Abstract
Intercellular communication can be mediated by extracellular small regulatory RNAs (sRNAs). Circulating sRNAs are being intensively studied for their promising use as minimally invasive disease biomarkers. To date, most attention is centered on exosomes and microRNAs as the vectors and the secreted species, respectively. However, this field would benefit from an increased understanding of the plethora of sRNAs secreted by different cell types in different extracellular fractions. It is still not clear if specific sRNAs are selected for secretion, or if sRNA secretion is mostly passive. We sequenced the intracellular sRNA content (19–60 nt) of breast epithelial cell lines (MCF-7 and MCF-10A) and compared it with extracellular fractions enriched in microvesicles, exosomes and ribonucleoprotein complexes. Our results are consistent with a non-selective secretion model for most microRNAs, although a few showed secretion patterns consistent with preferential secretion. On the contrary, 5′ tRNA halves and 5′ RNA Y4-derived fragments of 31–33 were greatly and significantly enriched in the extracellular space (even in non-mammary cell lines), where tRNA halves were detected as part of ∼45 kDa ribonucleoprotein complexes. Overall, we show that different sRNA families have characteristic secretion patterns and open the question of the role of these sRNAs in the extracellular space.
INTRODUCTION
Since the discovery that extracellular vesicles (EVs) can act as vehicles for the exchange of microRNAs (miRNAs) between cells (1), extracellular small regulatory RNAs (sRNAs) circulating in body fluids have attracted great attention both from a physiological point of view and because of their promising use as minimally invasive disease biomarkers (2). The potential of plasma or serum miRNA profiling for an earlier cancer diagnosis and to predict prognosis and response to therapy has been recently reviewed by Schwarzenbach et al. (3).
In order to persist in the extracellular environment and participate in cell-to-cell communication, cell-secreted sRNAs must be protected from ubiquitous extracellular RNases. To date, circulating sRNAs have been described in a variety of RNase-insensitive protein or lipid complexes, or encapsulated inside different types of EVs (4). The vast majority (above 90%) of circulating miRNAs is reportedly present in stability-conferring ribonucleoprotein complexes (5,6). Different proteins such as the catalytic core of the miRNA-induced silencing complex, Argonaute 2 (5,7) or nucleophosmin 1 (8), have been associated with circulating miRNAs in plasma and cell culture media. Small RNAs have also been described in plasma in association with lipoprotein particles (9).
However, EV-associated sRNAs have received special attention, in part because of the perceived potential of EVs to interact with and deliver cargo to specific target cells. EVs include microvesicles, exosomes and apoptotic bodies. Although the terms exosomes and microvesicles are often confounded in the literature (10), these EVs differ both in their physicochemical parameters (size, density) and in their biogenesis. Exosomes are typically described as having diameters of 40–100 nm and as being released into the extracellular space by fusion of multivesicular bodies (late endosomes containing intraluminal vesicles) with the cell membrane (11). Microvesicles are considered to be larger in size, more irregular in shape and derived from outward blebbing of the plasma membrane. Finally, apoptotic bodies are derived from cells that have undergone programmed cell death and contain cellular components such as partially degraded nucleic acids, peptides and lipids (12).
The cellular mechanisms underlying secretion of sRNAs and their selection and sorting into different secretory routes are still not fully understood. It has been reported that miRNA secretion is an active ATP-dependent process (8), and that the exporting cell selects specific miRNAs for export (8,13). Other authors have proposed that extracellular miRNA levels mirror their intracellular abundances, and that secretion occurs mostly in a non-selective manner. Extracellular sRNAs might even be nothing more than a consequence of necrotic cell death, releasing highly stable Argonaute/miRNA complexes (7). The different hypotheses regarding selective or non-selective miRNA export from cells and their roles in cell-to-cell communication have been reviewed elsewhere (14,15) and remain a matter of debate.
The advent of high throughput sequencing has revealed that the sRNA populations found inside a cell go far beyond the classical small interfering, piwi-associated and miRNA families. Non-protein-coding functional RNAs such as rRNA, tRNA, snRNA and snoRNA show terminal and asymmetric processing to yield smaller RNA entities, involved (or predicted to be involved) in specific regulatory pathways (16). In particular, tRNAs are cleaved near the anticodon loop to yield tRNA halves, some of which were shown to inhibit cap-dependent translation initiation and to activate a cytoprotective stress-response reprogramming protein translation (17). Smaller tRNA-derived regulatory fragments (18) were also shown to enter the RNA interference pathway or regulate protein synthesis by a variety of mechanisms [reviewed in (19,20)]. Interestingly, recent reports have highlighted the presence of 5′ tRNA halves (21–23) and 5′ YRNA-derived fragments (21,24) as abundant vesicle-free soluble complexes in plasma and serum. Moreover, these species were also detected in extracellular vesicles derived from immune cells (13) and human semen (25). Thus, although most of what is known about small RNA secretion is still restricted to miRNAs, non-canonical small RNA families are also secreted by certain cell types.
Herein, we sequenced the intracellular small RNA content of serum-free cultures of breast epithelial cell lines (MCF-7 and MCF-10A), and compared it with the small RNA content of different extracellular fractions following conventional protocols for microvesicle, exosome and ribonucleoprotein complex enrichment. Our results showed that most miRNAs are consistent with a non-selective secretion model, with few exceptions. In contrast, 5′ tRNA halves and 5′ RNA Y4-derived fragments of 31–33 nt showed secretion patterns consistent with preferential secretion. More than half of the reads mapped to the human genome in the ribonucleoprotein-associated fraction from both cell lines were 5′ tRNA halves from Gly and Glu isoacceptors. This corresponded to an approximately 20-fold increase in their relative abundance in comparison with the intracellular compartment. High abundance of tRNA halves was also observed in the ribonucleoprotein-enriched fraction of HeLa and NCI-H1299 conditioned media, suggesting that secretion of tRNA halves is a rather general mechanism, common to cell lines obtained from different sources. Overall, we show that different small RNA families have differential and characteristic secretion patterns, raising questions about the biological meaning of these non-canonical small RNAs in the extracellular space.
MATERIALS AND METHODS
Cell cultures
Cells were routinely incubated in a humidified chamber at 37°C with 5% CO2 without antibiotics. MCF-7, MCF-10A and NCI-H1299 cells were purchased from ATCC and used at low passage (<10). MCF-7 cells were cultured in EMEM (ATCC) + 0.01 mg/ml recombinant human insulin + 10% FBS (Gibco) until the desired cell number was obtained. After media removal and rinsing with phosphate buffered saline (PBS), cells were changed to defined media [modified from (26)] consisting of EMEM plus 3.8 μg/ml recombinant human insulin, 0.8 ng/ml EGF, 5 μg/ml transferrin, 475 pg/ml PGF2α and 20 μg/ml fibronectin. After an adaptation period of 48 h, the media was completely removed, cells were rinsed with PBS and new defined media was added. After further incubation for 48 h, conditioned media was collected. Cells (70% confluent) were detached using a trypsin-EDTA solution, counted and lysed. The same procedure was followed for HeLa and NCI-H1299 cells. MCF-10A cells were cultured in MEGM (Lonza) according to ATCC recommendations, but addition of cholera toxin was omitted. After the desired cell number was obtained, cells were detached using trypsin-EDTA (which was later neutralized with soybean trypsin inhibitor) and 1×106 viable cells were plated in 75 cm2 flasks. After 24 h, the media was removed; cells were rinsed with PBS and incubated in MEGM without the addition of bovine pituitary extract (MEGM–BPE media). After an adaptation period of 48 h, the media was completely removed, cells were rinsed with PBS and new defined media (MEGM–BPE) was added. Conditioned media corresponding to t = 48 h was collected, and cells (at a density of ∼70%) were detached, counted and lysed. Cell death at the time of conditioned media collection was below 5% in all cases as performed by trypan blue staining.
Purification of extracellular fractions
Conditioned serum-free media (t = 48 h) was harvested from culture flasks and immediately centrifuged at 300 g to remove detached cells and stored at −20°C. Once thawed, the media was centrifuged at 2000 g and 4°C for 30 min to remove cell debris and apoptotic blebs. The supernatant was centrifuged for 0.5 h at 16 000 g and 4°C. The pellet (p16 fraction) was washed twice in PBS and resuspended in a desired volume of PBS or directly subjected to RNA extraction. The supernatant was gently filtered by 0.22 μm and centrifuged for 2.5 h at 100 000 g and 4°C in a Beckman Coulter Optima XPN-90 ultracentrifuge using an SW40 Ti rotor. The pellet (p100 fraction) was washed with 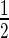 initial volume of PBS before resuspension in a desired volume of PBS and protein/RNA extraction. The supernatant of the first 100 000 g centrifugation (S100 fraction) was concentrated to ∼250 μl with 10 000 MWCO ultrafiltration units (Vivaspin 20, Sartorious Stedim Biotech) and subjected to RNA extraction. Purified fractions p16 and p100 were characterized by nanoparticle tracking analysis (NTA) using a NanoSight NS500 (Malvern Instruments), as described in the supplementary materials section.
initial volume of PBS before resuspension in a desired volume of PBS and protein/RNA extraction. The supernatant of the first 100 000 g centrifugation (S100 fraction) was concentrated to ∼250 μl with 10 000 MWCO ultrafiltration units (Vivaspin 20, Sartorious Stedim Biotech) and subjected to RNA extraction. Purified fractions p16 and p100 were characterized by nanoparticle tracking analysis (NTA) using a NanoSight NS500 (Malvern Instruments), as described in the supplementary materials section.
