Abstract
Ewing Sarcoma is a biologically aggressive bone and soft tissue malignancy affecting children and young adults. Ewing Sarcoma pathogenesis is driven by EWS/Ets fusion oncoproteins, of which EWS/Fli1 is the most common. We have previously shown that microRNAs (miRs) regulated by EWS/Fli1 contribute to the pro-oncogenic program in Ewing Sarcoma. Here we show that miR-22, an EWS/Fli1-repressed miR, is inhibitory to Ewing Sarcoma clonogenic and anchorage-independent cell growth, even at modest overexpression levels. Our studies further identify the H3K9me1/2 histone demethylase KDM3A (JMJD1A/JHDM2A) as a new miR-22-regulated gene. We show that KDM3A is overexpressed in Ewing Sarcoma, and that its depletion inhibits clonogenic and anchorage-independent growth in multiple patient-derived cell lines, and tumorigenesis in a xenograft model. KDM3A depletion further results in augmentation of the levels of the repressive H3K9me2 histone mark, and downregulation of pro-oncogenic factors in Ewing Sarcoma. Together, our studies identify the histone demethylase KDM3A as a new, miR-regulated, tumor promoter in Ewing Sarcoma.
Keywords: Ewing sarcoma, microRNA, epigenetics, histone demethylase
INTRODUCTION
Ewing Sarcoma is an aggressive malignancy of bone and soft tissues affecting children and young adults.1 Ewing Sarcoma pathogenesis is driven by potent fusion oncoproteins arising from recurrent chromosomal translocations, most commonly between the EWS gene and one of a number of Ets transcription factor genes.2 By far, the most common fusion oncoprotein in Ewing Sarcoma is EWS/Fli1, accounting for 80–90% of cases. The EWS/Fli1 fusion oncoprotein is a gain-of-function aberrant regulator of gene expression.2 A variety of downstream pathways of EWS/Fli1 action have been described,2 but many remain unknown.
MicroRNAs (miRs), identified recently as a new class of regulators of gene expression, are often aberrantly expressed in cancer and other disease states.3,4 MiRs can exert wide-ranging effects on disease phenotypes, and have attracted attention as potential new therapeutic agents or/and targets.5 We6 and others7–10 have recently identified miRs with altered expression in Ewing Sarcoma, and shown that miRs contribute to the sarcoma phenotype. However, understanding of miR-mediated pathways in Ewing Sarcoma pathogenesis remains limited.
In the present manuscript, we demonstrate that miR-22, a miR normally repressed by the EWS/Fli1 oncoprotein, is growth inhibitory in multiple Ewing Sarcoma cell lines, and we identify the H3K9me1/2 histone demethylase KDM3A as a new miR-22 target. Epigenetic modifiers have recently emerged as playing key roles in cancer,11,12 with particular importance in the pathogenesis of pediatric tumors.13,14 However, current understanding of the role of histone demethylases in cancer is limited, as are the effects of modulation of the H3K9 histone mark. We show that KDM3A, an epigenetic modifier not previously implicated in sarcomagenesis, is overexpressed in Ewing Sarcoma, and that this results in enhanced oncogene expression and promotion of the tumorigenic phenotype. Together, our studies reveal a new miR-regulated, epigenetic, tumor-promotional pathway in Ewing Sarcoma, downstream of the EWS/Fli1 oncoprotein.
