The authors developed and assessed an innovative course-based undergraduate research experience that emphasized collaboration among students and focused on data analysis.
Abstract
We present an innovative course-based undergraduate research experience curriculum focused on the characterization of single point mutations in p53, a tumor suppressor gene that is mutated in more than 50% of human cancers. This course is required of all introductory biology students, so all biology majors engage in a research project as part of their training. Using a set of open-ended written prompts, we found that the course shifts student conceptions of what it means to think like a scientist from novice to more expert-like. Students at the end of the course identified experimental repetition, data analysis, and collaboration as important elements of thinking like a scientist. Course exams revealed that students showed gains in their ability to analyze and interpret data. These data indicate that this course-embedded research experience has a positive impact on the development of students’ conceptions and practice of scientific thinking.
INTRODUCTION
Learning science means learning to do science.
—Vision and Change: A Call to Action (American Association for the Advancement of Science, 2011, p. 14)
Incorporating research experiences into the undergraduate curriculum is a major goal of national reform efforts (American Association for the Advancement of Science [AAAS], 2011; National Research Council [NRC], 2003). Research involving undergraduate students has been called the “purest form of teaching” (NRC, 2003, p. 87), and research experiences for undergraduates have been identified as “an integral component of biology education for all students” (AAAS, 2011, p. xiv). Despite these recommendations, the practical implications of involving students in research remain daunting. Historically, the integration of research into the undergraduate biology curriculum has primarily been in the form of research apprenticeships in faculty research labs (Russell et al., 2007), but there are not enough of these positions available at most institutions to give all students the opportunity to participate in authentic research.
One solution to this problem is to integrate research experiences into traditional high-enrollment lab courses (Handelsman et al., 2004; Sundberg et al., 2005; Auchincloss et al., 2014). Such course-based undergraduate research experiences, or CUREs, have five defining characteristics: 1) There is an element of discovery, so that students are working with novel data. 2) Iteration is built into the lab. 3) Students engage in a high level of collaboration. 4) Students learn scientific practices. 5) The topic is broadly relevant so that it could potentially be publishable and/or of interest to a group outside the class (Auchincloss et al., 2014). One of the major goals of these CUREs is that they reflect a real research experience in order to give students a more accurate conception of how scientific research is done.
In response to calls for reform, a diverse range of CUREs have been developed (for a list, see www.curenet.franklin.uga.edu). Some of these courses are extensions of faculty members’ research projects (e.g., Brownell et al., 2012). In this format, students have the opportunity to work on a research problem in a formal course, and questions they explore can contribute to a faculty member’s research program (Kloser et al., 2011; Brownell and Kloser, 2015). An alternative model is a stand-alone lab course in which students conduct research that is only peripherally related, if at all, to a local faculty member’s research program. This approach is exemplified by the Howard Hughes Medical Institute (HHMI) SEA PHAGES program (Jordan et al., 2014) and the Genetics Education Partnership (Shaffer et al., 2010, 2014), both of which are multi-institution programs. These courses have been packaged so they can be implemented at a diverse range of institutions, and inter-institutional collaboration is encouraged.
While many CUREs currently exist, most of these have been small-sized classes taught to students who volunteer to participate. However, volunteer students and nonvolunteer students have previously been shown to have different affective gains from a CURE (Brownell et al., 2013), indicating that findings from volunteer populations may not be generalizable to students in required CUREs. Additionally, assessment of CUREs has been primarily in the form of student self-report surveys (e.g., CURE survey; Lopatto et al., 2008). While student self-reporting can be useful if one is interested in affective measures such as confidence or interest, it is not as effective at determining students’ abilities to interpret data or how similar their thinking processes are to expert scientists. Different means of assessment need to be used to further probe the impact of CUREs on students (Brownell and Kloser, 2015; Corwin et al., 2015).
We developed a CURE that replaced our “cookbook” introductory biology course; it is required of all undergraduate biology majors and is not directly related to any faculty member’s research at this institution. The primary purpose of this course is to engage students in research to shift their “thinking like a scientist” from novice to expert. This construct has been defined in previous literature (Druger et al., 2004; Hunter et al., 2007; Hurtado et al., 2009; Etkina and Planinši, 2014). We solicited responses from expert scientists who hold a PhD in their discipline to further articulate this construct; our definition is outlined in Table 1.
Table 1.
Our definition of the construct “thinking like a scientist” based on prior studies and consensus of an expert panel of PhD-level scientists
| Element | Description | Agreement in prior literature |
|---|---|---|
| Make discoveries | Scientists formulate questions, make observations, collect data, analyze and interpret data, test hypotheses, and draw conclusions. | Druger et al., 2004; Hunter et al., 2007; Hurtado et al., 2009; Etkina and Planinši, 2014 |
| Make connections between seemingly unconnected phenomena | Scientists are able to think in multiple ways and design multiple types of experiments to test the same idea. New ideas often result from thinking differently. Science is not a linear process. | Hunter et al., 2007 |
| Critically evaluate data with skepticism | Scientists critique both their own experiments and the experiments of others. There is the need to repeat experiments to see whether more evidence backs up a claim; one experiment is not enough. | Druger et al., 2004; Hunter et al., 2007 |
| Seek opportunities to share their findings and communicate with others | Scientists present their work to others in the form of scientific posters, oral presentations, and written reports. Communication of their interpretations to the broader community is important, because scientists are working toward common goals. | Hurtado et al., 2009 |
The purpose of this study was to assess the impact of this required high-enrollment CURE on 1) student conceptions of what it means to think like a scientist and 2) student ability to analyze and interpret scientific data, one of the key components of what it means to think like a scientist.
The novelty of this particular study is in the curriculum, how the course is required for a large population of introductory students, specific aspects of the course that promote and benefit from large-scale collaboration, and how we assessed the impact of the course curriculum through coded open-ended written responses and student learning gains in data interpretation. We hope that this can be a model for others interested in developing and assessing CUREs that are designed to give all students graduating with a biology degree the experience of doing research.
CURRICULUM DESCRIPTION
Course Content: Investigating Human p53 Mutants Using Yeast as a Model System
We sought to identify an unsolved scientific problem that would engage student interest in human biology and could be investigated using molecular and cell biology techniques accessible to students with no previous lab experience. We reasoned that analysis of a human disease–related protein would satisfy the first criterion and that use of budding yeast as an experimental system would satisfy the second. We chose the human tumor suppressor gene p53 as the basis for study. p53 is a transcription factor that promotes DNA repair, cell cycle arrest, and apoptosis (Levine, 1997; Sionov and Haupt, 1999). p53 is mutated in more than 50% of cancers (Hollstein et al., 1991; Soussi et al., 1994; Whibley et al., 2009). We note that Gammie and Erdeniz (2004) used a similar rationale in creating a smaller lab course based on the human mismatch repair protein MSH2 (Gammie and Erdeniz, 2004).
The specific scientific research question in our course is to characterize mutant versions of p53 identified in human tumors. Each mutant version of p53 contains a single point mutation that changes one amino acid in the p53 protein sequence. Although much is known about p53 function, the specific functional defects of many mutant p53 proteins are uncharacterized. We used yeast as a model system because it is inexpensive to culture, grows rapidly, and is easily genetically manipulated. Using standard yeast genetics and an array of basic molecular and cell biology techniques, students can explore the phenotype of their p53 mutant over the course of a 10-wk quarter and come to an initial conclusion about the molecular nature of the defect in their mutant (Schärer, 1992; Mager and Winderickx, 2005; Figure 1).
