Abstract
Loss of a base in DNA leading to creation of an abasic (AP) site leaving a deoxyribose residue in the strand, is a frequent lesion that may occur spontaneously or under the action of various physical and chemical agents. Progress in the understanding of the chemistry and enzymology of abasic DNA largely relies upon the study of AP sites in synthetic duplexes. We report here on interactions of diastereomerically pure metallo–helical ‘flexicate’ complexes, bimetallic triple-stranded ferro-helicates [Fe2(NN-NN)3]4+ incorporating the common NN–NN bis(bidentate) helicand, with short DNA duplexes containing AP sites in different sequence contexts. The results show that the flexicates bind to AP sites in DNA duplexes in a shape-selective manner. They preferentially bind to AP sites flanked by purines on both sides and their binding is enhanced when a pyrimidine is placed in opposite orientation to the lesion. Notably, the Λ-enantiomer binds to all tested AP sites with higher affinity than the Δ-enantiomer. In addition, the binding of the flexicates to AP sites inhibits the activity of human AP endonuclease 1, which is as a valid anticancer drug target. Hence, this finding indicates the potential of utilizing well-defined metallo–helical complexes for cancer chemotherapy.
INTRODUCTION
An abasic (apurinic or apyrimidinic, AP) site represents one of the most abundant, although rapidly repaired, DNA lesions in cells. AP sites are generated by the breakage of the N-glycosidic bond in DNA, which may occur spontaneously, chemically or enzymatically during base excision repair mechanism (1,2). It is estimated that under physiological conditions,∼10 000 apurinic sites and 500 apyrimidinic are formed per human cell per day (1,3). Therefore, AP site represents an attractive target for diagnostic and therapeutic applications. Selective ligands capable of changing their optical properties upon binding to an AP site could be used for a sensitive detection of these lesions (4–9). Many different structures have been developed to target AP sites; DNA base-intercalator conjugates (10,11), metalloinsertors (12,13), naphthalene derivatives (14), biaryl derivatives (15), an isoquinoline alkaloid berberine (16) or cyclobisacridine (17). It has been suggested that molecules selectively binding to AP sites could assist antitumour drugs by blocking the binding site of the AP endonucleases (10,18). These specialized enzymes are able to cleave DNA strands at AP sites and initialize the first stage of the repair of such lesions by a cascade of specific enzymes. Several molecules have been shown to inhibit AP endonucleases (19,20) and potentiate cytotoxicity of antitumour alkylating agent bis(chloroethyl)nitrosurea (BCNU) in vitro and in vivo (10,21,22).
We have recently reported (23) that [Fe2L1a3]Cl4 helical complexes (Figure 1A) can recognize and stabilize some unusual DNA structures such as Y-shaped three-way junctions, three-way junctions with unpaired nucleotides, the so-called T-shaped three-way junctions and DNA bulges containing one and more unpaired nucleotides. These water stable, optically and diastereochemically pure bimetallic structures with flexible linkers have been created via self-assembly from monometallic complexes containing functionalized pyridine/imine units (24–27). Since the stereoselectivity in these complexes does not rely on the helicate concept of mechanical coupling they are described as flexicates.
Figure 1.
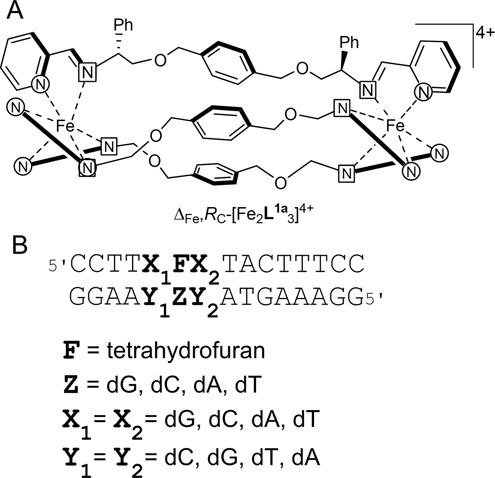
(A) Diastereomerically pure metallo–helical ‘flexicate’ complexes of chiral ligands L1a. Only one ligand shown in full. (B) Oligonucleotide duplex containing an abasic site mimicked by tetrahydrofuran and flanked by various bases X1 and X2. Y1 and Y2 correspond to complementary bases to X1 and X2. The letter Z indicates a base opposite an abasic site.
In the present study, we explored interactions of [Fe2L1a3]Cl4 flexicates with short DNA duplexes containing an AP site in different sequence contexts. We employed the following techniques: DNA melting temperature (Tm) measurements, fluorescence spectroscopy, gel electrophoresis, DNase I footprinting and isothermal titration calorimetry (ITC). Moreover, we examined if the binding of the flexicates to an AP site can affect the activity of human AP endonuclease 1 (APE1), since APE1 inhibitors have demonstrated potentiation of cytotoxicity of alkylating agents in preclinical models implying translational applications in cancer therapy (28). The results show that the flexicates can bind with high affinity to AP sites in a shape-selective manner and inhibit APE1 activity.
MATERIALS AND METHODS
Chemicals
The iron(II) flexicates [Fe2Ln3]Cl4 were synthesized as previously described (25). The synthetic oligodeoxyribonucleotides containing abasic furan (dSpacer) to mimic an abasic site were purchased from TriLink BioTechnologies (San Diego, CA, USA). The remaining synthetic oligodeoxyribonucleotides were from VBC-Biotech (Vienna, Austria). The quoted molar concentrations are related to the single strands. Stoichiometric amounts of oligonucleotides were mixed to form a duplex. T4 polynucleotide kinase and human AP endonuclease 1 (APE1) were purchased from New England Biolabs (Beverly, MA). [γ-32P]-ATP was from MP Biomedicals, LLC (Irvine, CA, USA). Acrylamide and bis(acrylamide) were from Merck KgaA (Darmstadt, Germany). Amiloride was purchased from Sigma (Prague, Czech Republic). Deoxyribonuclease I (DNaseI) was from Roche (Mannheim, Germany).
UV melting experiments
The stability of DNA duplexes in the absence or in the presence of the flexicates was monitored by measuring the absorbance at 260 nm as a function of temperature (Supplementary Figures S1–S4). The absorbances plotted in Supplementary Figures S1–S4 represent the values from which the absorbance determined for each sample at 15°C was subtracted. The experiment was run simultaneously on six masked 1 cm pathlength cuvettes of 1.2 ml volume using a Peltier controlled 6-sample cell-changer in a Varian Cary 4000 UV/vis spectrophotometer (1 nm bandwidth, average time: 10 s, heating rate 0.4°C/min). Melting temperature (Tm) was calculated within the thermal heating program by applying a first derivative calculation (Supplementary Figure S5). The melting curve for each duplex was recorded at least three times. The first derivative profiles exhibited in all cases a clearly defined maximum (Supplementary Figure S5) which made it possible to determine the Tm value accurately. The Tm values could be thus determined with an accuracy of ±0.5°C. The concentration of oligodeoxyribonucleotides was 3 × 10−6 M per strand. The buffer conditions were sodium phosphate buffer (10 mM, pH 7.0) and 100 mM NaCl.
