Abstract
FOXA1 (also known as hepatocyte nuclear factor 3α, or HNF-3α) is a protein of the FKHD family transcription factors. FOXA1 has been termed as a pioneer transcription factor due to its unique ability of chromatin remodeling in which the chromatin can be de-compacted to allow genomic access by nuclear hormone receptors, including androgen receptor (AR) and estrogen receptor (ER). In this review, we discuss our current understanding of FOXA1 regulation of prostatic and non-prostatic AR-chromatin targeting. We present an updated model wherein FOXA1:AR equilibrium in the nuclei defines prostatic AR binding profile, which is perturbed in prostate cancer with FOXA1 and/or AR de-regulation. Finally, we discuss recent efforts in exploring new horizons of AR-independent functions of FOXA1 in prostate cancer and interesting directions to pursue in future studies.
Keywords: Androgen receptor, Chromatin accessibility, EMT, FOXA1, Pioneering factor, Prostate cancer
Introduction
The forkhead box A1 (FOXA1; previously termed as hepatocyte nuclear factor 3α, HNF-3α) protein belongs to a superfamily of winged helix transcription factors.1, 2 The name of “forkhead box” gene family is originally derived from a prominent phenotypic feature of developmental defects observed in Drosophila with the fork head gene mutant, which manifests in the foregut and hindgut being replaced by ectopic head structures.3 Like other forkhead (FKHD) family proteins, FOXA1 controls gene transcription by directly binding to its consensus sequence, the FKHD motif. In addition, FOXA1 has been shown capable of opening surrounding chromatin and subsequently allowing other transcription factors, such as androgen receptor (AR), to come in close proximity to their target sites and thus exert transcriptional control of gene expression.4, 5, 6, 7 Although this transcription regulatory effect of FOXA1 is quite well understood, important new developments have been made recently concerning the functional roles of FOXA1 in prostate cancer. This review thus discusses current literature regarding the delicate mechanisms by which FOXA1 regulates AR signaling and the deregulation and implication of FOXA1 in prostate cancer progression.
FOXA1 in development
FOXA1 was initially discovered approximately 25 years ago as an important liver-enriched transcriptional regulator of hepatic differentiation, since it was found to occupy the promoters of liver genes α1-antitrypsin and transthyretin.8 Subsequent mouse studies have shown that Foxa1 expression can be observed in endoderm-, mesoderm- and ectoderm-derived tissues of adult mice.9 It has been reported that detectable Foxa1 mRNA could first be observed at E7 in the late primitive streak stage in the midline endoderm of mouse embryos, following that the expression could be seen in the notochord, neural plate and floor plate of the neural tube, indicating that Foxa1's roles can range from establishment of definitive endoderm to formation of neural tube patterning.10, 11, 12
Although Foxa1 null mice don't exhibit discernible morphological defects, they display severe growth retardation and die between postnatal days 2 and 14 (P2 and P14), which is resulted from a combination of phenotypes including dehydration and hypoglycemia.13, 14 Therefore, these observations indicate that FOXA1 plays a pivotal role in the maintenance of glucose homeostasis and pancreatic islet function. Tissue-specific deletion of Foxa1 in the pancreas shows that FOXA1 and FOXA2 jointly regulate the expansion of pancreatic primordial, specification of endocrine and exocrine compartments, and maturation of islet cells.15 Similarly, there is also evidence that FOXA1 is important for lung development by regulating respiratory epithelial differentiation,16 and that it acts in a complementary manner with FOXA2 to ensure proper branching morphogenesis of the lung.17 Moreover, it has been demonstrated that both FOXA1 and FOXA2 in conjunction are required for initiating the onset of hepatogenesis and hepatic specification.18 More recently, a study utilizing conditional knockout of Foxa1 and Foxa2 in dopamine neurons reports that both factors are required for dopamine neuron maintenance and that their loss can give rise to locomotor deficits resembling the manifestations of Parkinson's disease.19 Taken together, mice studies corroborate the notion that FOXA1 has critical influence on organogenesis.
