Abstract
Hepatitis C virus (HCV) represents a major health burden with more than 170 million individuals currently infected worldwide, equaling roughly 3% of the world’s population. HCV preferentially infects hepatocytes and is able to persist in up to 70% of infected individuals. It is estimated that up to 30% of chronically infected individuals will go on to develop progressive liver disease as a result of HCV infection, making the virus the leading cause of liver transplantation in the world. Currently there is no vaccine for HCV. In this study, we have taken a multi-step approach to develop a novel genotype 1a/1b consensus HCV NS3/NS4A DNA vaccine able to induce strong cellular immunity. We show that this construct is able to induce strong anti-NS3/NS4A T cell responses in C57BL/6 mice, as well as, in Rhesus Macaques. Our data suggest that DNA vaccines encoding HCV proteins NS3/NS4A merit further study in the context of future prophylactic and therapeutic HCV T cell based vaccines.
Keywords: HCV, NS3, NS4A, DNA vaccine, consensus
1. Introduction
Hepatitis C (HCV) is a small enveloped, positive stranded RNA virus that represents a major health burden worldwide with more than 170 million individuals currently infected [1]. One of the most successful of all human viruses, HCV preferentially infects heptocytes and is able to persist in the livers of up to 70% of all infected individuals [2]. It is estimated that up to 30% of chronically infected individuals will develop progressive liver disease, including cirrhosis and heptocellular carcinoma (HCC) during their lifetime making HCV infection the leading causes of liver transplantation in the world [1]. In addition, HCV and HBV infections are implicated in 70% of all cases of HCC, which is the third leading cause of cancer deaths worldwide [3].
Due to the persistent nature of the virus, HCV infection can be extremely difficult and expensive to treat. Most infected individuals do not receive treatment. However, those that do, pay on average 17,700 to 22,000 dollars for standard treatment protocols [4]. Genotype 1 infection, the most prevalent in Europe and North America, has the poorest prognosis with as little as 42% of individuals responding to standard treatments [5].
Therefore, the high prevalence of infection, lack of effective treatments and economic burden of chronic HCV, illustrates the urgent need for the development of novel immune therapy strategies to combat this disease. Currently there is no prophylactic or therapeutic vaccine for HCV, however there is evidence that natural and protective immunity to HCV exists [6–8]. In the majority of cases, convalescent humans are not protected against acute HCV infection, but rather, they are protected from the progression of infection to a chronic state [9]. Since it is the chronic state of infection that is mainly associated with HCV pathogenicity, this argues for the feasibility of a vaccine approach to control or treat this infection.
Understanding the adaptive immunity to this virus is critical for designing strategies, such as DNA vaccines, to combat viral infection. Although virus-specific antibodies are detected within 7–8 weeks post HCV infection [10] they do not protect against reinfection [11, 12] and can be completely absent following the resolution of infection [13, 14]. Instead, infected individuals that mount an early, multi-specific, intrahepatic CD4+ helper and CD8+ cytotoxic T-cell response can eliminate HCV infection [15–18]. In fact, it has been shown that an important correlate to resolution of acute infection is a strong T cell response against the structural proteins of the virus, in particular the NS3 protein [19, 20]. The correlation of NS3-specific T cell responses to resolution of acute infection, in addition to its low genetic variably and relative large size makes the NS3 protein of HCV an attractive target for T-cell based DNA vaccines.
DNA vaccines have many conceptual advantages over more traditional vaccination methods, such as live attenuated viruses and recombinant protein-based vaccines. DNA vaccines are safe, stable, easily produced, and well tolerated in humans with preclinical trials indicating little evidence of plasmid integration [21, 22]. In addition, DNA vaccines are well suited for repeated administration due to the fact that efficacy of the vaccine is not influenced by pre-existing antibody titers to the vector [23]. However, one major obstacle for the clinical adoption of DNA vaccines has been a decrease in the platforms immunogenicity when moving to larger animals [24]. Recent technological advances in the engineering of DNA vaccine immunogen, such has codon optimization, RNA optimization and the addition of immunoglobulin leader sequences have improved expression and immunogenicity of DNA vaccines [25–28], as well as, recently developed technology in plasmid delivery systems such as electroporation [29–31]. In addition, studies conducted in our laboratory and in others, have suggested that the use of consensus immunogens may be able to increase the breadth of the cellular immune response as compared to native antigens alone [27, 32, 33].
Therefore, we have combined these various techniques in order to create a novel construct encoding a HCV genotype 1a/1b consensus immunogen of proteins NS3/NS4A, which we will show is expressed in mammalian cells and is able to elicit strong cellular responses both in mice and Rhesus Macaques.