RESULTS AND DISCUSSION
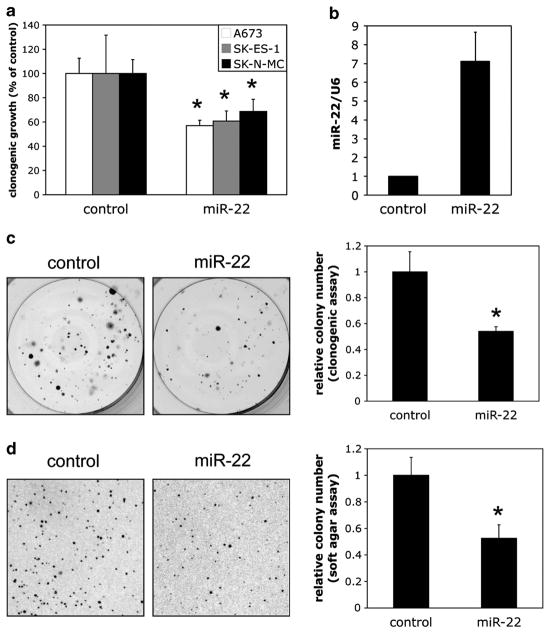
Our previous studies identified a group of candidate tumor suppressive miRs normally repressed by EWS/Fli1 in Ewing Sarcoma.6 Of these, miR-22 was of particular interest to us, as it had been shown to manifest tumor suppressive properties in many other cancers,15–22 thus representing an attractive candidate for potential miR replacement therapy. We thus sought to better characterize the effects of miR-22 replacement in Ewing Sarcoma. To simulate a potential therapeutic model, we introduced miR-22 mimics into three different, validated, patient-derived Ewing Sarcoma cell lines, and examined the effects on clonogenic growth. As shown (Figure 1a), this resulted in inhibition of colony formation in all the cell lines, compared with control (non-targeting) mimic. In order to further probe the function of miR-22 in Ewing Sarcoma, we generated A673 cells stably overexpressing miR-22 using a retroviral miR expression system. This system yielded approximately sevenfold overexpression of mature miR-22 (Figure 1b). Stable miR-22 overexpression, even at such relatively modest levels, resulted in inhibition of A673 colony formation in a clonogenic assay and a soft agar assay for anchorage-independent growth (Figures 1c and d). Thus, miR-22 overexpression in Ewing Sarcoma cells is inhibitory to clonogenic and anchorage-independent growth.
Figure 1.
MiR-22 is inhibitory to clonogenic and anchorage-independent growth in Ewing Sarcoma. (a) The indicated Ewing Sarcoma cell lines (all described previously,6 and authenticated by short tandem repeat (STR) profiling) were transfected with 25 nM miR-22 mimic or non-targeting negative control mimic, as previously described,6 harvested the following day and plated at 500 cells per well in six-well plates. Colonies were stained 14 days later with 0.1% crystal violet in 25% MeOH, and quantified using NIS-elements imaging software. Results represent the mean and standard deviation of triplicate platings, with the control set to 100%. (b) For stable overexpression of miR-22, the miR-22 genomic locus, including approximately 250 bp of upstream and downstream flanking sequence, was PCR amplified from Ewing Sarcoma A673 cells, and cloned into the pMSCV-Puro retroviral expression vector, using standard molecular techniques and verification by sequencing. Replication-incompetent, vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped, infectious retrovirus was prepared essentially as previously described,6 using a retroviral packaging system. Transduction of Ewing Sarcoma cells with viral supernatant was performed as previously described.6 Control cells were infected with empty pMSCV-Puro. Following selection with Puromycin (2 μg/ml), quantification of mature miR-22 levels was performed on whole Trizol-extracted RNA using the Qiagen miScript SYBRgreen quantitative reverse transcription-PCR system, with U6 as the internal control, as previously described.6 MicroRNA primers were obtained from Qiagen. MiR-22 level in control cells is set to 1. (c) Clonogenic assays on control and miR-22 stably overexpressing cells were performed as in a, in the presence of Puromycin (2 μg/ml). Quantifications represent the mean and standard error of the mean (s.e.m.) of two independent experiments, each performed in at least triplicate; representative images from one experiment are also shown. (d) Soft agar assays were performed as described,6 using 1 ×104 cells. Quantifications represent the mean and s.e.m. of two independent experiments, each performed in at least triplicate; representative images from one experiment are also shown. *P<0.05 relative to control, using a two-way Student’s t-test with unequal variance.