Gel electrophoresis of DNA duplexes containing an AP site
Stoichiometric amounts of oligonucleotides at the concentration of 1 × 10−5 M per strand were mixed together in the buffer to form a duplex. One of the strands was 5’-end labelled using T4 polynucleotide kinase and [γ-32P]ATP. Flexicates were then added to the mixture so that the final concentration of the oligonucleotides in the samples was 5 × 10−6 M. The samples were analysed by electrophoresis on 15% polyacrylamide gels in buffered solutions consisting of Tris(hydroxymethyl)amino methane (89 mM), borate (89 mM, pH 8.3) and ethylenediaminetetraacetic acid (1 mM) run at 5°C.
Fluorescence spectroscopy
A 1 μM solution of the oligonucleotide duplex was prepared in a 1 cm quartz cuvette in a total volume of 2.5 ml. The buffer was composed of sodium phosphate buffer (10 mM, pH 7.0) and NaCl (100 mM). Small volumes (2.5 μl) of flexicates were added to the solution to obtain the desired concentration and thoroughly mixed by pipetting. The mixture was kept undisturbed for 3 min at room temperature. The fluorescence was measured by using Varian Cary Eclipse spectrofluorophotometer. The excitation and emission wavelengths were set to 310 and 365 nm, respectively, the excitation and emission slit widths were 5 nm, and the integration time was set to 5 s.
Amiloride displacement
A 1 μM solution of the oligonucleotide duplex and amiloride was prepared in a 1 cm quartz cuvette in a total volume of 2.5 ml. The buffer was composed of sodium phosphate buffer (10 mM, pH 7.0) and NaCl (100 mM). Small volumes (2.5 μl) of flexicates were added to the solution to obtain the desired concentration and thoroughly mixed by pipetting. The mixture was kept undisturbed for 3 min at room temperature. The fluorescence was measured by using Varian Cary Eclipse spectrofluorophotometer. The excitation and emission wavelengths were set to 380 and 415 nm, respectively, the excitation and emission slit widths were 5 nm and the integration time was set to 5 s.
Isothermal titration calorimetry (ITC)
Heat flow during isothermal titration was measured at 20°C with a VP-ITC microcalorimeter (MicroCal Inc., Northampton, MA, USA). The standard titration buffer for these studies contained 100 mM NaCl with 10 mM phosphate buffer (Na2HPO4/NaH2PO4 [pH 7.0]). In a typical experiment, 6 μM DNA duplex solution (1.4 ml) was titrated with 80 μM flexicate solution, using 300 μl syringe rotating at 490 rpm. A titration consisted of 28 injections of 10 μl each, with 20 s duration and 240 s between injections. Samples were equilibrated thermally prior to a titration until the baseline has levelled off. The peaks produced during titration were converted to heat output per injection by integration and correction for the cell volume and sample concentration. Data from individual titrations were analysed by using Origin 5.0 software package (Origin, Northampton, MA, USA) and fitted to a one-site (or n identical sites), two independent sites, or sequential binding or more binding model (29) to extract the relevant thermodynamic parameters.
DNase I footprinting
Top or bottom strands of the oligonucleotide duplexes were 5’-end labelled using T4 polynucleotide kinase and [γ-32P]ATP and hybridized with the complementary bottom or top strands. A total of 5 μl solutions containing 1.11 × TKMC buffer (10 mM Tris pH 7.9, 10 mM KCl, 10 mM MgCl2, and 5 mM CaCl2), 3 × 10−4 M DNA (per nucleotide) and various concentrations of the flexicates were incubated for 15 min at 25°C. Cleavage was initiated by the addition of 1 μl of DNase I diluted in the precedent experiment to the concentration that was sufficient to achieve partial cleavage of the DNA duplexes. Samples were allowed to react for 10 min at room temperature before quenching with 6 μl of 2× concentrated formamide loading buffer followed by incubation at 90°C for 3 min. A total of 2 μl of the mixture containing DNA cleavage products were then withdrawn and resolved by polyacrylamide (PAA) gel electrophoresis under denaturing conditions (8%/8 M urea PAA gel). The autoradiograms were visualized and quantified by using the bio-imaging analyser.
Inhibition of APE1
Top strands of the oligonucleotide duplexes were 5’-end labelled using T4 polynucleotide kinase and [γ-32P]ATP and hybridized with the complementary bottom strands. Reaction mixtures (5 μl, total volume) containing DNA duplexes at the concentration of 2 × 10−7 M per strand in 50 mM potassium acetate, 20 mM Tris-acetate (pH 7.9), 10 mM magnesium acetate and 1 mM dithiothreitol (DTT) were preincubated in the absence or in the presence of various concentrations of the flexicates for 15 min at 25°C. Then 1 unit of APE1 in 1 μl was added into each reaction mixture and incubated for 10 min at 37°C. A total of 2 μl of the reaction mixture were then withdrawn, mixed with 2× concentrated formamide loading buffer followed by incubation at 90°C for 3 min and resolved by PAA gel electrophoresis under denaturing conditions (24%/8 M urea PAA gel). The autoradiograms were visualized and quantified by using the bio-imaging analyser.
RESULTS AND DISCUSSION
UV melting studies
The melting temperature (Tm) was determined to compare binding affinities of the flexicates for DNA duplexes containing an AP site flanked by various bases and with various bases placed opposite an AP site. The Tm corresponds to the midpoint of a smooth transition obtained by recording the absorbance at 260 nm as a function of the temperature (Supplementary Figures S1–4). The changed Tm values of the duplexes [Tm of the duplex modified by the flexicate—Tm of the control (unmodified duplex) (ΔTm)] are generally affected by the oligonucleotides’ dissociation constants and the stability of their own secondary structures. The stronger the interaction between DNA and [Fe2L1a3]Cl4, the more the Tm value increases (30,31).
The thermal stabilities of an AP site containing oligonucleotide duplexes flanked by G·C pairs on both sides and with dG, dC, dA and dT in opposite orientation to an AP site (X1 = X2 = G, Y1 = Y2 = C, Z = G, C, A and T; Figure 1B) in the presence of the flexicates were determined and results are summarized in Table 1. The fully matched duplex GGG/CCC of identical sequence to GFG/CCC duplex in which an AP site was replaced by guanine was used as a control and had a melting temperature of 53.1°C. The presence of an AP site markedly reduced thermal stabilities of the duplexes. The melting temperatures of the duplexes with purines (dG or dA) opposite an AP site were slightly higher than those with pyrimidines (dC or dT): 36.0 and 38.2°C for GFG/CGC and GFG/CAC, respectively, versus 35.5 and 34.1°C for GFG/CCC and GFG/CTC, respectively. The results in Table 1 show that the Tm values of an AP site containing duplexes in the presence of Λ- and Δ-[Fe2L1a3]Cl4 were increased by ∼14–18°C and ∼5–12°C, respectively. The maximum stabilization effect was observed for the duplexes with pyrimidines in opposite orientation to AP sites. In all cases, the Λ-[Fe2L1a3]Cl4 was more effective in increasing the thermal stability of an AP site containing DNA duplexes. Increasing the flexicate:duplex ratio from 1:1 to 2:1 had little impact on the melting temperature of the duplexes (0.1–1.3°C) which is consistent with the presence of a single dominant binding site for the flexicates. The value of Tm of the fully matched duplex GGG/CCC was negligibly affected in the presence of the flexicates.
