Abstract
DNA vaccines are a promising technology. Historically, however, the ability of DNA vaccines to induce high response rates and strong immune responses, especially antibody responses, in non-human primates and human clinical trials has proven suboptimal. Here, we performed a pilot study in rhesus macaques to evaluate whether we could improve the immunogenicity of DNA vaccines through the use of adjuvant technology and improved delivery systems. The study consisted of four groups of animals that received: DNA by intramuscular (IM) injection, DNA with plasmid-encoded IL-12 by IM injection, DNA by IM injection with in vivo electroporation (EP), and DNA with IL-12 by IM EP. Each group was immunized three times with optimized HIV gag and env constructs. Vaccine immunogenicity was assessed by IFNγ ELISpot, CFSE proliferation, polyfunctional flow cytometry, and antibody ELISA. Similar to previous studies, use of IL-12 as an adjuvant increased the gag and env-specific cellular responses. The use of EP to enhance plasmid delivery resulted in dramatically higher cellular as well as humoral responses. Interestingly, the use of EP to administer the DNA and IL-12 adjuvant combination resulted in the induction of higher, more efficient responses such that a 10 fold increase in antigen-specific IFNγ+ cells compared to IM DNA immunization was observed after a single immunization. In addition to increases in the magnitude of IFNγ production in the initial and memory responses, the combined approach resulted in enhancements in the proliferative capacity of antigen-specific CD8+ T cells and the amount of polyfunctional cells capable of producing IL-2 and TNFα in addition to IFNγ. These data suggest that adjuvant and improved delivery methods may be able to overcome previous immunogenicity limitations in DNA vaccine technology.
Keywords: DNA vaccine, non-viral, plasmid, electroporation, HIV, cytokine, adjuvant
Introduction
DNA, as a vaccine platform, has been used in numerous studies related to infectious diseases. DNA vaccines are attractive because of their improved safety profile compared to live attenuated vaccines and the simplicity of plasmid engineering and production compared to the manipulation required for the development of live attenuated and killed viral vaccines.
However, there is a question concerning the potency of DNA vaccines in non-human primates and in human studies. The results reported in small animal models have yet to be translated to non-human primates and human clinical trials. The DNA platform has been expanded extensively in human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) models [1–4]. A large number of clinical studies have evaluated different delivery platforms and formulations [5]. DNA vaccines for HIV antigens appear very well tolerated as part of a prime boost strategy [6]. However, overall cytotoxic T lymphocytes (CTL) response rates and magnitude are lower than optimally desired. Although DNA immunization is able to induce cellular responses, the antibody responses induced by this methodology are particularly weak. Furthermore, DNA immunization produces lower than optimal T cell helper responses as determined in proliferation assays, and induces little T cell polyfunctionality on its own.
To this end, numerous strategies are being developed to enhance the immunogenicity of DNA vaccines, such as plasmid optimization, adjuvants, and improved plasmid delivery. Plasmid optimization includes codon optimization for expression in human cells [7, 8], RNA optimization [9, 10], the addition of leader sequences to enhance translation [11] and the creation of consensus immunogens to induce broad cross-reactive responses to different viral isolates [12, 13].
A number of studies have examined the use of molecular adjuvants to expand immune responses to HIV vaccines [14]. Co-immunization of DNA with plasmid-encoded molecular adjuvants can aid in the antigen-specific activation and proliferation of immune cells. For instance, the cytokines IL-12 [15, 16] and IL-15 [17, 18] have been demonstrated to increase the activation, proliferation, and function of T cells. Furthermore IL-12, as a DNA vaccine adjuvant, can improve CTL responses several fold in non-human primates and improve protection in a SHIV model system [15, 19].
Increasing the immunogenicity of DNA vaccines through enhanced physical delivery methods is another area of investigation. Electroporation (EP), a method traditionally used for enhancing the transfection efficiency of cells in vitro, has been studied to enhance the uptake and expression of both therapeutic and vaccine plasmids [20, 21] and increase immune responses in tumor animal models [22]. More recently, EP has been studied as a method to enhance the potency of DNA vaccines in primates with encouraging results [23, 24]. However these studies did not examine in detail the functional aspects of the induced response.
In this pilot study we compared the ability of optimized DNA constructs encoding HIV gag and env in rhesus macaques with or without plasmid IL-12 adjuvant to induce vaccine-specific immune responses. We observed that, as previously reported [25], plasmid IL-12 increased T cell responses 5-fold as determined by quantitative ELISpot assay, resulting in substantially more robust memory T cell responses. However, EP delivered DNA generated 2-fold higher effector and memory T cell responses compared to the IL-12 IM adjuvanted DNA vaccine. The best responses were observed in the combination arm of EP + IL-12 adjuvant. Memory responses in this arm were 10-fold higher than the IM DNA alone and almost 2-fold higher than EP alone. We also observed 4-fold better antigen-specific proliferation in response to ex vivo peptide stimulation as determined by CFSE proliferation assay in the EP + IL-12 arm compared to EP alone. Functional analysis of T cells from the EP + IL-12 group revealed the presence of polyfunctional T cells which have been demonstrated to be present in HIV-infected long-term non-progressors, suggesting these cells may be better equipped to control infection. Together our data demonstrates a significant improvement in the immunogenicity of DNA vaccines in primates and support the further study of electroporation and plasmid-encoded cytokine adjuvants to enhance DNA vaccine performance.
