SUMMARY
We provide evidence that the Unc-51-like kinase 1 (ULK1) is activated during engagement of the Type I IFN receptor (IFNR). Our studies demonstrate that the function of ULK1 is required for gene transcription mediated via IFN-stimulated response elements (ISRE) and IFNγ activation site (GAS) elements and controls expression of key IFN-stimulated genes (ISGs). We identify ULK1 as an upstream regulator of p38α MAPK and establish that the regulatory effects of ULK1 on ISG expression are mediated possibly by engagement of the p38 MAPK pathway. Importantly, we demonstrate that ULK1 is essential for antiproliferative responses and Type I IFN-induced antineoplastic effects against malignant erythroid precursors from patients with myeloproliferative neoplasms. Together, these data reveal a role for ULK1 as a key mediator of Type I IFNR-generated signals that control gene transcription and induction of antineoplastic responses.
INTRODUCTION
Type I Interferons (IFNs) are cytokines with important antitumor, antiviral, and immunomodulatory properties (González-Navajas et al., 2012; Bekisz et al., 2013, Platanias, 2005). These cytokines have clinical activity against viral infections, and several human malignancies (Hervas-Stubbs et al., 2011; Bekisz et al., 2013; Kotredes et al., 2013; Platanias, 2013; Stein et al., 2013). Despite continuing efforts to define the precise mechanisms by which IFNs generate antineoplastic responses, the sequence of events and the specific coordination of different IFN-activated signaling cascades required for such responses remain incompletely defined (Platanias, 2013).
All Type I IFNs bind to Type I IFN receptor (IFNR), the engagement of which activates JAK-STAT (Janus activated kinase-signal transducer and activator of transcription) signaling pathways (Platanias, 2005; Stark and Darnell, 2012; Ivashkiv and Donlin, 2014). Beyond these pathways, activation of several other IFN-signaling cascades occurs during engagement of IFN receptors, including the p38 mitogen-activated protein kinase (MAPK) pathway (Uddin et al., 1999; Li et al., 2004), the phosphatidylinositol 3-kinase (PI3K)-AKT pathway (Kaur et al., 2008a; Kaur et al., 2008b), and the mammalian target of rapamycin complex 1 (mTORC1) and mTORC2 signaling cascades (Kaur et al., 2007; Kaur et al., 2012; Kaur et al., 2014). The functions of these pathways are essential for optimal transcription and/or mRNA translation of various interferon-stimulated genes (ISGs) that are needed for the induction of IFN-responses (Kaur et al., 2007; Kaur et al., 2008b; Kaur et al., 2014).
Although the relevance and functional importance of mTORC1 signals in promoting functional IFN-responses is well established (Kaur et al., 2007), the precise mechanisms and distinct roles of downstream mTORC1 effectors in the process remain to be defined. Previous work has demonstrated that activated mTORC1 prevents autophagy by phosphorylation of serine 757 (Ser757) of Unc-51-like kinase 1 (ULK1), and by disrupting the interaction between ULK1 and AMP-activated protein kinase (AMPK) (Kim et al., 2011). ULK1 and ULK2 are the closely related mammalian homologs of the serine/threonine autophagy-related (ATG) protein kinase ATG1, the first identified ATG product in yeast, and both of which are involved in the regulation of autophagy (Alers et al., 2012). In the present study we examined whether ULK1 is engaged in IFN-signaling and what role it plays in the induction of Type I IFN-mediated responses. Our studies provide evidence implicating ULK1 in Type I IFN-signaling and transcriptional activation of ISGs, and define a mechanism by which such ULK1-mediated activity occurs in the IFN-system, possibly involving regulation/activation of p38 MAPK.
RESULTS
Type I IFN-induced phosphorylation of ULK1 on serine 757 is AKT-dependent
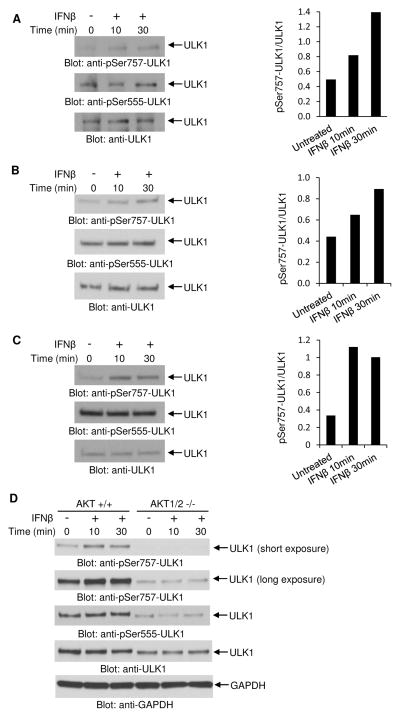
In initial studies we examined whether Type I IFN treatment induces phosphorylation of ULK1 in IFN-sensitive cells. Treatment of different IFN-sensitive malignant hematopoietic cell lines (U937, KT-1, and U266) with human IFNβ induced phosphorylation of ULK1 at the mTORC1 phosphorylation site (Kim et al., 2011), Ser757 (Figs. 1A–C). In contrast, there was no IFNβ-dependent induction of phosphorylation of ULK1 at Ser555 (Figs. 1A–C), the amino acid residue phosphorylated by AMPK (Bach et al., 2011). Previous studies have established that the serine/threonine protein kinase AKT is activated downstream of PI3K (Kaur et al., 2008a) and mTORC2 (Kaur et al., 2012) during engagement of the Type I IFNR and regulates downstream engagement of mTORC1 (Kaur et al., 2008b). We examined whether engagement of ULK1 in IFN-signaling requires upstream AKT activity. For this, we determined the effects of IFNβ treatment on the phosphorylation of ULK1 using Akt1/2 double knockout (Akt1/2−/−) mouse embryonic fibroblasts (MEFs) (Peng et al., 2003). Treatment of Akt1/2+/+ MEFs with mouse IFNβ resulted in phosphorylation of ULK1 on Ser757 (Fig. 1D). However, IFNβ-induced phosphorylation of ULK1 on Ser757 was defective in Akt1/2−/− MEFs (Fig. 1D). In contrast, there was no IFNβ-dependent induction of phosphorylation of ULK1 at Ser555 in both Akt1/2+/+ and Akt1/2−/− MEFs (Fig. 1D). Together, these data suggest that upstream AKT activity is essential for regulation of Type I IFN-induced phosphorylation of ULK1 on Ser757.
Figure 1. Engagement of the Type I IFN receptor results in phosphorylation of ULK1 at serine 757.
(A–C) Effects of IFNβ on the phosphorylation of ULK1 in (A) U937, (B) KT-1, and (C) U266 cell lines. (Left panels) Cells were left untreated or were treated with human IFNβ for 10 or 30 minutes, as indicated. Lysates were analyzed by SDS-PAGE and immunoblotted with an antibody against the phosphorylated form of ULK1 on Ser757. Equal amounts of cell lysates from the same experiment were resolved separately by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of ULK1 on Ser555 or ULK1. (Right panels) Bands were quantified by densitometry using ImageJ software, and data are expressed as ratios of pSer757-ULK1 over total ULK1. (D) Effects of IFNβ on the phosphorylation of ULK1 in Akt1/2+/+ and Akt1/2−/− MEFs. Cells were left untreated or were treated with mouse IFNβ for 10 or 30 minutes, as indicated. Lysates were analyzed by SDS-PAGE and immunoblotted with an antibody against the phosphorylated form of ULK1 on Ser757 and GAPDH. Short and longer exposures of p-Ser757 ULK1 from the same blot are shown. Equal amounts of cell lysates from the same experiment were resolved separately by SDS-PAGE and immunoblotted with antibodies against the phosphorylated form of ULK1 on Ser555 or ULK1.