Table 1. Thermal stability of a fully matched duplex GGG/CCC and corresponding duplexes containing an AP site flanked by guanines and with various bases opposite an AP site in the presence of Λ- and Δ-[Fe2L1a3]Cl4.
| Compound | ΔTm (°C) at 1:1a | ΔTm (°C) at 2:1b |
|---|---|---|
| GGG/CCC (Tm = 53.1°C) | ||
| Λ-[Fe2L1a3]Cl4 | 0.2 | 0.7 |
| Δ-[Fe2L1a3]Cl4 | 0.0 | 0.2 |
| GFG/CGC (Tm = 36.0°C) | ||
| Λ-[Fe2L1a3]Cl4 | 15.3 | 15.4 |
| Δ-[Fe2L1a3]Cl4 | 5.2 | 6.5 |
| GFG/CCC (Tm = 35.5°C) | ||
| Λ-[Fe2L1a3]Cl4 | 16.3 | 16.5 |
| Δ-[Fe2L1a3]Cl4 | 10.5 | 11.0 |
| GFG/CAC (Tm = 38.2°C) | ||
| Λ-[Fe2L1a3]Cl4 | 13.9 | 14.2 |
| Δ-[Fe2L1a3]Cl4 | 6.6 | 7.4 |
| GFG/CTC (Tm = 34.1°C) | ||
| Λ-[Fe2L1a3]Cl4 | 17.6 | 17.7 |
| Δ-[Fe2L1a3]Cl4 | 11.2 | 11.6 |
aHelicate:duplex was 1:1.
bHelicate:duplex was 2:1.
Table 2 presents data obtained for the duplexes containing an AP site flanked by A·T pairs on both sides and with various nucleotides placed opposite an AP site (X1 = X2 = A, Y1 = Y2 = T, Z = G, C, A and T; Figure 1B). Also in this case, the Tm values of an AP site containing duplexes in the absence of the helicates were a little higher if the base situated opposite an AP site was purine: 29.1 and 28.5°C for AFA/TGT and AFA/TAT, respectively, versus 25.9 and 27.6°C for AFA/TCT and AFA/TTT, respectively. The presence of the flexicates had little effect on the Tm of the fully matched duplex AGA/TCT (46.3°C). The Tm values of duplexes containing an AP site flanked by adenines in the presence of Λ- and Δ-[Fe2L1a3]Cl4 were increased by ∼11–15 and ∼7–14°C, respectively. The flexicates enhanced the thermal stability of duplexes with pyrimidines opposite an AP site more than those with purines. Λ-[Fe2L1a3]Cl4 was a better stabilizer of an AP site containing duplexes than Δ -[Fe2L1a3]Cl4. Doubling the flexicate:duplex ratio from 1:1 to 2:1 had small effect (0.7–1.4°C) on the stability of the duplexes.
Table 2. Thermal stability of a fully matched duplex AGA/TCT and corresponding duplexes containing an AP site flanked by adenines and with various bases opposite an AP site in the presence of Λ- and Δ-[Fe2L1a3]Cl4.
| Compound | ΔTm (°C) at 1:1a | ΔTm (°C) at 2:1b |
|---|---|---|
| AGA/TCT (Tm = 46.3°C) | ||
| Λ-[Fe2L1a3]Cl4 | 0.2 | 0.8 |
| Δ-[Fe2L1a3]Cl4 | 0.1 | 0.3 |
| AFA/TGT (Tm = 29.1°C) | ||
| Λ-[Fe2L1a3]Cl4 | 11.1 | 11.8 |
| Δ-[Fe2L1a3]Cl4 | 7.1 | 7.8 |
| AFA/TCT (Tm = 25.9°C) | ||
| Λ-[Fe2L1a3]Cl4 | 13.3 | 14.7 |
| Δ-[Fe2L1a3]Cl4 | 12.7 | 13.5 |
| AFA/TAT (Tm = 28.5°C) | ||
| Λ-[Fe2L1a3]Cl4 | 9.8 | 10.8 |
| Δ-[Fe2L1a3]Cl4 | 7.3 | 8.5 |
| AFA/TTT (Tm = 27.6°C) | ||
| Λ-[Fe2L1a3]Cl4 | 10.6 | 11.8 |
| Δ-[Fe2L1a3]Cl4 | 10.1 | 11.2 |
aHelicate:duplex was 1:1.
bHelicate:duplex was 2:1.
The Tm values of a fully matched duplex CGC/GCG and corresponding duplexes containing an AP site flanked by C·G pairs (X1 = X2 = C, Y1 = Y2 = G, Z = G, C, A and T; Figure 1B) are shown in Table 3. It can be seen, that the presence of a purine opposite an AP site enhanced thermal stability of the duplexes in the absence of the flexicates: 36.8 and 35.7°C for CFC/GGG and CFC/GAG, respectively, versus 31.3 and 33.0°C for CFC/GCG and CFC/GTG, respectively. The addition of Λ- and Δ-[Fe2L1a3]Cl4 increased the Tm values of duplexes by ∼6–15 and ∼4–12°C, respectively. In contrast, the Tm of the fully matched duplex CGC/GCG (54.4°C) was almost unaffected by flexicates. Similarly to the previous results, the duplexes containing pyrimidines (dC or dT) opposite an AP site were stabilized by flexicates more than those with purines (dG or dA). Λ-[Fe2L1a3]Cl4 was more efficient than Δ-[Fe2L1a3]Cl4 in stabilizing duplexes containing an AP site flanked by cytosines.
Table 3. Thermal stability of a fully matched duplex CGC/GCG and corresponding duplexes containing an AP site flanked by cytosines and with various bases opposite an AP site in the presence of Λ- and Δ-[Fe2L1a3]Cl4.
| Compound | ΔTm (°C) at 1:1a | ΔTm (°C) at 2:1b |
|---|---|---|
| CGC/GCG (Tm = 54.4°C) | ||
| Λ-[Fe2L1a3]Cl4 | 0.3 | 0.7 |
| Δ-[Fe2L1a3]Cl4 | -0.1 | 0.2 |
| CFC/GGG (Tm = 36.8°C) | ||
| Λ-[Fe2L1a3]Cl4 | 9.0 | 9.3 |
| Δ-[Fe2L1a3]Cl4 | 4.8 | 6.4 |
| CFC/GCG (Tm = 31.3°C) | ||
| Λ-[Fe2L1a3]Cl4 | 14.1 | 14.6 |
| Δ-[Fe2L1a3]Cl4 | 11.5 | 12.0 |
| CFC/GAG (Tm = 35.7°C) | ||
| Λ-[Fe2L1a3]Cl4 | 5.8 | 8.5 |
| Δ-[Fe2L1a3]Cl4 | 3.5 | 5.4 |
| CFC/GTG (Tm = 33.0°C) | ||
| Λ-[Fe2L1a3]Cl4 | 12.5 | 13.2 |
| Δ-[Fe2L1a3]Cl4 | 11.2 | 11.9 |
aHelicate:duplex was 1:1.
bHelicate:duplex was 2:1.
Finally, results listed in Table 4 show the impact of Λ- and Δ-[Fe2L1a3]Cl4 on the thermal stability of the fully matched duplex TGT/ACA and corresponding duplexes containing an AP site flanked on both sides by T·A pairs (X1 = X2 = T, Y1 = Y2 = A, Z = G, C, A and T; Figure 1B). In the absence of the flexicates, the melting temperatures of TGT/AGA and TGT/AAA duplexes having a purine base opposite an AP site were higher than those of TGT/ACA and TGT/ATA duplexes with a pyrimidine base in opposite orientation: 27.9 and 28.0°C versus 24.4 and 25.6°C, respectively. The presence of Λ- and Δ-[Fe2L1a3]Cl4 increased the Tm values of the duplexes by ∼5–14 and ∼5–15°C, respectively. Interestingly, in this case the Δ-enantiomer was slightly more potent in increasing the thermal stability of an AP site containing duplexes than the Λ-enantiomer.
