Abstract
Mutations in NLRP3 (CIAS1) are identified in a continuum of related inflammatory disorders, known as cryopyrinopathies since NLRP3 codes for the protein cryopyrin. Approximately 40% of patients with classic presentation lack mutations in the coding region of NLRP3 suggesting heterogeneity or epigenetic factors. Cryopyrin is a key regulator of proinflammatory cytokine release. Therefore, variations in the NLRP3 promoter sequence may have effects on disease state in patients with cryopyrinopathies and other inflammatory diseases. In this report, we confirmed three 5′-untranslated region splice forms with two separate transcriptional start sites, and identified potential promoter regions and six new DNA promoter variants. One variant is unique to a mutation negative cryopyrinopathy patient and increases in vitro gene expression. Additional studies can now be performed to further characterize the NLRP3 promoter and sequence variants, which will lead to better understanding of the regulation of NLRP3 expression and its role in disease.
Keywords: NLRP3, CIAS1, cryopyrin, NALP3, promoter
Introduction
The NLR proteins are a recently described family of immune regulatory proteins characterized by specific structural domains, including caspase recruitment domains, pyrin domains,1 nucleotide-binding domains, and leucine-rich repeat domains. Variations in these proteins have been associated with several human diseases including Crohn’s disease, Blau syndrome and vitiligo. One of these proteins known as cryopyrin (NALP3) is altered in patients with a spectrum of inflammatory syndromes, collectively referred to as cryopyrinopathies, which are characterized by fever, as well as cutaneous, joint and neurosensory symptoms.2,3 Cryopyrin is coded from NLRP3 (CIAS1), and heterozygous missense mutations in this gene are associated with a continuum of phenotypes that encompass three previously recognized syndromes: familial cold autoinflammatory syndrome (FCAS), Muckle-Wells syndrome (MWS) and neonatal onset multisystem inflammatory disease (NOMID). FCAS (MIM 120100), commonly known as familial cold urticaria, is an autosomal dominant systemic inflammatory disease characterized by intermittent episodes of rash, arthralgia, fever and conjunctivitis after generalized exposure to cold.4 Patients with MWS (MIM 191900) have similar episodes to those of FCAS, but symptoms are usually not associated with cold exposure. A significant percentage of patients with MWS also develop progressive sensorineural hearing loss (~60%) and systemic amyloidosis (~25%) leading to renal failure.5 NOMID (MIM 607115) is primarily sporadic, but dominant transmission has also been documented. These patients have chronic systemic inflammation involving the central nervous system, skin, eyes and joints, but also have potential noninflammatory symptoms including cartilage overgrowth and hearing loss.6 There is some correlation of specific NLRP3 nucleotide substitutions with phenotype; however, the same mutation has been associated with different phenotypes in different patients, suggesting additional genetic or environmental influences.7 In addition, there are several patients with classic cryopyrinopathy phenotypes that do no have readily detectable mutations in NLRP3-coding regions.
The expression of NLRP3 is highly regulated, being expressed only in certain cell types, such as neutrophils, monocytes8 and chondrocytes,2 and is affected by inflammatory processes9 and environmental factors. Recent expression studies show that NLRP3 expression is modified in several common and complex disease states, such as essential hypertension10 and osteoarthritis.11 We hypothesized that the promoter sequence of NLRP3 is a key component in its regulation, and that variations in promoter sequence may have significant effects on disease state in patients with cryopyrinopathies, as well as more common inflammatory diseases. Promoter regions of several genes have been implicated in contributing to a wide variety of diseases; from atopic eczema12 to Alzheimer’s disease.13 To date, no formal studies of the NLRP3 promoter region have been reported. Elucidating the regulation of expression of NLRP3 should allow for a better understanding of its role in innate immunity and in human diseases and could lead to novel targeted treatments.