In particular, a number of papers have demonstrated the significance of FOXA1 during development of the prostate and mammary glands. It has been said that the mammary ductal morphogenesis, but not the alveolar lineage, is dependent on FOXA1, and that while Foxa1-null glands can form milk-producing alveoli, they have lost ERα expression and functional activity, which ultimately result in compromised ductal lineage specification.20 Likewise, in the prostate, FOXA1 deficiency leads to abolished differentiation and maturation of luminal epithelial cells.21 Initially derived from the hindgut endoderm, the mouse prostate epithelium has persistent Foxa1 expression throughout the processes of prostate development, growth, and adult differentiation.22 The origin of the prostate is the urogenital sinus, which is a midline structure composed of an endoderm-derived epithelial layer and a mesoderm-derived mesenchymal layer.23 In the mouse, at approximately E17.5, prostatic morphogenesis starts to take place, prompted by responsiveness to circulating androgens and induction of AR activity.23 During the course of development, Foxa1 expression was characterized in all lobes of the murine prostate, and is specifically enriched in AR-expressing epithelial cells. FOXA1 plays a critical role in modulating AR-regulated transcriptional signaling in prostate epithelial cells,6 and concordantly Foxa1-deficient prostate has severely impaired ductal pattern formation, due to inhibition of ductal canalization and epithelial cytodifferentiation.21 As a consequence, the Foxa1-null prostate lacks structural maturity as well as secretory activities. Taken together, there is compelling evidence that FOXA1 is critically involved in growth and differentiation of prostatic cells and is required for prostate glandular morphogenesis.
FOXA1 deregulation in prostate cancer
As FOXA1 is highly involved in developmental processes and lineage specification in several organs, when expressed at aberrant levels it may disrupt normal physiological events and lead to formation of cancer. Molecular and genetic studies have shown that FOXA1 is often found to be abnormally expressed in a number of cancer types, including acute myeloid leukemia (AML), lung, esophageal, thyroid, breast and prostate cancers.24, 25, 26, 27, 28, 29, 30 At present, the prevailing views on FOXA1 expression in prostate cancer have not reached a consensus, with contrasting evidence seen in different cohorts of cancer patients. Analyses of human prostate cancer specimens have revealed that FOXA1 is overexpressed in metastatic as well as castration-resistant prostate cancer (CRPC) patients, but its expression is lower in normal and neoplastic transitional zone tissues.31 In addition, the level of FOXA1 may be positively correlated with conventional parameters indicative of cancer progression (including tumor stage and Gleason scores), and negatively correlated with relapse-free survival times.30, 31 In other words, high FOXA1 level is associated with poor prognosis. However, other studies have also demonstrated that low FOXA1 levels are found in metastatic and CRPC tumors and may in fact denote unfavorable prognostic outcome in advanced prostate cancer.32, 33 In order to reconcile these conflicting findings, the function of FOXA1 should be carefully dissected with respect to cellular context, taking into consideration the status of AR program and androgen responsiveness, to fully understand how FOXA1 may fit as a piece of jigsaw in the prostate cancer puzzle.
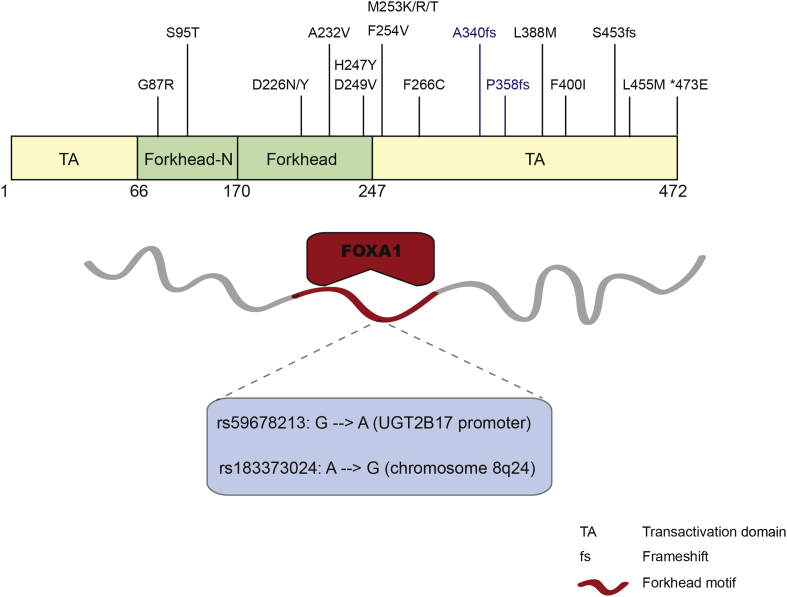
In addition to deregulation at expression level, mutations in the FOXA1 gene have also been uncovered in prostate tumors (Fig. 1), as reported in recent literature34 as well as in TCGA (The Cancer Genome Atlas). Recurrent FOXA1 gene mutations had recently been identified and characterized in 5 of 147 prostate cancers, including both localized as well as castration-resistant cases.35 Moreover, 4 of these 5 mutations are located in the C-terminal transactivation domain, and mutated FOXA1 was demonstrated to repress androgen signaling and augment tumor growth.35 Another independent study also reported 3 different non-silent mutations residing in or close to the forkhead domain in FOXA1,36 which are anticipated to disrupt DNA binding, but to what extent and how it may be related to prostate carcinogenesis will require further studies. Moreover, a recent study reported that, by adopting the methodology of 3D organoid culture system, the genetic heterogeneity of prostate cancer could be recapitulated, and FOXA1 was among the most mutated genes in the organoid CRPC lines.37
Figure 1.