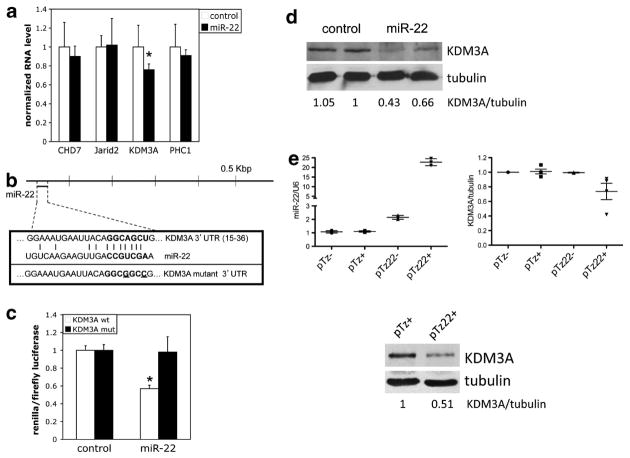
In order to better understand the mechanism of action of miR-22 in Ewing Sarcoma, we next sought targets that could potentially contribute to its growth-inhibitory phenotype. Examination of the predicted target profile of miR-22 disclosed a number of chromatin-modifying factors as candidate targets. Chromatin modifiers have recently emerged as critical modulators of oncogenesis,12 with a particularly prominent role in pediatric cancers.13 We thus explored the possibility that regulation of chromatin modifiers by miR-22 could promote Ewing Sarcoma oncogenesis. We selected four chromatin modifiers, CHD7, Jarid2, KDM3A and PHC1, as candidates of particular interest for analysis, based on the following criteria: identified as miR-22 targets by at least two of three prediction algorithms (TargetScan, PicTar and miRanda); predicted miR-22 site in proximal 1 kbp of 3′ untranslated region (UTR); targets implicated in promotion of oncogenesis in other systems. We first screened for changes in expression levels of these targets upon miR-22 overexpression in A673 cells, using quantitative reverse transcription-PCR. As shown in Figure 2a, miR-22 overexpression resulted in a roughly 20% decrease in the level of KDM3A (also known as JMJD1A and JHDM2A) mRNA, but not the other candidate targets. MiR-22 has a single, highly conserved, predicted target site in the proximal 50 base pairs of the KDM3A 3′UTR (Figure 2b). Using a 3′UTR reporter system, we verified that this site is responsive to regulation by miR-22 in A673 cells (Figure 2c). Finally, we asked whether miR-22 regulates KDM3A protein levels in Ewing Sarcoma. Using the same stable expression system as above (Figure 1), we found that KDM3A protein levels in A673 cells were lower in miR-22-overexpressing cells compared with controls (Figure 2d). To explore the regulation of KDM3A expression by miR-22 further, we performed similar analyses using an inducible miR-22 system in A673 cells. These experiments showed downregulation of KDM3A protein levels upon induction of miR-22 expression with doxycycline (Figure 2e, pMz22 + group), compared with controls, thus verifying our observation in miR-22 stably overexpressing cells. Moreover, using both miR expression systems, we demonstrated negative regulation of KDM3A expression by miR-22 in another Ewing Sarcoma cell line (TC71; Supplementary Figure 1). Interestingly, in both cell lines, the magnitude of negative regulation of KDM3A levels by miR-22 was variable from experiment to experiment, ranging from 10 to 60%. This suggests that miR-22 and KDM3A may be part of a more complex regulatory network, as is frequently true of miRs.23 Together, these studies support a role for miR-22 in the regulation of KDM3A expression in Ewing Sarcoma.
Figure 2.