Results and discussion
To identify the 5′ end of the NLRP3 promoter region, we determined the transcriptional start site through both rapid amplification of 5′cDNA ends (5′RACE) from peripheral leukocytes and by searching two online genome databases, NCBI (http://www.ncbi.nlm.nih.gov/) and UCSC (http://genome.ucsc.edu). As observed in over 20% of human genes,14 we identified alternative splicing in the 5′-untranslated region (5′UTR), and were able to confirm three different alternative splice forms and two transcriptional start sites (Figure 1a). The first splice form, labeled A, consists of one segment (1281 bp) from −1004 to +277 (the numbers correspond to position relative to the translational [ATG] start site, with +277 corresponding to the 3′ end of exon 1). Splice form B has the same transcriptional start site and consists of two segments, a 494 bp segment from −1004 to −511 and a 321 bp segment from −44 to +277. Splice form C has a different transcriptional start site and also consists of two segments, a 20 bp segment located from −2678 to −2608 and a 1025 bp segment from −748 to +277. A similar pattern is observed in the 5′ region of the interleukin-1 receptor antagonist and the NOD2 gene which have two or more 5′UTR alternative splice forms with different associated promoter regions with specific tissue expression.16,17 Both of these genes show a difference in context-dependent expression in that different cytokines result in a change in the ratio of expression of the two isoforms.
Figure 1.

NLRP3 promoter region. (a) Transcriptional start sites and 5′-untranslated region (UTR) splice forms. 5′ rapid amplification of 5′cDNA end (5′RACE) was performed using human leukocyte Marathon-Ready cDNA (BD Biosciences, San Jose, CA, USA). The First-Choice RLM-RACE kit (Applied Biosystems/Ambion, Austin, TX, USA) was used according to the manufacturer’s protocol with both the kit’s primers and the following gene-specific primers: ProckF2 (5′-CCAGAGCCTTCAGTTTGGAG-3′), F16dextR (5′-CCCTTCTGGGGAGGATAGTC-3′), ProckR2 (5′-GGTGAACAACCACTTCACGA-3′) and X2intR (5′-TTCCTGGCATATCACAGTGG-3′). These gene-specific primers were used to assist in the analysis of the RACE reactions according to the manufacturer’s protocol and also for their compatibility with annealing and optimization of the reactions. (b) Putative transcription factor binding sites. A total of 5000 bp of human genomic DNA, 5′ to the translational start site, was analyzed for putative transcription factor-binding sites using software from Transcriptional Element Search System (www.cbil.upenn.edu/tess). The search criteria and protocols were followed according to the website and Chapter 2.6 of Wiley’s Current Protocols in Bioinformatics.15 A single-nucleotide polymorphism (SNP) unique to one familial cold autoinflammatory syndrome (FCAS) patient is labeled by an asterisk. (c) Inserts for plasmid constructs. Construct labels refer to the distance from the translational start site. A primer at −690 was used because it included a portion of all three isoforms and was an optimal primer for cloning. Luciferase reporter constructs were created using polymerase chain reaction (PCR)-amplified fragments of the NLRP3 5′UTR and pGL3-Enhancer vector (Promega Corporation, San Luis Obispo, CA, USA). An unaffected human control was used for the template DNA. The PCR conditions and primers used to create constructs are described in Supplementary Table 1. PCR products were purified using the QIAquick PCR Purification kit (Qiagen, Valencia, CA, USA). The products were then digested using the restriction enzymes XhoI and NheI at 37°C for 18 h. The purified and digested PCR products were then directly cloned into the pGL3-Enhancer vector (Promega Corporation). All constructs were verified by direct sequencing using primers that annealed to the vector: RVP5 (5′-CCAGTGCAAGTGCAGGTGCCAGA-3′) and GLP3 (5′-TTTGGCGTCTTCCATGGTGGCTT-3′). DNA from an FCAS patient was used as a template for PCR to create the construct containing the SNP located at −1064 (Table 1).