RNA extraction and quantification
RNA purification was performed with Trizol LS (Invitrogen, Life Technologies) according to manufacturer's instructions with minor modifications to minimize small RNA lost in the precipitation step (duplication of isopropanol volume and washing with 80% ethanol). In p16 and p100 fractions, 10 μg RNAse-free glycogen was added as a carrier. RNA was quantified using a Qubit 2.0 fluorometer (Life Technologies) and a Qubit RNA high sensitivity kit, according to manufacturer's instructions.
Western blot analysis
Proteins were purified from cells and vesicles from the organic phase of the Trizol LS reagent used for RNA extraction, according to manufacturer's specifications. The protein pellet was resuspended in 1× RIPA buffer and quantified by the Bicinchoninic Acid assay. For western blots, 20 μg of samples were subjected to 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and electrotransferred to Hybond-P PVDF membranes (GE Healthcare, Amersham). After blocking, membranes were incubated overnight at 4°C with the following murine monoclonal antibodies: anti-TSG101 (4A10; Abcam; dilution 1/1000), anti-CD9 (clone C-4; Santa Cruz Biotechnologies; dilution 1/300) and anti-CD63 (MEM-259; Abcam; dilution 1/500). Blots were developed with horseradish-peroxidase-conjugated rabbit polyclonal secondary antibodies to mouse IgG – H&L (ab6728; Abcam).
Construction of libraries and high-throughput sequencing
Three biological replicates for each extracellular fraction and for each cell line and two biological replicates for intracellular fractions were sequenced. RNA inputs were on average 150 ng and 1.2 μg for extracellular and intracellular fractions, respectively. Intracellular RNA inputs were higher due to the low relative abundance of small RNAs and the predominance of larger RNA species (mostly rRNAs). DNA libraries were prepared using the NEBNext Small RNA Library Prep Set for Illumina (New England Biolabs) according to the manufacturer's instructions. Briefly, 3′ and 5′ adapters were ligated to input RNAs, reverse-transcribed and PCR amplified (15 cycles). Multiplexing indexes were included in the PCR reaction. PCR products were run on a 6% polyacrylamide gel and size-selected to include RNAs with an insert size < 60 nt. Libraries were analyzed using a 2100 Bioanalyzer (Agilent), quantified by the Qubit assay (Life Technologies) and submitted to sequencing (single-end) on either an Illumina Genome Analyzer IIx or an Illumina MiSeq desktop sequencer. Supplementary Figure S1 shows representative polyacrylamide gels and Bioanalyzer profiles of sequencing libraries obtained for MCF-7 p16, p100 and S100 fractions, compared to the concentrated non-conditioned media, which served as a negative control.
In the initial experimental design, reads of up to 72 nt were considered essential in order to reliably determine the 3′ ends of all sequenced RNAs. Once confirming lack of significant RNA populations above ∼40 nt in our libraries, the last biological replicates were sequenced with a 44-cycle format. The platform, number of cycles and sequencing depth in each data set are indicated in Supplementary Table S1.
Sequencing data analysis
After sample demultiplexing and adapter trimming (only sequences >15 bases which contained an identifiable 3′ adaptor were analyzed), FastQ files containing sequencing information were mapped to the human genome (hg19) with Bowtie (27), allowing one mismatch. Mapped reads were collapsed to unique sequences with their associated count numbers and aligned with Lastz (one mismatch allowance) to manually curated reference libraries containing unambiguous sequences corresponding to mature human miRNAs downloaded from miRBase release 20 (28); mature human tRNAs downloaded from the genomic tRNA database (http://gtrnadb.ucsc.edu/); and mature human ribosomal RNA, small nuclear RNA, small nucleolar RNA, vault RNA and YRNA sequences downloaded from GenBank (http://www.ncbi.nlm.nih.gov/nuccore/). Only one annotation per read was kept, prioritizing sense alignments with no mismatches on the entire query sequence. Since the Lastz algorithm uses seeds with a minimum length of 19, the size range of the RNAs under study was finally 19–60 nt (in 72-cycle format). Genomic coordinates corresponding to coding mRNA exons and tRNA 3′ trailers were retrieved from the UCSC Main table browser included in the Galaxy project online platform (29) and used to annotate mapped reads as CDS and tRF-1. Sequences annotated with more than one tag were manually resolved by considering the presence/absence of mismatches, length of the query sequence and abundance-based likelihood. The relative abundance of each unique sequence was expressed as reads per million (RPM) mapped reads by dividing its absolute count number by the total amount of mapped reads in the data set and multiplying by a million. Subsequent analysis was performed with in-house ad-hoc scripts. Statistical tests were performed using the software GraphPad Prism 5 and are detailed in the figure captions. Sequencing data were submitted to the sequence read archive database (http://www.ncbi.nlm.nih.gov/sra) under the BioProject ID: PRJNA270876
Stem-loop RT quantitative and droplet digital PCR (ddPCR)
We used a modification of the popular stem-loop RT miRNA quantification method (30) for small RNA quantification. For detection (in ddPCR) a universal hydrolysis probe (FAM-MGB-NFQ) was used (Life Technologies, USA) since this probe was shown to yield the same sensitivity and specificity as the original method (31). RNA samples were diluted to 10 ng/μl, and 2 μl were introduced into the retrotranscriptase mix (Applied Biosystems; Life Technologies) together with the folded (32) SL-RT primers at 10 nM final concentration (12 μl final volume). 20 μl DNAse-free water was added to each tube and 2 μl of diluted cDNA were used to yield 20 μl PCR reaction consisting of 1× ddPCR master mix (Bio-Rad), 1.5 μM forward primer, 0.7 μM reverse primer and 0.8 μM hydrolysis probe. Emulsification of the PCR reaction and droplet fluorescence readout was performed with a QX100 droplet digital PCR system (Bio-Rad) following manufacturer's protocols. For qPCR, a 2x SYBR green-based qPCR master mix (KapaBiosystems) was used with a Rotor-Gene 6000 qPCR system (Corbett Life Science). Quantification values (Cq) were considered relevant if at least 2 cycles below no-template or no-retrotranscriptase controls (whatever the least). Primer sequences and hydrolysis probes used are provided as supplementary material.
Nuclease and protease protection assays
For RNaseA protection assays, three replicates of p100 and S100 fractions were divided into two equal fractions, one of which received 4U/ml RNaseA (Sigma) while the other received an equal amount of PBS buffer. After 30 min. incubation at 37°C, an adequate volume of Trizol LS was added to denature the RNase and proceed to RNA isolation as previously indicated. For protease protection assays, three replicates of p100 and S100 fractions were divided into two equal fractions and proteinase K (Sigma) was added to both aliquots at 64μg/ml. After 30 min. incubation at 37°C, phenylmethylsulfonyl fluoride (PMSF; Sigma) was added at 5 mM final concentration. After protease inhibition, 4U/ml RNaseA was added to one of the aliquots (the other receiving an equal volume of buffer). Samples were incubated for 30 min at 37°C and subjected to RNA isolation. RNA from both assays was analyzed by SL-RT-qPCR specific to miR-21–5p [1–23nt], tRNAGluCUC [1–31] and tRNAGlyGCC [1–30]. The ΔCq values for paired samples were calculated (no RNase treatment – Rnase treatment) and taken together to determine the effect of RNase treatment versus no treatment (RNase protection), and of protease followed by RNase versus protease alone (protease protection).
RESULTS
Characterization and properties of extracellular vesicles isolated by differential centrifugation
We followed a broadly used conventional differential centrifugation-based protocol intended for the purification of different types of extracellular vesicles and the vesicle-free ribonucleoprotein fraction (33–36). However, since stepped ultracentrifugation procedures cannot achieve absolute separation by size (37), and because the exact centrifugation parameters needed to purify different EVs are still a matter of debate (38), we will avoid mechanistic definitions. Thus, we will simply refer to our extracellular samples as sequential fractionations of the extracellular medium, adopting the terms p16 (for 16 000 g pellet), p100 (for 100 000 g pellet) and S100 (for 100 000 g supernatant).
