Abstract
Homozygosity for a natural deletion variant of the HIV-coreceptor molecule CCR5, CCR5Δ32, confers resistance toward HIV infection. Allogeneic stem cell transplantation from a CCR5Δ32-homozygous donor has resulted in the first cure from HIV (‘Berlin patient’). Based thereon, genetic disruption of CCR5 using designer nucleases was proposed as a promising HIV gene-therapy approach. Here we introduce a novel TAL-effector nuclease, CCR5-Uco-TALEN that can be efficiently delivered into T cells by mRNA electroporation, a gentle and truly transient gene-transfer technique. CCR5-Uco-TALEN mediated high-rate CCR5 knockout (>90% in PM1 and >50% in primary T cells) combined with low off-target activity, as assessed by flow cytometry, next-generation sequencing and a newly devised, very convenient gene-editing frequency digital-PCR (GEF-dPCR). GEF-dPCR facilitates simultaneous detection of wild-type and gene-edited alleles with remarkable sensitivity and accuracy as shown for the CCR5 on-target and CCR2 off-target loci. CCR5-edited cells were protected from infection with HIV-derived lentiviral vectors, but also with the wild-type CCR5-tropic HIV-1BaL strain. Long-term exposure to HIV-1BaL resulted in almost complete suppression of viral replication and selection of CCR5-gene edited T cells. In conclusion, we have developed a novel TALEN for the targeted, high-efficiency knockout of CCR5 and a useful dPCR-based gene-editing detection method.
INTRODUCTION
Besides its physiological role, the chemokine receptor CCR5 plays an essential part during HIV infection, acting as the co-receptor for R5-tropic strains that usually mediate initial HIV infection. In the absence of CCR5 on T-helper cells, R5-HIV is unable to bind and thus cannot infect T lymphocytes (1). Approximately 10% of Caucasians are heterozygous, and 1% homozygous for a deletion within CCR5, named CCR5Δ32 (2). In CCR5Δ32-heterozygous individuals HIV infection is less efficient, whereas homozygosity essentially protects from HIV (3,4) making CCR5 an interesting target for HIV therapy (5).
Substances mediating pharmacological CCR5 blockade have successfully been introduced as part of combined antiretroviral therapy (cART) (6). However, cART has several limitations including long-term toxicity, resistance and high costs (7–9). In contrast, genetic disruption (‘knockout’) of the CCR5 gene by the means of genetic therapy would, in an ideal scenario, be effective as a one-time treatment. This hypothesis is not only based on the natural resistance seen for CCR5Δ32-homozygous individuals, but has also been proven in a case study (‘Berlin patient’). In that study, an HIV-patient transplanted with hematopoietic stem cells from an allogeneic CCR5Δ32-homozygous donor has not only been cured from his leukemia, but evidently also from HIV (2,10).
Recently, different approaches were developed for the genetic knockout of CCR5 using designer nucleases. The first designer nucleases broadly applied were zinc-finger nucleases (ZFNs). A CCR5-specific ZFN developed by Sangamo BioSciences has been tested in a phase-I clinical study using a recombinant adenoviral vector for delivery. That study provided proof of safety and feasibility, but also some indication for clinical efficacy of gene editing (11,12).
TAL effector nucleases (TALENs) represent second-generation designer nucleases. In direct comparison using identical target sequences, TALENs were shown to exert higher specificity and lower toxicity as compared to ZFNs (13,14). CCR5-specific TALENs were already described (13,15–16), but have not been translated toward clinical applications so far.
We designed a codon-optimized TALEN targeting the functionally relevant first intracellular loop of the CCR5 receptor (designated ‘CCR5-Uco-TALEN’). We show that CCR5-Uco-TALEN mediates CCR5-gene knockout at very high rates in primary T lymphocytes after mRNA transfection, and that gene-edited T lymphocytes are efficiently protected from infection with CCR5-tropic lentiviral vectors, but also fully replicating HIV-1.
MATERIALS AND METHODS
Cell culture
The T-cell line PM1 (17) was cultured in RPMI 1640 supplemented with 10% FCS, 2% L-Glutamine and 1% Sodium Pyruvate (all Life Technologies, Darmstadt, Germany). 293T cells (ATCC CRL-3216) were kept in DMEM, Glutamax (Life Technologies) supplemented with 10% FCS.
Peripheral blood mononuclear cells isolated from buffy coats were activated for 72 h with Dynabeads Human T-Activator CD3/CD28 (Life Technologies) following the manufacturer's instructions and maintained in X-VIVO 10 (Lonza, Basel, Switzerland) supplemented with 8% autologous plasma and 200 U/ml hIL-2 (18).
If not specified otherwise, cell culture was performed under standard conditions (37°C, 100% relative humidity, 5% CO2). Cell-culture material was purchased from Corning (Corning, NY), Greiner Bio One (Frickenhausen, Germany) and Sarstedt (Nümbrecht, Germany).
Design and cloning of TALE nucleases
CCR5-specific Uco-TAL effector nucleases were designed using the TALEN targeter (19). To identify potential off-target sites we applied the Paired Target Finder (19). Cloning was performed in accordance with the protocol developed by Sanjana et al. (20) with the TALE Toolbox Kit purchased from Addgene (Kit #1000000019). Sequences of TAL effector nucleases are displayed as the corresponding mRNA sequence in Supplementary Table S1.
In-vitro transcription of mRNA
In-vitro transcription (IVT) of mRNA was performed with T7-mScript Standard mRNA-Production System (Biozym, Hessisch-Oldendorf, Germany) and the RNeasy Kit (Qiagen, Hilden, Germany) as previously described (21). Prior to IVT, plasmids were linearized using restriction enzymes (AvrII for TALEN constructs, HindIII for control plasmid pJET-T7-eGFP) and purified using the QIAquick PCR-Purification Kit (Qiagen).
Electroporation of plasmid DNA and mRNA
Electroporation of mRNA into lymphocytes was performed as previously described (21). Shortly, cells were washed twice and resuspended at final densities of 2 x 106/600 μl for primary T cells and 1.5 x 106/600 μl for PM1 cells in Opti-MEM (Life Technologies). Electroporation was performed using the Gene-Pulser Xcell Total System (Biorad, Munich, Germany) in 4-mm cuvettes (BTX, Holliston, MA) using the following conditions: primary T cells (square-wave-pulse, 300V, 1 × 10 ms), PM1 cells (square-wave-pulse, 350V, 3 × 5 ms, pause between pulse: 0.1s). Cells were transferred into medium-containing 24- or 48-well plates and incubated at 32°C for 24 h as hypothermic conditions were shown to improve gene editing efficiency (15,22–23).