Requirement of ULK1/2 activity for transcriptional activation of Type I IFN-stimulated genes
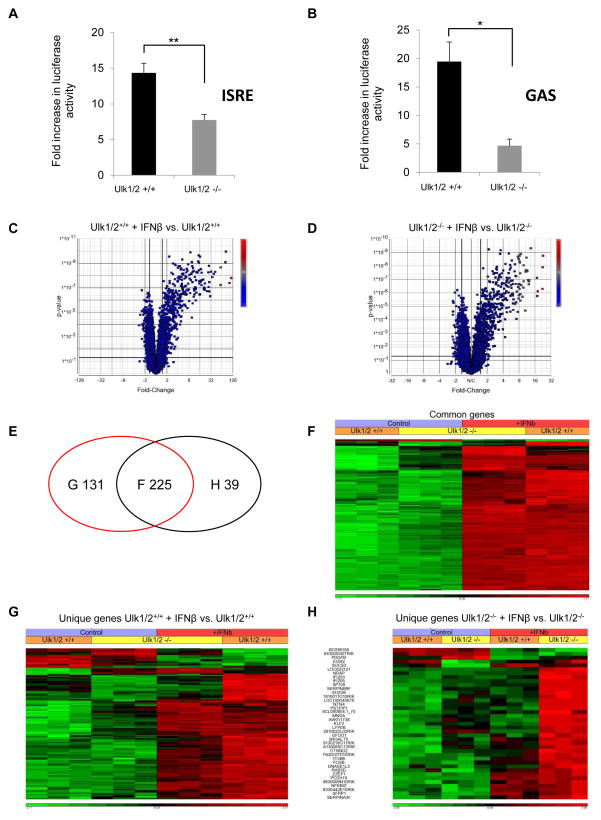
Our data establish that ULK1 is activated via the Type I IFNR. As the generation of IFN-responses depends on expression of ISGs and their protein products (Darnell et al., 1994; Stark and Darnell, 2012; Cheon et al., 2014), we initiated studies to determine whether ULK1 controls Type I IFN-dependent gene transcription. Initially, we determined whether ULK1/2 activity is required for transcriptional activation via IFN-stimulated response elements (ISRE) or IFNγ activation site (GAS) elements in luciferase reporter assays, using MEFs with targeted disruption of both the Ulk1 and Ulk2 genes. For these studies we used Ulk1/2+/+ and Ulk1/2−/− MEFs (Cheong et al., 2011), as ULK1 and ULK2 kinases were previously shown to have at least partially redundant functions in fibroblasts (Kundu et al., 2008; Lee and Tournier, 2011). IFNβ-dependent transcriptional activation via either ISRE or GAS elements was significantly reduced in the absence of Ulk1 and Ulk2 expression (Fig. 2A and 2B). To further define the role of ULK1/2 in ISG regulation, we sought to identify IFN-inducible genes differentially expressed in Ulk1/2+/+ and Ulk1/2−/− MEFs, using genome Illumina microarrays. Using principal component analysis (PCA) of differentially expressed genes we found that the three biological replicates of gene expression profiles cluster together, and that the control and IFNβ treated Ulk1/2+/+ and Ulk1/2−/− cells represent separated groups (Fig. S1A). Comparison of the transcriptomic profiles revealed IFN-inducible expression of 356 genes in Ulk1/2+/+ MEFs (Fig. 2C), whereas only 264 genes were inducible in Ulk1/2−/− MEFs (Fig. 2D). Notably, although 225 genes were induced in both Ulk1/2+/+ and Ulk1/2−/− MEFs (Figs. 2E and 2F), the expression of 84 of these genes was significantly higher in the Ulk1/2+/+ MEFs compared to the Ulk1/2−/− MEFs (Fig. 2F and Table S1, genes highlighted in red). 131 genes were found to be induced only in the Ulk1/2+/+ MEFs (Fig. 2E and 2G, Table S2), whereas 39 unique genes were induced in the Ulk1/2−/− MEFs (Fig. 2E and 2H, Table S3). The differentially expressed genes between Ulk1/2+/+ and Ulk1/2−/− MEFs were classified among biochemical pathways using the KEGG database (Tables S4–S6). Most of the genes whose transcriptional induction by IFNβ-treatment was defective or decreased in Ulk1/2−/− MEFs could be classified among biochemical pathways that regulate adaptive and innate immunity, as well as antiviral, antiproliferative, and pro-apoptotic responses (Table S4, genes highlighted in red and green; and Table S5). In contrast, genes induced by IFNβ only in Ulk1/2−/− MEFs could be classified among biochemical pathways that are involved in cell adhesion and DNA transcription (Table S6). A functional gene network, generated using IPA 2014 software, is shown in supplemental Fig. S1B and demonstrates relationships among the 215 genes the expression of which is defective or decreased in the absence of Ulk1/2.
Figure 2. Targeted disruption of Ulk1/2 gene expression impairs IFNβ-dependent gene transcription.
(A–B) Ulk1/2+/+ and Ulk1/2−/− MEFs were transfected with an ISRE-luciferase construct (A) or an 8X GAS-luciferase construct (B). 42 hours after transfection, the cells were incubated for 6 hours in the presence or absence of mouse IFNβ, and luciferase activity was measured. Data are expressed as fold increase of luciferase activity in response to IFNβ-treatment over control untreated samples for each condition. Bar graphs show means ± SE of four independent experiments for panel A and three independent experiments for panel B, using technical triplicates in each experiment. Statistical analyses were performed using Student’s t-test (*p < 0.05; **p < 0.01). (C–H) Differential expression of ISGs in Ulk1/2+/+ and Ulk1/2−/− MEFs. Cells were incubated in the presence or absence of mouse IFNβ for 6 hours. The gene expression profiles of untreated MEFs were compared with those of IFNβ-treated MEFs in three independent experiments, using MouseWG-6 v2.0 Expression BeadChips and Illumina iScan. (C–D) Volcano plots of differentially expressed genes after IFNβ treatment are shown for (C) Ulk1/2+/+ and (D) Ulk1/2−/− MEFs. 356 genes were differentially expressed between untreated and IFNβ-treated Ulk1/2+/+ cells (panel C), whereas 264 genes were differentially expressed between untreated and IFNβ-treated Ulk1/2−/− cells (panel D). (E) Venn diagram showing the gene expression overlap existing between differentially expressed genes in Ulk1/2+/+ MEFs (red ellipse) and in Ulk1/2−/− MEFs (black ellipse) after treatment with IFNβ. (F) Hierarchical clustering of differentially expressed genes in both Ulk1/2+/+ and Ulk1/2−/− MEFs upon IFNβ treatment. Differences in the effects of IFNβ treatment between Ulk1/2+/+ and Ulk1/2−/− MEFs are seen for 87 genes, 84 of these genes are characterized by a less efficient IFNβ-driven transcription in Ulk1/2−/− MEFs (see list of genes in Table S1). (G) Hierarchical clustering of differentially expressed genes only in Ulk1/2+/+ MEFs (see list of genes in Table S2). (H) Hierarchical clustering of differentially expressed genes only in Ulk1/2−/− MEFs (see list of genes in Table S3). All annotations presented here are based on statistical analyses and are presented with p values after FDR. Only annotations with p values < 0.05 are shown. See also Tables S4–S6 and Figure S1.