Materials and Methods
Animals
Rhesus macaques (Macaca mulatta) were housed at BIOQUAL, Inc. (Rockville, MD), in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care. Animals were allowed to acclimate for at least 30 days in quarantine prior to any experimentation.
Plasmids and plasmid products
Gag4Y contains an expression cassette encoding for a consensus sequence of the gag protein of HIV clades A, B, C, and D with several modifications including: the addition of a kozak sequence, a substituted IgE leader sequence, codon and RNA optimization for expression in mammalian cells. The Gag4Y gene was subcloned into the expression vector, pVax (Invitrogen, Carlsbad, CA), for further study. pEY-2E1-B contains an expression cassette encoding for a consensus sequence of the envelope of HIV clade B [26]. WLV104M is a plasmid encoding a rhesus IL-12 gene [27]. Plasmids were produced at Aldevron (Fargo, ND), and re-formulated at VGX Pharmaceuticals, Immune Therapeutics Division (The Woodlands, TX), in sterile water for injection with low molecular weight 1% (w/v) poly-L-glutamate sodium salt (LGS).
Immunization
Five rhesus macaques were immunized at weeks 0, 4, and 11 with 1.0mg of each pGag4Y and pEY2E1-B. Three of the macaques were electroporated following IM injection. Another group of five macaques were immunized at weeks 0, 4, and 8 with 1.0mg of each pGag4Y, pEY2E1-B, and WLV104 (1mL formulation, at a plasmid concentration of 3mg/mL) in one injection site. Of the five animals, two animals received the immunization by IM injection and three animals were electroporated following IM injection. All electroporation procedures were performed using the constant current Cellectra™ device (VGX Immune Therapeutics Division of VGX Pharmaceuticals, The Woodlands, TX). Electroporation conditions were 0.5 Amps, 3 pulses, 52 msec pulse length with 1 sec between pulses. This software-controlled device was designed to measure the tissue resistance immediately prior to plasmid delivery and generation of constant current square wave pulses, eliminating the risk of delivery outside the muscle tissue and potential plasmid loss [21, 28].
Blood Collection
Animals were bled every two weeks for the duration of the study. 10 mL of blood were collected in EDTA tubes. Peripheral blood mononuclear cells (PBMC) were isolated by standard Ficoll-hypaque centrifugation and then resuspended in complete culture medium (RPMI 1640 with 2mM/L L-glutamine supplemented with 10% heat-inactivated fetal bovine serum, 100 IU/mL penicillin, 100μg/mL streptomycin, and 55μM/L β-mercaptoethanol). Red blood cells (RBC) were lysed with ammonium chloride-potassium (ACK) lysis buffer (Cambrex Bio Science, East Rutherford, NJ).
CFSE of Cryo-preserved PBMCs
Cryo-preserved PBMCs were quick-thawed in a 37°C water bath and washed with complete media. Cells were incubated overnight in a 37°C incubator and cell counts were obtained the following day. Cells were pelleted and resuspended in 1 ml carboxyfluorescein diacetate, succinimidyl ester (CFDA SE) (Molecular Probes, Eugene, OR) in PBS (1:2000 dilution). Cells were incubated at 37°C for 10 min. Cells were washed with complete media and resuspended to a concentration of 1x106 cells/100μL and plated in 96 well round bottom plates with 100μL of 2 μg/mL recombinant HIV-1 p24 or gp120 (ImmunoDiagnostics, Woburn, MA) plus peptide pools. 5 μg/mL concavalin A (positive) and complete media (negative) were used as controls. Cultures were incubated for 5 days. Cells were first stained with Vivid dye violet, a live/dead cell marker, for 15 min on ice. Cells were washed once with PBS. Cells were then stained using anti-human CD3- phycoerythrin (PE) [clone SP34-2] (BD Pharmingen, San Jose, CA) and anti-human CD4- peridinin chlorophyll protein (PerCP) [clone L200], anti-human CD8- allophycocyanin (APC) [SK1] for 1 hour at 4°C. Cells were then washed twice with PBS and fixed with 1% paraformaldehyde. Data was collected using a LSRII flow cytometer (BD Biosciences, Franklin Lakes, NJ). Flow cytometry data was analyzed using FlowJo software (Tree Star, Ashland, OR), gating on CD3+ lymphocytes. Thirty to fifty thousand CD3+ lymphocytes were collected per sample.
Enzyme Linked Immunosorbant Assay (ELISA)
Ninety-six well plates were coated overnight with 100ng/well of recombinant HIV-1 IIIB p24 or gp120 (ImmunoDiagnostics) to determine HIV gag and env responses respectively. Plates coated with 100ng/well of bovine serum albumin served as a negative control. Plates were blocked with 3%BSA-PBST for 1 hour at 37°C. Plates were then incubated with four-fold serial serum dilutions for 1 hour at 37°C. Goat anti-monkey IgG horseradish peroxidase conjugated antibody was then added at a 1:10,000 dilution (MP Biomedicals, Aurora, OH) to the plates and incubated for 1 hour at 37°C. Tetramethylbenzidine (R&D systems, Minneapolis, MN) was used to develop the plates and reactions were stopped with 2N H2SO4. Optical densities (OD) were then measured.
IgG end-point titers were defined as the reciprocal serum dilution that resulted in OD values that were greater than twice the average OD value of the BSA wells.