Flow cytometry and sorting
Flow cytometry (FC) and fluorescence-activated cell sorting (FACS) were performed on LSRFortessa (405, 488, 561, 640-nm lasers) or CantoII (407, 488, 633-nm lasers), and on the FACSAria IIIu (407, 488, 561, 633-nm lasers, all BD Bioscience, Heidelberg, Germany), respectively. Data were analyzed using FACSDiva (BD Bioscience) and FlowJo (TreeStar, Ashland, OR) software. Surface staining was performed using the following anti-human antibodies: CD195-APC-Cy7 (BD, 557755); CD195-PerCP-Cy5.5 (BioLegend, San Diego, CA, 313716); CD3-Pacific Blue (BioLegend, 300431), CD4-FITC (BD, 550628), CD8-APC (BD, 557834), CD45-FITC (BD, 555482), CD34-PE (BD, 555822).
Standard, quantitative and gene-editing frequency digital PCR (GEF-dPCR)
Standard PCR to assess CCR5Δ32 status of T-cell donors was carried out as described (24).
To confirm gene editing on the molecular level we carried out quantitative (qPCR) and digital (droplet) PCR (dPCR). Sequences for CCR5-specific primer and probes are listed in Supplementary Table S2. qPCR was performed using the iTaq Universal Probes Supermix (Biorad) and the following program on the Mx3000P-cycler (Stratagene, La Jolla, CA): 60 s 95°C, 35x [5 s 95°C; 60 s 60°C]. For dPCR the ddPCR Supermix for Probes (Biorad), the Qx100 Droplet Generator (Biorad) and the Qx100 Droplet Reader (Biorad) were employed. PCR amplification was performed on the T-Gradient UNO (Biometra, Göttingen, Germany): 10 min 95°C, 40x [30 s 94°C; 60 s 60°C], 98°C 10 min.
Lentiviral vector production and transduction
Gibbon-ape-leukemia-virus-envelope (GALV-env) pseudotyped LeGO-S vectors encoding TSapphire were generated as previously described (25). For CCR5-tropic HIV-env (BaL-env) lentiviral particles encoding mCherry (LeGO-C) (26) viral supernatants were produced with 6 μg pcDNA3_BaL instead of GALV-env. Vector-particle containing supernatants were titrated on PM1 cells in the presence of 8 μg/ml DEAE-dextran as described (26).
HIV infection assay
At day 3 post activation, primary T cells were negatively selected for CD8 prior to HIV infection. To do so, CD3/CD28 Dynabeads were removed and CD8+ cells were depleted using the CD8 Microbeads Kit (Miltenyi Biotech) according to the manufacturer's instructions. CD4+ cells were electroporated as described above. At day 4 post electroporation, 2 × 106/ml CD8-depleted lymphocytes were resuspended in RPMI1640 supplemented with 1% FCS, 2 mM L-glutamine, 0.05mg/ml penicillin/streptomycin and 1 μg polybrene (Sigma-Aldrich) in 12-well plates. After addition of CCR5-tropic HIV-1 BaL-Luc (200 ng p24 antigen) or CXCR4-tropic HIV-1 NL4/3-Luc (20 ng p24), plates were centrifuged (730xg) for 90 min at 22°C. Following spinoculation, cells were maintained in RPMI1640 (10% FCS, 2mM L-glutamine, 0.05 mg/ml Penicillin/Streptomycin) supplemented with hIL-2 [500U]. At days 4 or 12 post-spinoculation, aliquots of 106 cells were washed twice in PBS (without Ca2+ and Mg2+) and lysed for luciferase assays performed according to the manufacturer's instructions (Luciferase Assay System from Promega) and for isolation of genomic DNA (gDNA) using the QIAmp-Blood DNA-Mini-Kit (Qiagen).
Next-generation sequencing (NGS) and bioinformatics analysis
For next-generation sequencing genomic regions of interest were amplified with primers containing the Nextera-transposase sequence. In a second PCR, ILMN indices and sequencing adapters were added to the fragments to unambiguously identify samples (for sequences of oligos please see Supplementary Table S2). PCR products were analyzed and purified by gel electrophoresis, isolated with QIAquick Gel-Extraction Kit (Qiagen) and pooled at equimolar ratios. Sequencing and bioinformatics analysis were performed by MicroSynth (Balgach, Switzerland). NGS data were corrected for background (Supplementary Figure S1).
[methyl-3H]-thymidine-incorporation assay
4 days after electroporation, primary T lymphocytes were seeded in 96-well plates at 2 × 105 cells/well in their standard medium. To determine proliferation capacity after CCR5 knock-out, T cells were reactivated with Dynabeads Human T-Activator CD3/CD28 for 72 h as recommended. Proliferation was determined by 3H-thymidine-incorporation assay as described (27,28).
RESULTS
CCR5-specific TALEN constructs
Gene editing of the CCR5 locus using different designer nucleases was reported, albeit often at relatively low efficiencies (13,16,29). To overcome this limitation we designed a novel, codon-optimized CCR5-specific TALEN (CCR5-Uco-TALEN) (Figure 1A). Both TALEN arms recognize 19-bp target sequences within the CCR5 gene corresponding to the very short first intracellular loop of CCR5 (Figure 1B), a region expected to be sensitive for amino-acid deletions or substitutions (30,31). A search for potential off-target sites using the Paired-Target Finder (19) identified the closest sequence (harboring six mismatches) within the CCR2 gene (Figure 1C) and a second one in the MUC16 gene (10 mismatches) (see below).
Figure 1.

CCR5-Uco-TALEN design. (A) Schematic representation of TALE-repeat variable di-residues used for recognition of the CCR5 locus. (B) Schematic representation of CCR5 conformation (modified from Dong et al. (31)), and the region targeted by CCR5-Uco-TALEN (red). TM = transmembrane region, EL = extracellular loop, ICL = intracellular loop. (C) TALEN binding sites in the genomic on-target (CCR5) and possible off-target (CCR2) loci.