FOXA1 mutations in prostate cancer. Somatic mutations of the FOXA1 gene that have been identified in localized and CRPC tumors, as well as in prostate cancer cell lines (the latter shown in blue, namely F266C, A340fs, P358fs). SNPs associated with prostate cancer risk have also been reported occurring within the consensus sequence of FOXA1 binding motifs.
Another level of FOXA1 deregulation in prostate cancer is reflected in somatic mutations of its cis-regulatory elements, which in turn affects FOXA1 transcriptional activity. It has been described that in prostate cancer there exists single nucleotide polymorphism (SNP) within the consensus forkhead motif, which is recognized and bound by the FOXA1 protein (Fig. 1). A prevalent SNP was identified in LNCaP prostate cancer cell line, which locates in the proximal promoter of the gene encoding UDP glucuronosyltransferase 2B17 (UGT2B17).38 This G to A polymorphism, UGT2B17 – 155 G/A, also appearing in NCBI SNP database as rs59678213, was shown to have a notable impact on FOXA1 binding, with the A-containing allele being 13-fold more active in luciferase assays.38 Moreover, another study revealed an SNP significantly associated with risk for prostate cancer in the chromosome 8q24 region (rs183373024), where it disrupts the FOXA1 recognition motif.39 As ChIP-seq data have reported AR and FOXA1 binding at this region in cell lines, it is predicted that this particular SNP in prostate cancer may cause disruption of FOXA1 and/or AR binding and thus lead to deregulation of some tumor suppressor genes. Several candidate genes were presented to be potential targets for this putative SNP-containing FOXA1 enhancer, however chromatin conformation capture (3C)-based techniques or CRISPR assays may be needed to assure the target gene.
FOXA1 defines prostate lineage-specific AR cistrome
Like other forkhead proteins, FOXA1 encompasses a winged helix domain that is composed of three α-helices, three β-sheets and two loops.40 This unique structure, which closely resembles that of linker histones,41 has imparted to FOXA1 the ability of binding to highly compacted chromatin and subsequently prying it open.42 In doing so, FOXA1 creates an open and easily accessible chromatin conformation to facilitate hormonal transcription factors such as estrogen receptor (ER) and AR to bind their cis-regulatory elements.43 Comparison of FOXA1 cistromes between breast and prostate cancer cells illustrates very distinct, lineage-specific profiles, where less than 40% of binding sites are shared in between.44 And yet, FOXA1 occupies a majority of the binding sites of AR and ER in prostate and breast cells, respectively, suggesting that FOXA1 may be critical in determining lineage-specific hormonal factor chromatin-targeting. It has been shown that FOXA1 is essential for AR-mediated prostatic gene activation, corroborated by the facts that FOXA1 regulatory elements are found in the core enhancer of the prostate-specific antigen (PSA) gene, a prototypical AR target, adjacent to the androgen response elements (AREs), and that perturbations in the FOXA1 motif can significantly abolish induction of PSA by androgens.6 Through bioinformatic and biochemical analyses, it has been discovered that the FKHD motif is enriched in AR cistromes,45, 46 and that FOXA1 can physically interact with AR.6 Thus, upon stimulation by androgens, AR translocates into the nucleus and preferentially binds cis-regulatory sequences that are largely pre-occupied by FOXA1, potentially under the recruitment by the FOXA1 protein. Expression profiling studies showed that FOXA1 indeed positively regulates prostatic gene expression induced by androgen.47