Isolated extracellular fractions were analyzed by transmission electron microscopy (TEM), NTA and western blot (Figure 1). Negative staining TEM showed vesicular content with size and morphology consistent with exosomes in p100, which was confirmed by NTA and immunoreactivity against the exosome markers TSG101, CD9 and CD63. Larger and irregular particles (with a diameter of 300–500 nm) were observed in p16. Even though this size is consistent with microvesicles, these larger particles co-purified with <100 nm spherical vesicles (Figure 1a, arrows). NTA analysis confirmed results obtained by TEM showing that the p100 fraction was mainly composed of particles in the expected size range for exosomes. On the contrary, p16 showed generally larger particles of widely varying size. This was confirmed by dynamic light scattering (DLS), which showed a much higher polydispersity index for p16 compared to p100 (0.43 versus 0.18), implying that p16 particles were relatively heterogeneous in size.
Figure 1.
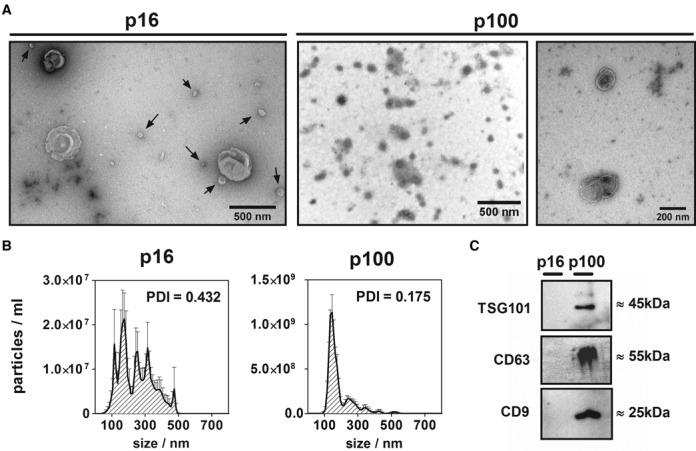
Characterization of p16 and p100 fractions. (A) Negative staining transmission electron microscopy of 16 000 g (p16; left) and 100 000 g (p100; middle: low magnification; right: high magnification) pellets of the conditioned medium of MCF-7 cells. Arrows in p16 show particles of <100 nm which co-purified with bigger (>300 nm) vesicles. (B) Size distribution (nanoparticle tracking analysis) of p16 (left) and p100 (right) fractions. Error bars correspond to the standard error of the mean for five repeated measurements of the same sample. The polydispersity index (PDI) of replicates of the same fractions measured by dynamic light scattering is also shown. (C) Western blot of exosomal markers: TSG101, CD63 and CD9 in equal protein loads of MCF-7 p16 and p100 fractions.
Different small RNA signatures characterize the intracellular and extracellular compartments of MCF-7 and MCF-10A cells
Three biological replicates for each extracellular fraction together with two biological replicates of the intracellular content of MCF-7 cells were sequenced. RNA yields in each preparation are shown in Supplementary Figure S2, together with a representative RNA size profile in the small RNA range. While the intracellular fraction showed the expected size distribution for intact intracellular RNAs (with a prominent 66 nt peak corresponding to full-length tRNAs) a broad peak at 30 nt characterized the S100 fraction. Most RNAs from EVs had a size above the upper limit of our libraries, although <60 nt RNAs were evident too. These characteristic size distributions are consistent with the major small RNA populations found in each extracellular fraction, as will be discussed later.
Main sequencing parameters and outputs for each data set are summarized in Supplementary Table S1. For MCF-7 cells, a total of 12.2, 11.4 and 23.7 million reads were obtained for p16, p100 and S100, respectively. Mapping to the human genome was 94.9% on average for intracellular (‘INTRA’) fractions, and the miRNA profiles from both replicates were highly correlated, with a Pearson's correlation coefficient of 0.927 (Supplementary Figure S3A).
Functional categorization of the small RNA reads was based on a >94% identity (1 mismatch allowance) to exonic regions of the genome and annotated ncRNAs. In the intracellular fractions, the majority of the reads ≥19 nt corresponded to mature miRNAs (43±9% of mapped reads, corresponding to 457 different miRNAs). They were followed by fragments derived from rRNAs (13.5%), snoRNAs (5.4%), tRNAs (2.2%), snRNAs (0.3%), YRNAs (0.3%), CDS (0.3%) and Vault RNAs (0.03%) (Figure 2a).
Figure 2.
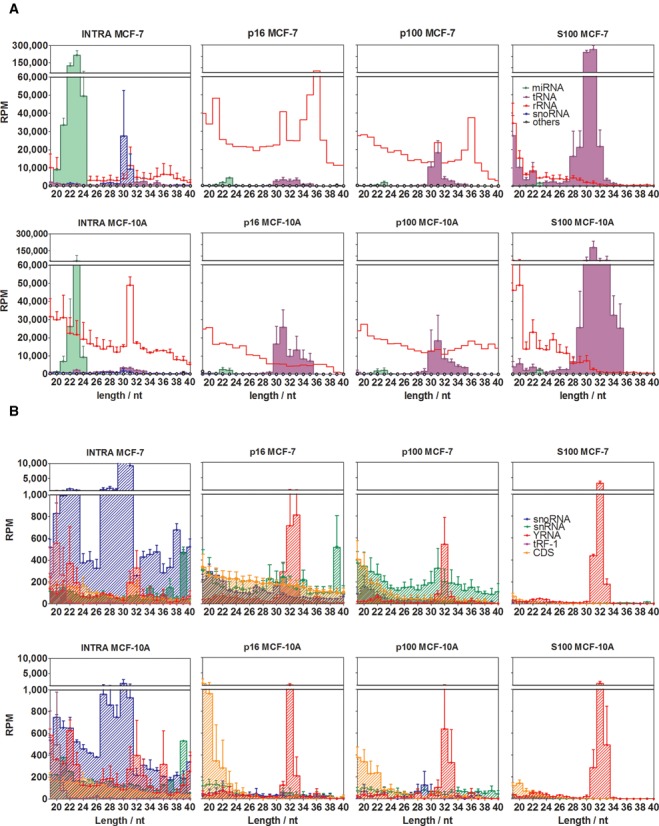
Different small RNA profiles characterize intracellular and extracellular fractions. Reads were annotated in one of the following functional categories: miRNAs (green area), rRNA-derived sequences (red line), snoRNA-derived sequences (blue diagonal pattern), tRNA-derived sequences (violet area), snRNA-derived sequences (green diagonal pattern), sequences derived from protein-coding exons (orange diagonal pattern), YRNA-derived sequences (diagonal red pattern) and 3′ trailer sequences from tRNA precursors (violet diagonal pattern). (A) and (B) are plots of the same data, but miRNAs, rRNA-derived and tRNA-derived sequences were omitted in (b) for reasons of scale. RPM: reads per million mapped reads. Error bars correspond to one standard error of the mean (n = 3 in p16, p100 and S100; n = 2 in INTRA), and were omitted for rRNA-derived sequences in p16 and p100 for reasons of clarity. The upper size limit of the graphs was set to 40 nt since no remarkable small RNA populations were detected beyond that size.
The extracellular fractions showed a completely different small RNA profile, characterized by very low miRNA relative abundances (< 1%) and supremacy of rRNA- and tRNA-derived fragments. This is in line with a recent report describing high levels of tRNA fragments with miRNA-like characteristics in EVs derived from MCF-7 cells (39). However, our data emphasize the predominance of tRNA halves (30–31 nt) rather than miRNA-like tRNA fragments (21–23 nt) in the extracellular space. In this regard, the S100 fraction showed a surprisingly high abundance of 5′ tRNA halves (62 ±1% of mapped reads that represented a 28-fold increase with respect to the intracellular compartment) and reproducible amounts of rRNA fragments (10 ± 1%). In contrast, miRNAs were only 0.26 ± 0.07% (164-fold decrease).
To address whether the observed differences in the small RNA profiles between extracellular fractions and the intracellular compartment were a singularity of the tumor-derived MCF-7 cell line, we repeated the same analysis in the non-malignant MCF-10A cell line. While miRNAs represented on average 11±7% of intracellular small RNAs, this number fell below 1% for the extracellular samples. In contrast, tRNA-derived fragments showed 3-, 2- and 18-fold increase in p16, p100 and S100, respectively, compared to the intracellular fraction. Similarly to MCF-7 cells, extracellular tRNA-derived sequences showed a narrow size distribution, with a mode at 30–31 nt (Figure 2a).
For reasons of scale, less-abundant small RNA categories were plotted separately (Figure 2b). Again, different signatures were observed for the intracellular and the extracellular fractions of each cell line, while corresponding samples from MCF-7 and MCF-10A cells were rather similar. Overall, our analysis showed that miRNAs and 30–31 nt snoRNA-derived species had significantly decreased relative abundances in the extracellular milieu compared to the intracellular compartment. Sequences mapping to tRNA genes showed exactly the opposite trend. Lastly, while YRNA-derived sequences showed relatively similar quantitative levels inside and outside the cells, a bias toward lengths of 31–33 nt was evident in the extracellular fractions. Of note, none of these changes were cell-line-specific.
