., Espinosa E., Hokamp K., Ussery D.W., Casadesus J., Dorman C.J. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2012;85:1072–1089. doi: 10.1111/j.1365-2958.2012.08162.x. [DOI] [PubMed] [Google Scholar]
- 68.MacQuarrie K.L., Fong A.P., Morse R.H., Tapscott S.J. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 2011;27:141–148. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hughes T.R., de Boer C.G. Mapping yeast transcriptional networks. Genetics. 2013;195:9–36. doi: 10.1534/genetics.113.153262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stergachis A.B., Haugen E., Shafer A., Fu W., Vernot B., Reynolds A., Raubitschek A., Ziegler S., LeProust E.M., Akey J.M., et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cortes T., Schubert O.T., Rose G., Arnvig K.B., Comas I., Aebersold R., Young D.B. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell. Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arnvig K.B., Comas I., Thomson N.R., Houghton J., Boshoff H.I., Croucher N.J., Rose G., Perkins T.T., Parkhill J., Dougan G., et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arnvig K.B., Young D.B. Identification of small RNAs in Mycobacterium tuberculosis. Mol. Microbiol. 2009;73:397–408. doi: 10.1111/j.1365-2958.2009.06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DiChiara J.M., Contreras-Martinez L.M., Livny J., Smith D., McDonough K.A., Belfort M. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res. 2010;38:4067–4078. doi: 10.1093/nar/gkq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamichhane G., Arnvig K.B., McDonough K.A. Definition and annotation of (myco)bacterial non-coding RNA. Tuberculosis (Edinb.) 2013;93:26–29. doi: 10.1016/j.tube.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Safi H., Lingaraju S., Amin A., Kim S., Jones M., Holmes M., McNeil M., Peterson S.N., Chatterjee D., Fleischmann R., et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-beta-D-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 2013;45:1190–1197. doi: 10.1038/ng.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dass B.K., Sharma R., Shenoy A.R., Mattoo R., Visweswariah S.S. Cyclic AMP in mycobacteria: characterization and functional role of the Rv1647 ortholog in Mycobacterium smegmatis. J. Bacteriol. 2008;190:3824–3834. doi: 10.1128/JB.00138-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stapleton M.R., Smith L.J., Hunt D.M., Buxton R.S., Green J. Mycobacterium tuberculosis WhiB1 represses transcription of the essential chaperonin GroEL2. Tuberculosis (Edinb.) 2012;92:328–332. doi: 10.1016/j.tube.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai G., Schaak D.D., McDonough K.A. cAMP levels within Mycobacterium tuberculosis and Mycobacterium bovis BCG increase upon infection of macrophages. FEMS Immunol. Med. Microbiol. 2009;55:68–73. doi: 10.1111/j.1574-695X.2008.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grainger D.C., Hurd D., Harrison M., Holdstock J., Busby S.J. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Picossi S., Flores E., Herrero A. ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genomics. 2014;15:22. doi: 10.1186/1471-2164-15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gordon B.R., Li Y., Wang L., Sintsova A., van Bakel H., Tian S., Navarre W.W., Xia B., Liu J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gordon B.R., Imperial R., Wang L., Navarre W.W., Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen J.M., Ren H., Shaw J.E., Wang Y.J., Li M., Leung A.S., Tran V., Berbenetz N.M., Kocincova D., Yip C.M., et al. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 2008;36:2123–2135. doi: 10.1093/nar/gkm1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gupta M., Sajid A., Sharma K., Ghosh S., Arora G., Singh R., Nagaraja V., Tandon V., Singh Y. HupB, a nucleoid-associated protein of Mycobacterium tuberculosis, is modified by serine/threonine protein kinases in vivo. J. Bacteriol. 2014;196:2646–2657. doi: 10.1128/JB.01625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharadamma N., Khan K., Kumar S., Patil K.N., Hasnain S.E., Muniyappa K. Synergy between the N-terminal and C-terminal domains of Mycobacterium tuberculosis HupB is essential for high-affinity binding, DNA supercoiling and inhibition of RecA-promoted strand exchange. FEBS J. 2011;278:3447–3462. doi: 10.1111/j.1742-4658.2011.08267.x. [DOI] [PubMed] [Google Scholar]
- 87.Hunt D.M., Sweeney N.P., Mori L., Whalan R.H., Comas I., Norman L., Cortes T., Arnvig K.B., Davis E.O., Stapleton M.R., et al. Long-range transcriptional control of an operon necessary for virulence-critical ESX-1 secretion in Mycobacterium tuberculosis. J. Bacteriol. 2012;194:2307–2320. doi: 10.1128/JB.00142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blasco B., Japaridze A., Stenta M., Wicky B.I., Dietler G., Peraro M., Pojer F., Cole S.T. Functional dissection of intersubunit interactions in the EspR virulence regulator of Mycobacterium tuberculosis. J. Bacteriol. 2014;196:1889–1900. doi: 10.1128/JB.00039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Akhter Y., Tundup S., Hasnain S.E. Novel biochemical properties of a CRP/FNR family transcription factor from Mycobacterium tuberculosis. Int. J. Med. Microbiol. 2007;297:451–457. doi: 10.1016/j.ijmm.2007.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