We first evaluated the potency of CCR5-Uco-TALEN in direct comparison with a previously published (13) CCR5 TALEN and a codon-optimized version (CCR5-Mco-TALEN) thereof (Supplementary Table S3). To do so, we quantified CCR5 knockout upon plasmid transfection into CCR5+ reporter cells (32). CCR5-Uco-TALEN outcompeted both the original CCR5 TALEN (13; data not shown) and CCR5-Mco-TALEN (69% versus 32.5% CCR5 knockout after delivery of 2 × 10 μg TALEN plasmids—both arms) (Supplementary Figure S2).
Efficient TALEN delivery, particularly into primary cells, remains a challenge (33,34). We recently introduced a high-efficiency, electroporation-based protocol for transfer of TALEN mRNAs into T lymphocytes (21). As mRNA electroporation has several advantages with respect to clinical applications, we here aimed at applying this technique to TALEN-mediated CCR5 knockout in primary T cells. To do so, we inserted the CCR5-Uco-TALEN in the same backbone successfully used for in-vitro transcription of TCR-TALENs (21). Based thereon, we were able to produce large amounts of mRNA by in-vitro transcription. It is of note that the only modifications introduced into the in-vitro transcribed mRNA are a cap-structure and a poly(A)-tail available in standard IVT-kits.
mRNA electroporation for TALEN delivery into the T-cell line PM1
To adapt the mRNA-electroporation protocol (21) for CCR5-Uco-TALEN, we first established a CCR5-positive reporter T-cell line susceptible toward electroporation with mRNA. To this aim we employed CD4+ PM1 cells widely used in HIV-infection assays (35). As the bulk culture of PM1 cells showed heterogeneous CCR5 expression, we derived single-cell clones expressing both, CD4 and CCR5, by FACS (Supplementary Figure S3). We applied eGFP mRNA to identify optimal electroporation conditions (Supplementary Figure S4) (21).
Using these conditions, we electroporated PM1 cells with CCR5-Uco-TALEN mRNA. We observed very high gene-editing frequencies (up to 94%) as determined by NGS (Figure 2A and B). In a kinetics study, we could demonstrate that on the molecular level CCR5 knockout was essentially completed three days after electroporation (Figure 2A). In a dose-effect experiment, we found that the rate of NHEJ-mediated mutations directly correlated with the loss of CCR5 expression as measured by FC (Figure 2B and C).
Figure 2.

Characterization of CCR5-Uco-TALEN in the HIV-susceptible T-cell line PM1. (A) NHEJ-induced mutations at the CCR5 locus over time. PM1 cells were electroporated with 5 μg mRNA of each CCR5-Uco-TALEN arm and genomic DNA was harvested at the indicated time points. NHEJ frequency was determined by next-generation sequencing (NGS). (B) Correlation between loss of CCR5 expression and accumulation of NHEJ-induced mutations in dependence on amounts of electroporated CCR5-Uco-TALEN mRNA. CCR5 expression was determined by flow cytometry (FC), NHEJ frequency was determined by NGS. (C) Loss of CCR5 expression after electroporation with increasing mRNA amounts of CCR5-Uco-TALEN determined by FC. (D) + (E) BL2-compatible HIV-infection assay. Cells were transduced with both, LeGO-S encoding TSapphire pseudotyped with GALV-env and LeGO-C2 encoding mCherry pseudotyped with CCR5-tropic gp160 (BaL). mCherry positivity representing HIV-susceptibility was normalized against GALV infectibility. (D) FC data of PM1 cells electroporated with either no TALEN-mRNA (mock) or the indicated amounts of mRNA of each Uco-TALEN-arm (L+R). (E) Quantification of CCR5-Uco-TALEN mediated protection against HIV infection in relation to mRNA doses, normalized against GALV.
We next wanted to determine the impact of CCR5 knockout on susceptibility toward HIV infection. We first established a biosafety-level-2 (BL2) compatible infection assay. To this aim we produced gp160 (BaL)-enveloped lentiviral vectors encoding mCherry (LeGO-pC2) (26), which, as HIV, require CCR5 for entry. As control, we simultaneously transduced the cells with TSapphire-encoding GALV-pseudotyped LeGO-S. That pseudotype is not dependent on CCR5, but utilizes the unrelated Pit-1 receptor (36). As shown in Figure 2D, susceptibility toward (CCR5-dependent) LeGO-pC2 transduction strongly decreased after treatment with Uco-TALEN-mRNA (see Q2, Q4), whereas transduction rates for CCR5-independent LeGO-S remained stable. Quantification of infectability (Figure 2E) showed protection rates of up to 86% for cells treated with 5 μg of each CCR5-Uco-TALEN arm (L+R) compared to homodimeric controls (5 μg CCR5-Uco-L, only).
Quantitative and digital PCR for detection of NHEJ-induced mutations
After successful high-level CCR5 knockout in PM1 cells, we proceeded with primary T cells. CCR5 expression is very variable among different donors (see below, Figure 4B) limiting the informative value of FC analysis. Despite some methodological constraints, NGS of designer-nuclease treated genomes can be a robust high-throughput assay to quantify mutation frequencies at the genomic level. However, this method is labor- and cost-intensive, in particular for small data sets. To overcome this limitation, we aimed at establishing an alternative, reliable detection method based on multiplex real-time PCR.
Figure 4.