The DNA regions upstream of the two different NLRP3 transcriptional start sites were analyzed for putative transcription factor-binding sites. Within these distinct regions, several potential transcription factor-binding sites were identified (Figure 1b). No typical TATA or CAAT boxes were found in these regions, however several other transcription factor sites were; including Sp1, c-Myb, AP-1 and c-Ets. All four of these transcription factors have been associated with myeloid-specific genes;18 however, they are not restricted to myeloid cells. These factors often work in combination for myeloid-specific gene expression and are also important for myeloid cell differentiation.19 Sp1 has been shown to be important in TATA-less promoters, because as seen with TATA boxes, Sp1 sites are able to recruit the TFIID complex resulting in transcription.20
After confirming the 5′ alternative splice forms and putative transcription factor-binding sites, luciferase reporter constructs of varying sizes were developed to assay the promoter activity of the region up to 3600 bp 5′ of the ATG site (Figure 1c). There was some promoter activity observed in all of the initial constructs tested; however, the region associated with the first two splice forms (within 2600 bp of the ATG site) had more significant promoter activity (Figure 2a) compared with the region associated with the third splice form (Figure 2b). The largest luciferase expression was observed in the two constructs containing the area from −690 to −1109 and −690 to −1463. In order to determine the essential region of promoter activity, a construct deleting the region from −690 to −1114 was generated and this construct showed no significant luciferase expression suggesting that this region is necessary for promoter activity. Regions encompassing the essential region but extending beyond −1463 had less promoter activity, suggesting there may be transcriptional inhibitor regions in the NLRP3 promoter. In addition to the pGL3-Enhancer vector without insert, two other negative controls were also studied: pRL-TK Vector alone (1.2±1.7%) and a transfection with no vectors added (3.7±2.1%) (data not shown).
Figure 2.
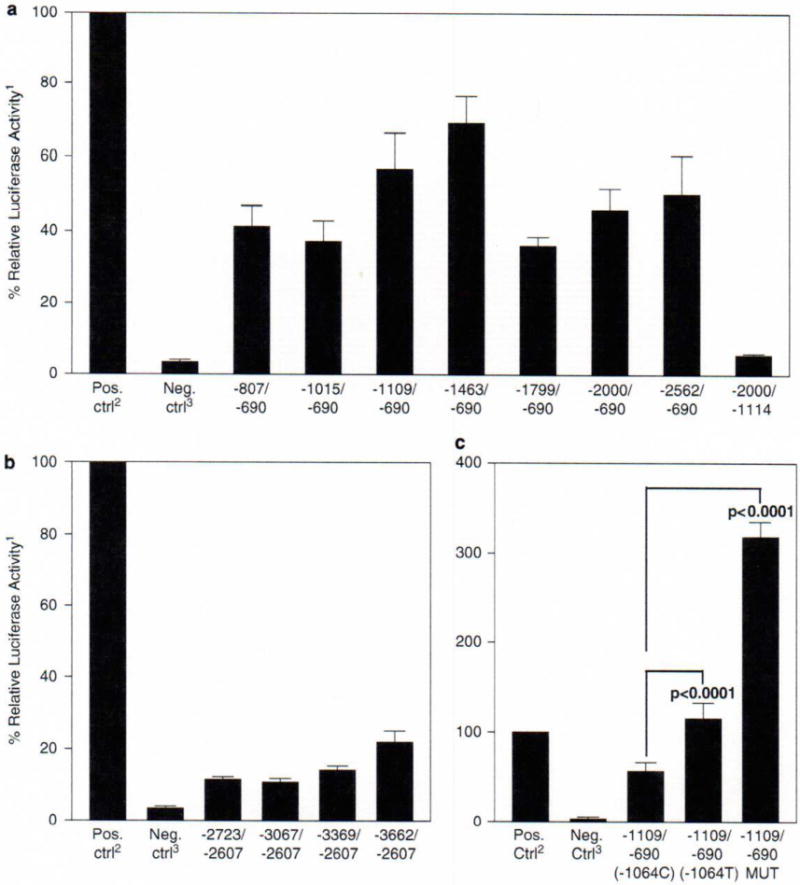
Luciferase assays. (a) Luciferase expression for transcriptional start site one constructs. A total of 5 × 104 HeLa cells were grown in 24-well plates in 0.5 ml media (Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum, glutamine, penicillin and streptomycin) overnight. The HeLa cells were then transfected with 400 ng of luciferase reporter constructs and 100 ng of pRL-TK Vector (Promega Corporation) using FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN, USA). After 48 h incubation, passive lysis of the cells was performed and the lysate was used to measure the luciferase expression using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer’s protocol. A GENios Pro (Tecan Systems Inc., San Jose, CA, USA) was used to measure the relative light units. (b) Luciferase expression for transcriptional start site two constructs. Cell culture, transfection and luciferase assays were performed as described in (a). (c) Luciferase expression of −1064C/T and Sp1 mutant. Cell culture, transfection and luciferase assays were performed as described in (a). To mutate the Sp1 site located at −1073 upstream of the translation start site, the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) was used according to the manufacturer’s protocol. This Sp1 site was mutated from 5′-CCGCCC-3′ to 5′-CCGAAC-3′ using the following primers: QCPro_mSP1F (5′-TCCTGACCTCAGGTGATCCGAACACCTCGGCCTCCCAAAGTG-3′) and QCPro_mSPlR (5′-CACTTTGGGAGGCCGAGGTGTTCGGATCACCTGAGGTCAGGA-3′). The -1109/-690 wild-type luciferase reporter construct was used for the template DNA. The Sp1 mutant construct was verified by direct sequencing. 1All luciferase activities are expressed as a percentage of the positive control, with the positive control as 100%. 2pGL3-Control Vector (Promega Corporation) was used as the positive control. 3pGL3-Enhancer Vector (Promega Corporation) without an insert was used as the negative control. P-values were determined by Student’s t-test; n = 12 for all assays except controls (n = 15) and −1109/−690 MUT (n = 6). Error bars represent the standard error of the mean.