Table 4. Thermal stability of a fully matched duplex TGT/ACA and corresponding duplexes containing an AP site flanked by thymines and with various bases opposite an AP site in the presence of Λ- and Δ-[Fe2L1a3]Cl4.
| Compound | ΔTm (°C) at 1:1a | ΔTm (°C) at 2:1b |
|---|---|---|
| TGT/ACA (Tm = 48.1°C) | ||
| Λ-[Fe2L1a3]Cl4 | 0.8 | 1.0 |
| Δ-[Fe2L1a3]Cl4 | 0.2 | 0.4 |
| TFT/AGA (Tm = 27.9°C) | ||
| Λ-[Fe2L1a3]Cl4 | 4.7 | 6.2 |
| Δ-[Fe2L1a3]Cl4 | 5.3 | 6.9 |
| TFT/ACA (Tm = 24.4°C) | ||
| Λ-[Fe2L1a3]Cl4 | 13.1 | 14.1 |
| Δ-[Fe2L1a3]Cl4 | 14.6 | 15.0 |
| TFT/AAA (Tm = 28.0°C) | ||
| Λ-[Fe2L1a3]Cl4 | 4.5 | 6.3 |
| Δ-[Fe2L1a3]Cl4 | 5.0 | 6.6 |
| TFT/ATA (Tm = 25.6°C) | ||
| Λ-[Fe2L1a3]Cl4 | 10.2 | 11.2 |
| Δ-[Fe2L1a3]Cl4 | 13.4 | 14.0 |
aHelicate:duplex was 1:1.
bHelicate:duplex was 2:1.
In the aggregate, results from ultraviolet (UV) melting studies demonstrate that the flexicates have higher affinity for AP sites in which a pyrimidine base (dC or dT) is in opposite orientation to the lesion. On closer inspection, data in Tables 1–4 reveal that duplexes containing AP sites flanked by G·C pairs, GFG/CTC and GFG/CCC, were the most stabilized and duplexes with AP sites flanked by T·A pairs, TFT/AAA and TFT/AGA the least stabilized in the presence of Λ- and Δ-[Fe2L1a3]Cl4. Increasing the flexicate:duplex ratios from 1:1 to 2:1 had little effect on the thermal stability of the duplexes which is consistent with the presence of one major binding site for the flexicates. On the other hand, it cannot be excluded that additional less affinity binding sites for flexicates are present in the duplexes with AP sites as it can be deduced from our ITC data (vide supra). The flexicates showed little affinity for the fully matched duplexes.
Electrophoretic mobility shift assay
We employed the electrophoretic mobility shift assay to examine whether the stability of the complex formed between flexicates and an AP site containing duplex is strong enough to withstand migration through a polyacrylamide gel. The autoradiograms of the gels run at 5°C (Figure 2) show interaction of Λ- and Δ-[Fe2L1a3]Cl4 with GFG/CTC and GFG/CAC duplexes. It can be seen that a new slower-migrating band indicating formation of the flexicate–DNA duplex complex appears for GFG/CTC in the presence of Λ-[Fe2L1a3]Cl4 (Figure 2A, lanes 1–6). The autoradiogram of the gel in Figure 2B (lanes 1–6) shows that the interaction of Λ-[Fe2L1a3]Cl4 with GFG/CAC duplex was not so strong as in the former case which is consistent with the previous results from the UV melting studies showing that flexicates prefer binding to an AP site with a pyrimidine placed opposite the lesion. We did not observe formation of the complexes between GFG/CTC and GFG/CAC duplexes and Δ-[Fe2L1a3]Cl4 (Figure 2A and B, lanes 7–12) which is also in agreement with the previous data demonstrating that Δ-[Fe2L1a3]Cl4 is a weaker binder than Λ-[Fe2L1a3]Cl4 to AP sites flanked by purines.
Figure 2.
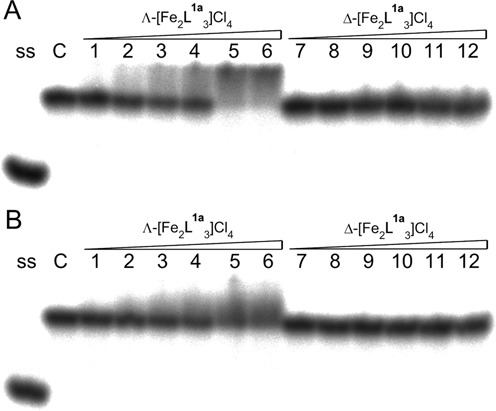
Autoradiogram of the gel run at 5°C, demonstrating interactions of the Λ- and Δ-[Fe2L1a3]Cl4 with GFG/CTC (A) and GFG/CAC (B) duplexes. Lane ss: control containing one strand in the buffer. Lane C: control containing duplex in the buffer in the absence of the flexicates. Lanes 1–6: duplex mixed with Λ-[Fe2L1a3]Cl4 at 0.25:1, 0.5:1, 0.75:1, 1:1, 1.5:1 and 2:1 (flexicate:duplex) ratios, respectively. Lanes 7–12: duplex mixed with Δ-[Fe2L1a3]Cl4 at 0.25:1, 0.5:1, 0.75:1, 1:1, 1.5:1 and 2:1 (flexicate:duplex) ratios, respectively.
2-aminopurine fluorescence studies
2-aminopurine (2AP), a fluorescent analogue of adenine, has been widely used as a probe of small molecule binding to DNA (32,33) or RNA (34,35). The fluorescence of 2AP is strongly quenched within the structure of double-stranded DNA or RNA, but is enhanced when the base stacking or base pairing is perturbed. Other factors that affect the fluorescence of 2AP are collisions with other bases and biomolecular interactions (36,37). We used AFA/T2APT duplex containing an AP site flanked by A·T pairs on both sides and with 2AP opposite an AP site to monitor the binding of Λ- and Δ-[Fe2L1a3]Cl4 (Figure 3). Titrations of AFA/T2APT by Λ- and Δ-[Fe2L1a3]Cl4 lead to a marked decrease of the fluorescence intensity of 2AP followed by a slight increase of the fluorescence when the concentration of flexicates reached ∼2 μM. Plots of the data with linear interpolations superimposed suggest that Λ- and Δ-[Fe2L1a3]Cl4 bind to AFA/T2APT at 1:1 flexicate:duplex ratio. The quenching of the 2AP fluorescence by [Fe2L1a3]Cl4 could be explained by enhanced base stacking interactions between the 2AP and adjacent bases in the presence of [Fe2L1a3]Cl4 or by direct interactions of the 2AP with the flexicates.
Figure 3.
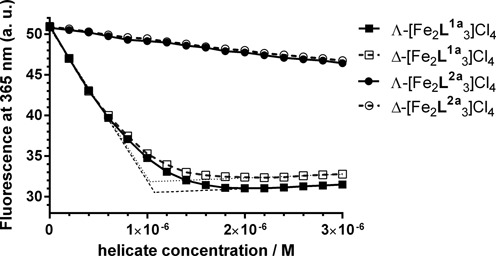
Fluorescence titrations of the 2AP-labelled duplex AFA/T2APT at 1 μM concentration with [Fe2L1a3]Cl4 and [Fe2L2a3]Cl4 helicates. The buffer conditions were 10 mM sodium phosphate buffer (pH 7) and 100 mM NaCl.