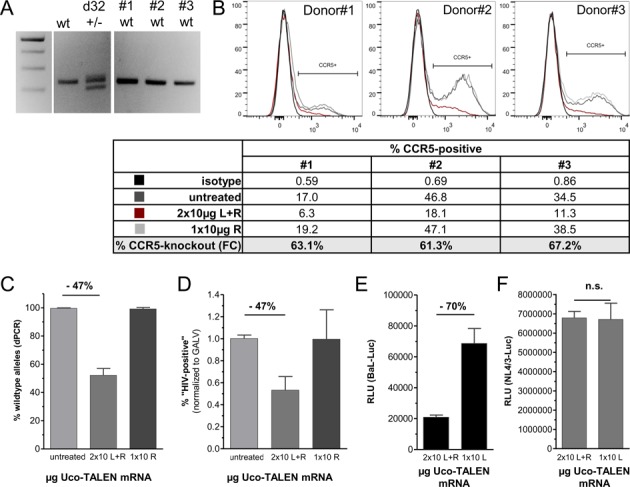
CCR5-Uco-TALEN enable high-level gene knockout leading to efficient protection against R5-tropic HIV independent of initial CCR5 expression. (A) Four blood donors were tested by qualitative PCR for presence of the CCR5Δ32 allele (24). One donor (d32) was heterozygous for CCR5Δ32, three donors (#1–3) were homozygous for wild-type CCR5. (B) Consistent gene editing by CCR5-Uco-TALEN despite high inter-individual variations in CCR5 expression levels. CCR5 expression in individual donors varied between 17.0% and 46.8% as assessed by FC (compare FACS plots and lane ‘untreated’ in the Table). Correspondingly, we observed large differences in CCR5 expression after CCR5-Uco-TALEN (lane ‘2 × 10 μg Uco L+R’ in the Table), whereas homodimeric treatment with one single TALEN arm (‘1 × 10 μg Uco R’) did not result in changes of CCR5-positive cells. Importantly, relative changes in CCR5 levels upon CCR5-Uco-TALEN were almost identical (61–67%) for all three individual T-cell donors, independent of initial CCR5 expression (lane ‘% CCR5 knockout (FC)’). (C) Digital PCR performed on gDNA samples isolated from cells treated in (B) indicated app. 50% gene editing on the DNA level (n = 6; P < 0.0005). (D) In line with dPCR data, transduction with gp160-pseudotyped lentiviral vectors was suppressed by 47% (n = 3; P < 0.0005). (E) Sample #2 was tested in a short-term infection assay with replication-competent CCR5-tropic luciferase expressing HIV-1 (BaL-Luc) after CD8 depletion. Viral replication measured based on luciferase activity was strongly suppressed as compared to the homodimeric control (n = 6, P < 0.0001). (F) No impact of CCR5 editing on infection by CXCR4-tropic HIV-1 (NL4/3-Luc) as estimated for the same sample (n = 6, P = 0.7425). ‘2 × 10 L+R' indicates transfection with both TALEN arms ( = functional TALEN), ‘1 × 10 L' indicates transfection with the right TALEN arm, only ( = homodimeric control).
We first amplified the CCR5 target locus by PCR using gDNA derived from CCR5-Uco-TALEN treated primary T cells. PCR products representing single alleles were analyzed by Sanger sequencing to identify indels (Figure 3A). Based on these data, we designed two differently labeled TaqMan probes specific for (i) the region not affected by TALEN cutting and subsequent NHEJ-mediated repair (‘HEX probe’) and (ii) the region affected by mutation in the analyzed allele (‘FAM probe’). We reasoned that this set of probes enables the simultaneous detection of total alleles and mutated alleles in one duplex-qPCR.
Figure 3.

Quantitative and digital PCR for detection of NHEJ-induced mutations. (A) Alignment of sequenced alleles of CCR5-Uco-TALEN treated primary T cells to define affected alleles. Based on the deletion pattern probes were designed to detect (i) an evidently unaffected region (HEX probe, green) and (ii) a region mutated by NHEJ in all analyzed alleles (FAM probe, blue). (B) ΔCt values for qPCR reactions performed on CCR5-Uco-TALEN treated primary T cells using the probes shown in (A). Cells were electroporated with the indicated amount of CCR5-Uco-TALEN mRNA and genomic DNA was isolated from polyclonal cultures at day 4 post electroporation. Although significant changes in ΔCt values were detected, absolute quantification of mutated alleles was not possible. qPCR was performed in triplicates. (C) GEF-dPCR on samples used in (B) results in two types of amplicons differing in their capability to bind the fluorescent probes displayed in (A). Resulting HEX+/FAM+-double positive droplets represent wild-type allels, whereas HEX+/FAM- droplets stand for edited alleles. Quantitative analysis of GEF-dPCR can be performed using a 2D dot plot as shown; thresholds used to distinguish GEF-dPCR-positive and –negative signals (dots) are indicated. (D) + (E) Comparison of relative knockout frequencies as determined by (D) dPCR versus flow cytometry (FC) or (E) dPCR versus NGS. For experimental settings, please refer to (B). ***P < 0.001, ****P < 0.0005 (tested by one-way-ANOVA).
To test this, we performed qPCR on gDNA from primary T cells treated with increasing amounts of CCR5-Uco-TALEN mRNA and indeed detected significant changes in ΔCt values compared to untreated cells (Figure 3B). However, despite the clear dose-dependency we were not able to absolutely quantify mutation frequencies in TALEN-treated cells based on qPCR. In contrast, digital PCR has been shown to allow precise quantification of target sequences, including allele frequencies (37–39). Hence, we characterized the same samples by duplex digital PCR designed to simultaneously detect wild-type and mutated alleles (Figure 3C). Here, unmodified (wild-type) CCR5 alleles could be expected to be HEX/FAM-double positive and therefore located in the upper right quadrant of a 2D dot plot (Figure 3C, Supplementary Figure S5). Consequently, numbers of FAM- and HEX-positive droplets in the upper right quadrant of the dot plot should be (nearly) identical. On the contrary, treatment with Uco-CCR5-TALEN was anticipated to impair binding of the NHEJ-sensitive probe and thereby result in a decrease or loss of the FAM signal without affecting the HEX signal. Thus, CCR5-gene edited alleles should fall out of the upper right quadrant—either as intermediate ‘raindrops’ not present in untreated samples or as completely FAM-negative droplets. As shown in Figure 3C and Supplementary Figure S5 this was exactly the case—numbers of FAM-negative droplets and intermediate ‘raindrops’ strongly increased in TALEN-treated samples in correlation to the applied amount of TALEN-mRNA. Accordingly, we proposed that numbers of unaffected alleles after TALEN treatment can be directly quantified based on remaining HEX+/FAM+ droplets in the upper right quadrant of the dot plot (Figure 3C, Supplementary Figure S5).
To validate this approach, we compared the obtained dPCR data with CCR5-knockout levels as assessed by FC and NGS. Indeed, we found almost perfect correlation between dPCR and FC (Figure 3D) and, more importantly, NGS results (Figure 3E). The highest variance between mutation frequencies measured by NGS versus dPCR was as low as 4% (Figure 3E—2×5 μg CCR5-Uco-TALEN mRNA).
Taken together these data clearly demonstrate that the dPCR technique developed here is a very dependable approach to determine mutation frequencies after Uco-CCR5-TALEN treatment. Notably, as compared to NGS dPCR is a fast and inexpensive method requiring low amounts of gDNA (<66ng/sample) and no bioinformatics.
