Abstract
Human papilloma-virus (HPV) infection is the major cause of cervical cancer. HPV 18 is the most prevalence high-risk HPV after type 16 that accounts for the largest number of cervical cancer cases worldwide. Currently, although prophylactic vaccines have been developed, there is still an urgent need to develop therapeutic HPV vaccines for targeting tumors post infection. In this study, we utilize a novel multi-phase strategy for HPV 18 antigen development with the goal of increasing anti-HPV18 cellular immunity. Our data show that this construct can induce strong cellular immune responses against HPV 18 E6 and E7 antigens in a murine model. Moreover, when applied to Rhesus monkeys, this construct is also able to elicit cellular immunity. These data suggest such DNA immunogens are candidates for further study in the eventual context of immunotherapy for HPV-associated cancers.
Keywords: HPV-18, E6 and E7 protein, DNA vaccine, consensus
1. Introduction
Cervical cancer is the second most common cancer among women worldwide, but the most common cancer in developing countries (1). Over 40 distinct HPV types have been identified to infect the genital tract, and 15 of them have been classified as high-risk in the development of cervical cancer (2). Among these high-risk HPV types, HPV16 is the predominant type in squamous cell carcinoma of the cervix, and HPV 18 is the second most common type associated with squamous cell carcinoma of the cervix, with prevalence ranging from 12.6% in Central/South America to 25.7% in South Asia (3). In addition, HPV 18 has been implicated in rapidly developing and potentially more aggressive cervical carcinomas (4, 5),
The development of effective therapeutic vaccines against HPV represents an opportunity to impact the pathogenesis of cervical cancer. Recently a FDA-approved HPV prophylactic vaccine has shown significant clinical efficacy in prevention of HPV infection, although it exhibits no efficacy in treatment of infected patients. Thus, development of therapeutic HPV vaccines through the induction of a strong cell-mediated response is still of great importance. The early HPV proteins, E6 and E7, represent tumor-specific antigens in cervical carcinoma and premalignant HPV-transformed cells. Integration of E6 and E7 genes into the host cell genome may lead to constitutive over expression of E6 and E7 proteins, mediating transformation of the cells to a malignant phenotype (6). These genes are also required for the maintenance of transformed phenotype. As a consequence, E6 and E7 are ideal immunotherapeutic targets to induce cellular immune responses against HPV-transformed cells (6, 7). Significant efforts have been made to develop HPV 16 therapeutic vaccines that target either E6 or E7 proteins (8–12). However, to date, there have been limited data on HPV 18 therapeutic vaccines. Since E6 and E7 proteins of different types of HPV are very variable and current HPV vaccines confer only type-specific immunity, novel immunotherapies for cervical cancer must consider that there is an urgent need to develop HPV 18 therapeutic vaccines.
DNA vaccines have conceptional advantages over traditional vaccines such as live attenuated virus and recombinant protein-based vaccines in the context of immune therapy. DNA vaccines appear to be very well tolerated in humans. Preclinical safety studies indicate that there was little evidence of plasmid integration (13, 14). DNA vaccines can also be used for repeat administration as the efficacy of plasmid vectors are not influenced by pre-existing neutralizing antibodies (15). The ability to target multiple antigenic components may be a particularly important characteristic of DNA vaccines since multi-component DNA vaccines can be engineered to include specific immunogens that can optimize and amplify desirable immunologic responses. Furthermore, DNA vaccines appear to be very stable and simple to produce. However, initial studies reported that DNA vaccines exhibited low potency in large animals and humans. Accordingly, improving their immuno-potency is critical. Recently, several strategies aimed at improving the magnitude of the cellular immune responses induced by DNA vaccines, such as codon optimization (16, 17), RNA optimization (18, 19) and the addition of immunoglobin leader sequences that have weak RNA secondary structure (20), have been studied and applied successfully to HIV and Influenza vaccine development (21, 22).
Improvements in delivery are also highly important to improve the magnitude of immune responses induced by DNA vaccines. Among the most striking advances for plasmid delivery in vivo the most potent has been that of electroporation (EP). Studies have showed that EP has been a feasible method for DNA vaccine delivery to enhance both cellular and humoral responses (23, 24).