We have recently demonstrated that flexicates based on the L2a ligand (Supplementary Figure S6) are weak and nonspecific DNA binders that have no stabilizing effect on unusual DNA structures (23). The experiment was repeated with [Fe2L2a3]Cl4 to compare the influence of the L1a and L2a based flexicates on the 2AP fluorescence. The plot in Figure 3 shows that titrations of AFA/T2APT by Λ- and Δ-[Fe2L2a3]Cl4 resulted in a slight linear decrease of the 2AP fluorescence.
Amiloride displacement
Amiloride has been previously reported to selectively recognize thymine situated opposite an AP site (38). It has been determined by ITC that amiloride preferentially binds to an AP site flanked by G·C pairs with Ka = 2.7 × 106 M-1 (7). We titrated GFG/CTC duplex mixed at 1:1 ratio with amiloride by Λ- and Δ-[Fe2L1a3]Cl4 to examine if the flexicates can displace amiloride from its preferential binding site. The plots of the fluorescence intensity of amiloride as a function of Λ- and Δ-[Fe2L1a3]Cl4 concentrations in Figure 4 show that the fluorescence signal of amiloride was growing with increasing concentration of the flexicates up to 1.1–1.2 μM and then it levelled off.
Figure 4.
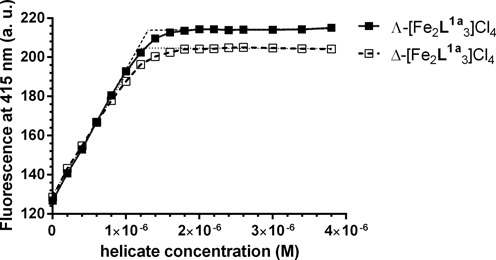
Displacement of amiloride from GFG/CTC duplex by Λ- and Δ-[Fe2L1a3]Cl4. The concentration of DNA duplex and amiloride was 1 μM and buffer conditions were 10 mM sodium phosphate buffer (pH 7) and 100 mM NaCl.
Since flexicates readily displaced amiloride from its binding site it can be concluded that Ka values of Λ- and Δ-[Fe2L1a3]Cl4 are much higher than that of amiloride. The linear interpolations of initial and final segments of the plots in Figure 4 intersect at ∼1 μM, which corresponds to 1:1 (flexicate:duplex) ratio. This result is consistent with the presence of one major binding site on GFG/CTC duplex for Λ- and Δ-[Fe2L1a3]Cl4, although the presence of an additional less affinity binding site cannot be excluded as it can be deduced from our ITC data (vide supra).
Isothermal titration calorimetry
In order to characterize the binding of the flexicates to an AP site containing duplexes we used ITC. Figure 5 shows ITC profiles resulting from the injections of Λ- (panels A and C) and Δ-[Fe2L1a3]Cl4 (panels B and D) into a solution of GFG/CTC (panels A and B) and GFG/CAC (panels C and D) duplexes. The plots of the heat evolved per mole of the flexicate added against the molar ratio of the flexicate to DNA duplex are shown below each ITC profile. In these plots, the data points correspond to the experimental injection heats, while solid lines represent the calculated fits of the data with the ‘two sets of sites’ binding model (39); the only one that yielded a reasonable fit of the experimental data. The calculated thermodynamic parameters are listed in Table 5. It can be seen that each ITC profile has two apparent phases suggesting two distinct binding events. The binding stoichiometry values estimated for the first binding event (N1) indicate that the flexicates bind to a higher-affinity site at 1:1 ratio. The steepness of the first of the two phases indicates relatively tight binding interaction. Indeed, fit of the ITC profile for the binding of Λ- and Δ-[Fe2L1a3]Cl4 to GFG/CTC duplex gives association constants for the first binding event (K1) of 6.8 × 107 M−1 and 4.7 × 107 M−1 (Table 5), respectively. These values are higher than the value reported previously for amiloride (2.7 × 106 M−1) which is in agreement with the observation that flexicates readily displaced amiloride from its binding site in GFG/CTC duplex. The binding affinity of Λ- and Δ-[Fe2L1a3]Cl4 to GFG/CAC duplex was not so high which also correlates with the results from the UV melting studies and electrophoretic mobility shift assay. The observed enthalpies for Λ- and Δ-[Fe2L1a3]Cl4 binding to GFG/CTC duplex, −41.4 and −21.9 kJ·mol−1, respectively, are more exothermic than those associated with binding to GFG/CAC duplex, −25.6 and −4.3 kJ·mol−1, respectively. In other words, the binding of the flexicates to an AP site with thymine placed opposite the lesion is enthalpically favourable. Taken together, the data demonstrate that the Λ-enantiomer binds to GFG/CTC and GFG/CAC duplexes with higher affinity than the Δ-enantiomer and that its binding is also associated with larger enthalpy changes. Unfortunately, we cannot currently provide an explanation and a binding model that would be consistent with the stoichiometry of the second binding event. The stoichiometry values N2 ≈ 0.26–0.37 (Table 5) indicate that the binding of the second flexicate molecule leads to the association of the duplexes. On the other hand, these results demonstrate that the first flexicate molecule binds to a unique high-affinity site on the target duplex.
Figure 5.
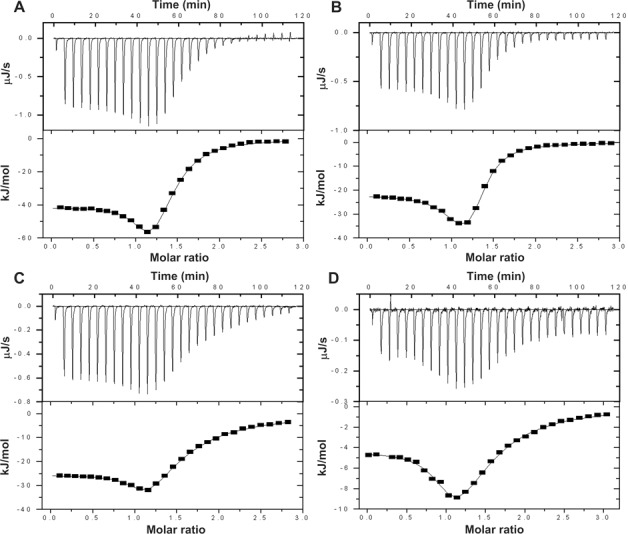
ITC binding curves for the titration of GFG/CTC (Aand B) and GFG/CAC (C and D) duplexes with Λ-[Fe2L1a3]Cl4 (A and C) and Δ-[Fe2L1a3]Cl4 (B and D) at 20°C in 10 mM phosphate buffer (pH 7.0) containing 100 mM NaCl. The upper panels in each Figure 5A–D show total heat released upon injecting of aliquots of the flexicate solution into a reaction cell containing an AP site containing duplex. The lower panels show the resultant binding isotherms (full squares) obtained by integrating the peak areas of each injection; the continuous lines represent the nonlinear least squares fit to a two sets of sites binding model. For other details, see the text.