CCR5-Uco-TALEN mRNA mediated high-level knockout of CCR5 resulting in suppression of HIV infection in primary T cells
We next aimed at testing effectiveness of CCR5-Uco-TALEN on primary T cells from different donors. First we tested T-cell samples from healthy individuals for presence of the CCR5Δ32 mutation using locus-specific PCR (24) (Figure 4A). T cells from three different donors with wild-type CCR5 were electroporated with 10 μg of each CCR5-Uco-TALEN arm after prestimulation. Three days after electroporation ( = d6 post activation) CCR5 expression of treated and untreated cells was determined by FC (Figure 4B). Expression levels of CCR5 varied between 17.0% (Donor#1) and 46.8% (Donor#2) (Figure 4B, Table). This is in good agreement with previous reports on intra-individual differences in CCR5 expression (40). In principle, it cannot be excluded that CCR5-expression levels impact accessibility for TALE nucleases. However, in our study treatment of T cells with CCR5-Uco-TALEN resulted in reduction of CCR5 surface expression by 61–67% in all three donors, independent of their initial CCR5-expression status (Figure 4B).
To absolutely quantify CCR5 knockout on the genome level we applied dPCR. In line with the FC results, we observed high-efficiency gene editing in CCR5-Uco-TALEN treated primary T lymphocytes (47%) (Figure 4C).
To verify the impact of CCR5 editing on HIV susceptibility of primary T cells, we again performed the BL2-infection assay using HIV-enveloped lentiviral vectors. Importantly, almost 50% of CCR5-Uco-TALEN treated cells showed resistance toward transduction with HIV-env-LeGO (Figure 4D), which was in excellent accord with the gene-editing frequencies.
Finally, we exposed the CCR5-edited T cells from one of the three donors to replication-competent laboratory strains of HIV, in which nef sequences were substituted by the gene encoding firefly luciferase (HIV-1 strains BaL-Luc and NL4/3-Luc). CCR5-edited T lymphocytes were efficiently (70%) protected from infection with the CCR5-tropic strain BaL in short-term assays (Figure 4E). This protection was mediated by entry inhibition, since infection with the CXCR4-tropic HIV strain NL4/3 was not impaired (Figure 4F).
T-cell viability and functionality, and TALEN off-target activity
Electroporation of primary T cells with mRNAs encoding functional proteins was previously shown to represent a comparatively gentle approach for transient gene delivery (41). In line with these data, we observed very good viability of T lymphocytes after electroporation with CCR5-Uco-TALEN, despite the fact that T cells were incubated at hypothermic conditions [32°C] for 24 h immediately after CD8+ T-cell depletion and electroporation (Supplementary Figure S6A).
To address whether mRNA treatment and/or CCR5 knockout had an immediate impact on TCR-signaling mediated T-cell proliferation, we re-stimulated gene-edited and control T lymphocytes from the above experiments (Figure 4) with CD3/CD28 beads and performed 3H-thymidine incorporation assays. We did not observe any evident impact of CCR5-Uco-TALEN on T-cell proliferation (Supplementary Figure S6B).
Another potential risk of genome editing is the off-target activity of designer nucleases (13,42–43). To address this point, we identified genomic sites with distinct homology to the CCR5 target region and thus a high likelihood for off-target binding. We reasoned that observed DNA editing frequencies at these sites would most probably be indicative for overall off-target activity. As expected, due to its pronounced sequence similarity (84%) the CCR2 gene contained the most probable off-target site (32/38 matched nucleotides). Another potential off-target site was located in the MUC16 gene (Supplementary Figure S7).
In order to quantify the frequency of off-target cutting in the CCR2 gene we PCR-amplified the target regions and performed deep sequencing (8000–16 000 individual sequences). In different samples, we observed sequence editing in 1.13–6.24% of total CCR2 alleles depending on the amount of transfected mRNA (Supplementary Figures S7 and 8). In relation to the number of edited CCR5 alleles, frequencies in the CCR2 locus corresponded to 2.4–11.63% with the highest relative off-target activity observed for high on-target rates (Supplementary Figure S7, Supplementary Table S4). No indel frequencies above background were found in the amplified target region of MUC16 (Supplementary Figure S7).
In order to establish an expedient assay for the detection of off-target activity in the CCR2 locus we set up a digital-PCR assay, in analogy to the one described above for CCR5 (primer sequences are provided in Supplementary Table S2). Again, the dPCR turned out to be very sensitive even at low gene-editing frequencies and showed good correlation with NGS (Supplementary Figure S8).
Selective advantage of CCR5-edited T cells from a CCR5Δ32-heterozygous individual during HIV infection
In the final set of experiments we tested primary T lymphocytes from a donor that eventually turned out to be heterozygous for the CCR5Δ32 allele (Figure 4A). CCR5-Uco-TALEN mediated knockout resulted in a reduction of CCR5 expression by app. 50% as assessed by FC and confirmed by dPCR (Figure 5A). In a short-term infection assay with CCR5-tropic replication-competent HIV-1 (BaL-Luc), infection was strongly (67%) suppressed in CCR5-Uco-TALEN-treated as compared to homodimeric-control cells (Figure 5B). These data were in perfect agreement with previous results obtained for homozygous wild-type T lymphocytes (compare Figure 4E).
Figure 5.

CCR5-Uco-TALEN treated T cells from a CCR5Δ32-heterozygous donor possess a selective advantage against HIV exposure. (A) High-rate CCR5 knockout in CCR5Δ32 heterozygous CCR5-Uco-TALEN treated T cells as assessed by FC (n = 3; P < 0.001 [‘2×10 L+R' versus mock], P < 0.01 [‘2×10 L+R' versus ‘1×10 L']) and dPCR (n = 3; P < 0.0001 [‘2×10 L+R' versus mock, ‘2×10 L+R' versus ‘1×10 L']). (B) CD8-depleted, CCR5Δ32 heterozygous, CCR5-Uco-TALEN treated T cells (‘2×10 L+R') efficiently suppress replication of CCR5-tropic HIV-1 (BaL-Luc) in short-term assay. HIV-1 de-novo infection was measured based on luciferase activity (n = 3; P < 0.01 [2 × 10 versus mock], P < 0.001 [2 × 10 versus 1 × 10]). (C) After incubation for 12 days, HIV replication was almost completely blocked in CCR5-Uco-TALEN treated, but not in homodimeric-treated control cells (n = 3; P = 0.0005). (D) At the end of the 12-day exposure to replication-competent HIV (as shown in (C)), a substantial increase in the percentage of CCR5-gene edited T cells was seen by NGS. On the contrary, no increase in frequencies of cells with mutations at the off-target locus CCR2 was noted.