MiR-22 negatively regulates KDM3A expression in Ewing Sarcoma. (a) Levels of the indicated mRNAs, normalized to U6 internal control, in control and miR-22 stably overexpressing A673 cells, were determined by quantitative reverse transcription-PCR (qRT-PCR) as in Figure 1. (b) Schematic of the KDM3A 3′UTR showing the location of the predicted conserved miR-22 target site; wild-type and mutant miR-22 target sequences used in reporter assays (mutated nucleotides are underlined). (c) A673 cells were cotransfected with a psiCHECK-2 reporter construct containing the wild-type KDM3A 3′UTR miR-22 site or the mutated site, introduced via standard cloning techniques followed by sequence verification, and miR-22 mimic or non-targeting negative control mimic. Reporter activity was determined as previously described.6 *P<0.05 relative to control, using a two-way Student’s t-test with unequal variance. (d) KDM3A levels in control and miR-22 stably overexpressing A673 cells, as determined by immunoblotting, with tubulin as loading control. Protein extracts were prepared in RIPA buffer (50 mM Tris pH 7.2, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) with protease/phosphatase inhibitors (1 × Roche protease inhibitor cocktail (#04-693-116-001), 20 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 100 μM sodium orthovanadate, 500 μM dithiothreitol). SDS–polyacrylamide gel electrophoresis, immunoblotting and enhanced chemiluminescence (ECL) detection were performed as previously described.6 KDM3A/tubulin ratios were determined using densitometric quantification, with the ratio in one of the controls set to 1. Primary antibodies used were: KDM3A (1:200, ProMab, #30134) and Tubulin (1:20,000, Sigma, T5168). (e) The miR-22 precursor was subcloned from pMSCV-Puro into the pTRIPz, tet-inducible, lentiviral expression vector44 and sequence verified. A673 cells were stably transduced with the resulting miR-22 expression construct (pTz22) or empty vector control (pTz), as described,44 and selected with Puromycin (2 μg/ml); miR expression was induced by treating the cells with doxycycline (2 μg/ml; ‘+’ groups) for 5–7 days; ‘−’ groups did not receive doxycycline. MiR-22 levels (left panel) were determined by qRT-PCR as in Figure 1. KDM3A levels, and KDM3A/tubulin ratios, were determined as above; right panel shows mean and s.e.m. from multiple experiments, each performed in duplicate; immunoblot data from one experiment are also shown below.
Although the precise cell of origin of Ewing Sarcoma remains to be defined, a number of studies have presented evidence for the mesenchymal stem cell as a candidate.24–26 Our immunoblotting analysis showed KDM3A levels in a panel of Ewing Sarcoma cell lines to be consistently higher compared with mesenchymal stem cells (Figure 3a), suggesting that KDM3A is upregulated in Ewing Sarcoma. To explore the question of KDM3A expression in Ewing Sarcoma further, we examined published gene expression profiling data for possible control of KDM3A levels by the EWS/Fli1 oncoprotein. This analysis revealed that KDM3A expression is positively regulated downstream of EWS/Fli1 (Supplementary Figure 2A). Moreover, similar data mining revealed that KDM3A is overexpressed in Ewing Sarcoma tumors (Supplementary Figure 2B). Taken together, these data indicate that KDM3A is upregulated in Ewing Sarcoma, and that this is at least in part a consequence of EWS/Fli1 expression. Moreover, our data above on negative regulation of KDM3A expression by miR-22, along with our previous observation of negative regulation of miR-22 by EWS/Fli1,6 suggest that downregulation of miR-22 contributes to the induction of KDM3A levels by EWS/Fli1.
Figure 3.