Enzyme Linked Immunospot Assay (ELISpot)
ELISpot were performed as previously described [15]. Antigen specific responses were determined by subtracting the number of spots in the negative control wells from the wells containing peptides. Results are shown as the mean value (spots/million splenocytes) obtained for triplicate wells.
Intracellular Cytokine Staining
Antibody Reagents
Directly conjugated antibodies were obtained from the following:BD Biosciences (San Jose, CA): IL-2 (PE), CD3 (Pacific Blue), IFN-γ (PE-Cy7), and TNF-α (Alexa Fluor 700), CD8 (APC) and CD4 (PerCP).
Cell stimulation and staining
PBMCs were resuspended to 1 x 106 cells/100μL in complete RPMI and plated in 96 well plates with stimulating peptides 100μL at 1:200 dilutions. An unstimulated and positive control (Staphylococcus enterotoxin B, 1μg/mL; Sigma-Aldrich, St. Louis, MO) was included in each assay. Cells were incubated for 5 hours at 37°C. Following incubation, the cells were washed three times in PBS and stained with surface antibodies. The cells were washed and fixed using the Cytofix/Cytoperm kit (BD PharMingen, San Diego, CA) according to manufacturer’s instructions. Following fixation, the cells were washed twice in the perm buffer and stained with antibodies against intracellular markers. Following staining, the cells were washed, fixed (PBS containing 1% paraformaldehyde), and stored at 4°C until analysis.
Flow cytometry
Cells were analyzed on a modified LSR II flow cytometer (BD Immunocytometry Systems, San Jose, CA). Fifty thousand CD3+ events were collected per sample. Data analysis was performed using FlowJo version 8.4.1 (TreeStar, San Carlos, CA). Initial gating used a forward scatter area (FSC-A) versus height (FSC-H) plot to remove doublets. The events were subjected to a lymphocyte gate by a FSC-A versus side scatter (SSC) dot plot. Following this, events are sequentially gated on CD3+, CD8+, and CD4− events versus IFN-γ to account for down-regulation. Following identification of CD8+ T cells, a gate was made for each respective function using combinations that provided optimal separation. After the gates for each function were created, we used the Boolean gate platform to create the full array of possible combinations, equating to 8 response patterns when testing 3 functions. Data are reported after background correction. Thresholds for positive responses were 10 events or 0.05%.
Results
Enhanced Cellular Responses with IL-12 co-immunization and electroporation
A number of studies have demonstrated the importance of TH1 mediated immunity in the control of viral replication in primary HIV infection [29, 30]. Given the importance of the HIV-specific cell-mediated immune (CMI) response in the early control of HIV replication during primary infection we evaluated the induction of CMI responses by IFNγ ELISpot after each immunization. After a single immunization (Figure 1A), the group receiving plasmid DNA by IM injection alone displayed weak cellular responses (74 ± 29 SFU/106 PBMCs), this response was modestly enhanced with the co-immunization of a rhesus IL-12 plasmid (136 ± 51.4 SFU/106 PBMCs). The EP group had an average response that was six times higher than the IM group (482 ± 181 SFU/106 PBMCs, p<0.1). The combination of IL-12 co-immunization with EP further doubled the number of IFNγ-producing cells (1030 ± 494 SFU/106 PBMCs, p<0.1).
Fig. 1.
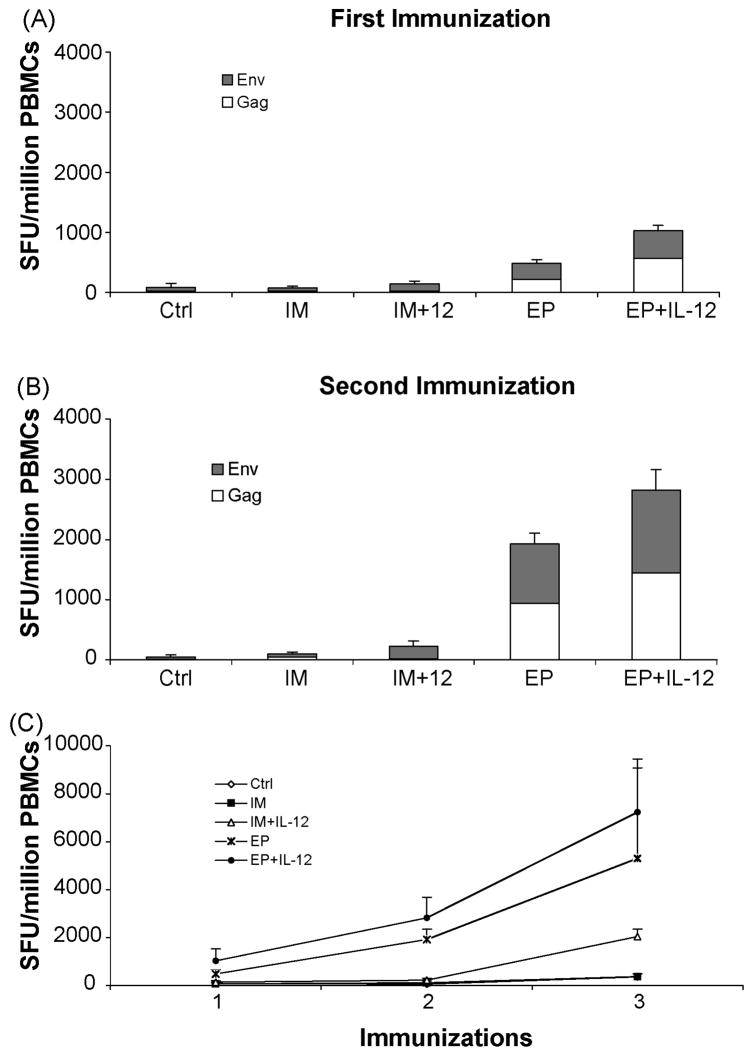
Enhanced cellular immune responses to HIV-1 consensus immunogens with IM co-injection of plasmid encoded IL-12 followed by EP. IFNγ ELISpots were performed two weeks after the (a) first immunization, (b) second immunization, and (c) third immunization (as seen in comparison to the other three). Responses to env are depicted as black bars and gag are depicted as white bars with the data shown as stacked group mean responses ± SEM.