2. Materials and Methods
2.1 Generation of HCV genotype 1a/1b consensus sequence of NS3/NS4A
The consensus sequence for NS3 was generated from fifteen different genotype 1a sequences and twenty-six different genotype 1b sequences. The consensus sequence for NS4A was generated from fifteen different genotype 1a sequences and nineteen different genotype 1b sequences. The sequences were obtained from GenBank, chosen from multiple countries to avoid sampling error and aligned using Clustal X (version 1.8) software to generate the final NS3/NS4A consensus sequence.
2.2 Additional Modification of the NS3/NS4A Consensus Sequence
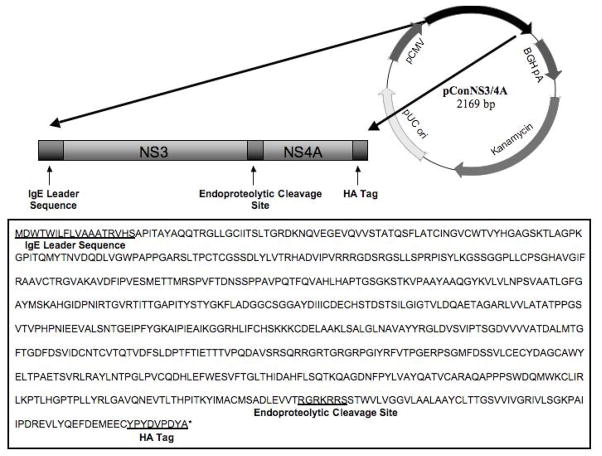
The consensus NS3/NS4A sequence was further modified through the addition of an IgE leader sequence, endoproteolytic cleavage site and a C-terminal HA tag, Fig. 1. GeneOptimizer™ (GENEART, Germany) was used to codon and RNA optimize the final sequence.
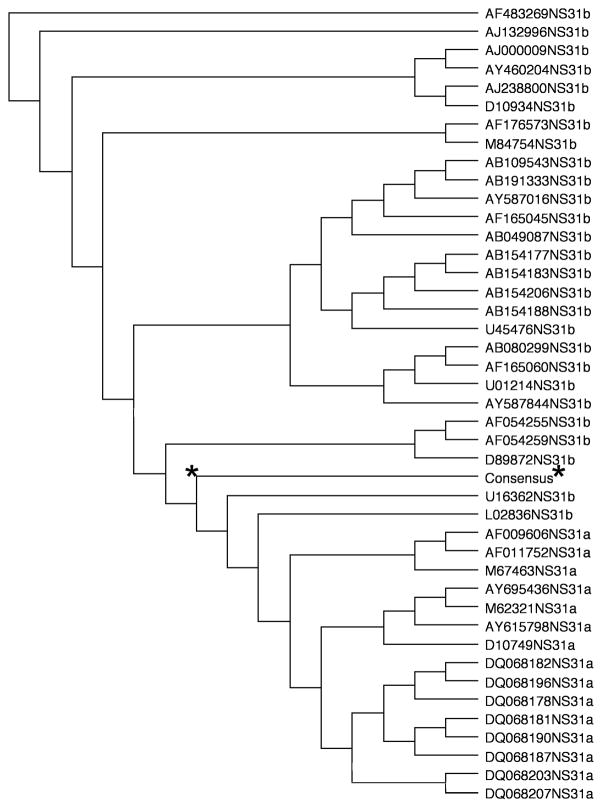
Fig. 1.
Phylogenetic analysis of pConNS3/NS4A’s genotype 1a/1b consensus sequence of NS3 as compared to individual genotype 1a and genotype 1b sequences for NS3. The genotype 1a/1b consensus sequence for NS3 was obtained from fifteen different HCV genotype 1a sequences and twenty-six different HCV genotype 1b sequences. The star represents the NS3 consensus sequence relative to its forty-one different component sequences.
2.3 HCV NS3/NS4A DNA Immunogen
The final consensus NS3/NS4A fusion gene (ConNS3/NS4A) was synthesized and sequence verified by GENEART (Germany). ConNS3/NS4A was digested with BamH1 and Not1, and subcloned in to the clinical expression vector pVAX (Invitrogen) under the control of the CMV promoter. The final construct was named pConNS3/NS4A.