Table 5. Thermodynamic parameters for the titration of GFG/CTC and GFG/CAC duplexes with Λ- and Δ-[Fe2L1a3]Cl4.
| GFG/CTC | GFG/CAC | |||
|---|---|---|---|---|
| Binding parameters | Λ-[Fe2L1a3]Cl4 | Δ-[Fe2L1a3]Cl4 | Λ-[Fe2L1a3]Cl4 | Δ-[Fe2L1a3]Cl4 |
| N1 | 1.03 ± 0.03 | 1.03 ± 0.09 | 1.03 ± 0.02 | 0.98 ± 0.05 |
| K1 (M−1) | (6.8 ± 1.8)·107 | (4.7 ± 2.2)·107 | (2.2 ± 0.4)·107 | (1.5 ± 0.5)·107 |
| ΔH1 (kJ·mol−1) | −41.4 ± 0.3 | −21.9 ± 0.4 | −25.6 ± 0.1 | −4.3 ± 0.1 |
| TΔS1 (kJ·mol−1) | 2.5 | 21.2 | 15.6 | 35.8 |
| N2 | 0.34 ± 0.05 | 0.26 ± 0.14 | 0.33 ± 0.07 | 0.37 ± 0.14 |
| K2 (M−1) | (1.8 ± 0.2)·106 | (3.2 ± 0.6)·106 | (4.5 ± 0.5)·105 | (5.4 ± 0.9)·105 |
| ΔH2 (kJ·mol−1) | −103.2 ± 13.8 | −75.3 ± 35.5 | −104.2 ± 22.3 | −29.5 ± 11.3 |
| TΔS2 (kJ·mol−1) | −68.3 | −38.8 | −72.5 | 2.6 |
DNase I footprinting
To further characterize the binding of the flexicates to an AP site we employed DNase I footprinting assay. Λ- and Δ-[Fe2L1a3]Cl4 were mixed with radioactively labelled GFG/CTC duplex at 0.5:1, 1:1, 2:1 and 3:1 (flexicate:duplex) ratios, respectively, followed by partial cleavage by DNase I. The autoradiograms of the DNA cleavage-inhibition patterns for the top and bottom strand labelled GFG/CTC duplex are shown in Figure 6A. As it can be seen, the inhibition of DNase I cleavage activity increases with increasing concentration of the flexicates. The maximum protection was observed when the flexicate:duplex ratio exceeded 1:1. Scheme in Figure 6B summarizing obtained results suggests that Λ- and Δ-[Fe2L1a3]Cl4 bind directly to an AP site or in its close proximity.
Figure 6.
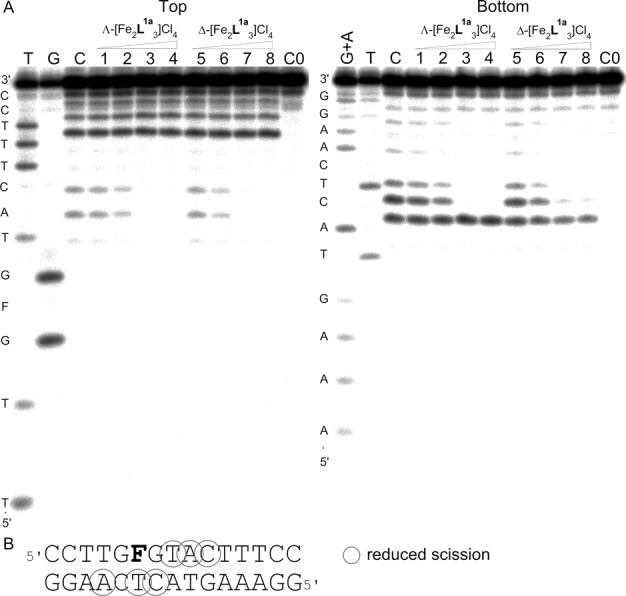
(A) Autoradiograms of DNase I footprint of 5’ end labelled top (left) or bottom (right) strand of the GFG/CTC duplex containing an AP site in the presence of increasing concentrations of Λ-[Fe2L1a3]Cl4 (lanes 1–4) and Δ-[Fe2L1a3]Cl4 (lanes 5–8). Lane C; GFG/CTC duplex in the absence of flexicates. Lanes 1–4; GFG/CTC duplex mixed with Λ-[Fe2L1a3]Cl4 at 0.5:1, 1:1, 2:1 and 3:1 (flexicate:DNA duplex) ratios, respectively. Lanes 5–8; DNA duplex mixed with Δ-[Fe2L1a3]Cl4 at 0.5:1, 1:1, 2:1 and 3:1 (flexicate:DNA duplex) ratios, respectively. Lane C0; DNA duplex in the absence of flexicates and DNase I. Lanes T, G and G + A correspond to T, G and G + A ladders. The nucleotide sequence of the oligonucleotide is shown on the left side of the gels. (B) Sequence of the DNA duplex containing an AP site indicated by letter ‘F’ showing sites (shown by circles) protected by Λ- and Δ-[Fe2L1a3]Cl4 from the cleavage by DNase I.
Inhibition of human AP endonuclease 1 (APE1)
An AP site containing duplexes GFG/CTC and GFG/CAC were treated with APE1 in the presence of various concentrations of Λ- and Δ-[Fe2L1a3]Cl4 to explore the influence of the flexicates on the cleavage activity of APE1. The autoradiogram of the gel in Figure 7A shows for the GFG/CTC duplex that APE1 activity was reduced by increasing concentrations of Λ- and Δ-[Fe2L1a3]Cl4. Gels for GFG/CTC and GFG/CAC duplexes were analysed by densitometry and the plots in Figure 7B demonstrate that the Λ-enantiomer was a more potent inhibitor of APE1 activity than the Δ-enantiomer and that the protection of an AP site from the cleavage by APE1 was higher if the base opposite an AP site was thymine rather than adenine. Both findings are in agreement with the previously obtained results. It might be argued that the flexicates inhibit APE1 by binding to this enzyme rather than by protecting the AP sites from the cleavage. If that would be the case then the cleavage of the duplexes should be independent of the base opposite the AP site. It is shown in Figure 7 that the cleavage activity of APE1 was distinctly different if the base opposite an AP site was thymine or adenine. Thus, the eventuality that the cleavage by APE1 of the duplexes containing AP sites was due to direct inhibition of APE1 appears unlikely. In addition, the latter conclusion has been also supported by the results of our competition experiments shown in the Supplementary Material and in Supplementary Figure S7.
Figure 7.
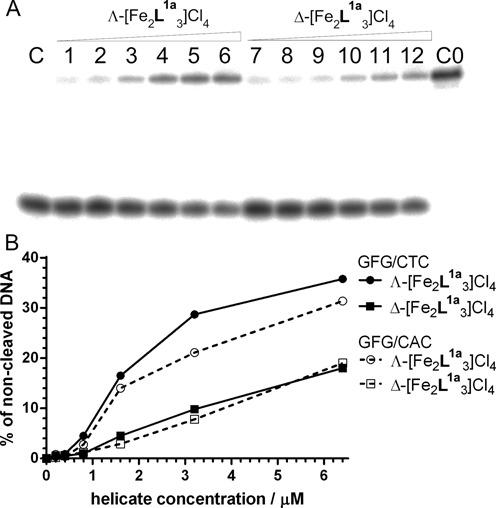
(A) Autoradiogram of the PAA gel showing cleavage of GFG/CTC duplex by APE1 in the presence of Λ- and Δ-[Fe2L1a3]Cl4. Lane C: GFG/CTC duplex treated with APE1 in the absence of flexicates. Lanes 1–6: GFG/CTC duplex treated with APE1 in the presence of 0.2, 0.4, 0.8, 1.6, 3.2 and 6.4 μM Λ-[Fe2L1a3]Cl4, respectively. Lanes 7–12: GFG/CTC duplex treated with APE1in the presence of 0.2, 0.4, 0.8, 1.6, 3.2 and 6.4 μM Δ-[Fe2L1a3]Cl4, respectively. Lane C0; GFG/CTC duplex in the absence of flexicates and APE1. The concentration of GFG/CTC duplex in the samples was 2.0 × 10−7 M. (B) The dependence of the cleavage of GFG/CTC and GFG/CAC duplexes by APE1 in the presence of increasing concentrations of Λ- and Δ-[Fe2L1a3]Cl4.