After two immunizations (Figure 1B), the IM and IM +IL-12 groups had a modest increase in ELISpot counts (104 ± 67.9 SFU/106 PBMCs and 223 ± 76.6 SFU/106 PBMCs, respectively). EP group had responses that were almost four fold higher (1924 ± 417 SFU/106 PBMCs, p<0.02) than the previous immunization and the EP+IL-12 group had again doubled the number of IFNγ-producing cells (2819 ± 872 SFU/106 PBMCs, p<0.04) compared to the EP arm alone.
After the third immunization (Figure 1C), the number of antigen specific cells in the EP group was more than a log higher than that of the IM group (5300 ± 3781 and 370 ± 110 SFU/106 PBMCs, respectively). The IM+IL-12 group also had a dramatic boost in cellular responses that were nearly a log higher than the previous immunization (2042 ± 311 SFU/106 PBMCs). As with the other two immunizations, the EP+IL-12 group was the most potent of all the vaccination groups (7228 ± 2227 SFU/106 PBMCs, p<0.05), consistent with a previous report [25]. Thus, the data indicate that the use of EP is sufficient to enhance vaccine-specific cellular immunity. Furthermore, the data demonstrates a clear enhancement of IFNγ ELISpot responses with the addition of plasmid IL-12.
Induction of cross-reactive envelope responses
A successful HIV vaccine will require the induction of cross-reactive immune responses. In this regard it was interesting to determine if EP + IL-12 could improve the magnitude of cross-reactivity to divergent peptide libraries. We compared the cross-reactive CTL responses induced by the env antigen using a peptide library from a consensus group M [12]. Cross-reactivity was observed in all groups. However the results displayed the same magnitude differences observed in the subtype B ELISpot analysis (Figure 2). After 3 immunizations, the IM group had the lowest response to the group M envelope peptides (222 ± SEM SFU/106 PBMCs). The addition of IL-12 doubled the response (540 ± SEM SFU/106 PBMCs). Higher group M envelope responses were induced with EP (830 ± SEM SFU/106 PBMCs), which were further enhanced with EP + IL-12 (1238 ± SEM SFU/106 PBMCs, p<0.01). Our data support the ability of the DNA vaccine to induce cross-reactive responses, in the adjuvant formulation and under optimized EP conditions.
Fig. 2.
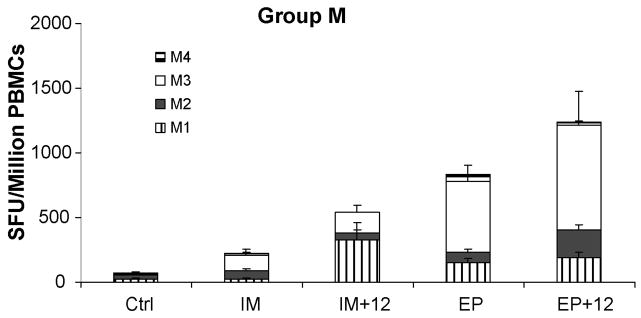
Enhanced cross-reactive cellular immune responses with intramuscular electroporation. After three immunizations, the total T-cell immune response in pEY2E1-B immunized macaques against four peptide pools of the HIV-1 group M peptides were determined by IFNγ ELISpot. The data are shown as stacked group means ± SEM.
Memory T cell Responses
An important issue is to be able to improve the generation of memory responses with the DNA platform. To assay for memory T cell responses we performed ELISpot analysis five months after the last DNA vaccination (Figure 3). In the IM groups, the addition of plasmid IL-12 resulted in nearly a 10-fold increase in memory cells (751 ± 11.1 and 78.6 ± 16.9 SFU/106 PBMCs). It is clear that IL-12 can positively impact this important T cell phenotype. The number of antigen-specific IFNγ producing cells was substantial in the EP group as well, however the IL-12 adjuvant + EP resulted in the most robust memory response (1231 ± 523.5 and 3795 ± 1336 SFU/106 PBMCs respectively, p<10−5 and p<0.002 vs. IM only group), indicating that the combined technology drives very strong T cell memory responses.
Fig. 3.