The mechanisms by which FOXA1 recognizes lineage-specific enhancers have also been investigated. It has been reported that FOXA1 binding can be guided by specific chromatin marks, namely mono- and di-methylation of histone 3 lysine 4 (H3K4me1, me2).44 Both are epigenetic signatures typically associated with enhancers, H3K4me1 and H3K4me2 genomic distribution was thought to provide a blueprint for directing differential FOXA1 binding in a lineage-specific manner. Furthermore, DNA methylation has also been shown to play a part in defining FOXA1 binding and enhancer activation. Genome-wide interrogation of DNA methylation reveals that FOXA1-bound enhancers are generally hypomethylated compared to juxtaposing genomic regions, in a pattern which is also correlated with cell type-specific FOXA1 binding.48 To better understand the sequence of events occurring at FOXA1-activated enhancers, kinetics study showed that binding of FOXA1 to chromatin could be detected prior to significant induction of H3K4me2 and DNA demethylation, which suggests that hypomethylation may be an epigenetic phenomenon succeeding FOXA1 binding rather than a pre-established mark.48 Although DNA hypomethylation does not seem to be required for FOXA1 binding, it is important for turning on FOXA1 transcriptional activity, as shown in luciferase reporter assays which exhibited reduced enhancer activation when constructs were methylated.48
FOXA1 prevents unrestricted, non-prostatic AR cistrome
Although FOXA1 pioneering of prostatic AR activities has been extensively studied, much less is known regarding aberrant AR programs in cancer cells, which often harbor AR amplification or overexpression, and FOXA1 loss or mutations. Whether and what cistrome AR binds in the absence of FOXA1 remained unaddressed for a while and have become questions of significance since the discoveries of FOXA1 mutations and loss in prostate cancer.33, 35, 36, 47 With more advanced sequencing technologies and scrupulous analytic efforts, a recent study demonstrated that FOXA1 is not only crucial for defining a prostate-specific AR program, but notably is also imperative for obstructing non-prostatic AR binding events.47 This latter feature is reflected in the observation that FOXA1 depletion in prostate cancer cells, instead of resulting in loss of total AR binding events, rather leads to substantial gain of new AR binding sites, reflecting AR reprogramming at the genome-wide level.7, 30, 33, 47, 49 It was reported that silencing of FOXA1 gave rise to a marked increase of over 2.5-fold in the number AR occupied sites.30 By grouping AR binding sites and target genes according to their relative responses to FOXA1 depletion, three classes of AR regulatory regions can be characterized: 1) sites that are independent of FOXA1 and remain unchanged regardless of FOXA1 depletion; 2) sites that require FOXA1 to open chromatin for AR recruitment (prostate-specific); 3) sites that only become bound by AR upon FOXA1 loss.30 Similar gain of AR binding sites upon FOXA1 depletion was also reported by an independent group.47 Our group performed motif analyses on endogenous as well as reprogrammed AR binding sites and showed that while in control cells a majority of the AR binding sites contained primarily the FKHD motif, upon FOXA1 knockdown this pattern became shifted towards binding sites that are dominated by the ARE motif.47 These gained ARE-mediated sites have not been pioneered by FOXA1 and are bound by AR through direct recognition of its consensus sequences, controlling a set of genes quite distinct from its FOXA1-pioneered, prostate-specific targets. This notion is further supported by the findings that in FOXA1-depleted cells, approximately half of the prostatic androgen-regulated genes lose their androgen-dependency, but more importantly, new sets of androgen-dependent genes not seen in parental cells are acquired.30 This set of new target genes may contribute to enhanced cell growth and metabolism potentially leading to prostate cancer progression.33 In addition, androgen-regulated genes in control cells are typically enriched for both AR and FOXA1, however in FOXA1-depleted cells, the acquired new sets of androgen-regulated genes only exhibited enrichment of binding sites unique to AR.30 Taken together, whether FOXA1 will facilitate or inhibit AR binding to specific genomic regions is context-dependent, depending largely on the co-occurrence of FKHD and ARE (or half ARE) motifs. It is clear that, in addition to its role in pioneering prostate-specific AR binding events, FOXA1 at the same time functions to inhibit AR binding to non-prostatic genes, the latter being equally important as the former in its ability to ensure prostate lineage differentiation.