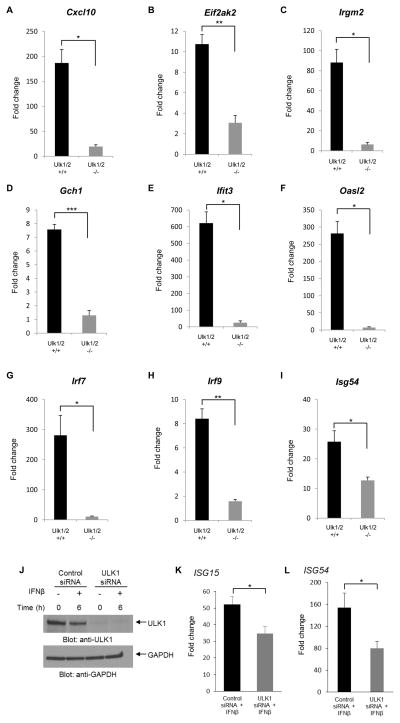
In further studies, we confirmed the requirement for ULK1/2 activity in the expression of several key ISGs, using quantitative RT-PCR (Fig. 3A–I). Among the genes the expression of which was found defective in the absence of Ulk1/2 were Cxcl10 (Zhang et al., 2005) and Eif2ak2 (García et al., 2006; McAllister and Samuel, 2009), both of which are involved in the induction of antiviral effects and control of apoptosis. The induction of several other genes the function of which is necessary for generation for IFN-biological responses was also defective in Ulk1/2−/− cells, including Irgm2 (Hunn et al., 2008), Gch1 (Rani et al., 2007; Alp and Channon, 2004); Ifit3 (Schmeisser et al., 2010; Liu et al., 2011); Oasl2 (Zhu et al., 2014); Irf7 (Sharma et al., 2003; Honda et al., 2005; Colina et al., 2008); Irf9 (Darnell et al., 1994; van Boxel-Dezaire et al., 2006); and Isg54/Ifit2 (Yang et al., 2012) (Fig. 3A–I). To determine whether ULK1 expression is required for transcriptional activation of IFN-induced genes in other cell types, studies were performed with human U937 cells in which ULK1 was knocked down using specific siRNAs (Fig. 3J). We found decreased IFN-inducible mRNA expression of ISG15 and ISG54 (Fig. 3K and L), genes with crucial roles in the induction of IFN-responses (Lenschow et al., 2007; Yang et al., 2012), further establishing a key role for ULK1 in Type I IFN-signaling.
Figure 3. Requirement of ULK1/2 activity for IFNβ-dependent transcription.
(A–I) Ulk1/2+/+ and Ulk1/2−/− MEFs were left untreated or were treated with mouse IFNβ for 6 hours. (J–L) U937 human leukemia cells were transfected with either control or ULK1 siRNAs. 24 hours after transfection, the cells were either left untreated or were incubated with human IFNβ for 6 hours. (J) Levels of ULK1 protein expression are shown, using Western immunoblotting, probing with ULK1-specific antibody. The immunoblot was also probed for GAPDH as a loading control. (A–I, K and L) Quantitative RT-PCR analyses of the relative mRNA expression of ISGs after IFNβ stimulation in (A–I) Ulk1/2+/+ and Ulk1/2−/− MEFs and in (K and L) U937 cells after siRNA transfection are shown. Expression levels of the indicated genes were determined using GAPDH for normalization. Data are expressed as fold change over untreated samples (A–I) or control siRNA untreated samples (K and L) and bar graphs represent means ± SE of three independent experiments for panels A, B, C, D, E, F, and L and four independent experiments for panels G, H, I, and K. Statistical analyses were performed using Student’s t-test between treated groups (*p < 0.05; **p < 0.01; ***p < 0.001).
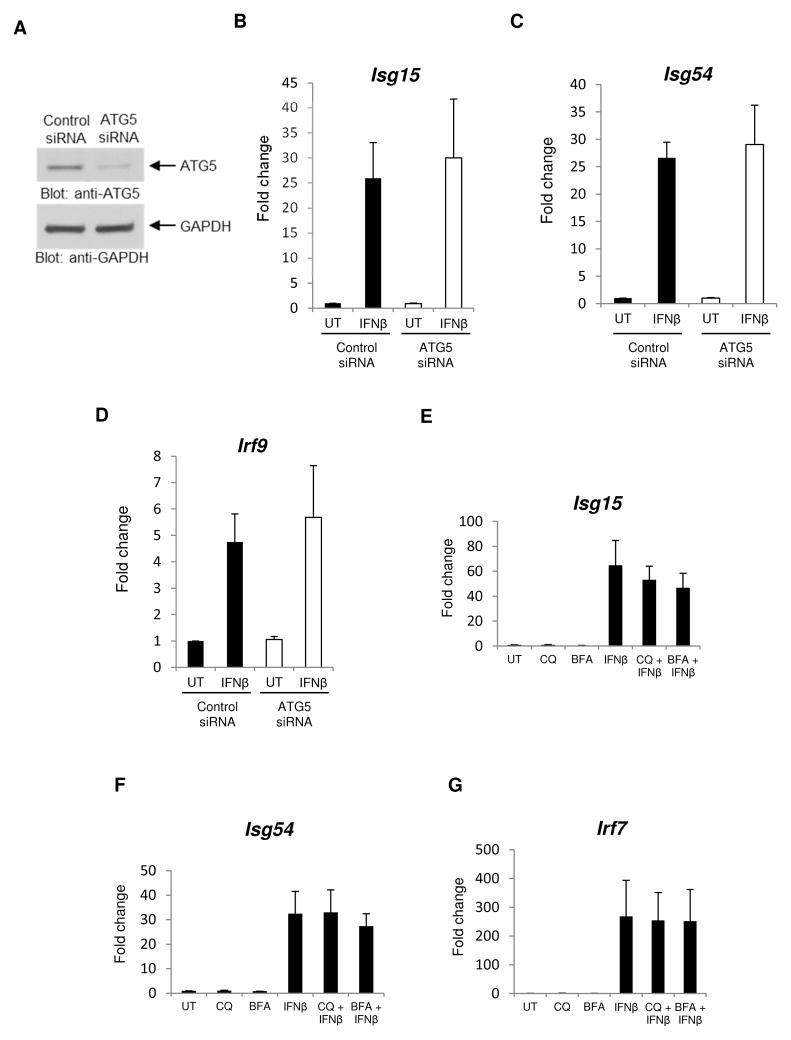
It has been extensively established that ULK1 regulates the induction of autophagy (Kim et al., 2011; Russel et al., 2013). In addition, there is also evidence for IFN-dependent induction of autophagy (Ambjørn et al., 2013; Schmeisser et al., 2014). We determined whether inhibition of autophagy modulates IFN-dependent transcriptional activation. The effects of siRNA-mediated knockdown of ATG5, a protein required in early stages of autophagosome formation (Mizushima et al., 2001), were initially determined. No significant differences on IFN-dependent Isg15, Isg54, and Irf9 mRNA expression were observed between control cells and cells in which ATG5 was knocked down (Figs. 4A–D). Consistent with this, treatment of cells with the autophagy inhibitors chloroquine or bafilomycin A1 (Klionsky et al., 2008) did not significantly affect ISGs mRNA expression (Figs. 4E–G), further establishing that ULK1 promotes Type I IFN-dependent transcriptional activation of key target genes in an autophagy-independent manner.
Figure 4. ULK1/2 activity regulates ISG transcription in an autophagy-independent manner.
(A) Total cell lysates from MEFs transfected with either control siRNA or ATG5-specific siRNA were resolved by SDS-PAGE and immunoblotted with anti-ATG5 or anti-GAPDH-specific antibodies, as indicated. (B–D) MEFs transfected with control siRNA or ATG5-specific siRNA were treated with mouse IFNβ for 6 hours, and mRNA expression for the indicated genes was assessed by quantitative RT-PCR, using GAPDH for normalization. Data are expressed as fold change over control siRNA untreated (UT) samples and bar graphs represent means ± SE of three independent experiments. (E–G) MEFs were treated with chloroquine (CQ), bafilomycin A1 (BFA) and/or mouse IFNβ. mRNA expression for the indicated genes was assessed by quantitative RT-PCR, using GAPDH for normalization. Data are expressed as fold change over control untreated (UT) cells and bar graphs represent means ± SE of 5 independent experiments for panels E and G and 3 independent experiments for panel F.