Sequences mapping to ribosomal RNAs were detected in high quantities in every data set, but their relative abundances and size distributions showed the highest variation between replicates. A preference for 5′ fragments was remarkable, except for 5S-derived sequences where the vast majority of the sequences started at position +80–100 (Supplementary Figure S4). Peaks of 31 and 36 bases were recurrently observed in the intracellular and EV fractions (Supplementary Figure S5). However, lack of reproducibility between biological replicates prevented us from further analyzing this population.
Small RNAs derived from the Vault ribonucleoprotein particle (i.e. Vault RNA fragments) were also studied, since they were shown to regulate gene expression by miRNA-like mechanisms (40). Furthermore, Vault RNA fragments were shown to be enriched in extracellular vesicles derived from immune cells (13). We found modest amounts of Vault RNA fragments (321 and 1303 RPM in MCF-7 and MCF-10A, respectively) in the intracellular compartment. In MCF-7, Vault RNA-derived sequences were predominantly produced from the 5′ end (+1 position) and from the region +[48–69] of their respective precursors. As previously observed by other authors (13), only the inner fragments were detectable in the extracellular space, suggesting uneven secretion of these sRNAs. Furthermore, Vault RNA-derived sequences were probably secreted in extracellular vesicles, since only 7 reads (out of 13.3 million mapped reads; 0.5 RPM) were detected in the MCF-7 S100 fraction, while they reached 131 and 282 RPM in p16 and p100, respectively.
Most secreted miRNAs are sorted to the extracellular space in a non-selective manner
To determine whether our data supported or not a selective model of miRNA secretion we studied the correlation of miRNAs abundances between intra- and extracellular fractions. For this we plotted the Log2 RPM (reads per million of mapped reads) values for individual miRNAs in each extracellular fraction as a function of their corresponding intracellular expression (Figure 3a–c). Interestingly, when intracellular expression values for individual miRNAs reached a certain threshold (>25), a direct correlation between intracellular and extracellular values was evident and the data adjusted to a line of slope = 1, strongly indicating that miRNA secretion followed a non-selective behavior. Due to the low relative proportion of miRNAs in the total of secreted small RNAs, as discussed above, those miRNAs with low intracellular expression (<25 RPM) tended to be undetected in the extracellular fractions and gave rise to the accumulation of points over the X axis of the graphs. Conversely, Figure 3d shows that miRNAs detected in the extracellular fractions corresponded to those with the highest intracellular expression. For instance, while only 40% of intracellular miRNAs had abundances >20 RPM, more than 90% of detectable extracellular miRNAs had intracellular abundances above that value.
Figure 3.
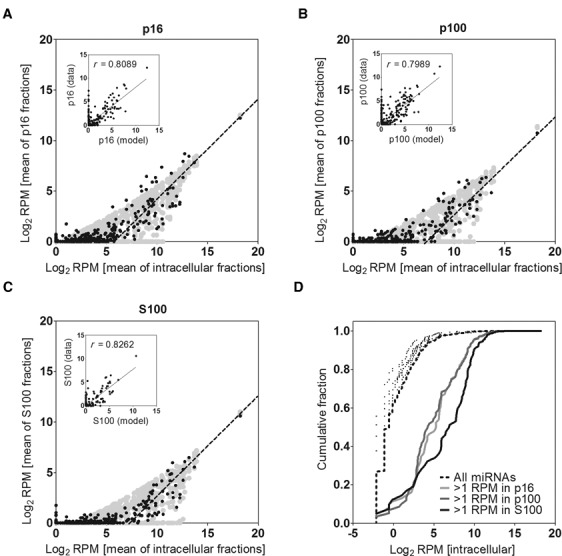
miRNA abundances in the extracellular fractions are consistent with a model of non-selective secretion for most miRNAs. Mean relative abundances (in Log2 scale) of each miRNA in the extracellular fractions p16 (A), p100 (B) and S100 (C) were plotted against their corresponding intracellular levels. Lines of slope = 1 were manually fitted to the data. A theoretical model which assumes passive secretion was overlaid (light gray area). Insets: correlation (Pearson's) between the model and the data. (D) Cumulative distribution of Log2 RPM values of intracellular miRNAs. The black dashed line includes all detected miRNAs in the intracellular fraction of MCF-7 cells (n = 456). The same analysis was repeated including only those miRNAs with >1 RPM in p16 (light gray line; n = 119), p100 (dark gray line; n = 114) or S100 (black line; n = 80). As a control, 5 subsamples of 80 randomly selected miRNAs were also analyzed (black dots).
To validate this conclusion, we constructed a theoretical model to predict the extracellular abundance of each miRNA based on its corresponding intracellular expression and assuming passive (i.e. non-selective) secretion. Briefly, extracellular abundances (in RPM) were predicted by multiplying intracellular RPM values for each miRNA by a sample-specific constant to make quantitatively comparable the intra- and extracellular compartments. This constant was calculated as the ratio of miRNA reads between the extracellular fraction and the intracellular compartment. Thus, under our model, the only variable that predicted the extracellular abundance of a given miRNA was its intracellular expression. After addition of sampling noise, we obtained a prediction of miRNA distribution under our model (gray area in Figure 3a–c) that adjusted well to the data (Pearson's correlation coefficients ≥0.8; Figure 3, insets).
It should be considered that our approach is empirical, not mechanistic. Thus, when we affirm that our data support a non-selective model of miRNA secretion we are not excluding the possibility that miRNAs were actually secreted through a controlled, regulated and selective process. What this means is that the extracellular abundance of most miRNAs is not high enough (or low enough) to separate from the ‘passive line’ beyond sampling noise and biological variability. In terms of hypothesis testing, our data do not allow us to reject the null hypothesis (i.e. non-selective secretion), given that the theoretical prediction of miRNA extracellular abundances under the null hypothesis and the observed values highly correlate.
Of note, correlation analysis serves to study the global behavior of miRNAs as a population but it does not allow determining if a small subset of individual miRNAs could be secreted selectively. As long as the number of exceptions is low, they would not significantly affect the global analysis supporting the non-selective model. To analyze this possibility, relative miRNA abundances between samples were normalized (i.e. we used reads per million total miRNA reads instead of RPM values), and the fold change between extracellular and intracellular fractions was calculated. Volcano plots showed significant (P < 0.05) differential (>4-fold) abundances for only four miRNAs in p16 (three increased and one decreased in the extracellular space), six miRNAs in p100 (five increased; one decreased) and none in S100 (Figure 4a). Consistent with the high correlation of miRNA expression between p16 and p100 (r = 0.96; Supplementary Figure S3), no miRNAs could serve to differentiate both fractions.
Figure 4.
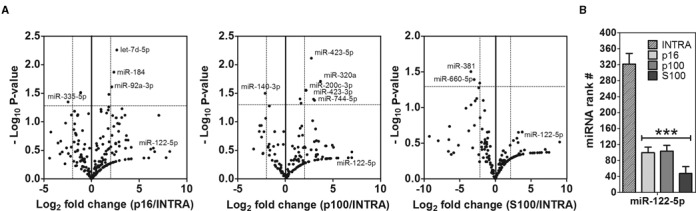
Detection of extracellular-enriched miRNAs. (A) Volcano plots showing miRNAs significantly enriched between extracellular and intracellular fractions. Sequence counts were normalized to the total sum of miRNAs in each data set and the corrected fold change was calculated and expressed as Log2. P-values were calculated by an unpaired two-tailed Welch's t test and expressed as −Log10. The horizontal dashed line shows P < 0.05. (B) Ranking analysis (i.e. position in a list of miRNAs ordered by abundance) of miR-122–5p. A significant (P < 0.001) lower ranking was found in the extracellular fractions (one-way ANOVA with Tukey's post hoc test).
Although miR-122–5p had the highest fold change between extracellular and intracellular samples (75-, 125- and 512-fold increase in p16, p100 and S100, respectively) the low absolute sequence counts did not provide enough statistical power to classify it as a candidate for selective secretion. Nevertheless, detection of this miRNA in the conditioned media of breast epithelial cells is remarkable, since miR-122–5p is the second-most specifically expressed miRNA in humans (41), with expression mostly restricted to the liver. Ranked by abundance, miR-122–5p varied from a very low average position of #320 in MCF-7 cell lysates to #100, #104 and #48 in p16, p100 and S100, respectively (P < 0.001; one-way ANOVA; Figure 4b).
To obtain sequencing-independent validation of these results we used stem-loop reverse-transcription (42) droplet digital PCR (SL-RT-ddPCR) because a typical problem in miRNA expression analysis by RT-qPCR is the selection of adequate methods for normalization (43). In our case, this issue was considered of paramount importance due to the lack of prior knowledge on adequate species for normalization in situations where both the query and the reference sequences are affected by the secretion process.