Once several areas with potential promoter activity and putative transcription factor-binding sites were identified, we sequenced the promoter regions (from −900 to −3700) in genomic DNA from 29 cryopyrinopathy patients (14 patients with and 15 patients without coding region mutations) looking for variations unique to these patients. For patients with mutations, we chose index cases from each of nine FCAS and four MWS families and one NOMID case in the UCSD cryopyrinopathy database. For patients without mutations, we studied one NOMID, one MWS and two FCAS cases from the UCSD database and nine NOMID and two MWS patients from the NIH database.21 This analysis identified six unreported sequence variants and confirmed five out of eight reported sequence variations in the NCBI SNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) (Table 1). One novel variant (−3242A) was found in a mutation negative African-American NOMID patient, however this variant was found in two out of twenty African-American controls. Another unique SNP (−1064T) was identified in a mutation negative FCAS patient that was not identified in over 200 matched controls (Table 1). This patient is a 68-year-old woman with classic FCAS symptoms and skin biopsy findings except for evidence of vasculitis. She has no family history and no mutations in the entire coding region.
Table 1.
Sequence variants in the 5′UTR of NLRP3
| Reference SNP ID | Varianta | Locationb | Caucasian controls | African-American controls | FCAS patients | MWS patients | NOMID patients |
|---|---|---|---|---|---|---|---|
| Rs2027432 | G/A | −3661 | Nonec | None | 2/4d | 1/2 | 2/16 |
| G/A | −3242 | 0/118 | 2/40 | 0/4 | 0/4 | 1/16 | |
| Rs3738448 | G/T | −2667 | 12/122 | None | 0/4 | 0/4 | 2/16 |
| Rs35728135 | G/-e | −2620 | 0/122 | None | 0/4 | 0/4 | 0/16 |
| C/G | −2392 | 18/120 | None | 1/4 | 0/4 | 4/16 | |
| A/G | −2353 | 18/120 | None | 1/4 | 0/4 | 4/16 | |
| Rs12079994 | G/A | −2194 | 12/120 | None | 0/4 | 0/4 | 2/16 |
| Δ63bpf | −2123 to −2060 | 20/120 | None | 0/4 | 0/4 | 3/16 | |
| Rs6426246 | A/C | −1921 | 0/200 | None | 0/22 | 0/14 | 0/22 |
| Rs4925648 | C/T | −1535 | 19/200 | None | 3/22 | 3/14 | 3/22 |
| T/C | −1293 | 19/200 | None | 3/22 | 3/14 | 3/22 | |
| Rs34912656 | −/Ag | −1136 | 0/200 | None | 0/22 | 0/14 | 0/22 |
| C/T | −1064 | 0/200 | None | 1/22 | 0/14 | 0/22 | |
| Rs12137901 | T/C | −894 | 38/200 | None | 7/22 | 5/14h | 7/22 |
Abbreviations: FCAS, familial cold autoinflaminatory syndrome; MWS, Muckle–Wells syndrome; NOMID, neonatal onset multisystem inflammatory disease; SNP, single-nucleotide polymorphism; 5′UTR, 5′ untranslated region.
Human genomic DNA from FCAS, MWS, and NOMID patients and from Caucasian and African-American controls were used for PCR and sequencing. Primers were designed using Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Variant frequency is represented as the variant allele over the total number of chromosomes sequenced. The PCR conditions and primers used to amplify and sequence the human genomic DNA are shown in Supplementary Table 2.