Figure 1.
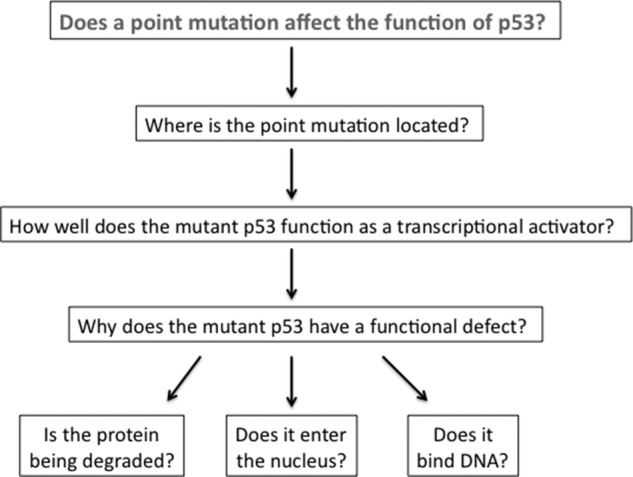
Research questions explored by students in the course over a 10-wk period. Students work through each of these questions to determine the functional defect of their mutant p53. The experimental protocols for each experiment have been developed; thus, the authenticity of the course stems from the data analysis on mutant alleles that have not previously been characterized, as well as the progressive refinement of student-articulated hypotheses and conclusions.
Course Goals: To Engage Students in a Research Experience to Encourage Scientific Thinking
This course’s primary goal was to shift students from novice to expert in their thinking as scientists in the context of a scientific research project. To achieve this, we incorporated the following features that align with the defining features of CUREs (Auchincloss et al., 2014): 1) discovery and relevance: students explored one longitudinal research question in depth for 10 wk, and neither instructors nor students knew in advance what the results of experiments would be; 2) collaboration: a high degree of collaboration required students to work with a partner on all aspects of the project, including experimentation, postlab assignments, and final presentations, and there were larger groups of students who shared data to achieve group conclusions; 3) iteration: multiple groups of students did the same experiments and compared data with one another; and 4) scientific practices: a) an emphasis on data interpretation and analysis and b) assessments that were representative of how scientists would evaluate one another, including a poster presentation and an oral presentation.
The course has a modular scalability in that five different p53 mutants were studied each term, and each lab section (10 students) studied the same five p53 mutants (one pair of students per mutant p53 allele). Even though the experimental procedures and the questions were mostly predetermined, the results and interpretations were unknown, creating a realistic research experience.
Course Organization, Student Population, and Instructional Team
Students enroll in this introductory lab course independent of an introductory biology lecture course. The course is intended for sophomore biology majors who are concurrently taking the introductory biology lecture series. All biology majors are required to take this course as their introductory biology lab course, and most premed students in nonbiology majors take this course to fulfill a medical school requirement; there were no other introductory biology lab course options available. Table 2 shows the demographic characteristics of students from Winter 2013, which is representative of the other terms.
Table 2.
Demographic characteristics of students in the lab course in Winter 2013 (n = 117)
| Class year | Gender | Ethnicity | Major | Prior research experience |
|---|---|---|---|---|
| Sophomore 53% | Male 40.2% | White 39.3% | Biology 37.6% | Yes 60.7% |
| Junior 31.6% | Female 59.8% | Asian 41% | Human biology 37.6% | No 39.3% |
| Senior 15.4% | Black 9.4% | Engineering 16.2% | ||
| Latin@ 7.7% | Other 8.5% | |||
| Pacific Islander 1.7% |
The course comprised one 75-min lecture/discussion section and one 4-h lab each week. The lecture/discussion section was used to introduce new material, to give students an opportunity for guided practice with some of the concepts of the course, and to compare and contrast data from different mutants. Three exams were also given during these discussion sections.
The 4-h lab was primarily dedicated to conducting experiments. Lab partners were randomly assigned at the beginning of the course, and students worked with the same lab partners each week to investigate their specific mutants.
A PhD-level lecturer taught each discussion session to 20 students; this lecturer taught two adjacent lab sessions, each with 10 students, simultaneously. Additionally, a graduate student teaching assistant helped coordinate the lab sessions, so there was an overall teacher-to-student ratio of 1:10. Two tenure-track faculty members with expertise in yeast genetics and four PhD-level instructors whose primary teaching responsibility was this course were responsible for designing and implementing the curriculum.
Course Design Elements Designed to Promote Scientific Thinking
We integrated the following design elements throughout the course to promote student thinking like a scientist.
Query as a Way to Structure Student Thinking.
QUERY is an acronym for “Question, Experiment, Results, and Your interpretation” (C. Anderson, unpublished observations). For each experiment students performed, they were asked to use the QUERY method to structure their thinking. This helped them to articulate the question they were trying to address, describe the experiment in detail, and differentiate results from their interpretation of the results. Specifically, students were asked about the question and experiment on each prelab assignment and the results and interpretation on each postlab assignment. Thus, we used QUERY as a way to scaffold the process of thinking like a scientist to introductory students. It required students to think about the “why” behind each experiment and distinguish the results from their interpretation of the results.
Hypothesis Testing and Making Predictions.
Although the instructors knew the order of the experiments, which was essential for planning and providing reagents to such a large class, the students did not know the order of the experiments in advance. Thus, after students completed the first set of analyses to determine whether their mutant p53s had transactivation defects, we gave students the opportunity to brainstorm about what might be causing the defects and how they might be able to test their hypotheses. Students engaged in a brainstorming session during which they used inductive logic to ask what next set of experiments they should design to answer their overarching question: “What is wrong with your p53 mutant?”
These brainstorming sessions provided opportunities for students to see the similarities and differences between experiments and experience the benefits of having multiple people working together to solve a problem. These sessions also prompted students to see the connections between individual experiments in answering the overall question (Figure 1).
Additionally, on each weekly postlab assignment, students were asked, based on data collected thus far, to develop hypotheses concerning possible molecular defects in their mutant p53s. This exercise could help students organize what they knew already and keep the big picture of the project in perspective. These activities were intended to help students see the project as one longitudinal project, even though they were completing a series of smaller experiments.
Data Interpretation.
Each week’s postlab assignment focused on the data analysis and interpretation of experiments conducted in the lab. Partners worked together on the data analysis and interpretation, and each pair of partners submitted a single, collaboratively prepared, postlab assignment. Thus, the collaboration between them was realistic of how scientists in a research lab would collaborate.
Building in Iteration and Comparing Data.
Within each lab room, each student pair worked on a different mutant, affording them ownership over their studies and independence in their interpretation of the data. However, students in different lab rooms examined the same set of five mutants. Three times during the 10-wk quarter, we held mutant group discussions, in which all the students working on the same mutant came together to compare their data and draw conclusions about their p53 mutants (Figure 2).
Figure 2.