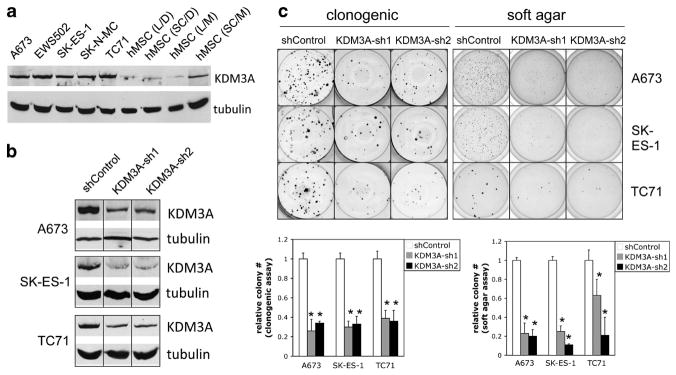
KDM3A is upregulated in Ewing Sarcoma, and promotes clonogenic and anchorage-independent growth. (a) KDM3A protein levels in the indicated Ewing Sarcoma cell lines, and mesenchymal stem cells (hMSCs), were determined by immunoblotting, with tubulin as loading control, as in Figure 2. MSCs were obtained from Lonza (L) and SciCell (SC), as previously,6 and grown in Dulbecco’s modified Eagle’s media (D) or proprietary stem cell media (M). (b) KDM3A protein levels in A673, SK-ES-1 and TC71 cells stably transduced with two different KDM3A-targeting lentiviral small hairpin RNAs (shRNAs) and non-targeting control, as determined by immunoblotting. ShRNA-mediated gene expression silencing via lentiviral delivery was performed as previously described.6 The control non-targeting shRNA consisted of a scrambled sequence (Addgene plasmid 186445). ShRNAs 1 and 2 for KDM3A correspond to TRCN0000021150 and TRCN0000021152 (all Sigma Mission shRNAs, distributed via the University of Colorado Cancer Center Functional Genomics Core Facility). (c) Clonogenic and soft agar assays were performed and analyzed as in Figure 1. Quantifications represent the mean and s.e.m. of two independent experiments, each performed in at least triplicate; representative images from one experiment are also shown. *P<0.05, determined using a two-way Student’s t-test with unequal variance.
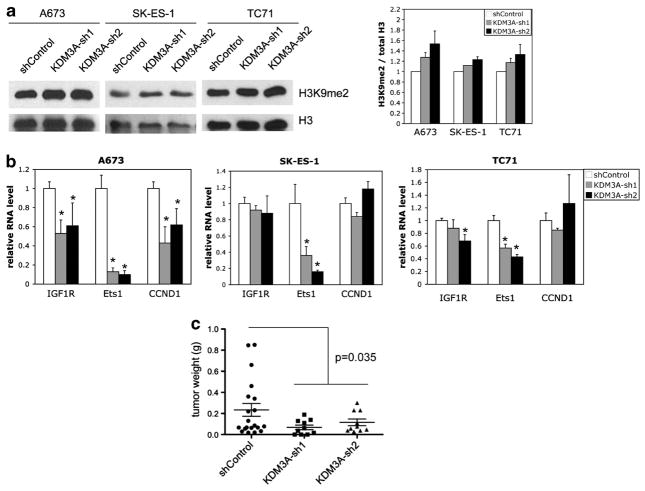
To determine the functional consequences of KDM3A upregulation in Ewing Sarcoma, we stably silenced KDM3A expression in three different Ewing Sarcoma cell lines, using two different lentivirally delivered small hairpin RNAs (Figure 3b), and examined the effects on clonogenic and anchorage-independent growth. Compared with non-targeting small hairpin RNA control, depletion of KDM3A protein levels resulted in robust inhibition of colony formation in both assays, in all three cell lines (Figure 3c). Thus, KDM3A is growth promoting in Ewing Sarcoma. Taken together with the above data on KDM3A expression, these findings identify induction of KDM3A as a new mechanism contributing to the EWS/Fli1-driven oncogenic program in Ewing Sarcoma.
KDM3A is an H3K9me1/2 histone demethylase.27 As H3K9 methylation in promoter regions is inhibitory to transcription,28,29 KDM3A demethylase activity tends to enhance gene expression. To begin to understand mechanisms by which KDM3A promotes the oncogenic phenotype in Ewing Sarcoma, we first verified that KDM3A depletion results in increased levels of H3K9 methylation (Figure 4a). Next, we examined the effects of KDM3A depletion on the expression levels of a panel of candidate targets: Cyclin D1 (CCND1), IGF-1R and Ets1. Cyclin D1, a known upregulated oncogene in Ewing Sarcoma,30 has been identified as a KDM3A-induced gene in bladder cancer.31 A recent study of KDM3A-regulated genes in bronchial epithelial cells32 identified IGF-1R, a key pro-oncogenic factor in Ewing Sarcoma, as a candidate KDM3A target. Last, Ets1, an established nuclear effector of receptor tyrosine kinase signaling and a known positive regulator of Cyclin D1 expression, is a KDM3A-upregulated gene in endothelial cells.33 We found robust positive regulation of Cyclin D1 by KDM3A in A673 cells, but not the other cell lines. IGF-1R was also robustly regulated in A673 cells, while data from the other cell lines were suggestive of possible weaker regulation, especially in TC71 cells. Interestingly, Ets1 was the most robustly regulated gene in all three cell lines. Taken together, these data indicate that KDM3A positively regulates the expression of multiple oncogenes in Ewing Sarcoma, and identify Ets1 as a possible candidate mediator of KDM3A pro-oncogenic effects in this cancer.