On the other hand, recent research from our lab and others have indicated that utilization of “centralized” immunogens such as consensus immunogens or ancestral immunogens could increase the breadth of cellular immune responses than native DNA immunogens (21, 22, 25, 26). Such consensus immungens in the context of HPV may appear more foreign and thus facilitate immune stimulation, which is an important issue with a slow growth that may induce peripheral tolerance. Hence, we present here a novel engineered HPV 18 consensus E6 and E7 fusion DNA immunogenic designed by utilizing a multi-phase strategy. We then tested the expression in vitro. Finally the immunogenic of this construct was examined in mice by quantitative T cell assays. Lastly, we observed that this construct was able to elicit important cellular immunity in Rhesus Macaques.
2. Materials and methods
2.1. Construction of HPV-18 E6 and E7 consensus sequences
To generate HPV type 18 consensus E6 and E7 sequences, twelve HPV-18 E6 and E7 gene sequences collected from different countries were selected from Embank to avoid sampling bias. The HPV type 18 E6 and E7 consensus nucleotide sequences were obtained after performing multiple alignments.
2.2. Modifications of HPV-18 consensus E6 and E7 sequences
Several modifications were performed after obtaining HPV-18 consensus E6 and E7 sequences (Fig. 1). Condon optimization and RNA optimization was performed by using GeneOptimizer™ (GENEART, Germany).
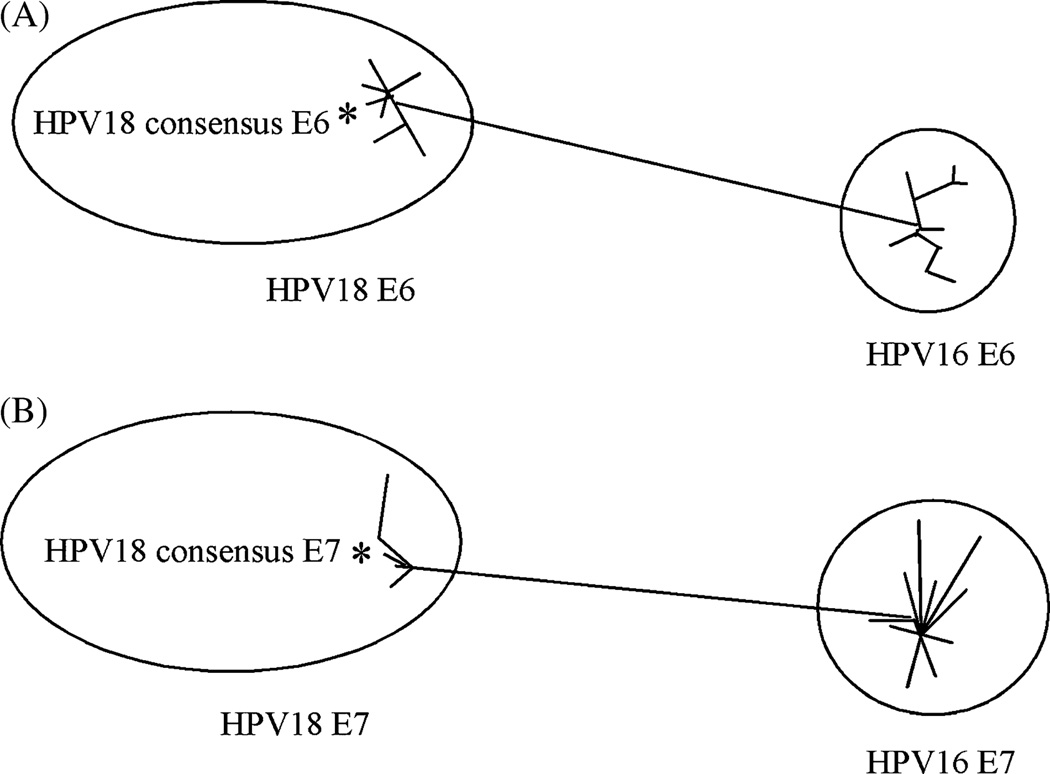
Fig. 1.