Variants are listed as the more common allele over the rarer allele.
Location of the variants represents the distance from the translational start site of NLRP3.
Evaluated in a population of atopic dermatitis patients and controls.22
One of the two FCAS patients sequenced was homozygous for the SNP listed, representing both of the rarer alleles.
This variant represents a single base pair deletion.
This variant represents a 63 base pair deletion.
This variant represents a single base pair insertion.
Two of the MWS patients sequenced were homozygous for the SNP listed, representing four out of the five rarer alleles.
As this SNP (−1064T) is unique to a mutation negative patient, not present in her unaffected brother, and is within an area of significant promoter activity, we generated a construct with this unique promoter variation and performed a luciferase analysis to determine if there is any change in expression. As this SNP is also located 9 bp downstream of a putative Sp1 site, we engineered a mutation in the consensus sequence of this Sp1 site to determine the relative importance of this Sp1 site. The analysis of the −1064T SNP showed a greater than twofold increase in luciferase expression, whereas mutating the Sp1 site showed a threefold increase in expression (Figure 2c). Although Sp1 typically increases expression of genes, Sp1 sites have also been shown to act as repressors.23–25 It has been suggested that coding region mutations in NLRP3 are ‘gain of function’ and result in a constitutively active cryopyrin; therefore, it is possible that this novel SNP (−1064T) could result in increased NLRP3 expression and a potentially new mechanism of disease causation in cryopyrinopathy patients.
In this report we have identified and confirmed (by sequencing) three 5′UTR splice forms with two separate transcriptional start sites and we identified regulatory promoter sequence in the region 5′ upstream of the NLRP3 gene. We also confirmed five out of eight previously reported variants and identified six new variants and at least one putative Sp1 site. We also showed that one unique SNP in a de novo mutation negative FCAS patient increases expression in vitro. Many genetic and epigenetic factors could be involved in disease expression in the cryopyrinopathies and this SNP may be yet another contributing factor in some patients. The initial characterization of the NLRP3 promoter and identification of new SNPs in the promoter region can be used to conduct further genetic studies in additional disease populations. In addition, these initial studies will allow for further characterization of the highly regulated expression of this important immune protein.
Supplementary Material
Footnotes
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/genes)
References
- 1.Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 2.Feldmann J, Prieur AM, Quarticr P, Berquin P, Certain S, Cortis E, et al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71:198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein cause familial cold autoinflammatory syndrome and Muckle–Wells syndrome. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman HM, Wanderer AA, Broide DH. Familial cold autoinflammatory syndrome: phenotype and genotype of an autosomal dominant periodic fever. J Allergy Clin Immunol. 2001;108:615–620. doi: 10.1067/mai.2001.118790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muckle TJ. The ‘Muckle–Wells’ syndrome. Br J Dermatol. 1979;100:87–92. doi: 10.1111/j.1365-2133.1979.tb03572.x. [DOI] [PubMed] [Google Scholar]
- 6.Prieur AM, Griscelli C, Lamport F, Truckenbrodt H, Guggenheim MA, Lovell DJ, et al. A chronic, infantile, neurological, cutaneous and articular (CINCA) syndrome. A specific entity analysed in 30 patients. Scand J Rheumatol Suppl. 1987;66:57–68. doi: 10.3109/03009748709102523. [DOI] [PubMed] [Google Scholar]
- 7.Neven B, Callebaut I, Prieur AM, Feldmann J, Bodemer C, Lepore L, et al. Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood. 2004;103:2809–2815. doi: 10.1182/blood-2003-07-2531. [DOI] [PubMed] [Google Scholar]