2.4 Immunofluorescence
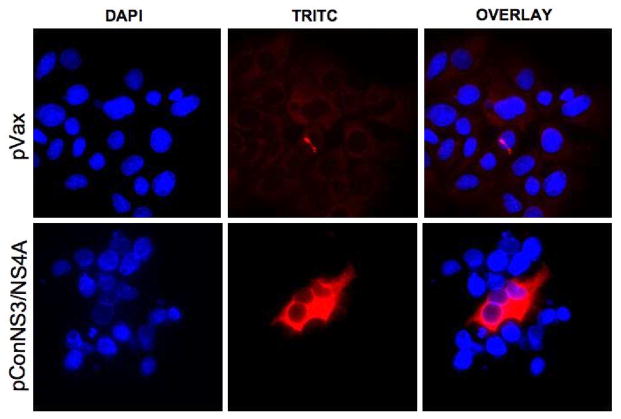
Huh7.0 cells were transiently transfected with pConNS3/NS4A using Lipofectamine™ (Invitrogen) according to the manufacturer’s guidelines. After 48 hours of transfection, the cells were permeabilized and expression of the proteins was determined using a mouse monoclonal antibody to the C-terminal HA tag of the fusion construct (Invitrogen) followed by a TRITC conjugated goat anti-mouse secondary antibody (Invitrogen).
2.5 Mouse Studies
2.5.1 Immunization/Electroporation
Female 6 to 8 week old C57BL/6 mice were purchased from Jackson Laboratories and were cared for in accordance with the National Institutes of Health and the University of Pennsylvania Institutional Care and Use Committee (IACUC) guidelines.
The mice were separated three mice per group and immunized with either pConNS3/NS4A or with the empty expression vector pVAX (negative control). Each mouse received three intramuscular injections, two weeks apart. Following each intramuscular injection, a three-electrode array made up of 26-gauge stainless steel electrodes was gently inserted into the muscle and a brief square-wave constant-current EKD was administered.
2.5.2 Splenocyte Isolation
The mice were sacrificed one week following the third immunization and the spleens were pooled according to group. The spleens were crushed using a Stomacher machine, and the resulting product was put through a 40μM cell strainer to isolate the splenocytes. The cells were treated 5min with ACK lysis buffer (Biosource) to clear the RBCs. Following lysis the splenocytes were resuspended in RPMI medium supplemented with 10% FBS. The cell number was determined with a hemocytometer.
2.5.3 IFN-gamma ELISpot
The mouse IFN-gamma ELISpot assays were conducted as previously described [32]. Splenocytes were stimulated with five different pools of 15mer peptides, overlapping by 8 amino acids and spanning the entire length of the pConNS3/NS4A protein. The peptides were synthesized by Invitrogen and pooled at a concentration of 2ug/ml/peptide. The splenocytes were plated at a concentration of 200,000 cells per well and the average number of spot forming units (SFU) was adjusted to 1×10^6 splenocytes for graphing purposes.
2.54 Epitope Mapping
The 15mer overlapping peptides were pooled into 21 separate pools, with each individual peptide represented in two pools of the 21 pools. Splenocytes were then stimulated with each pool in an IFN-gamma ELISpot assay as described above.
2.6 Rhesus Macaques Study
2.6.1 Study Design and Immunization
A total of five Rhesus Macaques of Indian origin were used and maintained in accordance with the Guide for the Care and Use of Laboratory Animals. Plasmids were prepared, HPLC purified (VGX Pharmaceuticals, Immune Therapeutics Division, The Woodlands, TX) and diluted in sterile water formulated with 1% (weight/weight) pol-L-glutamate sodium salt (MW=10.5 kDa) (Sigma).
The Macaques were intramuscularly injected followed by electroporation with the CELLECTRA™ adaptive constant current electroporation device and electrode arrays. Each monkey was injected with 1mg of pConNS3/NS4A in 0.75ml volume followed by three pulses of 0.5 Amp constant current, each 1 sec apart and 52 msec in length. Each monkey received two immunizations four weeks apart.
2.6.2 Blood Collection and PBMC Isolation
The Macaques were bled once before the first immunization and two weeks following each immunization. The animals were anesthetized with a mixture of ketamine (10mg/kg) and acepromazine (0.1mg/kg). Blood was collected in EDTA tubes and PBMCs were isolated using standard Ficoll-Hypaque density gradient centrifugation. The isolated PBMCs were resuspended in complete media (RPMI 1640 with 2mM/L L-glutamine supplemented with 10% heat inactivated FBS, 1X anti-biotic/anti-mycotic, and 55μM/L β-mercaptoethanol).