FOXA1:AR equilibrium and prostate cancer
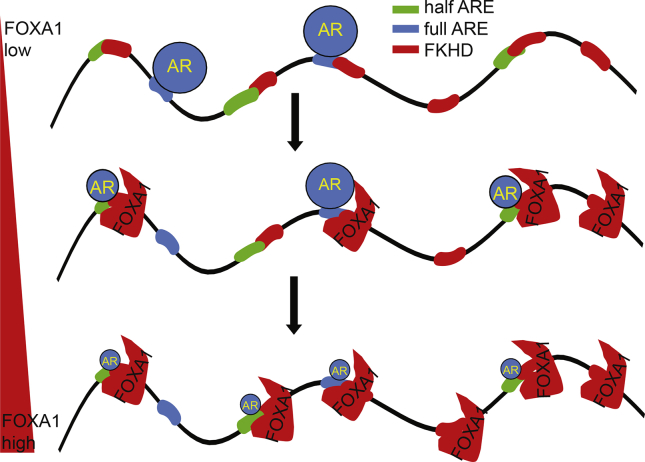
Previous studies have reported that even preceding androgen stimulation FOXA1 could bind to DNA and occupy a majority of AR-associated sites.30, 33, 44, 47 As demonstrated in one study, approximately 54% of AR binding sites were shared by FOXA1 in androgen-treated LNCaP cells, and 70% of these sites had FOXA1 pre-occupancy before hormone treatment.33 These studies have also revealed that on a global scale there exist a vastly more number of FOXA1 binding sites than AR; nearly 90% of FOXA1 binding sites are not co-occupied by AR. These data suggest that 1) FOXA1 may regulate many target genes independently of AR (which will be discussed in the next section) and 2) extensive FOXA1 binding events may be necessary to ensure AR binding to the prostate-specific genes. This latter point is supported by the observation that FOXA1 knockdown leads to AR re-distribution to many new sites.30, 33, 47 Using FAIRE-seq (formaldehyde-assisted isolation of regulatory elements followed by massively parallel sequencing), a study has reported that a significant proportion of FOXA1-bound sites exhibited low FAIRE signals along with repressive histone marks, reflective of a closed chromatin conformation, implying that FOXA1 binding is insufficient for functional activity of its bound enhancers.50 Moreover, we recently delineated that FOXA1 opens up large number of chromatin regions, only a portion of which, likely due to the co-occurrence of a half or full ARE within, are bound by AR.47 Our data suggest that, instead of directly fetching AR to the desired sites, FOXA1 binds to FKHD sites to increase the competence of any nearby half or full ARE sites for AR binding, compared to those AREs located at less accessible chromatin regions (Fig. 2).
Figure 2.
FOXA1:AR equilibrium determines the AR binding profile. The schematic model depicts that AR binds to ARE motifs in the absence of FOXA1, is recruited to FKHD sites with co-occurring full or half AREs when at equilibrium with FOXA1, and is diluted across the chromatin to form insignificant binding by excessive FOXA1.
One drawback of this system is that, when the amount of FOXA1 is in great excess to AR in the nuclei, it will inevitably dilute AR across the chromatin forming insignificant binding. This was demonstrated using the Du145 model system by manipulating overexpression of FOXA1 and AR in different levels, since these cells have negligible amounts of both endogenous proteins. As a result, moderate amount of FOXA1 shifts AR binding from ARE to FKHD sites. However, when FOXA1 is abundantly available, it could open up chromatin immensely, significantly increasing the number of open chromatin regions detected by FAIRE-seq, to the extent that AR binding becomes diluted across the entire genome. Consequently, total specific AR binding events are drastically reduced, since FOXA1 is able to direct AR to numerous open sites which may not be accessible had FOXA1 not been so abundantly present.47 Therefore, an equilibrium between nuclear FOXA1 and AR levels is critical for the maintenance of the prostatic AR program.
The FOXA1:AR equilibrium may be disrupted in various pathological conditions in prostate cancer. For example, in patients being treated with hormone deprivation therapy, there is markedly reduced nuclear AR level, leading to excessive FOXA1 and inhibition of residual AR binding to DNA. Therefore, FOXA1 knockdown in androgen-deprived LNCaP cells ultimately allows AR to form some significant binding events. Consistent with the changes in AR binding profile, FOXA1 depletion was also found to increase sensitivity to lower androgen levels, as measured by level of prostate-specific gene transcription.30 It was observed that androgen-induced genes become upregulated and androgen-repressed genes downregulated simply following FOXA1 knockdown, despite the lack of hormone stimulation.47 In other words, FOXA1 loss turns on androgen-independent AR transcriptional program, which might highlight a potential route for the development of castration resistance in prostate cancer. These studies are in good agreement with recent observations of FOXA1 loss-of-function mutations in prostate cancer and downregulation in CRPC.33, 35 In addition, studies have shown that AR primarily functions to promote G1-S cell cycle progression in androgen-dependent prostate cancer requiring the chromatin remodeling actions of FOXA1.51, 52, 53 On the contrary, in CRPC, AR function digresses to regulate G2-M transition instead, by modulating expression of new target genes such as UBE2C, a critical M phase checkpoint inhibitor.54 Importantly, the differential activities of AR are brought about by FOXA1 recruitment to regulatory sites of these genes prior to AR binding. Therefore, FOXA1 plays stage-specific and context-dependent functions in its regulation of particular AR program in either androgen-supplemented or -deprived conditions.