Absolute expression values were normalized to RNA input and expressed as Log2 (copies/ng RNA). A subset of miRNAs spanning different intracellular expression levels was selected. As expected, a strong correlation was found for these miRNAs between any extracellular fraction and MCF-7 cell lysates (Supplementary Figure S6A). Furthermore, experimental values could be adjusted to a regression line of slope = 1, which was predicted based on our theoretical model assuming passive secretion. This result, based on a random subset of moderately or highly expressed intracellular miRNAs, confirmed that, at least for these species, extracellular levels were mainly a function of their corresponding intracellular abundances. Analysis of miR-122–5p showed increased, yet not statistically significant abundance in S100. However, once miR-21–5p, miR-16–5p and let-7i-5p were validated as non-selectively secreted miRNAs in extracellular fractions, they were used as references for normalization in SL-RT-qPCR, supporting the selective secretion of miR-122–5p (Supplementary Figure S6B).
Overall, our results support both the non-selective secretion hypothesis for the vast majority of miRNAs and the selective export of a few candidates (11 out of 182 extracellular detected miRNAs) in specific extracellular compartments. This dual model might explain some of the apparent contradictions found in the literature regarding miRNA secretion. We also note that lack of biological replicates in many sequencing reports might have conduced to a generalized overestimation of the extent of specific miRNA sorting to the extracellular space.
The S100 fraction is greatly enriched in ribonucleoprotein complexes of 40–50 kDa containing 5′ halves from tRNAGly and tRNAGlu
As described above, while tRNA fragments represented fractional amounts of mapped reads in MCF-7 and MCF-10A cell lysates, they constituted an abundant small RNA category in all extracellular samples. This is in agreement with previous reports in which tRNA-derived sequences were detected in EVs from immune cells (13) and human semen (25). Interestingly, our study showed that tRNA-derived sequences were significantly enriched (P < 0.001 in MCF-7; P < 0.05 in MCF-10A) in the S100 fraction (Figure 5a), strongly suggesting their secretion as protein/RNA complexes. Again, this is in agreement with the recent finding of tRNA halves as abundant vesicle-free nucleoprotein complexes in mouse and human serum (21,22).
Figure 5.
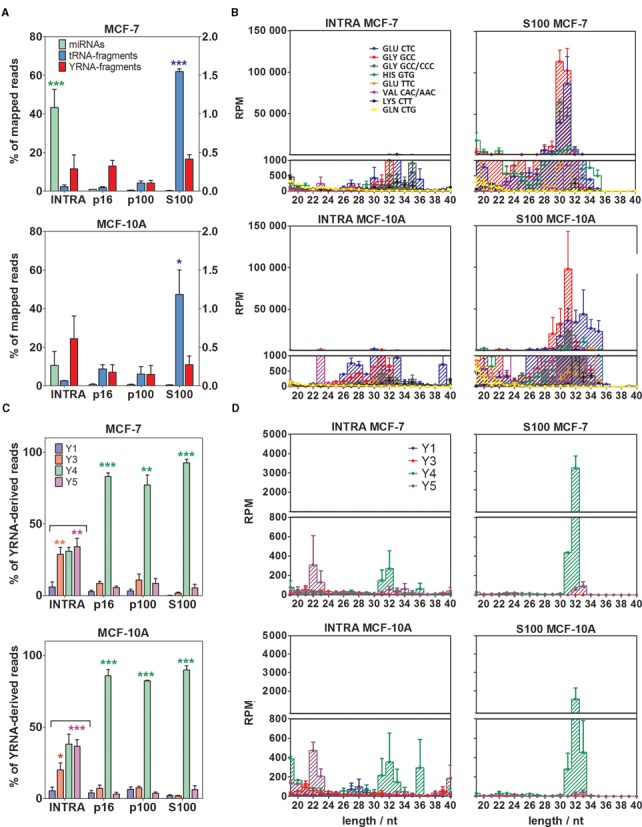
The extracellular milieu is enriched in 5′ tRNA halves and Y4 RNA-derived 5′ fragments, especially in the S100 fraction. (A) Comparison of the percentage of mapped reads corresponding to miRNAs (green; left axis), tRNA-derived sequences (blue; left axis) and YRNA-derived sequences (red; right axis) in the intracellular compartment and the extracellular fractions. Top: MCF-7 cells. Bottom: MCF-10A cells. (B) Size distributions and relative abundances of top eight tRNA-derived sequences. Left: intracellular. Right: S100 fraction. Top: MCF-7 cells. Bottom: MCF-10A cells. (C) Comparison of the percentage of YRNA-derived sequences corresponding to Y1 (blue), Y3 (red), Y4 (green) and Y5 (magenta). (D) Size distributions and abundances of YRNA-derived sequences. Distribution of the panels is the same as in (B). The upper size limit of both panels was set to 40 nt since no remarkable small RNA populations were detected beyond that size. Significantly different samples are shown with stars (one-way ANOVA with Tukey's post hoc test). For this and subsequent figures, *P < 0.05; **P < 0.01; ***P < 0.001.
In addition to RPM values of tRNA-mapping sequences that were orders of magnitude higher in S100 than in the intracellular fraction, qualitative differences were also evident (Figure 5b and Supplementary Figure S7A). In general terms, there was a correlation between the most abundant tRNA fragments found inside the cells and in the supernatants of the conditioned media. However, diversity was greatly restricted in extracellular fractions, and particularly in S100. For instance, sequences derived from only three different tRNAs (tRNAGluCUC and two different tRNAGlyGCC isodecoders) accounted for 89±3% and 88±3% of tRNA-derived reads in the S100 of MCF-7 and MCF10-A, respectively. These were also the top ranked tRNA sequences in the cell lysates, but accounted for only 44±9% and 63±9%, respectively.
Loss of diversity in tRNA-derived sequences in S100 was manifested not only by a restricted profile of tRNA fragments. The size distributions of these species were also notably condensed. In MCF-7, 80±5% of tRNA fragments had a length of 30–31 nt in S100 versus 9±2% in the cell lysates (P < 0.01; Welch's t-test). The profile was not so tight in MCF-10A cells supernatants, but a condensation was also evident and centered at 31–32 nt. With few exceptions, most tRNA-derived sequences corresponded to 5′ end fragments, irrespective of the cell line and extracellular fraction.
It has been claimed that tRNA-derived small RNAs can also be produced by RNase Z-mediated cleavage of the 3′ trailer sequence of pre-tRNAs, yielding the so-called tRF-1 or 3′ U tRFs (18). We detected modest quantities (1700±600 RPM) of tRF-1 sequences in the MCF-7 intracellular compartment. The majority of these sequences (74% in MCF-7, 63% in MCF-10A) corresponded to the proliferation-associated tRF-1001 (18), a 20 nt sequence derived from the 3′ trailer of tRNASerUGA. In contrast to tRNA halves, tRF-1 extracellular detection was rare. In MCF-7 S100, tRF-1 reads were below 20 RPM. Overall, from the diversity of tRNA fragments found in the cell lysates, only 5′ tRNA halves from Glu and Gly isoaceptors dominated the S100 fraction.
Validation by SL-RT-ddPCR and SL-RT-qPCR is shown in Figure 6a and b. As expected, 5′ halves derived from tRNAGluCUC and tRNAGlyGCC did not adjust to the ‘passive line’ defined by non-selectively secreted miRNAs, particularly in S100. On the contrary, their abundance was much higher than predicted by their intracellular levels (Figure 6a), implying either preferential secretion over miRNAs or differential stability in the extracellular space. Since RNA abundances were normalized to RNA input, systematic errors in RNA quantization obtained from different samples could have affected the results. Thus, we also normalized the abundances of 5′ tRNA halves using validated non-selectively secreted miRNAs as references. Irrespectively of the reference sequence, tRNA-halves from Glu and Gly isoaceptors were found to be significantly enriched in S100 versus intracellular fractions, both in ddPCR-based (Figure 6b) and qPCR-based assays (Figure 6c).
Figure 6.