IFN-gamma ELISpot assays were performed as previously described [34] using detection and capture antibodies from MabTech, Sweden.
3. Results
3.1 Construction of a Novel HCV Genotype 1a/1b Consensus Fusion Immunogen of HCV Structural Proteins NS3/NS4A
Due to the high mutational rate of HCV, designing immunogens with multiple immune target sites is important not only for protection against various strains of the virus, but for maintaining control in chronically infected individuals by guarding against viral escape mutants. Previous findings in our laboratory, as well as in others, have suggested that the use of consensus immunogens in the context of vaccines is able to elicit a more broad immune response as compared to vaccination with the native immunogens alone [27, 32, 33]. Based on these findings, we designed a construct encoding the NS3/NS4A consensus sequence for HCV genotypes 1a/1b in hopes of increasing the breadth of the immune response against the NS3/NS4A proteins. The consensus sequence was generated from a total of seventy-five different sequences obtained from GenBank. Clustal X (version 1.8) software was used to create multiple alignments needed to generate a single consensus sequence, Fig 1.
As depicted in Fig. 2, the consensus immunogen was further modified to increase expression and immunogenicity. NS4A was included within the construct due to its reported ability to enhance the stability, expression and immunogenicity of the NS3 protein [35–37]. An endoproteolytic cleavage site was introduced between the two protein sequences to ensure proper folding, as well as, better CTL processing. In addition, to further enhance expression of the construct, an IgE leader sequence was fused upstream of the start codon for the NS3 protein and the entire construct was codon and RNA optimized for expression in Homo sapiens. The final synthetically engineered ConNS3/NS4A gene was 2169 bp in length and was subcloned into the clinical expression vector pVAX using the BamH1 and Not1 restriction sites.
Fig. 2.
Plasmid map and sequence of pConNS3/NS4A. The sequences for the IgE leader, endoproteolytic cleavage site and C-terminal HA tag are underlined.
3.2 Expression of pConNS3/NS4A
Expression of pConNS3/NS4A was confirmed through transient transfection of a Huh7.0 cell line with pConNS3/NS4A, Fig.3. The translated proteins were detected with a monoclonal antibody against the C-terminal HA tag of the construct and were visualized using immunofluorescence. As a negative control, cells were also transiently transfected with the empty pVAX vector and stained with a monclonal anti-HA antibody.
Fig. 3.
Detection of pConNS3/NS4A expression via immunofluorescence (400X). Huh7.0 were transiently transfected with pConNS3/NS4A and expression of the gene product was detected using a monoclonal antibody against the C-terminal HA tag.
3.3 Immunization of C57BL/6 Mice with pConNS3/NS4A Induces Strong Cellular NS3-and NS4A- Specific Immune Responses
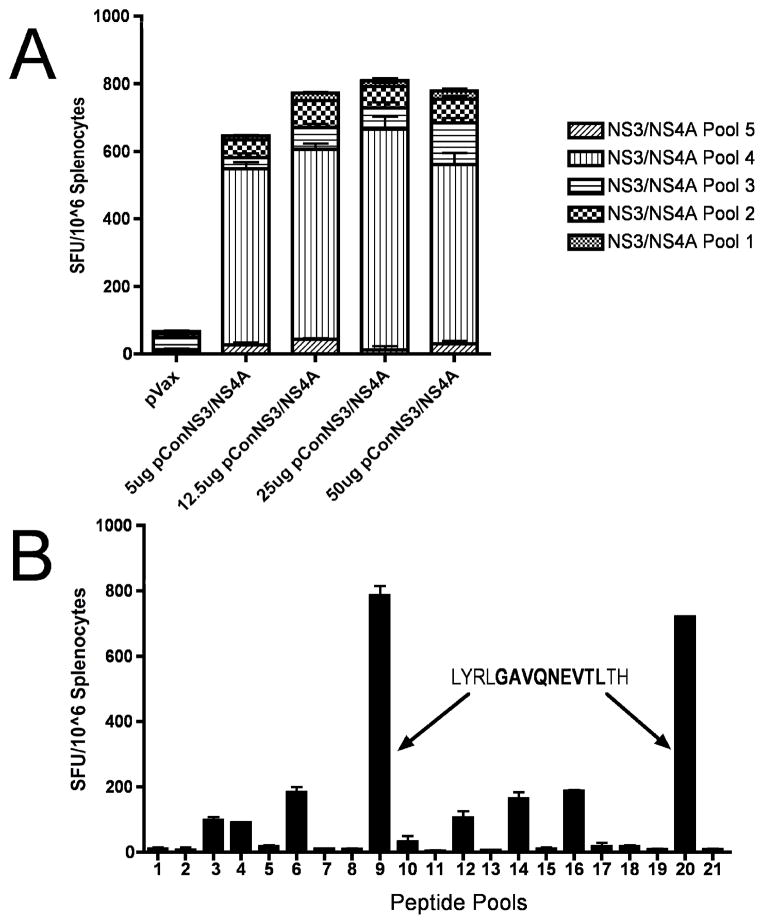
Following confirmation of the expression of pConNS3/NS4A, mice were immunized intramuscularly with the construct, followed by electroporation, in order to determine whether pConNS3/NS4A could induce cellular immune responses in mice. C57BL/6 received three immunizations using four different doses of pConNS3/NS4A. The mice were sacrificed one week following the third immunization and cellular immune responses to the construct were determined using IFN-gamma ELISpot assays. Splenocytes from vaccinated mice were stimulated with five pools of 15mer peptides overlapping by eight amino acids and spanning the sequence of pConNS3/NS4A. As shown in Fig. 4A, pConNS3/NS4A is able to induce potent cellular immune responses regardless of dose.