Role of FOXA1 in regulating prostate cancer cell growth and motility
Overall, both oncogenic and tumor-suppressive roles have been reported for FOXA1, which suggests that its precise contribution to cancer development or progression may be depended on disease stage, context, and treatment histories. In conjunction with AR signaling in the presence of androgen, FOXA1 is known to promote prostate cancer proliferation by inducing expression of cell cycle genes.32, 51 However, under androgen-depleted conditions, FOXA1 was shown to rather inhibit cell proliferation and its loss led to androgen-independent prostate cancer cell growth, being consistent with its regulation of AR signaling.47 In support with this tumor suppressor role, in mice with prostate-specific Foxa1 gene deletion, progressive hyperplasia can be observed, and Foxa1 knockout epithelial cells exhibit increased proliferation and altered morphology.55 Further, following castration, the number of Foxa1-positive cells was significantly reduced, supporting Foxa1 loss as a potential mechanism to castration resistance. Thus, like its modulation of AR signaling, FOXA1 regulation of prostate cancer cell growth is context-dependent.
Through analyses of genome-wide gene expression profiling, it has been discovered that FOXA1 may also possess AR-independent functions in inhibiting cell motility and epithelial-to-mesenchymal transition (EMT).32 In prostate cancer cells lacking AR expression, ectopic introduction of FOXA1 is sufficient to impede cell invasion and migration.32 On the other hand, loss of FOXA1 in LNCaP cells increases cell invasiveness, both in androgen-containing and -deprived conditions. Both cases demonstrated the AR-independent function of FOXA1 in inhibiting prostate cancer cell motility. Meanwhile, it is also found that FOXA1 can negatively regulate EMT, and loss of FOXA1 in LNCaP cells results in an astrocyte-like, fusiform, or fibroblastic phenotype characteristic of mesenchymal and neuroendocrine cells. Further analysis revealed that among direct transcriptional targets of FOXA1, SLUG was identified to be a key repressed gene that confers the anti-motility properties associated with FOXA1.32 Similar functions of FOXA1 in preventing metastasis have been reported in other forms of cancer as well, such as lung cancer and pancreatic cancer,56, 57 supporting the idea that this anti-EMT role is AR-independent. Being concordant with these functionalities, expression profiling datasets of prostate tumors confirm that FOXA1 is upregulated from benign tissue to localized tumor, but downregulated in metastatic CRPC tumors compared to localized ones.32, 33 However, there exists contrasting histological evidence that FOXA1 level is high in metastatic prostate cancer.58 FOXA1 expression level may need to be more carefully looked at taking into considerations of disease stage, hormone deprivation treatment history, and relative AR level.
Future directions
Despite these significant advances in our understanding of FOXA1 regulation of AR signaling and prostate cancer, many questions remain to be answered. Various mutations to the FOXA1 gene have been reported in prostate cancer. It will be critical in future studies to determine how these mutants differ from the wildtype FOXA1 in their regulation of AR signaling and prostate cancer progression. Further, the controversies regarding FOXA1 expression in primary specimens and its association with prostate cancer prognosis are yet to be carefully addressed. Since FOXA1 promotes cell growth in an AR-dependent manner, its expression level may be associated with poor clinical outcome only in patients with primary prostate cancer that is not castration-resistant. On the other hand, as FOXA1 is able to inhibit cell motility in an AR-independent manner, its expression level would predict good clinical outcome in patients who have been subjected to androgen-deprivation therapies. Importantly, due to the significance of the equilibrium between FOXA1 and AR levels, it will be imperative to take into consideration the FOXA1/AR ratio when predicting disease outcome. As FOXA1 loss has been shown to activate androgen-independent AR signaling and contribute to CRPC progression, a low FOXA1/AR ratio may be indicative of poor prognosis. Moreover, very recently it is reported that FOXA1 has a novel immunosuppressive function in T cells.59 It was shown that FOXA1 could be induced by interferon-β (IFN-β), which is used in treatment of relapsing-remitting multiple sclerosis (RRMS), an autoimmune disease where regulatory T (Treg) cells are defective. Additionally, generation of FOXA1+ Treg cells upon IFN-β treatment was found to associate with favorable clinical outcome in RRMS patients. Thus, these data indicate that it would be interesting to study whether the role of FOXA1 in T cells may be implicated in prostate cancer and tumor microenvironment. This will represent a new research direction of FOXA1 quite distinct from its well-established function in the AR context.