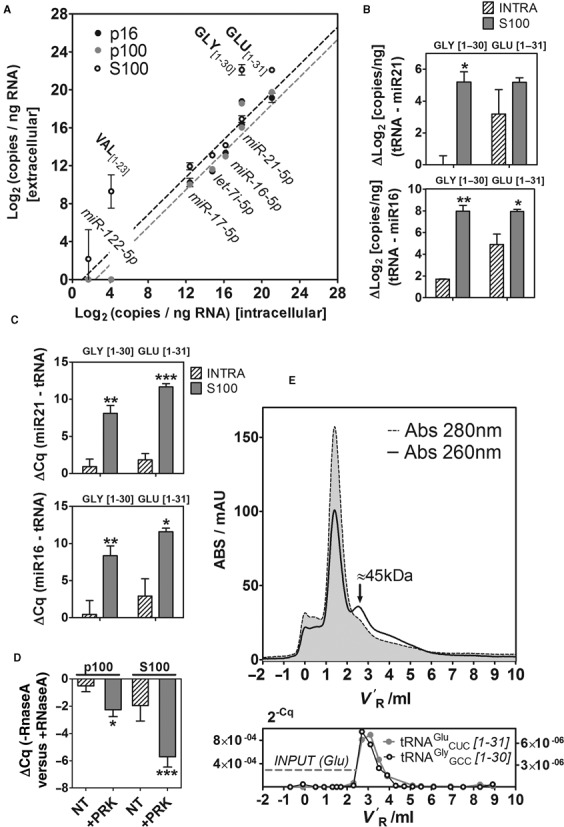
Validation of the sequencing results by SL-RT-ddPCR and SL-RT-qPCR, and purification of ribonucleoprotein complexes containing 5′ tRNA halves. (A) 2D-plots showing the relative abundance (Log2 copies per nanograms of input RNA) of selected miRNAs and tRNA-derived fragments between extracellular and intracellular fractions, measured by droplet digital PCR (ddPCR). Light gray, dark gray and white dots correspond to p16, p100 and S100, respectively. (B and C) Increased relative abundance (measured by ddPCR (B); qPCR (C)) of 5′ tRNA halves derived from tRNAGlyGCC (left; 1–30 nt) and tRNAGluCUC (right; 1–31 nt) in S100 (gray bars) with respect to the intracellular compartment (diagonal pattern). Abundances of tRNA halves were normalized to miR-21–5p and miR-16–5p. Significantly higher normalized abundances of tRNA halves in S100 versus INTRA are shown (Student t test; one-tailed). (D) Protease and RNase protection assays of the p100 and S100 fractions. ΔCq values were calculated between samples not treated and treated with RNaseA. The other experimental variable was treatment with Proteinase K (NT: not treated; +PRK: treated). Three biological replicates of each condition were analyzed, and each replicate was assayed for miR-21–5p and 5′ halves of tRNAGlyGCC and tRNAGluCUC (thus, each bar corresponds to nine different ΔCq values). (E) Top: The S100 fraction from MCF-7 cells was injected in a Superdex 75 10/300 column (size-exclusion chromatography) and fractions (200 μl) were eluted in 1× PBS using an ÄKTA purifier FPLC system (GE Healthcare, Amersham). The chromatograms correspond to the absorbance at 280 nm (gray area with dashed contours) and 260 nm (black line). V′R: adjusted retention volume (VR - Vm). Bottom: Eluted fractions were assayed for the presence of 5′ halves from tRNAGluCUC (gray; left Y axis) and tRNAGlyGCC (black; right Y axis) by SL-RT-qPCR. The horizontal gray dashed line corresponds to the input (before injection) 2−Cq value for tRNAGluCUC. The input 2−Cq value for tRNAGlyGCC was 7.5×10−8.
To address whether the small RNAs detected in S100 were present in genuine ribonucleoprotein complexes or in very small vesicles remaining in solution after 100 000 g centrifugation, RNase and protease protection assays of the p100 and S100 fractions were performed (Figure 6d). The p100 fraction was almost insensible to RNaseA (ΔCq < 0.5), as expected for RNAs encapsulated inside extracellular vesicles. Addition of 1% Nonidet P-40 prior to RNaseA (but not the detergent alone) made the RNAs completely sensitive to degradation (data not shown). Treatment with Proteinase K prior to RNaseA showed a moderate effect, reducing the levels of selected tRNA halves and miR-21–5p by ∼4 fold (ΔCq = −2.2). On the contrary, the S100 fraction was partially sensible to RNaseA (ΔCq = −1.96), but pretreatment of the sample with Proteinase K dramatically reduced the levels of the selected species (ΔCq = −5.71; different from zero with P < 0.001; ∼52 fold reduction). Taken together, these results are strong indicators that the RNAs present in the 100 000 g pellet and supernatant mostly correspond to different RNA-containing structures. This does not imply that the S100 fraction is totally vesicle-free or that p16 and p100 are completely depleted of ribonucleoprotein complexes. In particular, the observation that the p100 fraction was partially sensible to proteolysis might indicate that ribonucleoprotein complexes could co-purify with vesicles to a certain extent, although proteolytic destabilization of vesicular structures is another possibility.
To gain further insights into the nature of putative tRNA halves containing ribonucleoprotein complexes, the S100 fraction of MCF-7 and MCF-10A cells were analyzed by size-exclusion chromatography (Figure 6e and data not shown, respectively). The chromatogram obtained from a Superdex 75 10/300 column (GE Healthcare, Amersham) showed a prominent 280 nm peak at an adjusted retention volume (V′R) of 1.5ml, corresponding to the transferrin present in the defined media formulation. While absorbance at 260 nm followed the 280 nm profile, a 260 nm peak was observed at V′R = 2.5 ml which gave a 260/280 ratio >1 and was therefore indicative of the presence of nucleic acids. Eluted fractions were heat-denatured and analyzed by SL-RT-qPCR. The aforementioned 5′ halves from tRNAGluCUC and tRNAGlyGCC were detectable above input levels in the fractions which corresponded to the 260 nm peak (Figure 6e, bottom). Mild digestion with trypsin lowered the intensity and increased the Cq values of tRNA halves in the 260 nm peak, indicative of protease sensitivity (data not shown). Same-day calibration of the column determined that the hydrodynamic volume of the tRNA halves containing ribonucleoprotein complexes corresponded to that of a ∼45 kDa globular protein.
Extracellular-enriched RNA Y4-derived 5′ fragments
YRNAs are RNA polymerase III-transcribed small RNAs of 84–113 nt which associate with Ro60 and La proteins to yield ribonucleoprotein complexes whose molecular function are not yet completely understood. Under certain circumstances, these ncRNAs can be cleaved to yield 22–25 nt and 27–36 nt small RNAs in a Dicer-independent fashion (44). Interestingly, the presence of 5′ fragments from YRNAs was recently reported in extracellular vesicles from immune cells and human semen (13,25) and in serum and plasma (24). Of the four known human YRNA genes (Y1, Y3, Y4 and Y5), the authors found that the majority of circulating YRNA-derived sequences corresponded to fragments of RNA Y4 and its pseudogenes.
In our study, extracellular YRNA-derived sequences were found in every data set, with relative abundances that varied between 1000 and 10 000 RPM. In general, all sequences started at the +1 position of their respective precursors (5′-end fragments). In contrast to what was observed for miRNAs or tRNA-derived sequences, 5′ YRNA fragments did not show significant quantitative differences between intracellular and extracellular fractions (Figure 5a). However, when reads were sorted according to their corresponding YRNA precursors, dramatic changes in the intracellular and extracellular profiles became obvious (Figure 5c). While the intracellular compartments showed similar levels of Y3, Y4 and Y5 RNA-derived sequences, >85% of extracellular YRNA-derived reads corresponded to RNA Y4. Furthermore, while the size distributions of intracellular RNA Y4-derived reads showed peaks at 19, 26–29, 31–33 and 36 nt, the 31–33 nt population was predominant in the extracellular fractions (Figure 5d and Supplementary Figure S7B).
Abundance of 5′ tRNA halves in S100 is independent of culture conditions and not restricted to breast cell lines
It has been shown that tRNAs halves are produced in humans by ribonuclease 5 (angiogenin), and that angiogenin-mediated tRNA cleavage is upregulated under certain types of cellular stress (45–47). Thus, it is possible that the increased levels of tRNA halves in the S100 fraction were a consequence of a stress imposed by culturing cells under serum-free conditions. To evaluate this, MCF-7 cells were grown either in a different defined serum-free medium (MEGM; same used for MCF-10A) or following standard ATCC recommendations (EMEM plus insulin and 10% FBS). In both situations, ΔCq values for tRNA halves (normalized to non-selectively secreted miRNAs: miR21–5p or miR-16–5p) were higher in S100 compared to the intracellular compartment, especially for tRNAGluCUC (P < 0.05; Student t test; Supplementary Figure S8A-B). This shows that secretion of 5′ tRNA halves is not dependent on culture media and that the behavior is reproducible under standard growth conditions. It should be considered that both miR-16–5p and 5′ halves from tRNAGluCUC (but not from tRNAGlyGCC) were detected with low Cq values in the 100 000 g supernatant of non-conditioned EMEM plus 10% FBS. This observation highlights the importance of working with defined serum-free media when studying sRNAs present in ribonucleoprotein complexes because conventional exosome-depleted serum would still contribute to the S100 sRNA pool. Nevertheless, the increased abundance of tRNA halves in the S100 fraction of serum-containing cultures cannot be explained by serum contribution alone (Supplementary Figure S8B).