Fig. 4.
pConNS3/NS4A induces strong NS3- and NS4- specific T cell responses in C57BL/6 mice. The number of NS3- and NS4- specific IFN-gamma spot forming units (SFU) per million splenocytes was determined through IFN-gamma ELISpot assays. (A) Five groups of mice, three mice per group, were immunized intramuscularly with either pVAX (negative control) or four different doses of pConNS3/NS4A: 5ug, 12.5ug, 25ug or 50ug followed by electroporation. Splenocytes were isolated from each mouse, pooled according to group and stimulated with five different pools of overlapping peptides spanning the entire length of the pConNS3/NS4A protein sequence. (B) Matrix epitope mapping of pConNS3/NS4A. Splenocytes were isolated from C57BL/6 mice immunized with 12.5ug of pConNS3/NS4A and stimulated with 21 different pools of overlapping pConNS3/NS4A peptides with each peptide represented in two of the 21 pools. The dominant epitope was identified using IFN-gamma ELISpot assays as described above.
Next, using a matrix epitope mapping technique, the dominant epitope of pConNS3/NS4A was identified in IFN-gamma ELISpot assays. The dominant epitope of pConNS3/NS4A in C57BL/6 mice mapped to peptide 89 (LYRLGAVQNEVTLTH) located near the C-terminus of the NS3 protein, Fig. 4B. This is supported by previous research which identified the nine amino acid sequence of GAVQNEVTH, contained within peptide 89, as a dominant H-2b CTL epitope [35]. This finding argues for normal and effective processing of our consensus immunogen.
3.4 Immunization of Rhesus Macaques with pConNS3/NS4A Induces Strong Cellular NS3- and NS4A- Specific Immune Responses
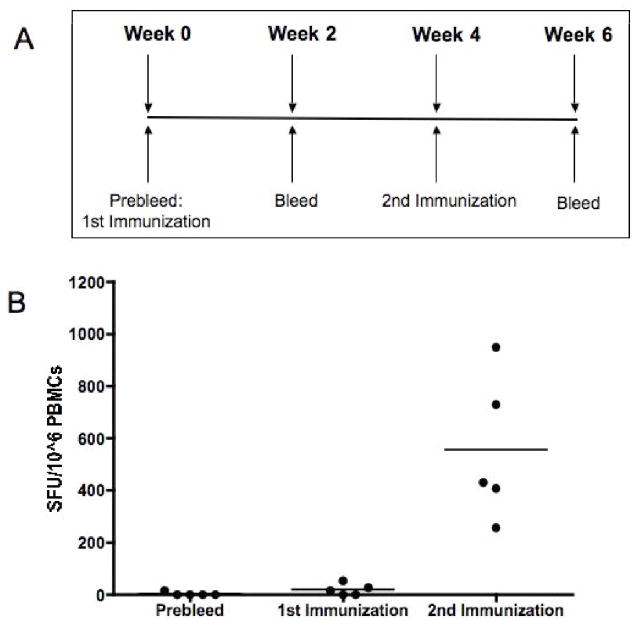
Although immunogenic in small animal models, DNA vaccines have typically lost potency when moved into larger animals. However, encouraged by the strong cellular immune responses elicited by pConNS3/NS4A in mice, we decided to test the immunogenicity of the construct in a larger animal model. Rhesus Macaques were immunized IM/EP with pConNS3/NS4A two times, four weeks apart, Fig. 5A. The animals were bled once before the first immunization and two weeks following each immunization. The animals’ immune responses to pConNS3/NS4A were determined with IFN-gamma ELISpot assays. Figure 5B depicts the sum of each monkey’s response to all five peptide pools, as well as, the average group response for each of the three time points. Cellular immune responses were not detectable following the prebleed and the first immunization, however, after the second immunization the responses increased dramatically to an average of 555 ± 280 SFU/10^6 PBMCs. Therefore, following only two immunizations, pConNS3/NS4A was able to elicit clear NS3- and NS4- specific cellular immune responses.