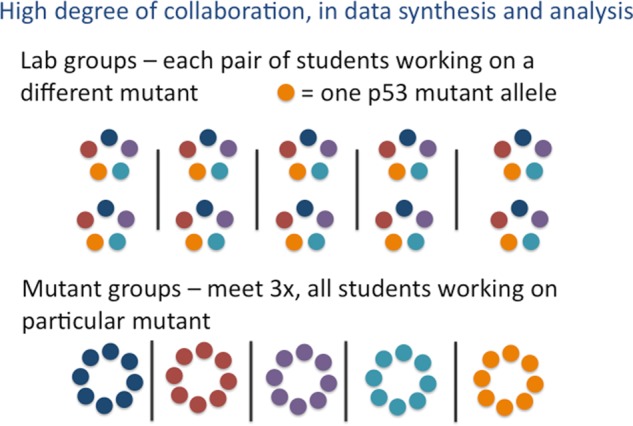
Collaboration among students in the course. Students work with one partner on a specific mutant allele of p53 for the whole 10 wk. In each 20-student combined lab section, two pairs of partners work on each mutant. For mutant group discussions, students from different lab sections who all work on the same mutant p53 compare data.
During one of these meetings, the group assigned to a specific mutant collectively designed an experiment, deciding how many different variables they wanted to test and weighing that against the benefit of having a higher number of replicates. These mutant group discussions allowed students to see, through the lens of their own p53 mutants, the inherent variability of biological data and, consequently, the importance of having multiple replicates of each experiment. When experiments did not yield an interpretable result, students engaged in a process of troubleshooting to determine what might have gone wrong. Differences between student results provided an opportunity to brainstorm possible sources of error and ways to make the data more reliable in the future; students also decided among themselves whether to include data points that could be considered outliers. Although instructors and teaching assistants facilitated these discussions, students were encouraged to lead the discussions, and student participation was integral.
We emphasized that the results of only one experiment are not enough to draw a conclusion; experiments must be repeated multiple times, which is a concept that students have been shown to have difficulty understanding (Brownell et al., 2014). Experiments must be repeated when they fail and when they work—one replicate is not enough to draw a conclusion in a biological experiment.
If an experiment did not yield interpretable results in a given week, students were expected to come in during another time before the next lab to repeat the experiment; this encouraged students to be diligent in their experimentation and also gave them a more realistic experience of what it feels like to have to repeat failed experiments, a necessary component of research.
Situating Student-Generated Data within What Is Known: Accessing Primary Literature and p53 Database.
Students read and discussed a primary scientific paper that was relevant to their investigation of p53 mutants. This exercise challenged students to think critically about published data and encouraged them to think about what other experiments they would need to do before their own work could be published (e.g., more replicates, additional experimental approaches to answer each question). The discussion also had students draw a connection between their work with yeast and the therapeutic implications in humans.
Students also accessed an online p53 database to explore research that had previously been done on their p53 mutants and the number of human tumors that had been identified with their p53 mutations. This allowed them to recognize what elements of the research project were novel and how their experiments were situated within the growing body of scientific knowledge.
Assignments Representative of How Scientists Would Present Data.
Because student results were preliminary, we sought to mimic what scientists in a research lab would do with preliminary data: present them to their colleagues in lab meeting and present them in a poster venue. For the lab meeting presentation, all students who studied the same mutant in the lab section presented one set of representative data and their interpretations from all the experiments. As they presented, other students and instructors asked questions concerning data analysis and interpretation, often comparing them with other sets of data that had been presented. For the final poster presentation, each partner pair created a scientific poster, which they took turns presenting during the poster session, a large venue with more than 50 posters. Students were required to visit other posters and compare other students’ results with their own results, completing graded worksheets that summarized their observations.
Building Community through Pass/Fail Grading.
The course was offered on a pass/fail basis, primarily because we wanted to create a community of collaborators rather than competitors. Additionally, we felt that having students strive for a letter grade, and thus focus on the final answers and point values, would detract from them learning how to think like scientists. This perspective was confirmed by students enrolled in the course: more than 50% of students preferred that the course was offered pass/fail, and the most common reason cited was the desire to avoid having to worry about points on each assignment (unpublished data). It has also been shown that offering courses on a pass/fail basis rather than assigning a letter grade can lead to improvements in psychological well-being (Bloodgood et al., 2009) and group cohesiveness (Rohe et al., 2006); we were most interested in group cohesiveness, because we wanted the students to view themselves as collaborators.
Accountability.
Because motivation can be a problem for pass/fail courses (Gold, 1971), we implemented a policy that required students to earn above a 70% on the average of three exams that focused on data interpretation and experimental design. These exams were completed individually. Students also needed to earn an average of 70% on their postlab assignments and final posters. These standards encouraged students to take the course seriously and held them accountable for what they learned; each year, only one to two students failed the course due to low exam scores.
Steps Taken to Implement a High-Enrollment Required CURE
Before this new course was implemented, the existing introductory biology lab course was a standard “cookbook” course in which the topics and organism studied changed every 2 wk and students worked through predetermined protocols to get a known “right” answer. The impetus for redesigning the course was a history of poor student evaluations for this course and a desire among faculty members to improve the course, in conjunction with national calls for biology lab course reform (NRC, 2003; AAAS, 2011).
Because we were uncertain how students would react to this new course or whether this type of course would be possible for a large, introductory population of students, we decided to gradually scale up the course. The new lab course was introduced in 2 yr of pilot versions of the course. In Winter 2010, a pilot version of the research-based course was taught to ∼20 students who volunteered to take it rather than the older “cookbook” course. Based on the positive reaction to the new course expressed in interviews and attitudinal and self-efficacy surveys (unpublished data), the course was scaled up to a larger group of students the next year. The course was taught in Winter 2011 to ∼40 students who were chosen at random (i.e., nonvolunteers) into either the pilot course or the existing cookbook lab course, and evaluation of the pilot course was largely positive (unpublished data). In Winter 2012, the new course was implemented to 250+ students, and four PhD-level instructors taught different sections of the course. Increased collaboration between students within the lab sections and in mutant groups was added to the course, as was a poster session at the end of the course. The curriculum that we are assessing is the most recent curriculum, which has now been taught in approximately the same way for five quarters (Winter 2012, Fall 2012, Winter 2013, Fall 2013, and Winter 2014) to a large-enrollment population of nonvolunteer students, giving us confidence that implementing this type of course in a large introductory setting as a required course is possible.
EVALUATION
Methods
To assess the course, we used an approach of analyzing pre- and postcourse open-ended written responses and course exams. Because time for assessment was limited in the 10-wk quarter, we have collected different types of data from the two times the course has been offered as a required component of the introductory curriculum (Fall 2012 and Winter 2013).
Students were given pre- and postcourse surveys that included open-ended questions about their conception of what it meant to think like a scientist. Question prompts were designed through a series of think-aloud interviews with students. Student responses to open-ended questions were coded using a combination of content analysis and grounded theory (Glaser, 1978; Glaser et al., 1968) to identify themes, from which specific categories were chosen. Two independent raters scored a subset of student responses, and they came to a consensus when they disagreed. The frequency of each student response was calculated for each category. Student responses could include more than one idea, so responses do not sum to 100%. All students enrolled in the course completed the surveys, and a random subset of student responses was used in the analysis; lab sections were chosen at random, and all student responses in those sections were included in the analysis. Post hoc, we made sure that student responses were included for each instructor and time slot (day and time) to minimize the bias that may be caused by only examining student responses from one instructor or from one section time. Data used are from Fall 2012 (n = 60) and Winter 2013 (n = 117). Statistical analysis was done using paired t tests (p < 0.05).