Figure 4.
KDM3A depletion results in upregulation of H3K9me2 levels, downregulation of oncogene expression and impaired tumorigenesis in Ewing Sarcoma. (a) Control and KDM3A-depleted A673, SKES1 and TC71 cells were harvested, and resuspended in lysis buffer (PBS containing 0.5% Triton X-100 and 2 mM phenylmethylsulfonyl fluoride) at 1 ×108 cells/ml and incubated for 10 min on ice. Lysates were centrifuged for 10 min at 14 000 r.p.m. and 4 °C. The supernatant was discarded, the pellet washed with ½ volume of lysis buffer and centrifuged again. The pellet was then resuspended in of the original lysis buffer volume of 0.2 N HCL, and placed in a rotator overnight at 4 °C to extract histones. 20 μg of the resulting extract protein were subjected to SDS–polyacrylamide gel electrophoresis, immunoblotting and ECL detection as in Figure 2. Primary antibodies used were: H3K9me2 (1:500, Abcam, #1220) and H3 (1:10,000, Abcam, #1791). The ratio of H3K9me2 to H3 was determined by densitometry. Quantification represents the mean and s.e.m. of two independent experiments, each performed in duplicate; the ratio in the control group is set to 1. (b) RNA transcript levels of the indicated genes in control and KDM3A-depleted Ewing Sarcoma cells, determined by qRT-PCR with U6 as internal control; data, normalized to control (set to 1), represent the mean and s.e.m. of two or more independent experiments, each performed in duplicate or triplicate. *P<0.05 relative to control, using a two-way Student’s t-test with unequal variance. (c) Tumor studies were performed according to an institutionally approved animal protocol. 1 ×106 cells, mixed 1:1 with Matrigel, were injected subcutaneously into the flank of immunocompromised (NCR Nu/Nu) mice. When the largest tumor reached 2 cm3, all animals were euthanized, and the tumors were excised and weighed. Tumor weights in each group are shown as individual data points, and mean and s.e.m.; P-value was determined using a two-way Student’s t-test with unequal variance.
Last, to demonstrate that the KDM3A pro-oncogenic effects are relevant to tumorigenesis, we examined the consequences of KDM3A depletion on tumor formation in an animal xenograft model. As shown (Figure 4c), stable depletion of KDM3A in Ewing Sarcoma A673 cells resulted in smaller tumors in a murine flank injection model. Thus, KDM3A promotes Ewing Sarcoma tumorigenesis.