To evaluate if secretion of 5′ halves from tRNAGluCUC and tRNAGlyGCC in S100 is a general phenomena, we extended the analysis to other epithelial cell lines derived from lung (NCI-H12299) and cervix (HeLa) tissues. In both cell lines, 5′ halves from tRNAGlyGCC were significantly enriched in the S100 fraction, while enrichment in 5′ halves from tRNAGluCUC was only observed in NCI-H1299 (Supplementary Figure S8C-D). Considering sequencing of non-malignant MCF-10A cells also supported the selective secretion (or increased stability) of tRNA halves in S100, increased abundance in tRNA halves seems to be a hallmark of the S100 fraction and is not restricted to a certain tissue or to a malignant phenotype.
DISCUSSION
To date, despite considerable effort invested in the field, circulating miRNA research has generally not yet resulted in highly specific, validated disease markers (48). A possible explanation, at least in non-hematological cancer research, is the great dilution of tumor-derived RNAs in the total pool of circulating miRNAs, most of which have a blood or endothelial cell origin (49). Altered miRNA profiles in the circulation of cancer patients and donors are usually reported, but discordant results appear across publications of different authors (50). Altered profiles of circulating miRNAs in early-stage cancer patients might be a consequence of nonspecific systemic responses rather than a true sampling of tumor RNA in the bloodstream (48). An alternative strategy is to focus in specific extracellular fractions instead of whole plasma or serum, especially if tumor-derived vesicles can be purified. To do this, a comprehensive profiling of sRNAs associated with different extracellular fractions is needed, together with a better understanding on sRNA secretion and sorting into these fractions. Our approach was also based on the inclusion and sequencing of biological replicates of each fraction in order to differentiate selective from non-selective secretion patterns with statistical confidence.
The small RNA content of three different extracellular vesicle subtypes (microvesicles, A33-positive and EpCAM-positive exosomes) secreted by a human colon cancer cell line has been recently reported (51). The authors reported selective enrichment of several miRNAs in EVs, and a subset of six miRNAs could discern exosomes from microvesicles. This implied selective export of miRNAs in specific types of vesicles, although lack of biological replicates did not allow assessment of the statistical significance of these results. In contrast, other authors have shown evidence supporting a general pattern of non-selective miRNA secretion (7). Due to the high degree of dispersion observed between biological replicates in high-throughput sequencing (HTS) data (52), and in accordance with the guidelines of the International Society for Extracellular Vesicles (53), we sequenced three biological replicates of each extracellular fraction of MCF-7 cells in order to address whether miRNA expression values statistically support either the selective or the non-selective secretion hypothesis in this cell line. Biological replicates were prepared at different times, even months apart, in order to minimize the effect (often underestimated) of cross-contamination during HTS library preparation (54).
Our results showed that the general pattern of miRNA secretion in MCF-7 cells was mainly non-specific, with a good correlation between the intracellular levels of each miRNA and their corresponding extracellular abundances. This was the case for the three extracellular fractions assayed: p16, p100 and S100. It has been recently suggested that miRNA sorting to exosomes may be a passive mechanism to dispose miRNAs in excess of their cellular targets (55). Nevertheless, a subset of miRNAs was consistently found in higher than expected levels based on their representation in the intracellular space. Thus, it is possible that both the non-selective (7) and the selective secretion hypotheses may be correct (i.e. most miRNAs present in the cytoplasm are randomly sampled into the extracellular space, but certain miRNAs might be actively directed to the export pathway). An intriguing case was miR-122–5p, a well-known liver-specific miRNA, which was observed in relatively high levels in two out of three replicates of the S100 fraction, despite being almost undetectable in the intracellular space (both in the sequencing data and in qPCR replicates). Although present in the ‘noise region’, increased levels of circulating miR-122–5p were recently reported in patients with breast cancer and predicted clinical outcome (56). Whether this was a consequence of a non-specific liver response or the specific secretion of a ‘rare’ miRNA by breast cancer cells, the observation deserves further attention.
A vast universe of small RNAs exists beyond miRNAs, and one of the advantages of HTS-based approximations is that different small RNA families can be studied simultaneously in a comprehensive manner. In contrast to miRNAs, we have shown that the profiles of other small RNAs are completely altered between the intracellular compartment and the extracellular fractions. Small nucleolar RNA-derived sequences were almost depleted in the extracellular milieu, despite being one of the most abundant small RNA categories in the cell lysates (albeit far less abundant than miRNAs). On the contrary, 5′ tRNA halves were significantly enriched in the extracellular space. These results are in good agreement with a previous report comparing immune cells and immune cell-derived EVs (13). Interestingly, we found that YRNA-derived sRNAs in the extracellular space showed a complete bias toward 5′-end fragments from RNA Y4, the most abundant fragments in plasma and serum (24).
Considering NanoSight and ddPCR quantification and RNA yields in the p100 fraction, we estimate that one copy of miR-21–5p, miR-16–5p and let-7i was obtained from 98, 780 and 2058 exosomes, respectively. The levels of tRNA halves were higher, but still far below the 1:1 stoichiometry (1 copy per 7 and 16 exosomes for tRNAGluCUC and tRNAGlyGCC 5′ halves, respectively). This is in accordance with a recently published report showing that exosomes carry an average of one given miRNA per 128 particles (6).
Besides extracellular vesicles, protein/RNA complexes emerge as potentially relevant vectors for RNA secretion and transport in the extracellular space. In the blood, 5′ tRNA halves and 5′ YRNA-derived fragments were also preferentially associated with ribonucleoprotein complexes (22,24). Our sequencing of 100 000 g supernatants from the conditioned media of MCF-7 cells showed a near 20-fold increase (P < 0.001) in the abundance of 5′ tRNA halves with respect to the intracellular compartment. As observed for RNA Y4-derived sRNAs, only 5′ tRNA fragments of 30–31 nt seemed to show preferential secretion, implying that the cells are able to discriminate between the varieties of tRNA-derived sequences of different lengths found in the intracellular compartment. Size selection makes sense if 5′ tRNA halves are effectively secreted in complex with proteins. Indeed, we have isolated by chromatography the protease-sensitive protein/tRNA-halves complex from the S100 fraction, which showed a smaller size than previously reported in serum (22). Such difference in size can correspond to complexes with different proteins in conditioned media and serum, a difference in oligomerization state, or the presence/absence of auxiliary proteins in the complexes. Identification of the presumptive tRNA half-binding protein/s is ongoing.
A second mammary epithelial cell line (MCF-10A), immortalized but not tumorigenic, was also sequenced, and almost identical results were obtained. This means that the observed secretion biases were not simply a peculiarity of MCF-7 cells. An additional advantage of the MCF-10A cell line was its routine culture under serum-free defined conditions. Since miR-16–5p and 5′ halves from tRNAGluCUC were detectable by qPCR even in 100 000 g supernatants of medium containing 10% fetal bovine serum, we strongly advise the use of serum-free conditions is when studying the ribonucleoprotein-associated small RNA profile in cell culture. The observation that tRNA halves were secreted above miRNA levels by lung and cervix cell lines is indicative that the secretion pathways underlying our sequencing results are common to different cell types.
Overall, we have presented evidence supporting different patterns of secretion for various small RNA families and their sorting into different extracellular compartments. Our results highlight the importance of the ribonucleoprotein fraction as a biologically relevant and understudied compartment in which to seek small RNAs of different sources. A mixed model of miRNA secretion was presented. We do not wish to speculate as to what role the predicted secretion-selected miRNAs might play in the biology of breast cancer, but the possibility deserves further research. For the majority of cases, miRNA secretion conformed better to a non-selective secretion model. In contrast, YRNA- and tRNA-derived sequences showed clear signs of being secreted in a selective manner, though differential stability is an alternative explanation.
Extracellular processing of secreted tRNAs is another possibility which for the moment cannot be discarded. Extracellular processing in cancer-cell-derived exosomes was recently reported for miRNAs (36), and the ribonuclease presumably involved in the biogenesis of tRNA halves (angiogenin) is known to be secreted (57). As previously suggested (7), extracellular RNA-containing structures might be no more than by-products of necrotic cell death, though this does not exclude a possible role in intercellular communication. In this regard, a recent report from our group demonstrated that EVs containing tRNA halves could be transferred from parasites to susceptible mammalian cells and that this process is involved in Trypanosoma cruzi pathogenesis (58). Furthermore, secretion of tRNA halves in ribonucleoprotein complexes might transfer a stress response to surrounding cells, acting as dominant negative extracellular regulators of protein synthesis. However, 5′ halves of tRNAGluCUC do not bear the terminal oligoguanine motif involved in this process (59). Independently of sequencing motifs, the predicted stem-loop structure of tRNA halves (which includes the D-loop of their parental tRNAs) might confer aptameric-like properties to these biomolecules, serving as scaffolds for protein assembly or promoting conformational changes in cell-signaling proteins (even mediating their secretion).