Phylogenetic analysis based on a neighbor-joining (Kimura two-parameter distances) evaluation of E6 (A) and E7 (B) alignments. The star represents the consensus sequence, relative to its compoment viruses.
2.3. HPV-18 E6 and E7 DNA immunogens
The fusion gene encoding modified HPV type 18 consensus E6/E7 fusion protein (18ConE6E7) was synthesized and sequence verified by GENEART. The synthesized 18ConE6E7 was digested with EcoRI and NotI, cloned into the expression vector pVAX (Invitrogen) under the control of the cytomegalovirus immediate-early promoter and this construct was named as p18ConE6E7.
2.4. In vitro expression
Expression of p18ConE6E7 was detected by utilizing TNT® Quick Coupled Transcription/Translation System containing 35S-methionine (Promega. Madison, WI). The synthesized gene product was immunoprecipitated using an HPV-18 E6-specific monoclonal antibody (Invitrogen). The immunoprecipitated protein was electrophoresed on the SDS-PAGE gel (12%), and subsequently fixed and dried. The synthesized protein with incorporation of radioactive 35S was detected by autoradiography.
2.5. Mice Studies
2.5.1. Mice and Immunization/Electroporation
Female 6–8-week-old C57BL/6 mice were purchased from the Jackson laboratory. Their care was in accordance with the guidelines of the National Institutes of Health and the University of Pennsylvania Institutional Care and Use Committee (IACUC).
For DNA immunizations, the mice were separated into two groups of 5 mice each and immunized by electroporation with pVAX (control group) and p18ConE6E7, respectively. Each mouse was immunized with three times, each of 10 ug of DNA at biweekly intervals. Briefly, Square-wave pulses were used and delivered with the constant-current EKD that was designed and tested in our laboratory. A three electrode array (3–EA) was used and it consists of three 26-gauge solid stainless steel electrodes in an isosceles triangle formation, with the two long sides 0.5mm in length and short side 0.3 mm in length, held together with a nonconductive plastic. The sequence of events for plasmid administration/EP was as follows: place a disposable electrode array in the receptacle of the handle, press initiation button on handle and enter animal experimental group number, inject 10ug DNA plasmid using insulin syringe, immediately place the array into area surrounding the injection site, press initiation button on handle, and after 4 second countdown, pulses will be delivered. Five seconds following electroporation, the array is gently removed from muscle. All electrodes were completely inserted into the muscle during all treatments.
2.5.2.Splenocyte purification
Mice were sacrificed one week after the third immunization and the spleens were removed and pooled from each experimental group. The pooled spleens were placed in a sterile plastic bag and crushed using Stomacher. The cells were then put through a 40 µm cell strainer, collected and resuspended in ACK lysing buffer (Biosource) to remove erythrocytes. After lysis, the splenocytes were resuspended in RPMI 1640 medium with 10% FBS. Cells were counted using a hemocytometer.
2.5.3. IFN-γ ELISpot assay
Mouse IFN-γ ELISpot assay was performed as described previously (21). A set of peptides each containing 15 amino acid residues overlapping by 8 amino acids representing the entire consensus E6/E7 fusion protein sequence of HPV-18 was synthesized from Invitrogen. This set of peptides was pooled at a concentration of 2 ug/ml/peptide into 2 pools as antigens for specific stimulation of the IFN-γ release. The average number of spot forming cells (SFC) was adjusted to 1 × 106 splenocytes for data display.
2.5.3. Epitope mapping in mice
In order to map the reactive epitopes, individual synthesized HPV-18 E6 and E7 peptides were used as stimulator pools to perform IFN-γ ELISpot assay as described above.