Fig. 5.
pConNS3/NS4A induces strong NS3- and NS4- specific T cell responses in Rhesus Macaques. (A) Immunization schedule. Five Rhesus Macaques were immunized intramuscularly with 1mg pConNS3/NS4A following by electroporation. The monkeys received two immunizations, four weeks apart. (B) Responses were measured once before the first immunization and two weeks following each immunization. The number of NS3- and NS4- specific IFN-gamma spot forming units (SFU) per million PBMCs was determined through IFN-gamma ELISpot assays.
3.5 pConNS3/NS4A Elicits a Broad Cellular Immune Response in Rhesus Macaques
Unlike immunization in C57BL/6 mice, where the majority of the immune response was directed at one dominant epitope of pConNS3/NS4A contained within a single peptide pool, the majority of immunized monkeys (4 out of 5) were able to elicit strong cellular immune responses to at least two or more peptide pools of pConNS3/NS4A, Fig. 6. Although further epitope mapping studies are planned, this suggests that the Rhesus Macaques were able to elicit cellular immune responses against multiple sites within the NS3/NS4A proteins. Therefore, the consensus sequence of pConNS3/NS4A is able to elicit both strong and broad cellular responses against the NS3/NS4A proteins in Rhesus Macaques.
Fig. 6.
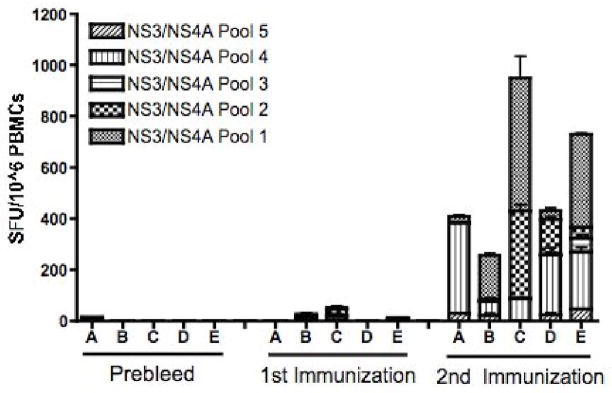
pConNS3/NS4A induces broad cellular immune responses in Rhesus Macaques. Shown are the individual responses of the five monkeys to each of the five peptide pools, before immunization and two weeks following each immunization. The number of NS3-and NS4- specific IFN-gamma spot forming units (SFU) per million PBMCs was determined through IFN-gamma ELISpot assays.
Discussion
It is known that HCV infected individuals who recover from acute infection are able to mount early, multi-specific, CD4+ helper and CD8+ cytotoxic T-cell responses [15–18]. It has also been reported that HCV-specific CTL responses are important for control of viral replication in chronically infected individuals [38, 39]. More specifically, a strong T cell response against the structural proteins of the virus, in particular the NS3 protein, has been reported to be an important correlate to clearance of acute infection [19, 20]. Due to the HCV virus’s high mutation rate, the relatively conserved structural proteins of the virus, such as NS3, are attractive candidates for T cell based vaccines.
DNA vaccines are well suited for eliciting strong T cell responses. Unlike immunization with recombinant proteins, which tend to produce Th2 responses, DNA vaccines are able to induce strong Th1 responses due to the ability of antigens to be delivered and processed intracellularly. In addition, DNA vaccines theoretically have unlimited boosting capability and can be readministered as often as desired without fear of inducing neutralizing antibodies to the plasmid, as is the case with recombinant viral vectors.
Due to the importance of NS3 specific T cell responses in clearance of acute infection and control of chronic infection, as well as, the clear advantages of DNA immunization, we have focused our efforts on creating a novel HCV DNA vaccine encoding the consensus sequence of genotype 1a and 1b for the HCV proteins NS3/NS4A (pConNS3/NS4A). Although DNA vaccines are able to elicit strong cellular immune responses in small animals, the major obstacle for adoption of DNA vaccine into a clinical setting has been the reduced immunogenicity of the platform when introduced in larger animal models. Therefore, in order to improve the potency of our construct, we took a multi-step approach in both its design and administration, including codon and RNA optimization, addition of an IgE leader sequence and administration of the construct with electroporation; modifications that have been shown to increase the expression and immunogenicity of DNA vaccines [25–31].