Conflicts of interest
The authors report no conflicts of interest in this work.
Acknowledgement
JY acknowledges funding from the NIH (R01CA172384), the U.S. Department of Defense (W81XWH-13-1-0319 and W81XWH-14-1-0023), and the American Cancer Society (RSG-12-085-01). Y.A.Y. was supported by the NIH/NCI training grant T32 CA09560.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Carlsson P., Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 2.Benayoun B.A., Caburet S., Veitia R.A. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27:224–232. doi: 10.1016/j.tig.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Weigel D., Jurgens G., Kuttner F., Seifert E., Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 4.Augello M.A., Hickey T.E., Knudsen K.E. FOXA1: master of steroid receptor function in cancer. EMBO J. 2011;30:3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll J.S., Liu X.S., Brodsky A.S. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Gao N., Zhang J., Rao M.A. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol. 2003;17:1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- 7.Sahu B., Laakso M., Pihlajamaa P. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer Res. 2013;73:1570–1580. doi: 10.1158/0008-5472.CAN-12-2350. [DOI] [PubMed] [Google Scholar]
- 8.Costa R.H., Grayson D.R., Darnell J.E., Jr. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besnard V., Wert S.E., Hull W.M., Whitsett J.A. Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expression Patterns. 2004;5:193–208. doi: 10.1016/j.modgep.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Monaghan A.P., Kaestner K.H., Grau E., Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- 11.Ang S.L., Wierda A., Wong D. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki H., Hogan B.L. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- 13.Kaestner K.H., Katz J., Liu Y., Drucker D.J., Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih D.Q., Navas M.A., Kuwajima S., Duncan S.A., Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc Natl Acad Sci USA. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao N., LeLay J., Vatamaniuk M.Z., Rieck S., Friedman J.R., Kaestner K.H. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besnard V., Wert S.E., Kaestner K.H., Whitsett J.A. Stage-specific regulation of respiratory epithelial cell differentiation by Foxa1. Am J Physiol Lung Cell Mol Physiol. 2005;289:L750–L759. doi: 10.1152/ajplung.00151.2005. [DOI] [PubMed] [Google Scholar]
- 17.Wan H., Dingle S., Xu Y. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 18.Lee C.S., Friedman J.R., Fulmer J.T., Kaestner K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 19.Domanskyi A., Alter H., Vogt M.A., Gass P., Vinnikov I.A. Transcription factors Foxa1 and Foxa2 are required for adult dopamine neurons maintenance. Front Cell Neurosci. 2014;8:275. doi: 10.3389/fncel.2014.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernardo G.M., Lozada K.L., Miedler J.D. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao N., Ishii K., Mirosevich J. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–3443. doi: 10.1242/dev.01917. [DOI] [PubMed] [Google Scholar]
- 22.Mirosevich J., Gao N., Matusik R.J. Expression of Foxa transcription factors in the developing and adult murine prostate. Prostate. 2005;62:339–352. doi: 10.1002/pros.20131. [DOI] [PubMed] [Google Scholar]
- 23.Marker P.C., Donjacour A.A., Dahiya R., Cunha G.R. Hormonal, cellular, and molecular control of prostatic development. Dev Biol. 2003;253:165–174. doi: 10.1016/s0012-1606(02)00031-3. [DOI] [PubMed] [Google Scholar]
- 24.Neben K., Schnittger S., Brors B. Distinct gene expression patterns associated with FLT3- and NRAS-activating mutations in acute myeloid leukemia with normal karyotype. Oncogene. 2005;24:1580–1588. doi: 10.1038/sj.onc.1208344. [DOI] [PubMed] [Google Scholar]
- 25.Lin L., Miller C.T., Contreras J.I. The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 2002;62:5273–5279. [PubMed] [Google Scholar]
- 26.Sano M., Aoyagi K., Takahashi H. Forkhead box A1 transcriptional pathway in KRT7-expressing esophageal squamous cell carcinomas with extensive lymph node metastasis. Int J Oncol. 2010;36:321–330. [PubMed] [Google Scholar]
- 27.Nucera C., Eeckhoute J., Finn S. FOXA1 is a potential oncogene in anaplastic thyroid carcinoma. Clin Cancer Res. 2009;15:3680–3689. doi: 10.1158/1078-0432.CCR-08-3155. [DOI] [PubMed] [Google Scholar]