We next performed a long-term infection assay with replication-competent CCR5-tropic HIV-1 (BaL-Luc). Following infection and 12 days of incubation, viral replication was almost completely abrogated in the CCR5-Uco-TALEN group, but not in the homodimeric-control group (Figure 5C). CCR5-Uco-TALEN treated cells were analyzed by NGS for the presence of indels before and after exposure to the replication-competent HIV-1 strain. Notably, at final analysis we observed a substantial increase in the percentage of cells harboring CCR5 indels, strongly indicating a selective advantage of gene-edited cells during infection (Figure 5D). In contrast, a slight decrease was found in the proportion of cells edited at the CCR2 off-target site (Figure 5D).
DISCUSSION
Virus-entry inhibition is the most promising strategy for preventing or suppressing HIV infection (44). The HIV-coreceptor CCR5 is a particularly interesting target for genetic knockout for several reasons: (i) the pronounced resistance toward HIV infection associated with natural CCR5Δ32 homozygosity, (ii) the documented absence of health problems in several million individuals homozygous for CCR5Δ32 and (iii) the possibility to cure HIV by transferring resistance (‘Berlin patient’) from an allogeneic donor (2,10). Importantly, due to the lack of matched CCR5Δ32-homozygous donors for most patients, but also given its high treatment-related toxicity allogeneic stem cell transplantation is not a realistic therapy option.
Alternatively, designer-nuclease mediated knockout of CCR5 in autologous cells from HIV patients has been suggested. Recently, proof-of-concept was provided by a phase-I study demonstrating safety and feasibility of infusing large numbers of T cells ex-vivo edited with a CCR5-ZFN. Notably, a profound and long-lasting effect on viral load was observed in one of six patients who underwent a treatment interruption. This patient was heterozygous for CCR5Δ32 and showed the highest CCR5-knockout level (28%). These data suggest that sufficiently high knockout rates are obligatory to ensure clinical efficacy (11,12). At the same time, the ‘Berlin patient' (10,45) indicates that efficient and early protection from CCR5-tropic HIV may prevent the switch toward CXCR4-tropic HIV able to infect CCR5-negative cells (46).
Besides ZFNs, alternative designer nucleases with high potential for clinical applications have become available, namely TAL effector nucleases (TALEN), CRISPR/Cas9 nucleases and Cre-recombinase based enzymes (i.e. Tre-recombinase) (47). In spite of differences in their structure and function, all genome-editing systems encounter common challenges. With regard to clinical application the latter include efficient delivery into the given target cells and the necessity to combine high on-target with limited off-target activity.
Here we have introduced a novel, highly active CCR5-specific designer nuclease, CCR5-Uco-TALEN. Due to their size and highly repetitive sequences, transfer of TALENs is particularly difficult (32). To overcome this problem we made use of a recently developed protocol for the efficient transfer of TALENs into T lymphocytes, which is based on mRNA electroporation (21). mRNA transfection has important advantages with regard to clinical application: (i) In certain regulatory environments, mRNA can be considered a pharmaceutical product, whereas virus-based vectors are considered as gene-modified organisms. This might ease requirements for its production and application depending on national regulations. (ii) mRNA-mediated TALEN expression is definitely transient. (iii) mRNA transfection requires only short-term activation of target cells. (iv) mRNA-electroporation technology has already been established under large-scale good manufacturing practice (GMP) conditions (41). (v) In contrast to viral or plasmid vectors, there is no risk of accidental integration potentially leading to insertional mutagenesis and undesired long-term expression.
CCR5-Uco-TALEN was designed to target the very short and hydrophilic first intracellular loop of CCR5 upstream from the natural Δ32 deletion, closely to the site targeted by the ZFN used in the clinical trial (11,30). After plasmid transfection the novel target site facilitated superior activity in direct comparison with a codon-optimized version of a previously published TALEN (13). More importantly, in primary T cells from different donors we regularly obtained gene-editing rates of app. 50%. These rates are similar to those observed after adenoviral gene transfer of the above mentioned CCR5-specific ZFN (30). As indicated by the results of the recent clinical CCR5/ZFN trial, such gene-editing rates might be sufficient to mediate HIV control (11). In line with that prediction, we observed profound protection of gene-edited T cells from infection with CCR5-tropic HIV-1 during short-term exposure to HIV, independently of the initial CCR5 status. We performed one experiment with ‘long-term’ (12 days) exposure to replication-competent virus, where we detected almost complete suppression of viral replication and substantial enrichment of CCR5-edited cells indicating their selective advantage. Subsequent genotyping revealed that the T-cell donor for that particular experiment was heterozygous for the Δ32 deletion, which might have contributed to the fact that only 67% of the CCR5 alleles were found to be edited by NGS.
Interestingly, CCR5-Uco-TALEN mediated CCR5-gene editing resulted in an unexpectedly high proportion of cells that completely lost CCR5 expression and were protected from HIV infection. It is tempting to speculate that deletion of hydrophilic amino acids in the region targeted by CCR5-Uco-TALEN may lead to changes in the protein structure and thus decreased CCR5 expression on the cell surface. In consequence, functional CCR5 knockouts would even occur in case of in-frame indels.
Clinically relevant off-target effects may hinder therapeutic application of designer nucleases. For CCR5-Uco-TALEN only one off-target site with less than 10 mismatches was identified by in-silico analysis. The latter is located in the CCR2 gene and displays high sequence identity (32/38 nucleotides; i.e. 84%), in line with the overall homology between CCR2 and CCR5. Using deep sequencing we observed indels in the CCR2 off-target region at frequencies between 1.1% and 6.2%. Available data points to a disproportionately high increase of off-target activity in relation to CCR5-editing rates. This might indicate some extravagation effect at saturating doses of TALENs supporting the strategy to limit expression strength and duration of designer nucleases in target cells. A second potential off-target site predicted by in-silico analysis was located in the MUC16 gene. For that locus, we did not observe off-target cutting by CCR5-Uco-TALEN. Of course, this does not preclude the presence of further off-target sites for our TALEN. At the same time, it seems reasonable to suggest that off-target activity at the most likely locus, i.e. the CCR2 gene, will be positively correlating with and thus indicative of overall off-target activity.