Additionally, open-ended questions on the postcourse surveys asked students whether their thinking like a scientist had changed as a result of the course and, if so, how their own thinking like a scientist had changed. Data used are from Fall 2012 (n = 60). Students were also asked specific Likert-scale questions about what components of the course were important for their understanding of thinking like a scientist. The average Likert score for each question and the SD were calculated. Data used are from Winter 2013 (n = 117).
Students’ ability to design experiments and interpret data were measured by assessing their scores on three exams. We determined the composition of questions focused on data analysis by having two independent raters review exam questions. We found that 50% of the total points on the exams asked students to read graphs or interpret figures and tables and 21% of the total points on the exams were questions that verbally described experimental results or conditions, so 71% of the exam points were specifically directed at eliciting student understanding of data analysis and interpretation. Thus, the composition of the exam questions was predominantly data analysis or interpretation questions, giving us confidence that this could be a measure of student ability to analyze and interpret data.
We characterized the cognitive level of each question on the exams (Crowe et al., 2008) and calculated a weighted Bloom average for each exam adapted from Freeman and colleagues (2011). We also calculated a difficulty score for each question, using a rubric that was developed by the course instructors (Table 3). To do this, we scrambled all the exam questions so the rater would not know which question came from which exam. We had two former instructors for this course blindly score the questions according to Bloom’s level and difficulty. They achieved greater than 80% interrater reliability on the Bloom’s level and 100% interrater reliability on the difficulty score. These former instructors did not teach the course in the term that used these particular exams, so they did not have knowledge about specific questions on specific exams. Additionally, these former instructors had an expert understanding of the course content and familiarity with what was explicitly covered in the course, so they knew how the data were typically presented to students in the course and what specific experiments students conducted. Scores were compared and discussed until consensus was achieved. Data used are from Winter 2013.
Table 3.
Difficulty rubric for exam questions
| Easy (1) | Definitions/explanations of what was previously presented in lab (e.g., purpose of a particular step of an experiment or fact about cancer) |
| Medium (2) | Students need to apply their knowledge to a situation that they experienced in lab or analyze the results of one graph/figure in the same way they analyzed it in lab (e.g., students have to predict what went wrong when given an experimental result). |
| Difficult: complex data or near transfer (3) | Students need to apply their knowledge to either a complex set of data (more than one figure at once) or to data presented in a novel way (e.g., students interpreting an unfamiliar graphical representation of data). |
While a common method of assessment in biology education is a pre–post test format, we chose not to give students a pretest focused on data analysis at the beginning of the course, because there was too much content-dependent information that students would not have known (e.g., how to interpret specific assays). We also chose not to give students a pre–post test on their content knowledge, because that was not one of our course goals. We were using yeast genetics to explore p53 and cancer as a model system; students had to learn certain details about the system to be able to pose hypotheses and analyze data, but we focused on content information only to the extent that it was necessary for them to learn the process skills. It would be interesting to see how students perform on a pre–post test of their ability to interpret data in a content-independent way, but at the time of our study, no such assessment tool existed.
Results
Finding 1: Students Show a More Expert-Like Conception of What It Means to Think Like a Scientist at the End of the Course and Perceive That Their Own Thinking Has Changed.
We found that student understanding of what it means to think like a scientist became significantly more nuanced and similar to expert scientists’ thinking when comparing their postcourse and precourse answers to the question: “What do you think it means to think like a scientist?” (Table 4). Specifically, students at the beginning of the course mentioned being curious, being critical or logical, developing hypotheses, or using the scientific method. However, at the end of the course, the responses were more grounded in their lab experience, including a focus on collaboration and data analysis. Specifically, students at the end of the course mentioned needing to be skeptical of data, the need to repeat experiments, how scientists can learn from failed experiments, and how there are multiple ways to approach a problem (Table 4).
Table 4.
Student responses to the open-ended question “What does it mean to think like a scientist?”a
| Percentage of responses categorized under this theme | |||
|---|---|---|---|
| Theme | Precourse | Postcourse | Example student responses |
| Involves collaboration | 0 | 20.5* | “This quarter has taught me the importance of working collaboratively with others in order to more fully understand the topic of research.” “Be willing to collaborate.” |
| Requires analyzing and interpreting data | 6.0 | 31.6* | “Thinking like a scientist requires lots of analyzing of data and asking so what? Why is this? What is next?” “One has to analyze the results and try to interpret them and then draw up other experiments that can confirm the results.” |
| Being skeptical of data | 0 | 18.8* | “It also means to be skeptical and critical of data, and to never trust just one set of data but try to continuously strive for accurate and less variable results.” “To not be stubborn and ignore results that contradict with your hypothesis.” |
| Need to repeat experiments | 2.6 | 12.8* | “Test and retest.” “Additionally, I have learned the importance of repeating experiments and testing hypotheses in a variety of ways in order to gain more significant data.” |
| Learn from mistakes/failed experiments | 1.7 | 9.4* | “Troubleshooting experiments that don’t go as planned, i.e., designing experiments to figure why the original experiment wasn’t working.” “Identify why errors occurred.” |
| QUERY as a way to organize thinking for each experiment | 0 | 15.4* | “It means asking a question, designing an informed hypothesis based on background knowledge, creating an experiment to test this question and interpreting the results. (QUERY).” “QUERY is a huge part in being able to think like a scientist.” |
| Using multiple approaches to answer a question; many ways of thinking | 9.4 | 17.9* | “As a scientist, you want to approach a topic or research points from multiple angles. There may be one experiment that shows X, but you always want to verify that result with other experiments. As a scientist, you want to realize that experiments have limitations and by having multiple experiments to support one another, you can synthesize a conclusion from all the data.” “Do a series of experiments to try and answer it while using both qualitative and quantitative methods.” |
| Critical, logical thinking | 25.6 | 21.4 | “Thinking like a scientist means thinking critically and thinking through all possibilities in order to best devise controls and alternate experiments.” “You have to make a conscious effort to not jump to conclusions you want to be true and only take what the data gives [sic] you.” |
| Developing hypotheses | 21.4 | 29.1* | “You try to learn more about the world around you by creating experiments and testing hypothesis [sic].” “Form educated hypotheses.” |
| Using the scientific method | 16.2 | 1.7* | “Think like a scientist means to constantly employ the scientific method—observe, hypothesize, experiment, conclude—in all aspects of life in order to reach better understandings of different topics.” “Thinking like a scientist means to think in a logical and organized method, in particular adhering to the scientific method. In such a method, research begins with an observation, hypothesis, question prediction, followed by the research and data and results.” |
| Generalized vague statement about being curious | 23.9 | 16.2* | “To be curious about how and why things work and to then pursue those curiosities with experimentation.” “To be inquisitive. A desire to learn how/why things work and how things break down/what causes them to malfunction.” |
*p < 0.05, paired t tests.
aData from Winter 2013 (n = 117).