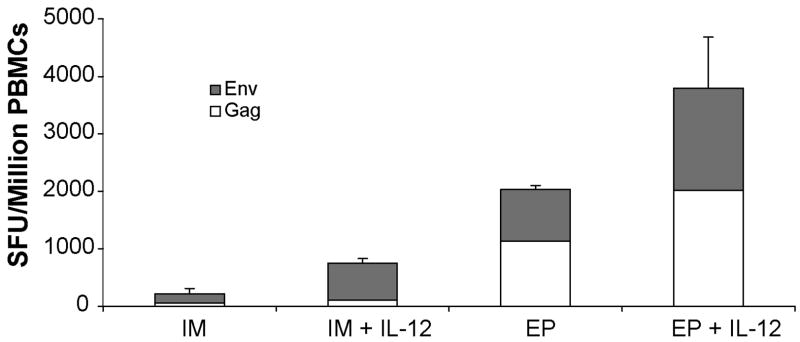
Enhanced memory responses to HIV-1 immunogens with IM electroporation and plasmid IL-12. Five months after the last immunization, ELISpot assays were performed to determine antigen-specific memory responses to gag and env in the IM and EP immunized groups with and without co-immunization with the IL-12 plasmid. The data are shown as group mean responses ± SEM.
Humoral immune responses to DNA vaccines
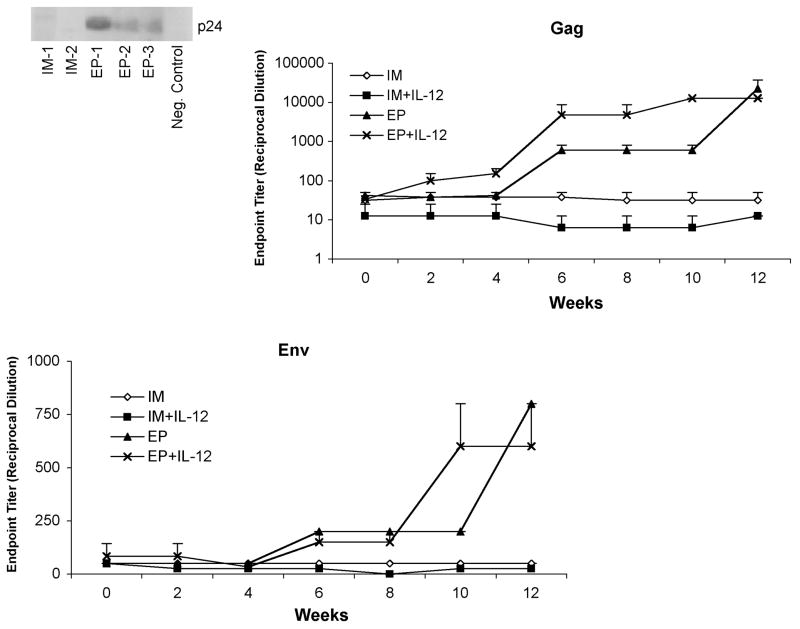
A historical weakness of DNA vaccine technology lies in its inability to induce clear antibody responses in non-human primates and in human clinical studies. We evaluated each group’s ability to induce both HIV-1 gag and env specific antibody titers to recombinant p24 and gp160 antigens in an ELISA format (Figure 4). For both antigens, the IM and IM + IL-12 groups did not show significant antibody titers (<1:50 endpoint titer). The electroporated groups exhibited dramatically higher gag antibody titers that were able to bind to recombinant p24 (Figure 4a). Although both the EP and the EP + IL-12 groups had similar endpoint titers at week 12 (22,400 and 12,800 respectively), the EP + IL-12 group generated a more vigorous antibody response. Furthermore, the EP + IL-12-induced response appeared earlier in the immunization scheme and rose to the maximum level more rapidly than other groups. The env antibody responses also reflected the results we observed with the gag antigen, albeit with lower endpoint titers.
Fig. 4.
Induction of p24 and gp160 antibodies. Serum was collected every two weeks over the course of the study. For each time point, gag and env antibody titers were determined by p24 and gp160 ELISA, respectively, in the IM and EP immunized groups with and without co-immunization with the IL-12 plasmid. The data are shown as group mean responses ± SEM.
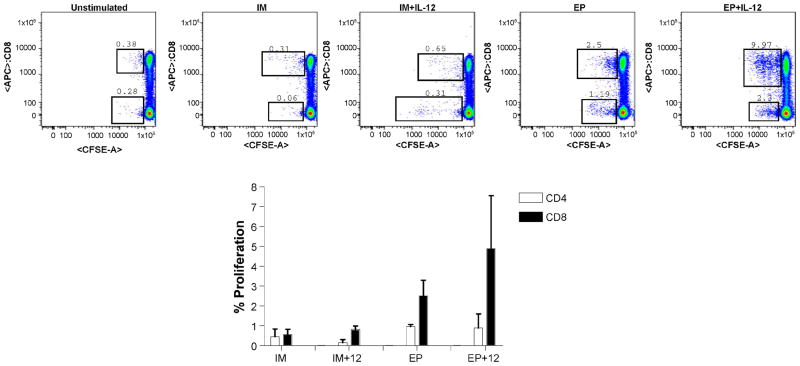
CD4+ and CD8+ T cell proliferation
Studies comparing HIV infected non-progressors with progressors have indicated that non-progressors have CD8+ T cells that maintain a higher proliferative capacity. Therefore, we examined the ability of gag specific CD4+ and CD8+ T cells to proliferate ex vivo following peptide stimulation among the different immunization arms. Cryo-preserved samples, collected two weeks after the final immunization, were stimulated and analyzed by a CFSE proliferation assay (Figure 5). The average CD4+ response increased similar to that observed in the ELISpot assay. By comparison, the CD8+ proliferation induction was much more dramatic in magnitude. We observed that IL-12 increased CD8+ T cell proliferation over IM alone and EP was substantially higher. The EP + IL-12 group had the highest percentage of CD8+ cells that were able to proliferate after ex vivo stimulation (2.51 ± SEM % and 4.88 ± SEM %, respectively). Obvious CD8+ T cell proliferation bands were observed in the EP + IL-12 arm, demonstrating the potent proliferative potential of this combined immunization.