DISCUSSION
The results show that [Fe2L1a3]Cl4 flexicates bind to an AP site containing DNA duplexes in a shape-selective manner as in the case of the binding of phenanthrenequinone diimine complexes of rhodium(III) to DNA (40,41). Flexicates preferentially bind to AP sites flanked by purines on both sides and their binding is enhanced when a pyrimidine is placed in opposite orientation to the lesion. This binding preference could be explained by the previously reported observation using nuclear magnetic resonance spectroscopy (42) that pyrimidines opposite an AP site stack poorly and are extrahelical while adenine (A) is stacked in an intrahelical conformation and guanine (G) can exist in both intra- and extrahelical conformations. Apart from an AP site flanked by thymines, the Λ-enantiomer was binding to all tested AP sites with higher affinity than the Δ-enantiomer.
Nevertheless, the binding mode of the flexicates to an AP site remains to be determined. The results indicate that the first flexicate molecule binds to a unique high-affinity site on the target duplex. We do not know if the flexicates bind directly to an AP site or in a close proximity to the lesion. The binding preference of the flexicates to AP sites having a poorly stacked pyrimidine base placed opposite the lesion suggests that the flexicate molecule could insert into the hollow cavity formed in the DNA double helix by the AP site and the looped-out base in opposite orientation. Another possibility is that the flexicate sticks to an AP site from the side, probably from the major groove.
In the aggregate, our results show that flexicates constitute a completely new group of ligands capable of selective binding to AP sites. Their structure is totally different from that of the previously reported molecules designed to bind to an AP site. In addition, the ability of the flexicates to inhibit DNA cleavage by human APE1 and that APE1 has been shown to have higher activity in tumour compared to normal tissue (43) suggests that the flexicates might be applied along with antitumour alkylating agents to potentiate their toxic activity in tumour cells (28).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors acknowledge that their participation in the EU COST Action CM1105 enabled them to exchange regularly the most recent ideas in the field of metallodrugs with several European colleagues.
FUNDING
Czech Science Foundation [P205/11/0856]. Funding for open access charge: Czech Science Foundation [P205/11/0856].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen H., Krokan H.E. Base excision repair in a network of defence and tolerance. Carcinogenesis. 2001;22:987–998. doi: 10.1093/carcin/22.7.987. [DOI] [PubMed] [Google Scholar]
- 3.Tropp B.E. Molecular Biology: Genes to Proteins. Sudbury, MA: Jones & Bartlett Publishers; 2012. p. 455. [Google Scholar]
- 4.Wu F., Sun Y., Shao Y., Xu S., Liu G., Peng J., Liu L. DNA abasic site-selective enhancement of sanguinarine fluorescence with a large emission shift. PLoS One. 2012;7:e48251. doi: 10.1371/journal.pone.0048251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu S., Shao Y., Wu F., Liu G., Liu L., Peng J., Sun Y. Targeting DNA abasic site by myricetin: sequence-dependent ESIPT emission. J. Lumin. 2013;136:291–295. [Google Scholar]
- 6.Wang X., Wang X., Cui S., Wang Y., Chen G., Guo Z. Specific recognition of DNA depurination by a luminescent terbium(III) complex. Chem. Sci. 2013;4:3748–3752. [Google Scholar]
- 7.Rajendran A., Zhao C., Rajendar B., Thiagarajan V., Sato Y., Nishizawa S., Teramae N. Effect of the bases flanking an abasic site on the recognition of nucleobase by amiloride. Biochim. Biophys. Acta. 2010;1800:599–610. doi: 10.1016/j.bbagen.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Sato Y., Nishizawa S., Yoshimoto K., Seino T., Ichihashi T., Morita K., Teramae N. Influence of substituent modifications on the binding of 2-amino-1, 8-naphthyridines to cytosine opposite an AP site in DNA duplexes: thermodynamic characterization. Nucleic Acids Res. 2009;37:1411–1422. doi: 10.1093/nar/gkn1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato Y., Zhang Y., Seino T., Sugimoto T., Nishizawa S., Teramae N. Highly selective binding of naphthyridine with a trifluoromethyl group to cytosine opposite an abasic site in DNA duplexes. Org. Biomol. Chem. 2012;10:4003–4006. doi: 10.1039/c2ob25513h. [DOI] [PubMed] [Google Scholar]
- 10.Belmont P., Jourdan M., Demeunynck M., Constant J.F., Garcia J., Lhomme J., Carez D., Croisy A. Abasic site recognition in DNA as a new strategy to potentiate the action of anticancer alkylating drugs. J. Med. Chem. 1999;42:5153–5159. doi: 10.1021/jm9901428. [DOI] [PubMed] [Google Scholar]
- 11.Berthet N., Constant J.F., Demeunynck M., Michon P., Lhomme J. Search for DNA repair inhibitors: Selective binding of nucleic bases acridine conjugates to a DNA duplex containing an abasic site. J. Med. Chem. 1997;40:3346–3352. doi: 10.1021/jm970225t. [DOI] [PubMed] [Google Scholar]
- 12.Zeglis B.M., Boland J.A., Barton J.K. Targeting abasic sites and single base bulges in DNA with metalloinsertors. J. Am. Chem. Soc. 2008;130:7530–7531. doi: 10.1021/ja801479y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeglis B.M., Boland J.A., Barton J.K. Recognition of abasic sites and single base bulges in DNA by a metalloinsertor. Biochemistry. 2009;48:839–849. doi: 10.1021/bi801885w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fakhari A., Rokita S.E. A new solvatochromic fluorophore for exploring nonpolar environments created by biopolymers. Chem. Commun. 2011;47:4222–4224. doi: 10.1039/c0cc04917d. [DOI] [PubMed] [Google Scholar]
- 15.Benner K., Bergen A., Ihmels H., Pithan P.M. Selective stabilization of abasic site-containing DNA by insertion of sterically demanding biaryl ligands. Chem. Eur. J. 2014;20:9883–9887. doi: 10.1002/chem.201403622. [DOI] [PubMed] [Google Scholar]
- 16.Wu F., Shao Y., Ma K., Cui Q., Liu G., Xu S. Simultaneous fluorescence light-up and selective multicolor nucleobase recognition based on sequence-dependent strong binding of berberine to DNA abasic site. Org. Biomol. Chem. 2012;10:3300–3307. doi: 10.1039/c2ob00028h. [DOI] [PubMed] [Google Scholar]