When we asked students at the end of the course whether their own thinking like a scientist had changed during their investigation of mutant p53, we found that 100% of students (n = 60) thought that their thinking like a scientist had changed (Table 5). In response to an open-ended question of how their thinking like a scientist changed, 83% of student responses could be classified as thinking scientifically, as follows: 1) collaboration is important (20%), 2) be skeptical of data and make only tentative conclusions (38%), 3) repeat experiments (27%), 4) learn from making mistakes/failed experiments (13.3%), 5) use multiple approaches to answer a problem (13.3%), and 6) controls are important. Thus, not only did students have a clearer idea of what it meant to think like a scientist, they felt as though they had acquired these skills as a result of taking this course.
Table 5.
Student responses at the end of the course to the open-ended question “How has your own thinking like a scientist changed during your investigation of mutant p53?”a
| Theme | Percentage of responses categorized under this theme | Example student responses |
|---|---|---|
| Collaboration | 20 | “I learned that science is collaborative work.” “I have learned it is essential to work together with the other groups in order to critically look at our experimental results before coming to a general conclusion.” |
| Being skeptical of data | 38 | “You can’t always trust your results because the procedure may need to be optimized.” “I learned not to get too attached to the results of any one experiment.” “Before I would have been more prone to quickly accept the results from science experiments as being always correct. But because this course taught us to question our results and look for possible sources of errors, I developed a more critical eye when interpreting experimental results.” |
| Need to repeat experiments | 27 | “I also realized that one set of data are not enough to make a conclusion. Many repeated attempts must give a similar result for the conclusion to be valid and consistent.” “Requiring multiple confirming data results to be able to conclude anything definitive about the mutant (rather than jumping to conclusions after a single experimental result)” |
| Learn from mistakes/failed experiments | 13.3 | “I also learned how to think like a scientist not only when my experiments worked, but when they didn’t work as well.” “Mistakes in my assay helped me think more critically about how an experiment is performed.” “I also am happy that errors occurred in the process, because troubleshooting them really helped me develop greater critical thinking skills, instead of just following the protocol.” |
| Using multiple approaches to answer a question; many ways of thinking | 13.3 | “That web-like thinking is how a scientist thinks – not a straight line.” “I learned how to attack problems from many different angles.” |
| Need to use controls | 15 | “Scientists must use positive and negative controls (I used to think a positive control would be enough)” “I understand the necessity of both positive and negative controls. Prior to this course, I only ever thought about having negative controls.” |
aData from Fall 2012 (n = 60).
Finding 2: Students Indicated That Specific Aspects of the Course Focused on Data Analysis and Collaboration, Including the Mutant Group Discussions, Were the Most Useful for Their Learning How to Think Like a Scientist.
At the end of the course, students were asked in an open-ended question what specific aspects of the course were most useful in helping them to think like a scientist. The most frequent themes that emerged after analyzing the data that captured 92% of student responses included (Table 6): 1) mutant group discussions (27%), 2) data analysis aspect of postlab assignments (26%), 3) performing different experiments on one longitudinal question (24%), 4) predicting results and comparing actual results with predicted ones (15%), 5) troubleshooting failed experiments (8%), and 6) collaborating with other students in the class (8%).
Table 6.
Student-generated ideas about what specific aspects of the course were most useful for their thinking like a scientista
| Theme | Percentage of responses that were categorized under this theme | Example student responses |
|---|---|---|
| Mutant group discussions | 26.5 | “I enjoyed mutant group discussions because it gave us an opportunity to combine data and come up with more accurate conclusions about our mutant. It showed if our data agreed and if it didn’t, we could explore possible reasons why this was so.” “Getting such varied results even within same mutant group was eye-opening.” |
| Data analysis and future directions aspects of postlabs | 25.6 | “Postlab, b/c they always emphasized analysis of data and trying to hypothesize why some results occurred.” “Parts of the postlab asking about future directions or possible causes; understanding reasons for doing experiments.” |
| Performing different experiments on one longitudinal question | 23.9 | “The fact that we used the whole quarter to answer one research question about p53 was helpful because it showed the time and complexity involved in scientific thinking.” “The multiple experiments aimed at only elucidating a small part of the puzzle that slowly built to an overall picture was [sic] a good technique.” |
| Brainstorming experiments, predicting results, and comparing actual results with predicted ones | 15.4 | “A specific example that I enjoyed was when we were presented our results which did not match up with our predictions and we had to determine why we didn’t get our derived results.” “The assignment where we had to come up with our own experiments was the most difficult since we had to think as scientists. We had to find an issue we wanted to investigate and come up with a procedure to do it.” |
| Troubleshooting failed experiments | 7.7 | “Tests that failed. Inconclusive data from group. Sooo [sic] many errors possible.” “Learning about the details of the experiments and not just about the results helped me understand the sort of controls and errors and stuff I should look for.” |
| Collaborating with other students in the class | 7.7 | “Working with other groups—get a feel for the community of scientific research. Exploring potential problems on our own first—got us really thinking about what we were doing and why.” “Specifically, I really enjoyed working on the postlabs with a partner and being able to analyze the results with someone.” |
aData from Winter 2013 (n = 117).
In addition, we gave students a list of some of the design elements of the course and asked them to rate these items on a five-point Likert scale on how useful a particular component of the course was for improving their thinking like a scientist (1, not at all useful; 2, slightly useful; 3, moderately useful; 4, very useful; and 5, extremely useful). Once again, we found that components of the course that related to collaboration and data analysis were most highly rated (Table 7), with the one exception being “your instructor’s teaching.” However, the majority of instruction that occurred in the lab was focused on data analysis, so it could be that students were referencing the instructor teaching them this skill.
Table 7.
Aspects of the course that were most useful for improving your thinking like a scientista
| Component of course | Mean (SD) |
|---|---|
| Analyzing your own data | 4.35 (0.68) |
| Working with a partner on all aspects of the project | 4.30 (0.87) |
| Your instructor’s teaching | 4.23 (0.85) |
| Comparing your data with data from other groups working on the same mutant in your lab section | 4.16 (0.81) |
| Completing postlab assignments | 4.12 (0.77) |
| Creating a poster | 3.45 (1.16) |
| Mutant group discussions | 3.42 (1.24) |
| Reading a primary scientific paper | 3.37 (1.14) |
| Using the QUERY method | 3.32 (1.13) |
| Comparing your data with data from other groups working on a different mutant in your lab section | 3.27 (1.19) |
| Designing your own experiment | 3.18 (1.13) |
| Repeating experiments that did not work the first time | 3.12 (1.11) |
| Working through the handouts during lab | 3.07 (1.03) |
| Course exams | 3.05 (0.91) |
aStudents evaluated this question on a closed-ended Likert scale (1, not at all useful; 2, slightly useful; 3, moderately useful; 4, very useful; 5, extremely useful). Data from Winter 2013 (n = 117).
An interesting comparison between the open-ended data and the Likert-scale data is that mutant group discussions was the most frequent response for the open-ended question but was only rated in between moderately useful and very useful on the Likert scale. Additionally, the SD is large, indicating that this aspect of the course may be polarizing for students or that perhaps the quality of the different mutant group discussions was variable. Different instructors facilitated these discussions and may have emphasized different concepts.