Fig. 5.
CD4+ and CD8+ T cell proliferation. Cryo-preserved samples collected two weeks following the final immunization were stimulated with gag peptides and p24 protein in vitro for 5 days to determine the proliferative capacity of gag-specific T cells. Representative dot plots are shown for each group. Proliferative responses are shown as group mean responses ± SEM with CD4 depicted as white bars and CD8 are depicted as black bars.
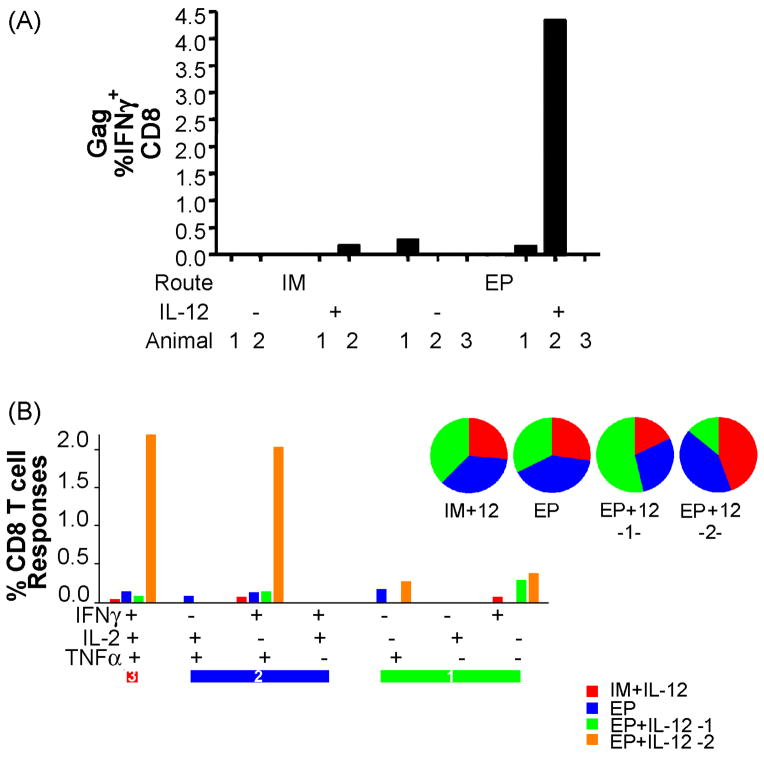
Polyfunctional CD8+ T cell responses
Although we have clearly observed the induction of a robust IFNγ effector response following EP and IL-12 co-immunization, IFNγ production is only one function that CD8+ T cells are capable of performing. To get a more detailed understanding of the vaccine-induced immune response in the various arms of the study we performed intracellular cytokine analysis by polychromatic flow cytometry to characterize the functional and phenotypic profile of CD8+ T cells (Figure 6). Samples collected three months following the final immunization were stimulated with gag peptides and stained for intracellular cytokine production of IFNγ, TNFα and IL-2. Out of all groups, only one animal in the IM + IL-12 and one animal in the EP only group had a detectable IFNγ response. Two out of the three animals in the EP + IL-12 immunized group had gag-specific IFNγ producing CD8+ T cells. The IM + IL-12 responder had a small percentage of polyfunctional cells that stained for all three cytokines as well as a population that had lost its ability to produce IL-12. The EP responder had slightly higher polyfunctional responses that were comprised of four different populations. The most dramatic response was seen in the second EP + IL-12 animal. More than 2% of its CD8+ T cells were able to produce all three cytokines and 2% were able to produce both IFNγ and TNFα. Clearly the number of animals in each group is low and requires additional primate studies to confirm these results, however collectively the trends observed appear encouraging.
Fig. 6.
Induction of gag-specific polyfunctional CD8+ T cells. PBMCs were stimulated in vitro with a gag peptide pool mix for 5 hours. Cells were then stained for intracellular production of IFNγ, TNFα and IL-2. Animals with detectable gag-specific IFNγ+ CD8+ T cells (a) were further analyzed for polyfunctional populations (b-d). Pie charts show the proportion of gag-specific CD8+ T cells that have 3 functions (red), 2 functions (green) or 1 function (blue). Cells incubated with complete media alone were used to determine background responses for each polyfunctional population and was subtracted from the gag-specific response. The threshold for detection was set at 10 events or 0.05%.
Discussion
We present herein a pilot macaque study to evaluate a DNA vaccine immune potency with or without plasmid IL-12 cytokine adjuvant and with or without EP as a delivery enhancer. We sought to determine the contributions of plasmid IL-12 and EP in augmenting vaccine induced responses. Despite the relatively small number of animals in each group, we observed substantial differences and common themes in all assays.
In this study we observed enhanced immune responses when animals were co-immunized with plasmid IL-12. In fact, after three IM immunizations we observed levels of IFNγ responses that have been reported for some viral vectors. However, the use of EP for plasmid delivery induced similar levels of responses after a single immunization, demonstrating the utility of EP in increasing the efficacy of DNA vaccines. Further enhancement of cellular immune responses was achieved when the two approaches were used together. These trends were also seen in the induction of memory and cross-reactive cellular immune responses.