- 8.Manji GA, Wang L, Geddes BJ, Brown M, Merriam S, Al-Garawi A, et al. PYPAF1, a PYRIN-containing Apaf1-like protein that assembles with ASC and regulates activation of NF-kappa B. J Biol Chem. 2002;277:11570–11575. doi: 10.1074/jbc.M112208200. [DOI] [PubMed] [Google Scholar]
- 9.Rosengren S, Hoffman H, Bugbee W, Boyle D. Expression and regulation of cryopyrin and related proteins in rheumatoid arthritis synovium. Ann Rheum Dis. 2005;64:708–714. doi: 10.1136/ard.2004.025577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omi T, Kumada M, Kamesaki T, Okuda H, Munkhtulga L, Yanagisawa Y, et al. An intronic variable number of tandem repeat polymorphisms of the cold-induced autoinflammatory syndrome 1 (CIAS1) gene modifies gene expression and is associated with essential hypertension. Eur J Hum Genet. 2006;14:1295–1305. doi: 10.1038/sj.ejhg.5201698. [DOI] [PubMed] [Google Scholar]
- 11.Mahr S, Burmester GR, Hilke D, Göbel U, Grützkau A, Häupl T, et al. Cis- and trans-acting gene regulation is associated with osteoarthritis. Am J Hum Genet. 2006;78:793–803. doi: 10.1086/503849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak N, Yu CF, Bussmann C, Maintz L, Peng WM, Hart J, et al. Putative association of a TLR9 promoter polymorphism with atopic eczema. Allergy. 2007;62:766–772. doi: 10.1111/j.1398-9995.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 13.Ge YW, Lahiri DK. Regulation of promoter activity of the APP gene by cytokines and growth factors: implications in Alzheimer’s disease. Ann NY Acad Sci. 2002;973:463–467. doi: 10.1111/j.1749-6632.2002.tb04684.x. [DOI] [PubMed] [Google Scholar]
- 14.Pesole G, Grillo G, Larizza A, Liuni S. The untranslated regions of eukaryotic mRNAs: structure, function, evolution and bioinformatic tools for their analysis. Brief Bioinform. 2000;1:236–249. doi: 10.1093/bib/1.3.236. Review. [DOI] [PubMed] [Google Scholar]
- 15.Schug J. Current Protocols in Bioinformatics. Wiley; 2003. Using TESS to predict transcription factor binding sites in DNA sequence; pp. 2.6.1–2.6.9. version 1. [DOI] [PubMed] [Google Scholar]
- 16.Gabay C, Smith MF, Eidlen D, Arend WP. Interleukin 1 receptor antagonist (IL-1Ra) is an acute-phase protein. J Clin Invest. 1997;99:2930–2940. doi: 10.1172/JCI119488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenstiel P, Huse K, Franke A, Hampe J, Reichwald K, Platzer C, et al. Functional characterization of two novel 5′ untranslated exons reveals a complex regulation of NOD2 protein expression. BMC Genom. 2007;8:472. doi: 10.1186/1471-2164-8-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke S, Gordon S. Myeloid-specific gene expression. J Leukoc Biol. 1998;63:153–168. doi: 10.1002/jlb.63.2.153. Review. [DOI] [PubMed] [Google Scholar]
- 19.Skalnik DG. Transcriptional mechanisms regulating myeloid-specific genes. Gene. 2002;284:1–21. doi: 10.1016/s0378-1119(02)00387-6. Review. [DOI] [PubMed] [Google Scholar]
- 20.Pugh BF, Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 21.Aksentijevich I, Putnam C, Remmers EF, Mueller JL, Le J, Kolodner RD, et al. The clinical continuum of cryopyrinopa-thies: novel CIAS1 mutations in North American patients and a new cryopyrin model. Arthritis Rheum. 2007;56:1273–1285. doi: 10.1002/art.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macaluso F, Nothnagel M, Parwez Q, Petrasch-Parwez E, Bechara FG, Epplen JT, et al. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp Dermatol. 2007;16:692–698. doi: 10.1111/j.1600-0625.2007.00589.x. [DOI] [PubMed] [Google Scholar]
- 23.Steinke JW, Barekzi E, Hagman J, Borish L. Functional analysis of −571 IL-10 promoter polymorphism reveals a repressor element controlled by spl. J Immunol. 2004;173:3215–3222. doi: 10.4049/jimmunol.173.5.3215. [DOI] [PubMed] [Google Scholar]
- 24.Pietrzak M, Puzianowska-Kuznicka M. p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol Chem. 2008;389:383–393. doi: 10.1515/BC.2008.039. [DOI] [PubMed] [Google Scholar]
- 25.Zhan M, Yu D, Liu J, Glazer RI, Hannay J, Pollock RE. Transcriptional repression of protein kinase Calpha via Sp1 by wild type p53 is involved in inhibition of multidrug resistance 1 P-glycoprotein phosphorylation. J Biol Chem. 2005;280:4825–4833. doi: 10.1074/jbc.M407450200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