To investigate this further, we asked students what was the single greatest strength of the mutant group discussions in an open-ended question (Table 8). Ninety percent of the responses for the greatest strength could be classified as 1) confirming whether their data agreed or disagreed with other students’ data, 2) achieving consensus about functional defect, 3) discussing possible sources of error, and 4) representative of a scientific lab’s discussion of data.
Table 8.
Single greatest strength of the mutant group discussionsa
| Theme | Percentage of responses categorized under this theme | Example student responses |
|---|---|---|
| Confirming that “their data” were similar to others by comparing results | 54.7 | “The single greatest strength of the mutant group discussion was to have the opportunity to compare our data with that of the other mutant groups.” “Being able to see that our results were not completely askew.” |
| Achieving consensus about functional defect of mutant p53 | 24.8 | “Verify/compare results to the same experiment to simulate conducting multiple trials.” “It was very beneficial to see some consensus or lack of consensus between teams; it put everything more in perspective.” |
| Discussing possible sources of error in experiments | 13.7 | “Discussing the results each group got and which were the possible correct results. Determining reasons for any outliers.” “An opportunity to discuss possible sources of variation and the strength of our controls.” |
| Representative of an authentic lab discussion about data | 6.0 | “A preview of what collaboration in a lab is actually like (people sometimes get completely different results and those differences need to be reconciled).” “Collaborating was always interesting and made our results feel much more real.” |
aData from Winter 2013 (n = 117).
Students in this class seemed to display one of two distinct mind-sets: 1) those who viewed “their data” as the data that they themselves had collected and 2) those who viewed “their data” as the collective set of data derived from their mutant group. Those ∼55% of responses that held onto the idea that their data were only the data that they had collected with their own hands used the mutant groups as a way to confirm that they had done the experiment correctly (i.e., they had a similar result as everyone else) or to determine whether they had made a mistake (i.e., they had a different result from everyone else), but they failed to understand the larger benefit of having multiple people working on the same mutant as part of a collective research team. This disconnect and apparent disappointment in the mutant groups was captured by the following responses:
“Sometimes we failed to reach a conclusion, since often we all had conflicting results.”
“[Mutant groups] were drawn out and inconclusive.”
“The greatest area for improvement for the mutant discussion group is reaching a more firm and assertive group conclusion.”
These students missed the idea that purpose of the mutant group discussions was to illustrate the inherent variability of scientific data and the inappropriateness of drawing conclusions based on one replicate of one experiment. These students wanted an answer, the “right” answer, and were frustrated when effort was spent and no conclusion could be reached.
However, the other group of students (∼25% of responses) seemed to understand that the data they had collected with their own hands were one set out of a larger collaborative data set that they would use to determine the functional defect of their mutant p53; thus, the mutant group discussions became paramount for these students’ understanding of the significance of repetition of experiments and in statistical analysis of results, and their responses captured this:
“Being able to compare your data to better understand what next steps to take and what was actually happening with our p53 mutation.”
“The opportunity to be away from the lab and have the time to sit and actually process and discuss what we’re doing in class. Also the opportunity to talk to other mutant groups and think about what could have gone wrong, what are the general trends etc. was really great in actually understanding the lab content. I think this was the best learning experience of the entire lab.”
Finding 3: Students Showed Improvement in Their Ability to Analyze and Interpret Data.
We designed three exams to test student ability to analyze and interpret data, including understanding the significance of biological variation, repetition of experiments, and how to analyze the data they had obtained during their investigations into the functional defects of p53. Some of the questions were based on specific experiments they had conducted, but others required students to analyze data in a novel way or to transfer their knowledge to a novel scenario (see Supplemental Material for sample questions).
Average student scores on these exams remained constant over the term; students’ average scores were 87.1% on exam 1, 86.6% on exam 2, and 88.2% on exam 3 (Table 9). Student performance on exams was not curved or normalized in any way. While the total score remained constant, the Bloom’s level of the average exam question increased significantly over the term (Kruskal-Wallis χ2 = 13.7, df = 2, p = 0.001). Average weighted Bloom levels were 2.69, 2.91, and 3.61 for exams 1, 2, and 3, respectively. Additionally, we designed a “difficulty scale” based on whether students were giving an explanation of what was already presented in the lab course, were examining data in the same way they had been presented the data in class, or were faced with a more complex set of data presented in a novel way. Based on this difficulty scale, exam 1 was 1.61, exam 2 was 1.82, and exam 3 was 2.41. Thus, the exams had a significant increase in the difficulty of the average exam question (Kruskal-Wallis χ2 = 10.35, df = 2, p = 0.006).
Table 9.
Student ability to analyze data improved as the exams got more difficult and their exam scores stayed constanta
| Average student score | Weighted average Bloom score for each exam | Weighted average difficulty score for each exam | |
|---|---|---|---|
| Exam 1 | 87.1% | 2.69 | 1.61 |
| Exam 2 | 86.6% | 2.91 | 1.82 |
| Exam 3 | 88.2% | 3.61 | 2.41 |
aData from Winter 2013 (n = 117).
DISCUSSION
In this study, we present an innovative curriculum that allows students to experience research in the context of an introductory lab course. One of the strongest aspects of this course is that it is required of all biology majors, thus giving all biology majors exposure to research at the undergraduate level. Many CUREs have only been offered to a small number of volunteers, which limits the ability to generalize conclusions drawn from the evaluation of these smaller courses. With the scale-up of this course, every biology major at this institution engages in a research project as part of his or her undergraduate curriculum. Additionally, it makes scientific research more inclusive by limiting the number of hurdles students have to jump through to engage in research and by removing the selection process that faculty members use to pick students for individual apprenticeships (Bangera and Brownell, 2014).
Gains in Student Conceptions of What It Means to Think Like a Scientist
The theme of thinking like a scientist has appeared in other studies of traditional independent research apprenticeships, although in contrast to these other studies, our definition did not include deeper conceptual knowledge about the topic (Seymour et al., 2004; Hunter et al., 2007). We limited our definition to what would be considered process skills (Coil et al., 2010) or core competencies (AAAS, 2011) and divorced it from conceptual understanding or affective benefits.
We found that students’ thoughts about what it means to think like a scientist transitioned as a result of the course. At the beginning of the course, students had a vague, general view of what it means to think like a scientist. Many student responses stated that scientists were critical thinkers, were curious about the world around them, and asked questions. However, not one precourse response included references to collaboration or being skeptical of data, and very few mentioned data analysis, the need to repeat an experiment, or learning from failed experiments—all of which were mentioned in the postcourse responses. We conclude that students’ conceptions of what it means to think like a scientist became more reflective of the views of practicing scientists (Table 1) and more grounded in students’ experiences of working closely with data. Additionally, students highlighted the importance of collaboration in thinking like a scientist, a trait that has been emphasized as a core competency in Vision and Change (AAAS, 2011).
Interestingly, while many students mentioned the scientific method as part of what it meant to think like a scientist in the beginning of the course, few students mentioned it at the end of the course. We interpret this to mean that students’ conceptions of science shifted away from the idea that science is a rigid, step-by-step method (e.g., “the scientific method”) to an understanding that it is a process that is responsive to experimental results and constantly evolving in the face of new data, new techniques, and new ideas. This emphasis on data was echoed when students were asked about the specific aspects of the course that contributed to an improvement in their thinking like a scientist: data-centered collaborative aspects of the course were highlighted. Students focused on mutant group discussions and postlabs as sources of scientific thinking. Building multiple opportunities for students to think critically about data, including having them repeat experiments, helped students grasp the variability that is inherent to experimental science and the need for multiple, independent observations to build convincing support for a hypothesis.