ULK1 mediates Type I IFN-dependent activation of p38 MAPK
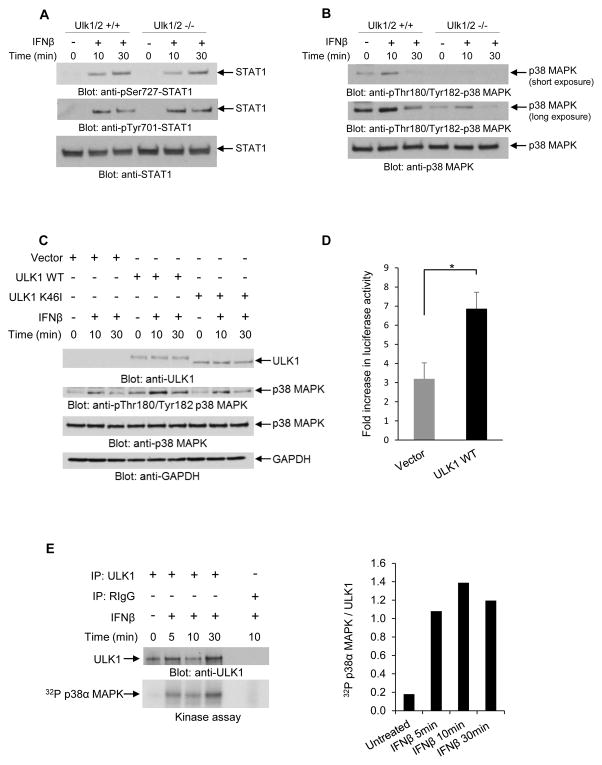
To define the mechanisms by which ULK1 activity may regulate Type I IFN-dependent transcriptional activation, we examined whether it is required for activation of pathways that control Type I IFN-dependent transcriptional activation of sensitive genes. As activation of Stat1 is essential for transcriptional induction of genes that contain ISRE or GAS elements in their promoters (Stark and Darnell, 2012), we first determined if phosphorylation/activation of Stat1 is Ulk1/2-dependent in MEFs. IFNβ-dependent phosphorylation of Stat1 on serine 727 and on tyrosine 701 was inducible in both Ulk1/2+/+ and Ulk1/2−/− MEFs (Fig. 5A), indicating that the functions of ULK1/2 are not required for Type I IFN-induced activation of Stat1. As Stat1 is a key Type I IFN-regulated protein involved in complexes that control both ISRE- and GAS-dependent transcription, these studies suggested that the effects of ULK1/2 on Type I IFN-inducible transcriptional activation are independent of modulation of the classical STAT pathways.
Figure 5. ULK1/2 activity is required for Type I IFN-dependent activation of the p38 MAPK.
(A) Ulk1/2+/+ and Ulk1/2−/− MEFs were treated with mouse IFNβ as indicated. Equal amounts of total cell lysates were resolved by SDS-PAGE and consecutively immunoblotted with antibodies against pSer727-STAT1, pTyr701-STAT1, and STAT1, as indicated. (B) Ulk1/2+/+ and Ulk1/2−/− MEFs were treated with mouse IFNβ for 10 or 30 minutes, as indicated. Equal amounts of total cell lysates were resolved by SDS-PAGE and consecutively immunoblotted with antibodies against the phosphorylated form of p38 MAPK on Thr180/Tyr182 and against p38 MAPK. Short and longer exposures of p-Thr180/Tyr182 p38 MAPK from the same blot are shown. (C) Ulk1/2−/− MEFs were transfected with pcDNA6.2 empty vector (Vector), or ULK1 WT, or ULK1-K46I (kinase inactive) plasmids, as indicated. 48 hours after transfection, the cells were treated with mouse IFNβ for 10 and 30 minutes, as indicated. Equal amounts of total cell lysates were resolved by SDS-PAGE and consecutively immunoblotted with the indicated antibodies. (D) Ulk1/2−/− MEFs were transfected with pcDNA6.2 empty vector (Vector), or ULK1 WT plasmids, as indicated. 24 hours after transfection, these cells were transfected with an 8X GAS-luciferase construct. 42 hours later, the cells were incubated for 6 hours in the presence or absence of mouse IFNβ, and luciferase activity was measured. Data are expressed as fold increase of luciferase activity in response to IFNβ-treatment over control untreated samples for each condition. Bar graphs show means ± SE of three independent experiments using technical triplicates in each experiment. Statistical analyses were performed using Student’s t-test (*p < 0.05). (E) U937 cells were starved overnight prior to IFNβ treatment, and then were treated with human IFNβ for 5, 10, and 30 minutes, as indicated. After cell lysis, equal amounts of protein were immunoprecipitated with either ULK1-specific antibody or control non-immune rabbit IgG (RIgG). In vitro kinase assays to detect ULK1 activity were subsequently performed on the immunoprecipitates, using p38α MAPK recombinant inactive protein as an exogenous substrate. (Left and top panel) Immunoblot demonstrating total immunoprecipitated ULK1 expression used in each condition for the in vitro kinase assay. (Left and bottom panel) Autoradiography film demonstrating ULK1-induced phosphorylation of p38α MAPK after IFNβ treatment is shown. Note: a lane between ULK1 and RIgG immunoprecipitates was loaded with 1x loading dye for best separation between the wells. (Right) Bands were quantified by densitometry using ImageJ software, and data are expressed as ratios of 32P p38α MAPK over total immunoprecipitated ULK1. See also Figure S2.
Previous studies have demonstrated that the p38 MAPK pathway complements the function of STAT pathways and plays a critical role in Type I IFN-induced transcriptional activation via both ISRE and GAS elements (Uddin et al., 1999; Uddin et al., 2000; Li et al., 2004). We examined the possibility that the effects of ULK1/2 on ISG transcription are mediated by effects on p38 MAPK activity. We found that IFNβ-induced phosphorylation of p38 MAPK was substantially decreased in Ulk1/2−/− MEFs as compared to Ulk1/2+/+ MEFs (Fig. 5B). Additionally, this defective p38 MAPK phosphorylation could be rescued by ectopic re-expression of wild-type ULK1 (ULK1 WT), but not a kinase-inactive ULK1 mutant (ULK1 K46I) (Egan et al., 2011) (Fig. 5C). Complementation of Ulk1/2−/− MEFs with ULK1 WT also restored IFN-induced transcriptional activation via GAS elements (Fig. 5D). Moreover, we found that p38α MAPK is phosphorylated by ULK1 kinase in in vitro assays (Figs. 5E and S2), further suggesting that p38 MAPK mediates the regulatory effects of Ulk1 in Type I IFN-dependent transcriptional activity.