Our analyses of the role of a single miR in EWS/Fli1-driven oncogenesis have thus uncovered a new tumor promoter in Ewing Sarcoma, the H3K9 histone demethylase KDM3A. To our knowledge, these studies represent the first demonstration of a role for histone demethylation in Ewing Sarcoma oncogenesis, and add to the growing list of epigenetic modifiers implicated in the pathogenesis of this cancer, including the H3K27 histone methyltransferase EZH234 and the Polycomb group gene BMI-1.35 A direct chromatin-modifying function for the EWS/Fli1 oncoprotein has also recently been suggested.36 Epigenetic modifications, as a class, are emerging as a powerful driving force in cancer.11,12 In pediatric cancers, which tend to harbor fewer mutations than their adult counterparts,37 epigenetic modifications may make a disproportionate contribution to initiation and maintenance of the oncogenic state, as suggested by recent studies in retinoblastoma,14 and pointed out by others.13 Encouragingly, targeting of proteins responsible for modulating and interpreting the cell’s epigenetic state appears to be feasible,38 and may represent a worthwhile alternative or complementary approach to ongoing efforts to target the EWS/Fli1 oncoprotein in Ewing Sarcoma.39,40
A substantial challenge in the development of effective epigenetically targeted therapies is understanding of the complex biology of the targets themselves. With respect to the H3K9 methylated state, overexpression or/and amplification of other histone demethylases with activity against H3K9 has been identified in other cancers, including the pediatric cancers neuroblastoma and medulloblastoma, as has inactivation of H3K9 methyltransferases.38 Of note, our preliminary analyses indicate that other sarcomas may express similar levels of KDM3A to Ewing Sarcoma (Supplementary Figure 3). Thus, diminution of H3K9 methylation may be generally favorable to oncogenesis, whereas its augmentation may be a desirable activity for an epigenetic therapeutic. However, the current state of knowledge of the precise roles and mechanistic consequences of H3K9 methylation, including differential properties of the mono, di and tri-methylated state, is limited,28,41 and in need of further study.
Our studies identify a new mechanism, active in at least some cell lines, for induction of two established oncogenes in Ewing Sarcoma, IGF-1R and Cyclin D1. We have previously shown that IGF-1R expression is in part regulated by miR-100 levels,6 while Cyclin D1 has also been shown to be directly induced by EWS/Fli1.42 Interestingly, Cyclin D1 expression is silenced via H3K9 methylation in differentiating cardiomyocytes,43 suggesting that modulation of the H3K9 methyl mark may be a more general mechanism of regulation of this pivotal oncogene. Importantly, identification of epigenetic mechanisms simultaneously controlling the expression of multiple oncogenes opens the door to potential new therapeutic strategies. Histone demethylases, including KDM3A, have an enzymatic activity that could be drugged with small molecule inhibitors. Further examination of the biology of KDM3A and related factors will help determine whether modulation of H3K9 methylation should be pursued as a therapeutic approach in Ewing Sarcoma, and potentially other cancers.
Supplementary Material
Acknowledgments
We thank Kathrin Bernt and Tobias Neff for advice on histone mark analysis, Steve Lessnick for retroviral packaging constructs, Heide Ford for critical reading of the manuscript, and the University of Colorado Cancer Center DNA Sequencing, Flow Cytometry, and Functional Genomics core facilities. This work was supported by the Boettcher Foundation’s Webb-Waring Biomedical Research Program, Department of Defense Discovery Award (W81XWH-12-1-0296), Alex’s Lemonade Stand Foundation for Childhood Cancer, and funds from the University of Colorado School of Medicine and Cancer Center (to PJ); and Merit Award from the US Department of Veterans’ Affairs and R01CA138528-2522717 (to RAW).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Ludwig JA. Ewing sarcoma: historical perspectives, current state-of-the-art, and opportunities for targeted therapy in the future. Curr Opin Oncol. 2008;20:412–418. doi: 10.1097/CCO.0b013e328303ba1d. [DOI] [PubMed] [Google Scholar]
- 2.Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene. 2010;29:4504–4516. doi: 10.1038/onc.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKinsey EL, Parrish JK, Irwin AE, Niemeyer BF, Kern HB, Birks DK, et al. A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene. 2011;30:4910–4920. doi: 10.1038/onc.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DN, et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene. 2011;30:2173–2180. doi: 10.1038/onc.2010.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Vito C, Riggi N, Suva ML, Janiszewska M, Horlbeck J, Baumer K, et al. Let-7a is a direct EWS-FLI-1 target implicated in Ewing’s sarcoma development. PLoS One. 2011;6:e23592. doi: 10.1371/journal.pone.0023592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, Delattre O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene. 2012;32:3915–3921. doi: 10.1038/onc.2012.403. [DOI] [PubMed] [Google Scholar]