However, we saw that many students still struggled in the mutant groups with not being able to find a “right” answer. This may indicate that we need to integrate more opportunities for mutant group discussions into the course and more explicit discussion of the importance of biological replicates in order to come to a conclusion.
Documented Learning Gains in Student Ability to Interpret Data
In pilot studies, we saw that students’ confidence in their ability to analyze data improved as a result of this course and through exam analysis (unpublished data). Confidence does not always correlate with ability, but here we demonstrated that their ability to analyze data and draw conclusions about experiments improved as a result of the course. As the difficulty of exams increased, students’ average scores on the exams remained constant. This is particularly encouraging, because these exams were not high-stakes exams. We wanted to establish a culture of collaboration rather than competition, so we chose to have the course be ungraded and taken only as pass/fail. Students were required to obtain 70% on the average of the three exams but were not rewarded for performing better than that. Despite the potential lack of motivation, we still saw student improvement in the ability to analyze data. This is important, because most assessments of CUREs rely on student self-reporting of their abilities rather than one measurement of their actual ability; we were able to document learning gains, and others interested in assessing CUREs could use a similar method.
Advantages of a Large-Enrollment Course
Finally, rather than a large course enrollment being an inconvenience, the specific format of the course actually benefits from having large numbers of students working on the same mutant alleles and sharing data. More students meant a larger number of replicates and higher probability of seeing a conclusive pattern in the data. We acknowledge that our enrollment of ∼200–300 students, while large by the standards of our institution, is not large by the standards of others that need to cater to more than 1000 introductory-level students. However, we believe that this course could be scaled up further and are interested in working with others to help adopt this course, or similar courses, at their institutions.
LIMITATIONS OF INTERPRETATIONS
Teacher Effect May Be Acting Independently of the Curriculum
This course was taught by PhD-level instructors, which likely contributes to an “instructor effect” that has an impact distinct from the curriculum itself on student learning. Our goal for this study was to examine the course curriculum rather than an individual instructor’s teaching, but the two by their very nature are entwined. It could be possible that the gains we saw are due to the synergistic effects of the research-based curriculum and having expert scientists as instructors. We specifically decided to have PhD-level instructors, because we wanted instructors with expertise in scientific thinking, but we hope that it is possible to replicate the same quality of high-level instruction by using more advanced graduate students or postdoctoral fellows.
It is also important to note that instructors all had very different teaching styles, ranging from a more traditional lecturing to more interactive active-learning teaching, and diverse training in biological sciences. While we do not see notable differences among instructors on most of the assessments, leading us to conclude that the major source of impact for the course is derived from the common curriculum, there was one instance in which particular instructors had significant differences. In Fall 2012, for two of the instructors, more than 90% of the student responses could be categorized as “thinking like a scientist,” whereas this was only 50% for one of the other instructors. For others interested in developing this type of course and having the course taught by instructors with diverse teaching experience and/ or styles, it will be important to determine whether instructors with limited teaching experience (e.g., first-year graduate students) can adequately facilitate this type of course to achieve the desired learning gains.
Student Population May Limit Generalizations
Another critical caveat arising from our evaluation of the benefits of this type of course for an introductory population of students is that the student population at Stanford includes a high proportion of high achievers. Even though this is the first biology lab experience for them in college, a significant percentage of our students have had previous research experience. Although we acknowledge that this is likely perceived as a limitation to generalizing our results to all universities, there are students in this cohort who have no prior research experience and a weak biology background. Furthermore, unlike many CUREs, students are required to take this course, so there is no self-selection bias.
Although our population is high achieving, it is considerably ethnically diverse. White students make up less than 40% of the class, with almost 20% of the students coming from historically underrepresented minority populations. Additionally, ∼15% of Stanford undergraduates are the first in their family to attend a 4-yr college.
Cost of Running This Type of Course
Although we encourage others to adopt a similar curriculum, this course does have an associated cost that may be a barrier for many institutions. We were fortunate to receive several grants to ensure ongoing commitment to the course. In addition to start-up costs (e.g., equipment), there are high costs for experimental materials and supplies (e.g., antibodies, the DNA-binding assay kits). Although there are ways to decrease the cost of running this course by not doing particular experiments, the cost of running a molecular course is inherently higher than other types of biological courses. As budgets become tighter, this does present a logistical challenge to those interested in developing this type of course. As a contrast to this high-cost course, we have developed another research-based lab course that integrates teaching and research; it focuses on ecological relationships and is a low-cost alternative that also gives students a research experience in the context of an introductory biology lab course. For an overview and assessment of that course, see Brownell et al. (2012) and Kloser et al. (2013).
CONCLUSION
We encourage others to consider developing this type of high-enrollment course; it allows students the opportunity to think about larger data sets than would be possible from a small 10–20 person class. Additionally, the opportunities for collaboration are extensive. However, a larger class is logistically more challenging. We encourage others to start small and then build from there; our first pilot year we had only 20 students and few of the experiments worked. In the second year, more of the experiments worked, but kinks still needed to be worked out. It took many iterations of the course to work out the logistical challenges associated with this type of course, and even after seven iterations, not all of the experiments work every time. The most frequent complaints from students the first two times the course was offered centered on course logistics rather than on content, curriculum, or instruction. While logistics may seem insignificant and often are less of a problem in a lecture-based course, they can be significant barriers to achieving course goals in a lab course.
This course is a way to expose all students, not a select few, to the joys and challenges of research, and it gives them a more realistic impression of how research is done. We urge others to consider adopting this type of course-embedded research experience as a way to make research more accessible to a larger population of students who likely would not pursue independent research on their own (Bangera and Brownell, 2014). This course, and others like it, can be instrumental for providing all students with the opportunity to learn how to do science.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this course came from a National Science Foundation TUES (0941984), a Hoagland Award for Innovations in Undergraduate Teaching, an HHMI education grant, and a generous gift from a private donor. We thank the 700+ students who have participated in this course, in particular the students who took the course during the initial pilot phases, and the graduate student teaching assistants who helped teach the course, particularly Jared Wegner and Dan Van de Mark. Charles Anderson (now at Penn State University) first coined “QUERY” as an educational concept, and it has now been incorporated into the course. We are especially grateful to Nicole Bradon for her technical assistance in preparing reagents and equipment for the weekly student laboratories. We also acknowledge the support of colleagues in the Department of Biology, including Shyamala Malladi, Matt Knope, Waheeda Khalfan, Tad Fukami, and the leadership of our former department chairman, Bob Simoni. Additionally, Rich Shavelson and Matt Kloser at the Stanford Graduate School of Education were instrumental in early discussions of course assessment.