ULK1/2 activity is required for induction of Type I IFN-dependent antiviral and antiproliferative effects
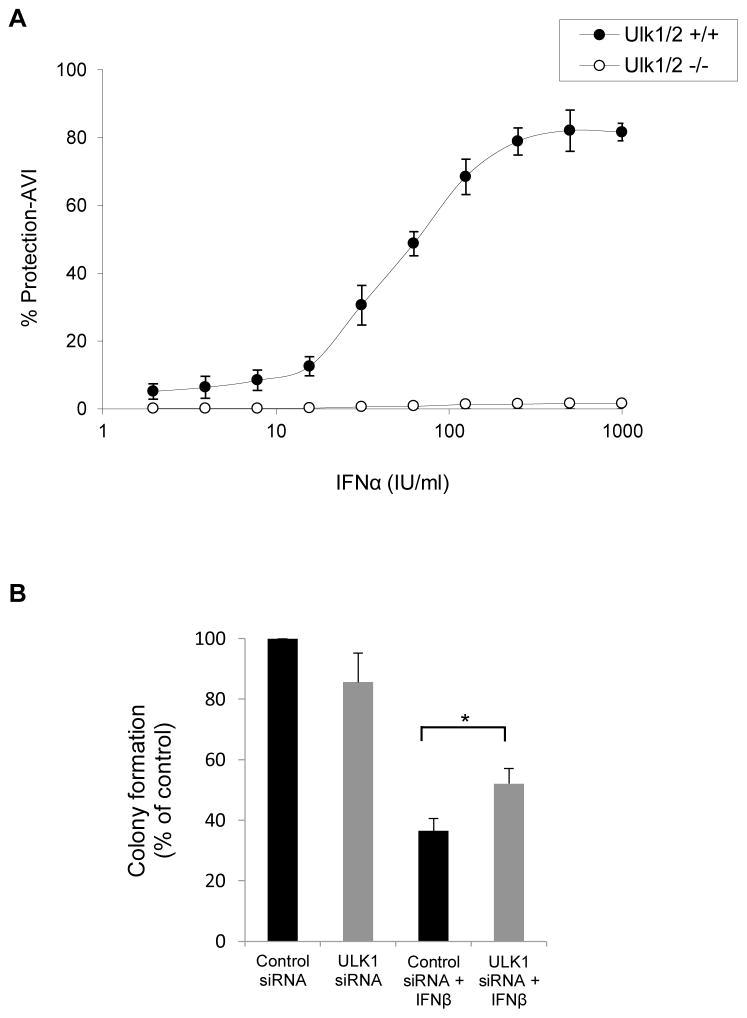
To define whether the defective Type I IFN-dependent gene transcription seen in Ulk1/2−/− MEFs has consequences in the generation of antiviral responses by Type I IFNs, the ability of mouse IFNα to protect cells from encephalomyocarditis virus (EMCV) infection was compared in Ulk1/2+/+ and Ulk1/2−/− MEFs. Ulk1/2−/− MEFs were much more sensitive to EMCV infection compared to Ulk1/2+/+ MEFs (Fig. S3). Specifically, at least a 50-fold reduction in infective dose was required to induce comparable EMCV-induced cytopathic effects (CPE) in the Ulk1/2−/− MEFs compared with the Ulk1/2+/+ MEFs (Fig. S3). Moreover, IFNα-induced antiviral dose-response data indicated that Ulk1/2−/− cells are also less responsive to mouse IFNα-treatment compared with the Ulk1/2+/+ cells (Fig. 6A). Together, these data show that Ulk1/2−/− MEFs are more sensitive to viral infection and less sensitive to the antiviral effects of IFNα compared with Ulk1/2+/+ MEFs, establishing that engagement of Ulk1/2 is required for the control of Type I IFN-generated antiviral responses in MEFs.
Figure 6. Regulation of Type I IFN responses by ULK1/2 activity.
(A) Ulk1/2+/+ and Ulk1/2−/− MEFs were seeded in quadruplicate in individual wells of 96 well plates, and then treated with mouse IFNα for 16 hours, as indicated. Ulk1/2+/+ cells were subsequently challenged with a 1:2×104 dilution of encephalomyocarditis virus (EMCV), and the Ulk1/2−/− cells with a 1:106 dilution of EMCV. EMCV-induced cytopathic effects (CPE) were determined 24 hours later. The data are expressed as percent protection from CPE adjusted to viral infective dose (% Protection-AVI). Values shown represent means ± SD of two independent experiments. See also Figure S3. (B) U937 cells were transfected with either control siRNA or ULK1 siRNA, and leukemic CFU-L colony formation was assessed in clonogenic assays in methylcellulose in the presence or absence of human IFNβ, as indicated. Data are expressed as percent colony formation of control siRNA-transfected untreated cells, and bar graphs represent means ± SE of five independent experiments. Statistical analysis was performed using Student’s t-test (*p < 0.05).
We also determined whether ULK1 is required for the generation of Type I IFN-antiproliferative responses. For this purpose, we performed studies involving siRNA-mediated knockdown of ULK1 in U937 cells, followed by assessment of IFNβ-inhibitory responses on leukemic CFU-L colony growth. As shown in Fig. 6B, inhibition of expression of ULK1 partially reversed suppression of CFU-L colony formation by IFNβ-treatment, implicating ULK1 as a signaling element required for the generation of Type I IFN-antiproliferative effects.
ULK1 is critical for IFN-regulation of normal hematopoiesis and the generation of IFN-responses in MPNs
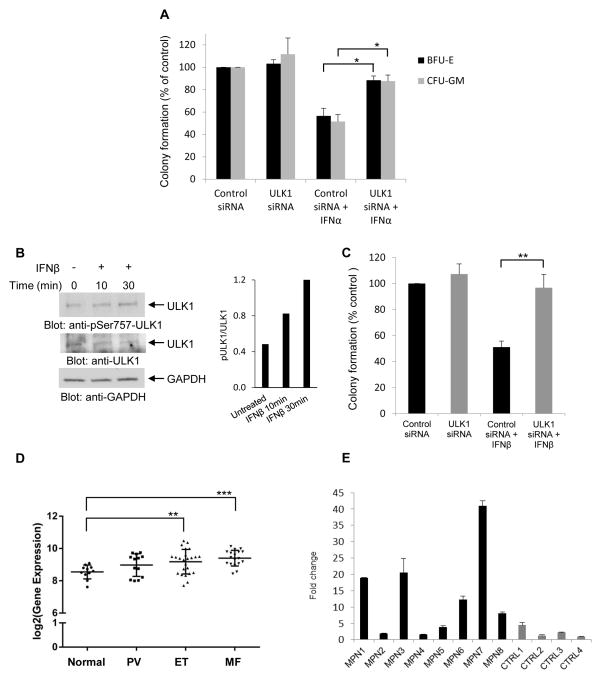
As Type I IFNs are potent regulators of normal hematopoiesis (Platanias, 2005), in subsequent studies we determined if engagement of ULK1 activity is necessary for the generation of growth inhibitory responses on normal CD34+-derived hematopoietic precursors. For this purpose, we used specific siRNAs to knockdown ULK1 expression in primary normal human bone marrow progenitors and examined the effects of this knockdown on the inhibitory effects of IFNα on CD34+-derived erythroid and myeloid precursors. As expected, treatment with human IFNα suppressed the growth of normal myeloid (CFU-GM) and early erythroid (BFU-E) progenitors in clonogenic assays in methylcellulose (Fig. 7A). However, these suppressive effects were reversed by ULK1 knockdown (Fig. 7A), indicating key and essential roles for ULK1 in the control of normal hematopoiesis by Type I IFNs.
Figure 7. Requirement of ULK1 for the regulatory effects of Type I IFNs on normal and malignant human hematopoiesis.
(A) Normal human bone marrow-derived CD34+ cells were transfected with either control siRNA or ULK1 siRNA, and then incubated in clonogenic assays in methylcellulose in the presence or absence of human IFNα, and myeloid (CFU-GM) and erythroid (BFU-E) progenitor colony formation were assessed. Data are expressed as percent colony formation of control siRNA-transfected untreated cells, and represent means ± SE of three independent experiments. Statistical analysis was performed using Student’s t-test (*p < 0.05). (B) (Left panel) Serum-starved circulating primary peripheral blood mononuclear cells from a patient with PV were treated with human IFNβ for 10 or 30 minutes, as indicated. Total cell lysates were resolved by SDS-PAGE and consecutively immunoblotted with p-S757 ULK1 and ULK1 antibodies. The immunoblot was also probed for GAPDH as a loading control. (Right panel) Bands were quantified by densitometry using ImageJ software, and data are expressed as ratios of p-ULK1/ULK1. (C) Peripheral blood mononuclear cells from patients with PV were transfected with either control siRNA or ULK1 siRNA and the effects of human IFNβ on malignant erythroid (BFU-E) colony formation were assessed by clonogenic assays in methylcellulose. Data are expressed as percent colony formation of control siRNA-transfected untreated cells, and represent means ± SE of five independent experiments, using cells from 5 different patients with Polycythemia Vera. (D) Box plot shows gene expression of ULK1 in neutrophils in a large independent cohort of normal individuals (normal, n = 11), and patients with PV (n = 13), ET (n = 24), and MF (n = 18). Statistical analyses were performed using Student’s t-test comparing expression in each MPN group to the normal group (**p < 0.01; ***p < 0.001). (E) Quantitative RT-PCR analysis for ULK1 mRNA expression in neutrophils isolated from different patients with MPNs (MPN1-8) and age-matched controls (CTRL1-4). Expression levels of the ULK1 gene was determined using GAPDH for normalization. Data are expressed as fold change over CTRL4 and bar graphs represent means ± SD for two independent assays.