- 10.Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S, et al. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol. 2012;226:796–805. doi: 10.1002/path.3007. [DOI] [PubMed] [Google Scholar]
- 11.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan RJ, Bernstein BE. Molecular biology. Genetic events that shape the cancer epigenome. Science. 2012;336:1513–1514. doi: 10.1126/science.1223730. [DOI] [PubMed] [Google Scholar]
- 13.Lawlor ER, Thiele CJ. Epigenetic changes in pediatric solid tumors: promising new targets. Clin Cancer Res. 2012;18:2768–2779. doi: 10.1158/1078-0432.CCR-11-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Liang S, Yu H, Zhang J, Ma D, Lu X. An inhibitory effect of miR-22 on cell migration and invasion in ovarian cancer. Gynecol Oncol. 2010;119:543–548. doi: 10.1016/j.ygyno.2010.08.034. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhang Y, Zhao J, Kong F, Chen Y. Overexpression of miR-22 reverses paclitaxel-induced chemoresistance through activation of PTEN signaling in p53-mutated colon cancer cells. Mol Cell Biochem. 2011;357:31–38. doi: 10.1007/s11010-011-0872-8. [DOI] [PubMed] [Google Scholar]
- 17.Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, et al. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 2010;24:447–463. doi: 10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel JB, Appaiah HN, Burnett RM, Bhat-Nakshatri P, Wang G, Mehta R, et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 2011;30:1290–1301. doi: 10.1038/onc.2010.510. [DOI] [PubMed] [Google Scholar]
- 19.Ting Y, Medina DJ, Strair RK, Schaar DG. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem Biophys Res Commun. 2010;394:606–611. doi: 10.1016/j.bbrc.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Xiong J, Du Q, Liang Z. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene. 2010;29:4980–4988. doi: 10.1038/onc.2010.241. [DOI] [PubMed] [Google Scholar]
- 21.Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y, et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277:1684–1694. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–1220. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582–3600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riggi N, Cironi L, Provero P, Suva ML, Kaloulis K, Garcia-Echeverria C, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 2005;65:11459–11468. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 25.Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68:2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 26.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 27.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22 :1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 30.Wai DH, Schaefer KL, Schramm A, Korsching E, Van Valen F, Ozaki T, et al. Expression analysis of pediatric solid tumor cell lines using oligonucleotide microarrays. Int J Oncol. 2002;20:441–451. [PubMed] [Google Scholar]
- 31.Cho HS, Toyokawa G, Daigo Y, Hayami S, Masuda K, Ikawa N, et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer. 2012;131:E179–E189. doi: 10.1002/ijc.26501. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Kluz T, Zhang R, Costa M. Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis. 2010;31:2136–2144. doi: 10.1093/carcin/bgq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mimura I, Nangaku M, Kanki Y, Tsutsumi S, Inoue T, Kohro T, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;32:3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci USA. 2009;106:5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douglas D, Hsu JH, Hung L, Cooper A, Abdueva D, van Doorninck J, et al. BMI-1 promotes ewing sarcoma tumorigenicity independent of CDKN2A repression. Cancer Res. 2008;68:6507–6515. doi: 10.1158/0008-5472.CAN-07-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD, et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res. 2012;22:259–270. doi: 10.1101/gr.125666.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene. 2012;31:3827–3844. doi: 10.1038/onc.2011.552. [DOI] [PubMed] [Google Scholar]
- 39.Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat Med. 2009;15:750–756. doi: 10.1038/nm.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C, et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst. 2011;103:962–978. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuma M, Okita H, Hata J, Umezawa A. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene. 2003;22:1–9. doi: 10.1038/sj.onc.1206055. [DOI] [PubMed] [Google Scholar]
- 43.Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, et al. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- 44.Cittelly DM, Dimitrova I, Howe EN, Cochrane DR, Jean A, Spoelstra NS, et al. Restoration of miR-200c to ovarian cancer reduces tumor burden and increases sensitivity to paclitaxel. Mol Cancer Ther. 2012;11:2556–2565. doi: 10.1158/1535-7163.MCT-12-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.