- 28.Hu X., Stern H.M., Ge L. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 29.Robbins C.M., Tembe W.A., Baker A. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahu B., Laakso M., Ovaska K. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerhardt J., Montani M., Wild P. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. Am J Pathol. 2012;180:848–861. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 32.Jin H.J., Zhao J.C., Ogden I., Bergan R.C., Yu J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013;73:3725–3736. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D., Garcia-Bassets I., Benner C. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J.L., Holmes K.A., Carroll J.S. FOXA1 mutations in hormone-dependent cancers. Front Oncol. 2013;3:20. doi: 10.3389/fonc.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grasso C.S., Wu Y.M., Robinson D.R. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbieri C.E., Baca S.C., Lawrence M.S. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao D., Vela I., Sboner A. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu D.G., Gardner-Stephen D., Severi G. A novel polymorphism in a forkhead box A1 (FOXA1) binding site of the human UDP glucuronosyltransferase 2B17 gene modulates promoter activity and is associated with altered levels of circulating androstane-3alpha,17beta-diol glucuronide. Mol Pharmacol. 2010;78:714–722. doi: 10.1124/mol.110.065953. [DOI] [PubMed] [Google Scholar]
- 39.Hazelett D.J., Coetzee S.G., Coetzee G.A. A rare variant, which destroys a FoxA1 site at 8q24, is associated with prostate cancer risk. Cell Cycle. 2013;12:379–380. doi: 10.4161/cc.23201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hannenhalli S., Kaestner K.H. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark K.L., Halay E.D., Lai E., Burley S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 42.Cirillo L.A., McPherson C.E., Bossard P. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 44.Lupien M., Eeckhoute J., Meyer C.A. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q., Li W., Liu X.S. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lupien M., Brown M. Cistromics of hormone-dependent cancer. Endocr Relat Cancer. 2009;16:381–389. doi: 10.1677/ERC-09-0038. [DOI] [PubMed] [Google Scholar]
- 47.Jin H.J., Zhao J.C., Wu L., Kim J., Yu J. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun. 2014;5:3972. doi: 10.1038/ncomms4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serandour A.A., Avner S., Percevault F. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L., Huang H., Dougherty G., Zhao Y., Hossain A., Kocher J.P. Epidaurus: aggregation and integration analysis of prostate cancer epigenome. Nucleic Acids Res. 2015;43:e7. doi: 10.1093/nar/gku1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eeckhoute J., Lupien M., Meyer C.A. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 2009;19:372–380. doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y., Chen S.Y., Ross K.N., Balk S.P. Androgens induce prostate cancer cell proliferation through mammalian target of rapamycin activation and post-transcriptional increases in cyclin D proteins. Cancer Res. 2006;66:7783–7792. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]
- 52.Knudsen K.E., Arden K.C., Cavenee W.K. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J Biol Chem. 1998;273:20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 53.Balk S.P., Knudsen K.E. AR, the cell cycle, and prostate cancer. Nucl Recept Signal. 2008;6:e001. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q., Li W., Zhang Y. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeGraff D.J., Grabowska M.M., Case T.C. FOXA1 deletion in luminal epithelium causes prostatic hyperplasia and alteration of differentiated phenotype. Lab Invest. 2014;94:726–739. doi: 10.1038/labinvest.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H., Meyer C.A., Fei T., Wang G., Zhang F., Liu X.S. A systematic approach identifies FOXA1 as a key factor in the loss of epithelial traits during the epithelial-to-mesenchymal transition in lung cancer. BMC Genomics. 2013;14:680. doi: 10.1186/1471-2164-14-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song Y., Washington M.K., Crawford H.C. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain R.K., Mehta R.J., Nakshatri H., Idrees M.T., Badve S.S. High-level expression of forkhead-box protein A1 in metastatic prostate cancer. Histopathology. 2011;58:766–772. doi: 10.1111/j.1365-2559.2011.03796.x. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y., Carlsson R., Comabella M. FoxA1 directs the lineage and immunosuppressive properties of a novel regulatory T cell population in EAE and MS. Nat Med. 2014;20:272–282. doi: 10.1038/nm.3485. [DOI] [PubMed] [Google Scholar]