However, due to the high mutation rate of HCV, we believe a construct able to elicit both strong and broad cellular immune responses targeting multiple sites within the immunogen, would be better able to drive immunity against various strains of the virus, as well as, allow for better possible control of the virus in infected individuals by guarding against viral escape mutants. Previous studies both in our laboratory and others, have suggested that incorporation of multiple sequences to create one consensus immunogen is able to produce broader immune responses [27, 32, 33]. Therefore, as part of our construct design we incorporated seventy-five different HCV genotype 1a/1b sequences of the proteins NS3/NS4A in order to create one consensus immunogen.
We have shown that the construct, pConNS3/NS4A, is expressed in cell culture and that it is able to induce strong NS3- and NS4- specific T cell responses in C57BL/6 mice following three immunizations and is able to elicit both strong and broad NS3- and NS4A- specific T cell responses in a larger animal model, Rhesus Macaques, following only two immunizations. In fact, our study is one of only two DNA vaccine studies looking at NS3- specific immune responses induced in Rhesus Macaques. While both studies used similar sequence optimization methods, plasmid delivery systems and identical vaccination schedules, this construct was able to induce much higher NS3 specific immune responses with fewer immunizations immunizations and one-fifth the amount of DNA as compared to a previous study in which animals received three immunizations of 5mg of DNA [40]. In addition, we have shown that pConNS3/NS4A is able to elicit broad responses in Rhesus Macaques. Unlike C57BL/6 mice, which responded to one dominant epitope contained within one peptide pool, the majority of the Rhesus Macaques were able to elicit strong cellular immune responses to multiple peptide pools, suggesting that these monkeys are able to mount a response to multiple epitopes within pConNS3/NS4A. Further study of the immune responses induced by this construct, including epitope mapping of the cellular immune responses in Rhesus Macaques, is currently in progress.
Acknowledgments
This ms was supported in part by funding from the NIH to DBW and from VGX pharmaceuticals. The laboratory notes possible commercial conflicts associated with this work, which may include; Wyeth, VGX, BMS, Virxsys, Ichor, Merck, Althea, & Aldeveron.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomson BJ, Finch RG. Hepatitis C virus infection. Clin Microbiol Infect. 2005;11(2):86–94. doi: 10.1111/j.1469-0691.2004.01061.x. [DOI] [PubMed] [Google Scholar]
- 2.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436(7053):946–52. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 3.Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25(27):3834–47. doi: 10.1038/sj.onc.1209562. [DOI] [PubMed] [Google Scholar]
- 4.Salomon JA, et al. Cost-effectiveness of treatment for chronic hepatitis C infection in an evolving patient population. Jama. 2003;290(2):228–37. doi: 10.1001/jama.290.2.228. [DOI] [PubMed] [Google Scholar]
- 5.Manns MP, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 6.Weiner AJ, et al. Intrahepatic genetic inoculation of hepatitis C virus RNA confers cross-protective immunity. J Virol. 2001;75(15):7142–8. doi: 10.1128/JVI.75.15.7142-7148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassett SE, et al. Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection. Hepatology. 2001;33(6):1479–87. doi: 10.1053/jhep.2001.24371. [DOI] [PubMed] [Google Scholar]
- 8.Lanford RE, et al. Cross-genotype immunity to hepatitis C virus. J Virol. 2004;78(3):1575–81. doi: 10.1128/JVI.78.3.1575-1581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houghton M, Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature. 2005;436(7053):961–6. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 10.Pawlotsky JM. Diagnostic tests for hepatitis C. J Hepatol. 1999;31(Suppl 1):71–9. doi: 10.1016/s0168-8278(99)80378-x. [DOI] [PubMed] [Google Scholar]
- 11.Farci P, et al. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258(5079):135–40. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 12.Lai ME, et al. Hepatitis C virus in multiple episodes of acute hepatitis in polytransfused thalassaemic children. Lancet. 1994;343(8894):388–90. doi: 10.1016/s0140-6736(94)91224-6. [DOI] [PubMed] [Google Scholar]
- 13.Cooper S, et al. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10(4):439–49. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 14.Post JJ, et al. Clearance of hepatitis C viremia associated with cellular immunity in the absence of seroconversion in the hepatitis C incidence and transmission in prisons study cohort. J Infect Dis. 2004;189(10):1846–55. doi: 10.1086/383279. [DOI] [PubMed] [Google Scholar]