In further studies, we examined whether the engagement of ULK1 by the Type I IFNR is essential for generation of antineoplastic responses. It is well established that polycythemia vera (PV) and other Philadelphia-chromosome negative MPNs are sensitive to Type I IFN therapy and Type I IFN-treatment is currently used for the treatment of such neoplasms (Tefferi and Vainchenker, 2011; Kiladjian et al., 2011; Cassinat et al., 2014). We determined whether ULK1 is required for generation of Type I IFN-dependent growth inhibitory effects on malignant erythroid progenitors from patients with PV. When primary peripheral blood mononuclear PV cells (PBMCs) were treated with IFNβ, we observed induction of ULK1 phosphorylation on serine 757 (Fig. 7B). Importantly, when the effects of Type I IFN-treatment on malignant erythroid progenitors from 5 different MPN patients were assessed, we found that siRNA-mediated targeted inhibition of ULK1 expression reversed the suppressive effects of IFNβ on primitive malignant erythroid precursors in vitro (Fig. 7C). Thus, ULK1 engagement via the Type I IFNR appears to be essential for generation of antineoplastic effects in MPNs.
We next determined whether ULK1 expression is upregulated in the peripheral blood of MPN patients. Specific analysis for ULK1 gene expression from a previously reported microarray profiling study in neutrophils from a cohort of patients with chronic MPNs (Rampal et al., 2014) showed that ULK1 expression is increased in different groups of MPN patients, including patients with PV, essential thrombocythemia (ET), and myelofibrosis (MF) (Fig. 7D). The upregulation of expression of ULK1 was also seen in another independent group of MPN patients (Fig. 7E) using RT-PCR analysis for ULK1 mRNA expression, establishing upregulation of ULK1 expression in MPNs and suggesting a mechanism to explain the unique sensitivity of these neoplasms to the effects of Type I IFNs.
DISCUSSION
Type I IFNs are cytokines with important biological effects in vitro and in vivo and have been used extensively in the treatment of various malignancies, viral syndromes, and autoimmune disease in humans (Borden et al., 2007; Cheon et al., 2014; Platanias, 2013). The important biological and therapeutic properties of Type I IFNs reflect the induction of expression of key genes via the Type I IFNR that mediate diverse biological responses, including antineoplastic, immune modulatory, and antiviral effects (Cheon et al., 2014; Kroczynska et al., 2014). The precise mechanisms accounting for the transcriptional activation and mRNA translation of such ISGs have been the focus of extensive work that led to the original discovery of JAK-STAT pathways (Darnell et al., 1994; Platanias, 2005; Stark et al., 2012). Besides the classical JAK-STAT signaling cascades, the Type I IFNR engages several other cellular pathways in normal and malignant cells (Platanias, 2005; van Boxel-Dezaire et al., 2006; González-Navajas et al., 2012), the functions of which are required for the generation of Type I IFN-dependent biological responses (Platanias, 2005). Among them, the p38 MAPK cascade appears to act as an auxiliary pathway, necessary for optimal transcription of ISGs, without modulating elements of the STAT-pathway (Platanias, 2005). Despite a better understanding of the mechanisms of Type I IFN-signaling in the past decades, several questions have remained unanswered, particularly the mechanisms that define signaling specificity. Although in some cases selective use of the Type I IFNR subunits may account for differential gene expression among distinct Type I IFNs (de Weerd et al., 2013), the precise effectors of such specific pathways and the potential interactions with other cytokine receptors remain to be defined (Kaur et al., 2013). Also, there is a need to identify cellular elements linking pathways that control IFN-dependent gene transcription to the ones that regulate subsequent mRNA translation of ISGs, as this should allow better understanding of the mechanisms that account for specificity of expression of ISG products.
In the present study we provide evidence that the kinase ULK1 is phosphorylated by engagement of the Type I IFNR at serine 757, a mTORC1 phosphorylation site known to inhibit ULK1 in pathways that control the initiation of autophagy (Kim et al., 2011). Furthermore, our data show that ULK1 is activated after engagement of the Type I IFNR and that its activated form can, either directly or through intermediate kinases, phosphorylate the p38 MAPK in immune complex kinase assays in vitro. This suggests that during Type I IFN-treatment, the pro-autophagic functions of ULK1 are blocked, and instead, ULK1 activity is possibly re-directed towards regulation of the p38 MAPK pathway. Consistent with this, Type I IFN-inducible activation of the p38 MAPK pathway is defective in cells with targeted disruption of the Ulk1/2 genes and appears to result in defective downstream ISG transcription. Moreover, Type I IFN-induced activation of p38 MAPK and transcriptional activation via GAS elements is restored by ectopic expression of ULK1 WT protein in Ulk1/2−/− MEFs. Hence, it is possible that for optimal activation of p38 MAPK pathway in the Type I IFN-system, the activities of both ULK1 and MKK3/6 (Li at al., 2005) are required. Notably, the effects of ULK1 on ISG expression appear to reflect selective regulation of the p38 MAPK pathway, as functional engagement of STAT1 is intact in Ulk1/2−/− cells.
In previous studies we had demonstrated that AKT is required for mRNA translation of ISGs but not ISG-transcription (Kaur et al., 2008b). In the present study we provide evidence that Akt1/2 activity is required for Type I IFN-induced ULK1 phosphorylation at serine 757. Furthermore, given that transcription of ISGs is defective in Ulk1/2−/− MEFs and in U937 cells in which ULK1 has been knockdown, it is possible that another Type I IFN-activated kinase(s) act(s) upstream of ULK1 during engagement of the Type I IFNR and concomitant regulation of ULK1 by such kinase may be necessary for the transcriptional activity of ULK1. Some examples of possible kinases are PKC-δ and ERK1 kinases, which have been identified as potential kinases of ULK1 (Mack et al., 2012) and are activated downstream of Type I IFNR (Platanias, 2005), but this remains to be determined in future studies.
The potential engagement of ULK1 in Type I IFN-signaling has important functional implications for the generation of the effects of Type I IFNs. Our studies provide evidence for an involvement of ULK1 in the induction of both Type I IFN-antiviral responses and growth inhibitory activities. They also suggest key and essential roles for ULK1 in the generation of the suppressive regulatory effects of Type I IFNs on normal hematopoiesis, by demonstrating that ULK1 knockdown reverses suppression of myeloid (CFU-GM) and early erythroid (BFU-E) hematopoietic progenitors. Most importantly, our data identify ULK1 as an essential element for the generation of the antineoplastic effects of Type I IFNs on primitive malignant hematopoietic precursors from patients with PV, an MPN where IFN-treatment has major clinical activity (Tefferi et al., 2011). Remarkably, when expression of ULK1 mRNA was specifically analyzed in a large cohort of patients with different MPNs, we found significant increases in ULK1 expression in different subtypes of MPNs, including PV, essential thrombocytosis, and myelofibrosis. There is prior evidence that the p38 MAPK pathway is involved in the generation of Type I IFN-antileukemic effects (Mayer et al., 2001), while other studies have shown that p38 MAPK activation is essential for the generation of the inhibitory effects of Type I IFNs on JAK2V617F-positive hematopoietic progenitor cells from MPN patients (Lu et al., 2010). Our findings suggest a mechanism by which ULK1 and p38 MAPK are engaged in Type I IFN-signaling in MPNs and, most importantly, provide an explanation for the unique sensitivity of these malignancies to the effects of Type I IFNs, due to overexpression of ULK1.