Although we have observed higher ELISpot responses to the Group M peptides in the EP and the EP+12 groups we cannot distinguish whether these differences reflect an enhancement in the breadth or in the magnitude of the immune response. Others showed that a DNA vaccine encoding an optimized version of the nonstructural region of hepatitis C virus delivered by EP induced substantially more potent, broad, and long-lasting CD4+ and CD8+ cellular immunity than IM DNA injection in mice and in rhesus macaques [31]. Thus, epitope mapping studies are needed to address this issue.
While DNA immunizations have performed poorly in the induction of humoral immunity, our data clearly demonstrates the ability of electroporation to stimulate vaccine-specific antibody responses, consistent with previous data [23]. All of the animals in the EP groups seroconverted, while IM immunization, with or without IL-12, were unable to induce significant antibody production. Although the EP and the EP + IL-12 groups had similar endpoint titers after three immunizations the kinetics of antibody induction was slightly faster in the EP + IL-12 group. We have not yet determined the ability of these antibodies to neutralize virus, however the env responses are still modest. Regardless of their ability to neutralize virus, antibodies may still contribute to the control of virus through antibody-dependent cellular cytotoxicity [22]. In addition, there are numerous reports suggesting antibody responses exert selective pressure on the virus and, therefore, may provide at least some support in viral protection or suppression.
In a recent study, Luckay and colleagues compared the use of EP in an HIV vaccine that included an IL-12 antigen in a rhesus macaque model [25]. They observed similar increases in antigen-specific IFNγ responses by ELISpot and antibody responses. Having seen dramatic increases in cellular and humoral immune responses in the animals that received the HIV antigens with plasmid IL-12 and delivered by EP, we wanted to further characterize the vaccine induced T cell response. An important aspect of a vaccine-induced immune response for HIV is the proliferative capacity of antigen-specific cells. The proliferative capacity of CD8+ T cells appeared to be enhanced with EP and plasmid IL-12. This data supports the memory expansion observed in the ELISpot assay where expansion of antigen specific T cells is likely the result of the enhanced proliferative potential of the EP+ IL-12 arm.
Studies of HIV-1 infected clinical non-progressors demonstrated the maintenance of virus-specific CD8 T cells that were capable of multiple functions [32]. This suggests that the induction of polyfunctional T cells will be an important factor for a successful vaccine. In this study we observed a large population of gag-specific CD8 T cells that were able to produce IFNγ, TNFα, and IL-2 in an animal in the EP + IL-12 group. The production of IL-2 may obviate the necessity for CD4+ help for the induction of CD8+ T cell memory [33], which may be particularly important in a therapeutic setting where functional CD4 T cells may be a limiting factor [34]. This encouraging pilot study will require follow-up studies to confirm and extend these results and test their ability to impact viral challenge. While the immune responses we observed here are significantly greater than previously reported for DNA vaccines, the polyfunctionality of the responses may play a more important role in viral clearance or suppression. Close monitoring of multiple aspects of the antiviral immune response must be monitored in order to determine the correlates of immune protection.
In summary, by a variety of parameters we observe that we can substantially improve the immune potency of an optimized plasmid vaccine in non-human primates by combining the adjuvant activity of plasmid IL-12 with improved plasmid delivery using electroporation. The increased magnitudes will allow for detailed investigation of the immune biology of the DNA approach in the non-human primate model in previously unexplored detail.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sun Y, et al. Virus-specific cellular immune correlates of survival in vaccinated monkeys after simian immunodeficiency virus challenge. J Virol. 2006;80(22):10950–6. doi: 10.1128/JVI.01458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Gegerfelt AS, et al. Long-lasting decrease in viremia in macaques chronically infected with simian immunodeficiency virus SIVmac251 after therapeutic DNA immunization. J Virol. 2007;81(4):1972–9. doi: 10.1128/JVI.01990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosati M, et al. DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J Virol. 2005;79(13):8480–92. doi: 10.1128/JVI.79.13.8480-8492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malkevitch N, et al. Evaluation of combination DNA/replication-competent Ad-SIV recombinant immunization regimens in rhesus macaques. AIDS Res Hum Retroviruses. 2004;20(2):235–44. doi: 10.1089/088922204773004969. [DOI] [PubMed] [Google Scholar]
- 5.Hokey DA, Weiner DB. DNA vaccines for HIV: challenges and opportunities. Springer Semin Immunopathol. 2006;28(3):267–79. doi: 10.1007/s00281-006-0046-z. [DOI] [PubMed] [Google Scholar]
- 6.Mulligan MJ, et al. Excellent safety and tolerability of the human immunodeficiency virus type 1 pGA2/JS2 plasmid DNA priming vector vaccine in HIV type 1 uninfected adults. AIDS Res Hum Retroviruses. 2006;22(7):678–83. doi: 10.1089/aid.2006.22.678. [DOI] [PubMed] [Google Scholar]