Functionally, CCR2 is involved in migration and potentially proliferation of immune cells (48). At the same time, given the relatively low numbers of CCR2-edited T cells in the graft, an impact on overall T-cell activity is not anticipated. Actually, we did not observe an evident loss in functionality of CCR5-Uco-TALEN treated T lymphocytes. As noted above, TALENs have been suggested to confer higher specificity with lower probability for off-target cutting and lower cytotoxicity as compared to ZFNs (13,14). To address this point, we calculated the off-target-to-on-target ratio for CCR5-Uco-TALEN in comparison with those CCR5-ZFNs, for which off-target data have been reported. The resulting figures support the proposed higher specificity of TALENs (Supplementary Table S4).
Finally, in this work we have introduced a novel, very sensitive digital-PCR technique for assessing the rate of NHEJ-mediated editing of the CCR5-Uco-TALEN target site in the CCR5 gene. Importantly, indel frequencies assessed by this method were in excellent agreement with respective data obtained by NGS. In contrast to NGS, dPCR is simple, fast, comparatively cheap, and does not require bioinformatics. As we have shown in this work for the off-target site in the CCR2 gene, the dPCR technique can easily be adapted to other gene loci targeted by designer nucleases and might therefore be useful to concurrently assess both their on- and off-target activities.
In conclusion, we have presented a highly efficient CCR5-specific TALEN that confers gene-editing rates so far not reached for this target with TALE nucleases. In the view of previous data with CCR5-specific ZFNs (11) we suggest that this new designer nuclease, CCR5-Uco-TALEN, in conjunction with the proposed gentle and efficient delivery method, i.e. mRNA electroporation, has a promising potential for clinical translation. Based on recent clinical experience (11,46), early protection of autologous T cells from CCR5-tropic HIV strains seems to be the most rational clinical strategy to efficiently suppress viral replication and prevent dissemination of HIV-1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors are indebted to Tanja Sonntag, Almut Uhde, Johannes Polke, Dagmara Nelson (UKE Isotope lab core facility), Melanie Lachmann, Susanne Roscher and Regine Thiele (all three: UKE FACS Core Facility) for expert technical assistance. We are also grateful to the Dept. of Transfusion Medicine of UKE for kind help. We would like to thank Tanja Stahl and Anita Badbaran for their valuable input in regard to dPCR and NGS, and Kristoffer Riecken for helpful discussions. Additionally, we thank Christian Hundsrucker and Sebastian Strempel from Microsynth for technical expertise. We also wish to thank Rainer Loew (EUFETS) for initially providing mRNA.
FUNDING
Deutsche Forschungsgemeinschaft [SFB841/SP2 to B.F., in part]; Forschungsförderung Medizin (FFM) program of the Medical faculty of the UMC Hamburg-Eppendorf [to U.M., in part]. Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG).
Author contribution: U.M. designed the study, performed most experiments, analyzed and interpreted data and wrote the manuscript; R.M. performed T-cell experiments, S.H. established cell lines and assays used in this study, P.A. performed proliferation assay, B.B. helped with establishment of mRNA transfection protocol; I.H. and J.H. performed experiments with infectious HIV; B.F. designed the study, analyzed data and wrote the manuscript. All authors have read the manuscript and confirmed their authorship.
Conflict of interest statement. U.M. and B.F. have submitted a patent application for CCR5-Uco-TALEN (decision pending). None of the other authors has declared competing interests.
REFERENCES
- 1.Dragic T., Litwin V., Allaway G., Martin S., Huang Y., Nagashima K., Cayanan C., Maddon P., Koup R., Moore J., et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 2.Hütter G., Nowak D., Mossner M., Ganepola S., Müssig A., Allers K., Schneider T., Hofmann J., Kücherer C., Blau O., et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 3.Liu R., Paxton W.A., Choe S., Ceradini D., Martin S.R., Horuk R., MacDonald M.E., Stuhlmann H., Koup R.A., Landau N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 4.Dean M., Carrington M., Winkler C., Huttley G.A., Smith M.W., Allikmets R., Goedert J.J., Buchbinder S.P., Vittinghoff E., Gomperts E., et al. Genetic restriction of HIV-1 Infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 5.Burke B.P., Boyd M.P., Impey H., Breton L.R., Bartlett J.S., Symonds G.P., Hütter G. CCR5 as a natural and modulated target for inhibition of HIV. Viruses. 2014;6:54–68. doi: 10.3390/v6010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fätkenheuer G., Pozniak A.L., Johnson M.A., Plettenberg A., Staszewski S., Hoepelman A.I.M., Saag M.S., Goebel F.D., Rockstroh J.K., Dezube B.J., et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat. Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 7.Schackman B.R., Gebo K.A., Walensky R.P., Losina E., Muccio T., Sax P.E., Weinstein M.C., Seage G.R., Moore R.D., Freedberg K.A. The lifetime cost of current human immunodeficiency virus care in the United States. Med. Care. 2006;44:990–997. doi: 10.1097/01.mlr.0000228021.89490.2a. [DOI] [PubMed] [Google Scholar]
- 8.Chen R.Y., Accortt N.A., Westfall A.O., Mugavero M.J., Raper J.L., Cloud G.A., Stone B.K., Carter J., Call S., Pisu M., et al. Distribution of health care expenditures for HIV-infected patients. Clin. Infect. Dis. 2006;42:1003–1010. doi: 10.1086/500453. [DOI] [PubMed] [Google Scholar]
- 9.Deeks S.G., Phillips A.N. HIV infection, antiretroviral treatment, ageing, and non-AIDS related morbidity. BMJ. 2009;338:a3172. doi: 10.1136/bmj.a3172. [DOI] [PubMed] [Google Scholar]
- 10.Allers K., Hütter G., Hofmann J., Loddenkemper C., Rieger K., Thiel E., Schneider T. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 11.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G., et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kay M.A., Walker B.D. Engineering cellular resistance to HIV. N. Engl. J. Med. 2014;370:968–969. doi: 10.1056/NEJMe1400593. [DOI] [PubMed] [Google Scholar]
- 13.Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mussolino C., Alzubi J., Fine E.J., Morbitzer R., Cradick T.J., Lahaye T., Bao G., Cathomen T. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014;42:6762–6773. doi: 10.1093/nar/gku305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J., et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 16.Kim H., Um E., Cho S.-R., Jung C., Kim H., Kim J.-S. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods. 2011;8:941–943. doi: 10.1038/nmeth.1733. [DOI] [PubMed] [Google Scholar]