It should be noted that in a recent study, ULK1 was shown to inhibit STING activity, leading to inhibition of IRF3, and consequent suppression of Type I IFN production (Konno et al., 2013). These events appear to function as a negative-feedback control mechanism to prevent sustained transcription of ISGs (Konno et al., 2013) and limit development of IFN-dependent autoimmune inflammatory disorders (Gall et al., 2012). The results of our studies, taken in context with the report of Konno et al. (2013), suggest a dual regulatory role for ULK1 in the control of Type I IFN-responses, acting as a “molecular switch” in the IFN-system that regulates the balance and duration of IFN-biological responses. In this model, ULK1 appears to regulate directly early signals that control ISG expression and induction of Type I IFN-responses. At the same time, a more delayed ULK1-mediated effect appears to be the suppression of Type I IFN production by suppressing STING activity, thus limiting/optimizing the response. The recognition of this unique role for ULK1 should have important clinical-translational implications, as modulation of the ULK1 activity may be used as an approach to selectively enhance the activity of Type I IFNs on MPN cells.
EXPERIMENTAL PROCEDURES
Materials and some of the methods can be found in the Supplemental Information.
Cells and cell culture
U937, U266, and KT-1 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. The immortalized Akt1/2+/+ and Akt1/2−/− MEFs were kindly provided by Dr. Nissim Hay (University of Illinois at Chicago) (Peng et al., 2003). The immortalized Ulk1/2+/+ and Ulk1/2−/− MEFs have been described previously (Cheong et al., 2011). MEFs were cultured in DMEM medium supplemented with 10% FBS and antibiotics. Peripheral blood from patients with PV was collected after obtaining consent approved by the Institutional Review Board of Northwestern University. Additionally, blood samples were collected from patients with MPNs and controls at Albert Einstein School of Medicine, under an Institutional Review Board approved study.
Immunoblotting
Cells were treated, transfected, and lysed as described in the supplemental information. Equal amounts of total cell lysates were resolved by SDS-PAGE and processed for immunoblotting essentially as in our previous studies (Uddin et al., 1999; Kaur et al., 2007; Kroczynska et al., 2009).
Luciferase Reporter Assays
Ulk1/2+/+ and Ulk1/2−/− MEFs were co-transfected with a β-galactosidase expression vector and either an ISRE-luciferase or 8X GAS-luciferase construct. Luciferase activities were measured and normalized to β-galactosidase activity as in previous studies (Uddin et al., 1999). See supplemental information for a detailed description.
Microarray Analysis
Total RNA was isolated from Ulk1/2+/+ and Ulk1/2−/− MEFs untreated or treated with IFNβ (n = 3), and labeled cRNA was hybridized to MouseWG-6 v2.0 Expression BeadChips. The GEO accession number for the microarray data is GSE60778. See supplemental information for a detailed description.
Quantitative RT-PCR
Quantitative RT-PCR was carried out using commercially-available FAM-labeled probes and primers (Applied Biosystems). The mRNA amplification was calculated as previously (Kaur et al., 2007), and the data were plotted as the increase of fold change as compared with control samples. See supplemental information for a detailed description.
Immunoprecipitations and in vitro kinase assays
In vitro kinase assays to detect ULK1 kinase activity in cells treated with IFNβ were performed essentially as in previous studies (Kroczynska et al., 2009). MAPK14 (p38α MAPK) recombinant human inactive protein was used as an exogenous substrate. See supplemental information for a detailed description.
Antiviral Assays
The antiviral effects of mouse IFNα on Ulk1/2+/+ and Ulk1/2−/− MEFs were determined in assays using ECMV as the challenge virus, as in previous studies (Kaur et al., 2012).
Hematopoietic Cell Progenitor Assays
The effects of ULK1 knockdown were assessed in leukemic (CFU-L), erythroid (BFU-E), or myeloid (CFU-GM) colony formation using clonogenic assays in methylcellulose (Stemcell Technologies) in the absence or presence of Type I IFNs as in previous studies (Mayer et al., 2001; Joshi et al., 2009; Kroczynska et al., 2012; Mehrotra et al., 2013; Kaur et al., 2014). See supplemental information for a detailed description.
Statistical Analyses
Student’s t-test was used for comparison of one observation between two groups. One-way analysis of variance (ANOVA) was used to compare more than two groups followed by Tukey’s test. Differences were considered statistically significant when p values were less than 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Nissim Hay (University of Illinois at Chicago, Chicago, IL) for the AKT1/2−/− and Akt1/2−/− MEFs. We also thank Dr. Reuben Shaw (The Salk Institute for Biological Studies, La Jolla, CA) for providing the ULK1 plasmids through Addgene. This work was supported by grants CA77816, CA155566, CA161196, and CA121192 from the National Institutes of Health and by a Merit review grant from the Department of Veterans Affairs, and by grant DPN/MOB109/II/2012. D.P.B. is an employee of BiogenIdec and owns BiogenIdec stock.
Footnotes
Supplemental Information includes three figures, six tables, and Supplemental Experimental Procedures.
AUTHOR CONTRIBUTIONS
Designed research: D.S., L.C.P.; performed research: D.S., S.M., B.K., T.D.B., B.L.S., B.M., J.K.A., B.M-K.; analyzed data: D.S., S.M., E.M.B., B.K., E.M.K, P.L., C.J., N.J, E.N.F., A.K.V., R.L.L.; L.C.P.; provided key materials: D.P.B., C.B.T; wrote the manuscript: D.S., L.C.P; conceived project: L.C.P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alers S, Löffler AS, Wesselborg S, Stork B. The incredible ULKs. Cell Commun Signal. 2012;10:7. doi: 10.1186/1478-811X-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alp NJ, Channon KM. Regulation of endothelial nitric oxide synthase by tetrahydrobiopterin in vascular disease. Arterioscler Thromb Vasc Biol. 2004;24:413–420. doi: 10.1161/01.ATV.0000110785.96039.f6. [DOI] [PubMed] [Google Scholar]
- Ambjørn M, Ejlerskov P, Liu Y, Lees M, Jäättelä M, Issazadeh-Navikas S. IFNB1/interferon-β-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy. 2013;9:287–302. doi: 10.4161/auto.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M, Larance M, James DE, Ramm G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011;440:283–291. doi: 10.1042/BJ20101894. [DOI] [PubMed] [Google Scholar]
- Bekisz J, Sato Y, Johnson C, Husain SR, Puri RK, Zoon KC. Immunomodulatory effects of interferons in malignancies. J Interferon Cytokine Res. 2013;33:154–161. doi: 10.1089/jir.2012.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassinat B, Verger E, Kiladjian JJ. Interferon alfa therapy in CALR-mutated essential thrombocythemia. N Engl J Med. 2014;371:188–189. doi: 10.1056/NEJMc1401255. [DOI] [PubMed] [Google Scholar]