REFERENCES
- American Association for the Advancement of Science. Vision and Change in Undergraduate Biology Education: A Call to Action. Washington, DC: 2011. [Google Scholar]
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, et al. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ. 2014;13:29–40. doi: 10.1187/cbe.14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangera G, Brownell SE. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sci Educ. 2014;13:602–606. doi: 10.1187/cbe.14-06-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood RA, Short JG, Jackson JM, Martindale JR. A change to pass/fail grading in the first two years at one medical school results in improved psychological well-being. Acad Med. 2009;84:655–662. doi: 10.1097/ACM.0b013e31819f6d78. [DOI] [PubMed] [Google Scholar]
- Brownell SE, Kloser MJ. Toward a conceptual framework for measuring the effectiveness of course-based undergraduate research experiences in undergraduate biology. Studies High Educ. 2015;40:525–544. [Google Scholar]
- Brownell SE, Kloser MJ, Fukami T, Shavelson R. Undergraduate biology lab courses: comparing the impact of traditionally based “cookbook” and authentic research-based courses on student lab experiences. J Coll Sci Teach. 2012;41:36–45. [Google Scholar]
- Brownell SE, Kloser MJ, Fukami T, Shavelson RJ. Context matters: volunteer bias, small sample size, and the value of comparison groups in the assessment of research-based undergraduate introductory biology lab courses. J Microbiol Biol Educ. 2013;14:176. doi: 10.1128/jmbe.v14i2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell SE, Wenderoth MP, Theobald R, Okoroafor N, Koval M, Freeman S, Walcher-Chevillet CL, Crowe AJ. How students think about experimental design: novel conceptions revealed by in-class activities. BioScience. 2014;64:125–137. [Google Scholar]
- Coil D, Wenderoth MP, Cunningham M, Dirks C. Teaching the process of science: faculty perceptions and an effective methodology. CBE Life Sci Educ. 2010;9:524–535. doi: 10.1187/cbe.10-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin LA, Graham MJ, Dolan EL. Modeling course-based undergraduate research experiences: an agenda for future research and evaluation. CBE Life Sci Educ. 2015;14:es1. doi: 10.1187/cbe.14-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe A, Dirks C, Wenderoth MP. Biology in Bloom: implementing Bloom’s taxonomy to enhance student learning in biology. CBE Life Sci Educ. 2008;7:368–381. doi: 10.1187/cbe.08-05-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druger M, Siebert ED, Crow LW (eds.) Teaching Tips: Innovations in Undergraduate Science Instruction, Arlington, VA. NSTA Press. 2004 [Google Scholar]
- Etkina E, Planinši G. “Thinking like a scientist.”. Physics World. 2014;27:48. [Google Scholar]
- Freeman S, Haak D, Wenderoth MP. Increased course structure improves performance in introductory biology. CBE Life Sci Educ. 2011;10:175–186. doi: 10.1187/cbe.10-08-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Erdeniz N. Characterization of pathogenic human MSH2 missense mutations using yeast as a model system: a laboratory course in molecular biology. Cell Biol Educ. 2004;3:31–48. doi: 10.1187/cbe.03-08-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser BG. Theoretical Sensitivity: Advances in the Methodology of Grounded Theory, vol. 2. Mill Valley, CA: Sociology Press; 1978. [Google Scholar]
- Glaser BG, Strauss AL, Strutzel E. The discovery of grounded theory; strategies for qualitative research. Nurs Res. 1968;17:364. [Google Scholar]
- Gold RM. Academic achievement declines under pass-fail grading. J Exp Educ. 1971:17–21. [Google Scholar]
- Handelsman J, Ebert-May D, Beichner R, Bruns P, Chang A, DeHaan R, Gentile J, Lauffer S, Stewart J, Tilghman SM, Wood SB. Scientific teaching. Science. 2004;304:521–522. doi: 10.1126/science.1096022. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Hunter AB, Laursen SL, Seymour E. Becoming a scientist: the role of undergraduate research in students’ cognitive, personal, and professional development. Sci Educ. 2007;91:36–74. [Google Scholar]
- Hurtado S, Eagan K, Sharkness J. Thinking and Acting Like a Scientist: Investigating the Outcomes of Introductory Science and Math Courses. Atlanta, GA: Association of Institutional Researchers Forum; 2009. [Google Scholar]
- Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SCR, et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio. 2014;5:e01051. doi: 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloser MJ, Brownell SE, Chiariello NR, Fukami T. Integrating teaching and research in undergraduate biology laboratory education. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001174. e1001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloser MJ, Brownell SE, Shavelson RJ, Fukami T. Effects of a research-based ecology lab course: a study of nonvolunteer achievement, self-confidence, and perception of lab course purpose. J Coll Sci Teach. 2013;42:90–99. [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lopatto D, Alvarez C, Barnard D, Chandrasekaran C, Chung H-M, Du C, Eckdahl T, Goodman AL, Hauser C, Jones CJ, et al. Genomics Education Partnership. Science. 2008;322:684–685. doi: 10.1126/science.1165351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager WH, Winderickx J. Yeast as a model for medical and medicinal research. Trends Pharmacol Sci. 2005;26:265–273. doi: 10.1016/j.tips.2005.03.004. [DOI] [PubMed] [Google Scholar]
- National Research Council. BIO2010: Transforming Undergraduate Education for Future Research Biologists. Washington, DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- Rohe DE, Barrier PA, Clark MM, Cook DA, Vickers KS, Decker PA. The benefits of pass-fail grading on stress, mood, and group cohesion in medical students. Mayo Clin Proc. 2006;81:1443–1448. doi: 10.4065/81.11.1443. [DOI] [PubMed] [Google Scholar]
- Russell SH, Hancock MP, McCullough J. Benefits of undergraduate research experiences. Science. 2007;316:548–549. doi: 10.1126/science.1140384. [DOI] [PubMed] [Google Scholar]
- Schärer E. Mammalian p53 can function as a transcription factor in yeast. Nucleic Acids Res. 1992;20:1539–1545. doi: 10.1093/nar/20.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour E, Hunter AB, Laursen SL, DeAntoni T. Establishing the benefits of research experiences for undergraduates in the sciences: first findings from a three-year study. Sci Educ. 2004;88:493–534. [Google Scholar]
- Shaffer CD, Alvarez C, Bailey C, Barnard D, Bhalla S, Chandrasekaran C, Chandrasekaran V, Chung HM, Dorer DR, Du C, et al. The Genomics Education Partnership: successful integration of research into laboratory classes at a diverse group of undergraduate institutions. CBE Life Sci Educ. 2010;9:55–69. doi: 10.1187/09-11-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer CD, Alvarez CJ, Bednarski AE, Dunbar D, Goodman AL, Reinke C, Rosenwald AG, Wolyniak MJ, Bailey C, Barnard D, et al. A course-based research experience: how benefits change with increased investment in instructional time. CBE Life Sci Educ. 2014;13:111–130. doi: 10.1187/cbe-13-08-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov RV, Haupt Y. The cellular response to p53: the decision between life and death. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- Soussi T, Legros Y, Lubin R, Ory K, Schlichtholz B. Multifactorial analysis of p53 alteration in human cancer: a review. Int J Cancer. 1994;57:1–9. doi: 10.1002/ijc.2910570102. [DOI] [PubMed] [Google Scholar]
- Sundberg MD, Armstrong JE, Wischusen EW. A reappraisal of the status of introductory biology laboratory education in US colleges & universities. Am Biol Teach. 2005;67:525–529. [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