- 17.Lusso P., Cocchi F., Balotta C., Markham P.D., Louie A., Farci P., Pal R., Gallo R.C., Reitz M.S. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehse B., Schade U.M., Li Z., Uhde A., Koch S., Goller B., Rüger R., Fehse N., Stockschläder M., Zander A.R. Highly-efficient gene transfer with retroviral vectors into human T lymphocytes on fibronectin. Br. J. Haematol. 1998;102:566–574. doi: 10.1046/j.1365-2141.1998.00785.x. [DOI] [PubMed] [Google Scholar]
- 19.Doyle E., Booher N., Standage D., Voytas D., Grendel V., VanDyk J., Bogdanove A. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanjana N.E., Cong L., Zhou Y., Cunniff M.M., Feng G., Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berdien B., Mock U., Atanackovic D., Fehse B. TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene Ther. 2014;21:539–548. doi: 10.1038/gt.2014.26. [DOI] [PubMed] [Google Scholar]
- 22.Doyon Y., Choi V.M., Xia D.F., Vo T.D., Gregory P.D., Holmes M.C. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat. Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 23.Carlson D., Tan W., Lillico S., Stverakova D., Proudfoot C., Christian M., Voytas D., Long C., Whitelaw B., Fahrenkrug S. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gade-Andavolu R., Comings D.E., MacMurray J., Rostamkhani M., Cheng L.S.-C., Tourtellotte W.W., Cone L.A. Association of CCR5 Δ32 deletion with early death in multiple sclerosis. Genet. Med. 2004;6:126–131. doi: 10.1097/01.gim.0000127274.45301.54. [DOI] [PubMed] [Google Scholar]
- 25.Mock U., Thiele R., Uhde A., Fehse B., Horn S. Efficient lentiviral transduction and transgene expression in primary human B cells. Hum. Gene Ther. Methods. 2012;23:408–415. doi: 10.1089/hgtb.2012.160. [DOI] [PubMed] [Google Scholar]
- 26.Weber K., Bartsch U., Stocking C., Fehse B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol. Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 27.Abramowski P., Steinbach K., Zander A.R., Martin R. Immunomodulatory effects of the ether phospholipid edelfosine in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2014;274:111–124. doi: 10.1016/j.jneuroim.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Abramowski P., Otto B., Martin R. The orally available, synthetic ether lipid edelfosine inhibits T cell proliferation and induces a type I interferon response. PLoS One. 2014;9:e91970. doi: 10.1371/journal.pone.0091970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badia R., Riveira-Muñoz E., Clotet B., Esté J.A., Ballana E. Gene editing using a zinc-finger nuclease mimicking the CCR5Δ32 mutation induces resistance to CCR5-using HIV-1. J. Antimicrob. Chemother. 2014;69:1–5. doi: 10.1093/jac/dku072. [DOI] [PubMed] [Google Scholar]
- 30.Perez E.E., Wang J., Miller J.C., Jouvenot Y., Kim K.A., Liu O., Wang N., Lee G., Bartsevich V. V, Lee Y.-L., et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong H.-F., Wigmore K., Carrington M.N., Dean M., Turpin J.A., Howard O.M.Z. Variants of CCR5, which are permissive for HIV-1 infection, show distinct functional responses to CCL3, CCL4 and CCL5. Genes Immun. 2005;6:609–619. doi: 10.1038/sj.gene.6364247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mock U., Riecken K., Berdien B., Qasim W., Chan E., Cathomen T., Fehse B. Novel lentiviral vectors with mutated reverse transcriptase for mRNA delivery of TALE nucleases. Sci. Rep. 2014;4:6409. doi: 10.1038/srep06409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holkers M., Maggio I., Liu J., Janssen J.M., Miselli F., Mussolino C., Recchia A., Cathomen T., Gonçalves M.A. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2012;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chuah M.K., Vandendriessche T. Optimizing delivery and expression of designer nucleases for genome engineering. Hum. Gene Ther. Methods. 2013;24:329–332. doi: 10.1089/hgtb.2013.166. [DOI] [PubMed] [Google Scholar]
- 35.Wilen C.B., Wang J., Tilton J.C., Miller J.C., Kim K.A., Rebar E.J., Sherrill-Mix S.A., Patro S.C., Secreto A.J., Jordan A.P.O., et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011;7:e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller A.D., Chen F. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sykes P.J., Neoh S.H., Brisco M.J., Hughes E., Condon J., Morley A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 38.Vogelstein B., Kinzler K.W. Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl T., Böhme M., Kröger N., Fehse B. Digital PCR to assess haematopoietic chimaerism after allogeneic stem cell transplantation. Exp. Hematol. 2015 doi: 10.1016/j.exphem.2015.02.006. doi:10.1016/j.exphem.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Wu L., Paxton W.A., Kassam N., Ruffing N., Rottman J.B., Sullivan N., Choe H., Sodroski J., Newman W., Koup R.A., et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almåsbak H., Rian E., Hoel H.J., Pulè M., Wälchli S., Kvalheim G., Gaudernack G., Rasmussen A.-M. Transiently redirected T cells for adoptive transfer. Cytotherapy. 2011;13:629–640. doi: 10.3109/14653249.2010.542461. [DOI] [PubMed] [Google Scholar]
- 42.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabriel R., Lombardo A., Arens A., Miller J.C., Genovese P., Kaeppel C., Nowrouzi A., Bartholomae C.C., Wang J., Friedman G., et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 44.Von Laer D., Hasselmann S., Hasselmann K. Gene therapy for HIV infection: what does it need to make it work. J. Gene Med. 2006;8:658–667. doi: 10.1002/jgm.908. [DOI] [PubMed] [Google Scholar]
- 45.Hütter G., Schneider T., Thiel E. Transplantation of selected or transgenic blood stem cells - a future treatment for HIV/AIDS. J. Int. AIDS Soc. 2009;12:10. doi: 10.1186/1758-2652-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kordelas L., Verheyen J., Beelen D.W., Horn P.A., Heinold A., Kaiser R., Trenschel R., Schadendorf D., Dittmer U., Esser S. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N. Engl. J. Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 47.Weber N.D., Aubert M., Dang C.H., Stone D., Jerome K.R. DNA cleavage enzymes for treatment of persistent viral infections: recent advances and the pathway forward. Virology. 2014;454–455:353–361. doi: 10.1016/j.virol.2013.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tylaska L., Boring L., Weng W., Aiello R., Charo I.F., Rollins B.J., Gladue R.P. CCR2 regulates the level of MCP-1/CCL2 in vitro and at inflammatory sites and controls T cell activation in response to alloantigen. Cytokine. 2002;18:184–190. doi: 10.1006/cyto.2002.1031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