- 7.Andre S, et al. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72(2):1497–503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deml L, et al. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol. 2001;75(22):10991–1001. doi: 10.1128/JVI.75.22.10991-11001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider R, et al. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71(7):4892–903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muthumani K, et al. Novel engineered HIV-1 East African Clade-A gp160 plasmid construct induces strong humoral and cell-mediated immune responses in vivo. Virology. 2003;314(1):134–46. doi: 10.1016/s0042-6822(03)00459-8. [DOI] [PubMed] [Google Scholar]
- 11.Yang JS, et al. Induction of potent Th1-type immune responses from a novel DNA vaccine for West Nile virus New York isolate (WNV-NY1999) J Infect Dis. 2001;184(7):809–16. doi: 10.1086/323395. [DOI] [PubMed] [Google Scholar]
- 12.Gao F, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol. 2005;79(2):1154–63. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scriba TJ, et al. Functionally-inactive and immunogenic Tat, Rev and Nef DNA vaccines derived from sub-Saharan subtype C human immunodeficiency virus type 1 consensus sequences. Vaccine. 2005;23(9):1158–69. doi: 10.1016/j.vaccine.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Calarota SA, Weiner DB. Enhancement of human immunodeficiency virus type 1-DNA vaccine potency through incorporation of T-helper 1 molecular adjuvants. Immunol Rev. 2004;199:84–99. doi: 10.1111/j.0105-2896.2004.00150.x. [DOI] [PubMed] [Google Scholar]
- 15.Boyer JD, et al. SIV DNA vaccine co-administered with IL-12 expression plasmid enhances CD8 SIV cellular immune responses in cynomolgus macaques. J Med Primatol. 2005;34(5–6):262–70. doi: 10.1111/j.1600-0684.2005.00124.x. [DOI] [PubMed] [Google Scholar]
- 16.Schadeck EB, et al. A dose sparing effect by plasmid encoded IL-12 adjuvant on a SIVgag-plasmid DNA vaccine in rhesus macaques. Vaccine. 2006;24(21):4677–87. doi: 10.1016/j.vaccine.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 17.Xin KQ, et al. IL-15 expression plasmid enhances cell-mediated immunity induced by an HIV-1 DNA vaccine. Vaccine. 1999;17(7–8):858–66. doi: 10.1016/s0264-410x(98)00271-0. [DOI] [PubMed] [Google Scholar]
- 18.Kutzler MA, et al. Coimmunization with an optimized IL-15 plasmid results in enhanced function and longevity of CD8 T cells that are partially independent of CD4 T cell help. J Immunol. 2005;175(1):112–23. doi: 10.4049/jimmunol.175.1.112. [DOI] [PubMed] [Google Scholar]
- 19.Chong SY, et al. Comparative ability of plasmid IL-12 and IL-15 to enhance cellular and humoral immune responses elicited by a SIVgag plasmid DNA vaccine and alter disease progression following SHIV(89.6P) challenge in rhesus macaques. Vaccine. 2007 doi: 10.1016/j.vaccine.2006.11.070. [DOI] [PubMed] [Google Scholar]
- 20.Widera G, et al. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J Immunol. 2000;164(9):4635–40. doi: 10.4049/jimmunol.164.9.4635. [DOI] [PubMed] [Google Scholar]
- 21.Prud’homme GJ, Draghia-Akli R, Wang Q. Plasmid-based gene therapy of diabetes mellitus. Gene Ther. 2007;14(7):553–64. doi: 10.1038/sj.gt.3302907. [DOI] [PubMed] [Google Scholar]
- 22.Ugen KE, et al. Regression of subcutaneous B16 melanoma tumors after intratumoral delivery of an IL-15-expressing plasmid followed by in vivo electroporation. Cancer Gene Ther. 2006;13(10):969–74. doi: 10.1038/sj.cgt.7700973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otten G, et al. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine. 2004;22(19):2489–93. doi: 10.1016/j.vaccine.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 24.Otten GR, et al. Potent immunogenicity of an HIV-1 gag-pol fusion DNA vaccine delivered by in vivo electroporation. Vaccine. 2006;24(21):4503–9. doi: 10.1016/j.vaccine.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Luckay A, et al. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J Virol. 2007;81(10):5257–69. doi: 10.1128/JVI.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan J, et al. Enhanced cellular immune responses elicited by an engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Mol Ther. 2007;15(2):411–21. doi: 10.1038/sj.mt.6300036. [DOI] [PubMed] [Google Scholar]
- 27.Egan MA, et al. Priming with plasmid DNAs expressing interleukin-12 and simian immunodeficiency virus gag enhances the immunogenicity and efficacy of an experimental AIDS vaccine based on recombinant vesicular stomatitis virus. AIDS Res Hum Retroviruses. 2005;21(7):629–43. doi: 10.1089/aid.2005.21.629. [DOI] [PubMed] [Google Scholar]
- 28.Prud’homme GJ, et al. Electroporation-enhanced nonviral gene transfer for the prevention or treatment of immunological, endocrine and neoplastic diseases. Curr Gene Ther. 2006;6(2):243–73. doi: 10.2174/156652306776359504. [DOI] [PubMed] [Google Scholar]
- 29.Borrow P, et al. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68(9):6103–10. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koup RA, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68(7):4650–5. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capone S, et al. Modulation of the immune response induced by gene electrotransfer of a hepatitis C virus DNA vaccine in nonhuman primates. J Immunol. 2006;177(10):7462–71. doi: 10.4049/jimmunol.177.10.7462. [DOI] [PubMed] [Google Scholar]
- 32.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmerli SC, et al. HIV-1-specific IFN-gamma/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc Natl Acad Sci U S A. 2005;102(20):7239–44. doi: 10.1073/pnas.0502393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lichterfeld M, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J Exp Med. 2004;200(6):701–12. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]