- Cheon H, Borden EC, Stark GR. Interferons and their stimulated genes in the tumor microenvironment. Semin Oncol. 2014;41:156–173. doi: 10.1053/j.seminoncol.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina R, Costa-Mattioli M, Dowling RJ, Jaramillo M, Tai LH, Breitbach CJ, Martineau Y, Larsson O, Rong L, Svitkin YV, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- de Weerd NA, Vivian JP, Nguyen TK, Mangan NE, Gould JA, Braniff SJ, Zaker-Tabrizi L, Fung KY, Forster SC, Beddoe T, et al. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat Immunol. 2013;14:901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. 2011;17:2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, Kaiser F, Zerrahn J, Martens S, Howard JC. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 2008;27:2495–2509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Kaur S, Redig AJ, Goldsborough K, David K, Ueda T, Watanabe-Fukunaga R, Baker DP, Fish EN, Fukunaga R, Platanias LC. Type I interferon (IFN)-dependent activation of Mnk1 and its role in the generation of growth inhibitory responses. Proc Natl Acad Sci USA. 2009;106:12097–12102. doi: 10.1073/pnas.0900562106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Lal L, Sassano A, Majchrzak-Kita B, Srikanth M, Baker DP, Petroulakis E, Hay N, Sonenberg N, Fish EN, Platanias LC. Regulatory effects of mammalian target of rapamycin-activated pathways in type I and II interferon signaling. J Biol Chem. 2007;282:1757–1768. doi: 10.1074/jbc.M607365200. [DOI] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Joseph AM, Majchrzak-Kita B, Eklund EA, Verma A, Brachmann SM, Fish EN, Platanias LC. Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling. J Immunol. 2008a;181:7316–7323. doi: 10.4049/jimmunol.181.10.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, Hay N, Fish EN, Platanias LC. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci USA. 2008b;105:4808–4813. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Majchrzak-Kita B, Baker DP, Su B, Fish EN, Platanias LC. Regulatory effects of mTORC2 complexes in type I IFN signaling and in the generation of IFN responses. Proc Natl Acad Sci USA. 2012;109:7723–7728. doi: 10.1073/pnas.1118122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Platanias LC. IFN-β-specific signaling via a unique IFNAR1 interaction. Nat Immunol. 2013;14:884–885. doi: 10.1038/ni.2686. [DOI] [PubMed] [Google Scholar]
- Kaur S, Kroczynska B, Sharma B, Sassano A, Arslan AD, Majchrzak-Kita B, Stein BL, McMahon B, Altman JK, Su B, et al. Critical roles for Rictor/Sin1 complexes in interferon-dependent gene transcription and generation of antiproliferative responses. J Biol Chem. 2014;289:6581–6591. doi: 10.1074/jbc.M113.537852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–850. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- Kiladjian JJ, Mesa RA, Hoffman R. The renaissance of interferon therapy for the treatment of myeloid malignancies. Blood. 2011;117:4706–4715. doi: 10.1182/blood-2010-08-258772. [DOI] [PubMed] [Google Scholar]
- Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotredes KP, Gamero AM. Interferons as inducers of apoptosis in malignant cells. J Interferon Cytokine Res. 2013;33:162–170. doi: 10.1089/jir.2012.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B, Kaur S, Katsoulidis E, Majchrzak-Kita B, Sassano A, Kozma SC, Fish EN, Platanias LC. Interferon-dependent engagement of eukaryotic initiation factor 4B via S6 kinase (S6K)- and ribosomal protein S6K-mediated signals. Mol Cell Biol. 2009;29:2865–2875. doi: 10.1128/MCB.01537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B, Sharma B, Eklund EA, Fish EN, Platanias LC. Regulatory effects of programmed cell death 4 (PDCD4) protein in interferon (IFN)-stimulated gene expression and generation of type I IFN responses. Mol Cell Biol. 2012;32:2809–2822. doi: 10.1128/MCB.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B, Mehrotra S, Arslan AD, Kaur S, Platanias LC. Regulation of interferon-dependent mRNA translation of target genes. J Interferon Cytokine Res. 2014;34:289– 296. doi: 10.1089/jir.2013.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011;7:689–695. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, García-Sastre A, et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sassano A, Majchrzak B, Deb DK, Levy DE, Gaestel M, Nebreda AR, Fish EN, Platanias LC. Role of p38alpha Map kinase in Type I interferon signaling. J Biol Chem. 2004;279:970–979. doi: 10.1074/jbc.M309927200. [DOI] [PubMed] [Google Scholar]
- Li Y, Batra S, Sassano A, Majchrzak B, Levy DE, Gaestel M, Fish EN, Davis RJ, Platanias LC. Activation of mitogen-activated protein kinase kinase (MKK) 3 and MKK6 by type I interferons. J Biol Chem. 2005;280:10001–10010. doi: 10.1074/jbc.M410972200. [DOI] [PubMed] [Google Scholar]
- Liu XY, Chen W, Wei B, Shan YF, Wang C. IFN-induced TPR protein IFIT3 potentiates antiviral signaling by bridging MAVS and TBK1. J Immunol. 2011;187:2559–2568. doi: 10.4049/jimmunol.1100963. [DOI] [PubMed] [Google Scholar]
- Lu M, Zhang W, Li Y, Berenzon D, Wang X, Wang J, Mascarenhas J, Xu M, Hoffman R. Interferon-alpha targets JAK2V617F-positive hematopoietic progenitor cells and acts through the p38 MAPK pathway. Exp Hematol. 2010;38:472–480. doi: 10.1016/j.exphem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack HI, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer IA, Verma A, Grumbach IM, Uddin S, Lekmine F, Ravandi F, Majchrzak B, Fujita S, Fish EN, Platanias LC. The p38 MAPK pathway mediates the growth inhibitory effects of interferon-alpha in BCR-ABL-expressing cells. J Biol Chem. 2001;276:28570–28577. doi: 10.1074/jbc.M011685200. [DOI] [PubMed] [Google Scholar]
- McAllister CS, Samuel CE. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem. 2009;284:1644–1651. doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Sharma B, Joshi S, Kroczynska B, Majchrzak B, Stein BL, McMahon B, Altman JK, Licht JD, Baker DP, et al. Essential role for the Mnk pathway in the inhibitory effects of type I interferons on myeloproliferative neoplasm (MPN) precursors. J Biol Chem. 2013;288:23814–23822. doi: 10.1074/jbc.M113.476192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Platanias LC. Interferons and their antitumor properties. J Interferon Cytokine Res. 2013;33:143–144. doi: 10.1089/jir.2013.0019. [DOI] [PubMed] [Google Scholar]
- Peng XD, Xu PZ, Chen ML, Hahn-Windgasse A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, et al. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, Bass AJ, Pretz J, Ahn J, Hricik T, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123:e123–e133. doi: 10.1182/blood-2014-02-554634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani MR, Shrock J, Appachi S, Rudick RA, Williams BR, Ransohoff RM. Novel interferon-beta-induced gene expression in peripheral blood cells. J Leukoc Biol. 2007;82:1353–1360. doi: 10.1189/jlb.0507273. [DOI] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser H, Mejido J, Balinsky CA, Morrow AN, Clark CR, Zhao T, Zoon KC. Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J Virol. 2010;84:10671–10680. doi: 10.1128/JVI.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser H, Bekisz J, Zoon KC. New function of type I IFN: induction of autophagy. J Interferon Cytokine Res. 2014;34:71–78. doi: 10.1089/jir.2013.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein BL, Tiu RV. Biological rationale and clinical use of interferon in the classical BCR-ABL-negative myeloproliferative neoplasms. J Interferon Cytokine Res. 2013;33:145–153. doi: 10.1089/jir.2012.0120. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vainchenker W. Myeloproliferative neoplasms: molecular patho-physiology, essential clinical understanding, and treatment strategies. J Clin Oncol. 2011;29:573–582. doi: 10.1200/JCO.2010.29.8711. [DOI] [PubMed] [Google Scholar]
- Uddin S, Majchrzak B, Woodson J, Arunkumar P, Alsayed Y, Pine R, Young PR, Fish EN, Platanias LC. Activation of the p38 mitogen-activated protein kinase by type I interferons. J Biol Chem. 1999;274:30127–30131. doi: 10.1074/jbc.274.42.30127. [DOI] [PubMed] [Google Scholar]
- Uddin S, Lekmine F, Sharma N, Majchrzak B, Mayer I, Young PR, Bokoch GM, Fish EN, Platanias LC. The Rac1/p38 mitogen-activated protein kinase pathway is required for interferon alpha-dependent transcriptional activation but not serine phosphorylation of Stat proteins. J Biol Chem. 2000;275:27634–27640. doi: 10.1074/jbc.M003170200. [DOI] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–32. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liang H, Zhou Q, Li Y, Chen H, Ye W, Chen D, Fleming J, Shu H, Liu Y. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012;22:1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Yuan J, Cheung P, Chau D, Wong BW, McManus BM, Yang D. Gamma interferon-inducible protein 10 induces HeLa cell apoptosis through a p53-dependent pathway initiated by suppression of human papillomavirus type 18 E6 and E7 expression. Mol Cell Biol. 2005;25:6247–6258. doi: 10.1128/MCB.25.14.6247-6258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, Ibsen MS, Schmid-Burgk JL, Schmidt T, Ganapathiraju MK, et al. Antiviral Activity of Human OASL Protein Is Mediated by Enhancing Signaling of the RIG-I RNA Sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.