- 15.Lechner F, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191(9):1499–512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlach JT, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117(4):933–41. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 17.Thimme R, et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194(10):1395–406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grakoui A, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302(5645):659–62. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 19.Missale G, et al. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest. 1996;98(3):706–14. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diepolder HM, et al. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet. 1995;346(8981):1006–7. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 21.Martin T, et al. Plasmid DNA malaria vaccine: the potential for genomic integration after intramuscular injection. Hum Gene Ther. 1999;10(5):759–68. doi: 10.1089/10430349950018517. [DOI] [PubMed] [Google Scholar]
- 22.Nichols WW, et al. Potential DNA vaccine integration into host cell genome. Ann N Y Acad Sci. 1995;772:30–9. doi: 10.1111/j.1749-6632.1995.tb44729.x. [DOI] [PubMed] [Google Scholar]
- 23.Chattergoon M, Boyer J, Weiner DB. Genetic immunization: a new era in vaccines and immune therapeutics. FASEB J. 1997;11(10):753–63. doi: 10.1096/fasebj.11.10.9271360. [DOI] [PubMed] [Google Scholar]
- 24.Liu MA, Ulmer JB. Human clinical trials of plasmid DNA vaccines. Adv Genet. 2005;55:25–40. doi: 10.1016/S0065-2660(05)55002-8. [DOI] [PubMed] [Google Scholar]
- 25.Andre S, et al. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72(2):1497–503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deml L, et al. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol. 2001;75(22):10991–1001. doi: 10.1128/JVI.75.22.10991-11001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laddy DJ, et al. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25(16):2984–9. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frelin L, et al. Codon optimization and mRNA amplification effectively enhances the immunogenicity of the hepatitis C virus nonstructural 3/4A gene. Gene Ther. 2004;11(6):522–33. doi: 10.1038/sj.gt.3302184. [DOI] [PubMed] [Google Scholar]
- 29.Hirao LA, et al. Intradermal/subcutaneous immunization by electroporation improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine. 2008;26(3):440–8. doi: 10.1016/j.vaccine.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 30.Luckay A, et al. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J Virol. 2007;81(10):5257–69. doi: 10.1128/JVI.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahlen G, et al. In vivo electroporation enhances the immunogenicity of hepatitis C virus nonstructural 3/4A DNA by increased local DNA uptake, protein expression, inflammation, and infiltration of CD3+ T cells. J Immunol. 2007;179(7):4741–53. doi: 10.4049/jimmunol.179.7.4741. [DOI] [PubMed] [Google Scholar]
- 32.Yan J, et al. Enhanced cellular immune responses elicited by an engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Mol Ther. 2007;15(2):411–21. doi: 10.1038/sj.mt.6300036. [DOI] [PubMed] [Google Scholar]
- 33.Rolland M, et al. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J Virol. 2007;81(16):8507–14. doi: 10.1128/JVI.02683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer JD, et al. SIV DNA vaccine co-administered with IL-12 expression plasmid enhances CD8 SIV cellular immune responses in cynomolgus macaques. J Med Primatol. 2005;34(5–6):262–70. doi: 10.1111/j.1600-0684.2005.00124.x. [DOI] [PubMed] [Google Scholar]
- 35.Frelin L, et al. Low dose and gene gun immunization with a hepatitis C virus nonstructural (NS) 3 DNA-based vaccine containing NS4A inhibit NS3/4A-expressing tumors in vivo. Gene Ther. 2003;10(8):686–99. doi: 10.1038/sj.gt.3301933. [DOI] [PubMed] [Google Scholar]
- 36.Wolk B, et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74(5):2293–304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanji Y, et al. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69(3):1575–81. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rehermann B, et al. Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J Clin Invest. 1996;98(6):1432–40. doi: 10.1172/JCI118931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson DR, et al. The role of hepatitis C virus-specific cytotoxic T lymphocytes in chronic hepatitis C. J Immunol. 1997;158(3):1473–81. [PubMed] [Google Scholar]
- 40.Capone S, et al. Modulation of the immune response induced by gene electrotransfer of a hepatitis C virus DNA vaccine in nonhuman primates. J Immunol. 2006;177(10):7462–71. doi: 10.4049/jimmunol.177.10.7462. [DOI] [PubMed] [Google Scholar]