- 17.Berthet N., Michon J., Lhomme J., Teulade-Fichou M.P., Vigneron J.P., Lehn J.M. Recognition of abasic sites in DNA by a cyclobisacridine molecule. Chem. Eur. J. 1999;5:3625–3630. [Google Scholar]
- 18.Barret J.M., Etievant C., Fahy J., Lhomme J., Hill B.T. Novel artificial endonucleases inhibit base excision repair and potentiate the cytotoxicity of DNA-damaging agents on L1210 cells. Anticancer Drugs. 1999;10:55–65. doi: 10.1097/00001813-199901000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Lefrancois M., Bertrand J.R., Malvy C. 9-Amino-ellipticine inhibits the apurinic site-dependent base excision-repair pathway. Mutation Res. 1990;236:9–17. doi: 10.1016/0921-8777(90)90027-3. [DOI] [PubMed] [Google Scholar]
- 20.Malvy C., Safraoui H., Bloch E., Bertrand J.R. Involvement of apurinic sites in the synergistic action of alkylating and intercalating drugs in Escherichia coli. Anticancer Drug Des. 1988;2:361–370. [PubMed] [Google Scholar]
- 21.Alarcon K., Demeunynck M., Lhomme J., Carrez D., Croisy A. Diaminopurine-acridine heterodimers for specific recognition of abasic site containing DNA. Influence on the biological activity of the position of the linker on the purine ring. Bioorg. Med. Chem. Lett. 2001;11:1855–1858. doi: 10.1016/s0960-894x(01)00310-9. [DOI] [PubMed] [Google Scholar]
- 22.Alarcon K., Demeunynck M., Lhomme J., Carrez D., Croisy A. Potentiation of BCNU cytotoxicity by molecules targeting abasic lesions in DNA. Bioorg. Med. Chem. Lett. 2001;9:1901–1910. doi: 10.1016/s0968-0896(01)00097-9. [DOI] [PubMed] [Google Scholar]
- 23.Brabec V., Howson S.E., Kaner R.A., Lord R.M., Malina J., Phillips R.M., Abdallah Q.M.A., McGowan P.C., Rodger A., Scott P. Metallohelices with activity against cisplatin-resistant cancer cells; does the mechanism involve DNA binding. Chem. Sci. 2013;4:4407–4416. [Google Scholar]
- 24.Howson S.E., Scott P. Approaches to the synthesis of optically pure helicates. Dalton Trans. 2011;40:10268–10277. doi: 10.1039/c1dt11423a. [DOI] [PubMed] [Google Scholar]
- 25.Howson S.E., Bolhuis A., Brabec V., Clarkson G.J., Malina J., Rodger A., Scott P. Optically pure, water-stable metallo-helical ‘flexicate’ assemblies with antibiotic activity. Nat. Chem. 2012;4:31–36. doi: 10.1038/nchem.1206. [DOI] [PubMed] [Google Scholar]
- 26.Howson S.E., Allan L.E.N., Chmel N.P., Clarkson G.J., van Gorkum R., Scott P. Self-assembling optically pure Fe(A-B)3 chelates. Chem. Commun. 2009:1727–1729. doi: 10.1039/b821573a. [DOI] [PubMed] [Google Scholar]
- 27.Howson S.E., Allan L.E.N., Chmel N.P., Clarkson G.J., Deeth R.J., Faulkner A.D., Simpson D.H., Scott P. Origins of stereoselectivity in optically pure phenylethaniminopyridine tris-chelates M(NN ‘)3n+ (M = Mn, Fe, Co, Ni and Zn) Dalton Trans. 2011;40:10416–10433. doi: 10.1039/c1dt10588d. [DOI] [PubMed] [Google Scholar]
- 28.Abbotts R., Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat. Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Freyer M.W., Lewis E.A. Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008;84:79–113. doi: 10.1016/S0091-679X(07)84004-0. [DOI] [PubMed] [Google Scholar]
- 30.Xi Z., Zhang R., Yu Z., Ouyang D., Huang R. Selective interaction between tylophorine B and bulged DNA. Bioorg. Med. Chem. Lett. 2005;15:2673–2677. doi: 10.1016/j.bmcl.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Bai L.P., Cai Z.W., Zhao Z.Z., Nakatani K., Jiang Z.H. Site-specific binding of chelerythrine and sanguinarine to single pyrimidine bulges in hairpin DNA. Anal. Bioanal. Chem. 2008;392:709–716. doi: 10.1007/s00216-008-2302-7. [DOI] [PubMed] [Google Scholar]
- 32.Sielaff A., Mackay H., Brown T., Lee M. 2-aminopurine/cytosine base pair containing oligonucleotides: fluorescence spectroscopy studies on DNA-polyamide binding. Biochem. Biophys. Res. Commun. 2008;369:630–634. doi: 10.1016/j.bbrc.2008.02.049. [DOI] [PubMed] [Google Scholar]
- 33.Patel N., Berglund H., Nilsson L., Rigler R., McLaughlin L.W., Graslund A. Thermodynamics of interaction of a fluorescent DNA oligomer with the anti-tumour drug netropsin. Eur. J. Biochem. 1992;203:361–366. doi: 10.1111/j.1432-1033.1992.tb16558.x. [DOI] [PubMed] [Google Scholar]
- 34.Bradrick T.D., Marino J.P. Ligand-induced changes in 2-aminopurine fluorescence as a probe for small molecule binding to HIV-1 TAR RNA. RNA. 2004;10:1459–1468. doi: 10.1261/rna.7620304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacourciere K.A., Stivers J.T., Marino J.P. Mechanism of neomycin and Rev peptide binding to the Rev responsive element of HIV-1 as determined by fluorescence and NMR spectroscopy. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 36.Holz B., Klimasauskas S., Serva S., Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rachofsky E.L., Osman R., Ross J.B.A. Probing structure and dynamics of DNA with 2-aminopurine: Effects of local environment on fluorescence. Biochemistry. 2001;40:946–956. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]
- 38.Zhao C.X., Dai Q., Seino T., Cui Y.Y., Nishizawa S., Teramae N. Strong and selective binding of amiloride to thymine base opposite AP sites in DNA duplexes: simultaneous binding to DNA phosphate backbone. Chem. Commun. 2006:1185–1187. doi: 10.1039/b516575j. [DOI] [PubMed] [Google Scholar]
- 39.ITC Data Analysis in OriginH. Tutorial Guide Version 5.0. 1998.
- 40.Pyle A.M., Morii T., Barton J.K. Probing microstructures in double-helical DNA with chiral metal complexes: recognition of changes in base-pair propeller twisting in solution. J. Amer. Chem. Soc. 1990;112:9432–9434. [Google Scholar]
- 41.Sitlani A., Long E.C., Pyle A.M., Barton J.K. DNA photocleavage by phenanthrenequinone diimine complexes of rhodium(III): shape-selective recognition and reaction. J. Am. Chem. Soc. 1992;114:2303–2312. [Google Scholar]
- 42.Cuniasse P., Fazakerley G.V., Guschlbauer W., Kaplan B.E., Sowers L.C. The abasic site as a challenge to DNA-polymerase - a nuclear-magnetic-resonance study of G, C and T opposite a model abasic site. J. Mol. Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- 43.Yoo D.G., Song Y.J., Cho E.J., Lee S.K., Park J.B., Yu J.H., Lim S.P., Kim J.M., Jeon B.H. Alteration of APEI/ref-1 expression in non-small cell lung cancer: The implications of impaired extracellular superoxide dismutase and catalase antioxidant systems. Lung Cancer. 2008;60:277–284. doi: 10.1016/j.lungcan.2007.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