In any case, new light will come from the identification of the protein components of extracellular tRNA halves containing ribonucleoprotein complexes. A recent report has shown that bacterial YRNAs mimic tRNAs and are substrates of the same processing machinery (60). These observations and the finding of these species in circulation (22,24) suggest that there may be a biological reason for cleavage of tRNAs and secretion of 5′ halves into the extracellular space.
ACCESSION NUMBERS
High throughput sequencing data will be available at the sequence read archive database (http://www.ncbi.nlm.nih.gov/sra) under the BioProject ID: PRJNA270876.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Gonzalo Greif and Carlos Robello; Natalia Rego; Melissa McAlexander; Grace V. Hancock and Gabriela Casanova; Agustín Correa; Santiago Botasini and Eduardo Méndez for their help with HTS library preparation, initial data analysis, digital PCR, TEM, SEC and DLS, respectively. J.P.T. and A.C. are researchers of the Sistema Nacional de Investigadores (ANII, Uruguay).
FUNDING
Agencia Nacional de Investigación e Inovación (ANII) [FCE 2011 I 6159]; Fondo para la Convergencia Estructural del Mercosur (FOCEM) [COF 03/11]; Comisión Sectorial de Investigación Científica (CSIC) [PAS2014 ID51]. National Institutes of Health (NIH) [R21 AI102659 to K.W.W.] (in part). Funding for open access charge: Agencia Nacional de Investigación e Inovación (ANII) [FCE 2011 I 6159]; Comisión Sectorial de Investigación Científica (CSIC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 2.Cortez M.A., Bueso-Ramos C., Ferdin J., Lopez-Berestein G., Sood A.K., Calin G.A. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 4.Mittelbrunn M., Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell. Biol. 2012;13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevillet J.R., Kang Q., Ruf I.K., Briggs H.A., Vojtech L.N., Hughes S.M., Cheng H.H., Arroyo J.D., Meredith E.K., Gallichotte E.N., et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turchinovich A., Weiz L., Langheinz A., Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K., Zhang S., Weber J., Baxter D., Galas D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gould S.J., Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20389. doi:10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Hristov M., Erl W., Linder S., Weber P.C. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood. 2004;104:2761–2766. doi: 10.1182/blood-2003-10-3614. [DOI] [PubMed] [Google Scholar]
- 13.Nolte-'t Hoen E.N., Buermans H.P., Waasdorp M., Stoorvogel W., Wauben M.H., t Hoen P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turchinovich A., Samatov T.R., Tonevitsky A.G., Burwinkel B. Circulating miRNAs: cell-cell communication function. Front. Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turchinovich A., Weiz L., Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 2012;37:460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Li Z., Ender C., Meister G., Moore P.S., Chang Y., John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787–6799. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson P., Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebetsberger J., Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhahbi J.M., Spindler S.R., Atamna H., Boffelli D., Martin D.I. Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and YRNA fragment expression associated with breast cancer. Biomark. Cancer. 2014;6:37–47. doi: 10.4137/BIC.S20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhahbi J.M., Spindler S.R., Atamna H., Yamakawa A., Boffelli D., Mote P., Martin D.I. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akat K.M., Moore-McGriff D., Morozov P., Brown M., Gogakos T., Correa Da Rosa J., Mihailovic A., Sauer M., Ji R., Ramarathnam A., et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc. Natl. Acad. Sci. U.S.A. 2014;111:11151–11156. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhahbi J.M., Spindler S.R., Atamna H., Boffelli D., Mote P., Martin D.I. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol. Genomics. 2013;45:990–998. doi: 10.1152/physiolgenomics.00129.2013. [DOI] [PubMed] [Google Scholar]
- 25.Vojtech L., Woo S., Hughes S., Levy C., Ballweber L., Sauteraud R.P., Strobl J., Westerberg K., Gottardo R., Tewari M., et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes D., Sato G. Growth of a human mammary tumour cell line in a serum-free medium. Nature. 1979;281:388–389. doi: 10.1038/281388a0. [DOI] [PubMed] [Google Scholar]
- 27.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19.10.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung U., Jiang X., Kaufmann S.H., Patzel V. A universal TaqMan-based RT-PCR protocol for cost-efficient detection of small noncoding RNA. RNA. 2013;19:1864–1873. doi: 10.1261/rna.040501.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer M.F. Stem-loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. 2011 doi: 10.1002/0471142727.mb1510s95. Chapter 15, Unit 15.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crescitelli R., Lasser C., Szabo T.G., Kittel A., Eldh M., Dianzani I., Buzas E.I., Lotvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20677. doi:10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucharzewska P., Christianson H.C., Welch J.E., Svensson K.J., Fredlund E., Ringner M., Morgelin M., Bourseau-Guilmain E., Bengzon J., Belting M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turchinovich A., Weiz L., Burwinkel B. Isolation of circulating microRNA associated with RNA-binding protein. Methods Mol. Biol. 2013;1024:97–107. doi: 10.1007/978-1-62703-453-1_8. [DOI] [PubMed] [Google Scholar]
- 36.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witwer K.W., Buzas E.I., Bemis L.T., Bora A., Lasser C., Lotvall J., Nolte-'t Hoen E.N., Piper M.G., Sivaraman S., Skog J., et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. doi:10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeppesen D.K., Hvam M.L., Primdahl-Bengtson B., Boysen A.T., Whitehead B., Dyrskjøt L., Orntoft T.F., Howard K.A., Ostenfeld M.S. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J. Extracell. Vesicles. 2014;3:25011. doi: 10.3402/jev.v3.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guzman N., Agarwal K., Asthagiri D., Saji M., Ringel M.D., Paulaitis M.E. Breast Cancer-Specific miR Signature Unique to Extracellular Vesicles includes ‘microRNA-like’ tRNA Fragments. Mol. cancer Res. 2015;13 doi: 10.1158/1541-7786.MCR-14-0533. doi:10.1158/1541-7786.MCR-14-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persson H., Kvist A., Vallon-Christersson J., Medstrand P., Borg A., Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 41.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2004;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts T.C., Coenen-Stass A.M., Wood M.J. Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One. 2014;9:e89237. doi: 10.1371/journal.pone.0089237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicolas F.E., Hall A.E., Csorba T., Turnbull C., Dalmay T. Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 2012;586:1226–1230. doi: 10.1016/j.febslet.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 45.Yamasaki S., Ivanov P., Hu G.-F.F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell. Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G.F., Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 48.Witwer K.W. Circulating MicroRNA Biomarker Studies: Pitfalls and Potential Solutions. Clin. Chem. 2014;61:56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 49.Duttagupta R., Jiang R., Gollub J., Getts R.C., Jones K.W. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leidner R.S., Li L., Thompson C.L. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS One. 2013;8:e57841. doi: 10.1371/journal.pone.0057841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji H., Chen M., Greening D.W., He W., Rai A., Zhang W., Simpson R.J. Deep Sequencing of RNA from Three Different Extracellular Vesicle (EV) Subtypes Released from the Human LIM1863 Colon Cancer Cell Line Uncovers Distinct Mirna-Enrichment Signatures. PLoS One. 2014;9:e110314. doi: 10.1371/journal.pone.0110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill A.F., Pegtel D.M., Lambertz U., Leonardi T., O'Driscoll L., Pluchino S., Ter-Ovanesyan D., Nolte-'t Hoen E.N. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.22859. doi:10.3402/jev.v2i0.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tosar J.P., Rovira C., Naya H., Cayota A. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA. 2014;20:754–757. doi: 10.1261/rna.044263.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Squadrito M.L., Baer C., Burdet F., Maderna C., Gilfillan G.D., Lyle R., Ibberson M., De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 56.Wu X., Somlo G., Yu Y., Palomares M.R., Li A.X., Zhou W., Chow A., Yen Y., Rossi J.J., Gao H., et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J. Transl. Med. 2012;10:42. doi: 10.1186/1479-5876-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skorupa A., King M.A., Aparicio I.M., Dussmann H., Coughlan K., Breen B., Kieran D., Concannon C.G., Marin P., Prehn J.H. Motoneurons secrete angiogenin to induce RNA cleavage in astroglia. J. Neurosci. 2012;32:5024–5038. doi: 10.1523/JNEUROSCI.6366-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Silva M.R., das Neves R.F., Cabrera-Cabrera F., Sanguinetti J., Medeiros L.C., Robello C., Naya H., Fernandez-Calero T., Souto-Padron T., de Souza W., et al. Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol. Res. 2014;113:285–304. doi: 10.1007/s00436-013-3655-1. [DOI] [PubMed] [Google Scholar]
- 59.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X., Sim S., Wurtmann E.J., Feke A., Wolin S.L. Bacterial noncoding Y RNAs are widespread and mimic tRNAs. RNA. 2014;20:1715–1724. doi: 10.1261/rna.047241.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