2.6. Rhesus monkey studies
2.6.1. Study design and Immunization
In this study, a total of 10 rhesus macaques (Macaca mulatta) of Indian origin were used and divided into two groups (5 monkeys/per group): negative control group and p18ConE6E7-immunized group. Macaques were maintained in accordance with the Guide for the Care and Use of Laboratory Animals. Plasmid preparations were prepared at concentrations higher than 5 mg/mL, diluted in sterile water and formulated with 1% (weight/weight) poly-L-glutamate sodium salt (MW=10.5 kDa average) (Sigma, St. Louis, MO), further HPLC purified at VGX Pharmaceuticals, Immune Therapeutics Division, The Woodlands, TX.
Macaques were vaccinated intramuscularly in the semi-membranosous muscle in combination with EP using CELLECTRA™ adaptive constant current EP device and electrode arrays. Immediately following injection, 3 pulses at 0.5 Amps constant current, 52 msec pulse length with 1 sec between pulses, were applied. Each monkey was immunized with 1mg DNA and the injection volume was 0.75mL. The first two immunizations were performed three weeks apart, followed by a third immunization at week 8, and animals were bled two weeks after each immunization to measure immune responses.
2.6.2 Sample collection and PBMC Isolation
Rhesus macaques were bled every 3 weeks for the duration of the study. Animals were anesthetized with ketamine (10 mg/kg) mixed with acepromazine (0.1 mg/kg). Blood were collected in EDTA tubes. PBMCs were isolated from whole blood by standard Ficoll-Hypaque density gradient centrifugation, resuspended in complete culture medium (RPMI 1640 with 2 mM/L L-glutamine supplemented with 10% heat-inactivated FBS, 100 IU/mL penicillin, 100 µg/mL streptomycin, and 55 µM/L β-mercaptoethanol).
2.6.3. IFN-γ ELISpot assay
Monkey IFN-γ ELISpot were performed as previously described (27). Anti-monkey IFN-γ capture and detection antibodies (MabTech, Sweden) were used. Antigen-specific responses were determined by subtracting the number of spots in the negative control wells from the wells containing peptides.
2.7. Statistical Analysis
Student paired t-test was used for comparison of the cellular immune responses. In this study, p<0.05 has been considered statistically significant.
3. Results
3.1. Construction of a novel HPV type 18 E6 and E7 consensus-based fusion immunogen
The consensus sequences of HPV 18 E6 and E7 proteins were generated from 12 sequences retrieved from GenBank. The multiple alignment procedure applied in the phylogenetic study included the application of Clustal X (version 1.8). Fig 1A and B depicted phylogenic analyses of HPV 18 consensus E6 and E7 proteins. There were some differences among the HPV strains belonging to the same type in their E6 and E7 proteins. However, the genetic distances could go up to 47% in the E6 protein and 59% in the E7 protein between HPV16 and 18. The phylogenic analyses indicated that there might be advantages in using a type-specific E6/E7 consensus DNA vaccine.
As summarized in Fig. 2, several modifications were conducted after generating the consensus E6/E7 fusion sequence. A highly efficient leader sequence was fused in frame upstream of the start codon to facilitate the expression. The codon usage of this fusion gene was adapted to the codon bias of Homo Sapiens genes for better expression in humans. In order to further increase the expression level, RNA optimization (18, 19) was also performed: regions of very high (>80%) or very low (<30%) GC content and the cis-acting sequence motifs such as internal TATA boxes, chi-sites and ribosomal entry sites were avoided. Considering the safety issue, the domains of the E6 protein required for p53 binding/degradation were mutated or deleted (28, 29). The binding site of the E7 protein to the cellular retinoblastoma (Rb) protein was mutated (30). An endoproteolytic cleavage site was introduced between E6 and E7 protein for proper protein folding and better CTL processing. The synthetic engineered 18ConE6E7 gene was 860 bp in length. The 18ConE6E7 gene was subcloned into the pVAX expression vector at the EcoRI and NotI sites for further study.
Fig. 2.
The modifications and amino acid sequence of 18ConE6E7. The IgE leader sequence and endoproteolytic cleavage site are underlined. The residues in boxed regions are deleted. The * denotes mutation sites.
3.2. In Vitro Expression of 18ConE6E7
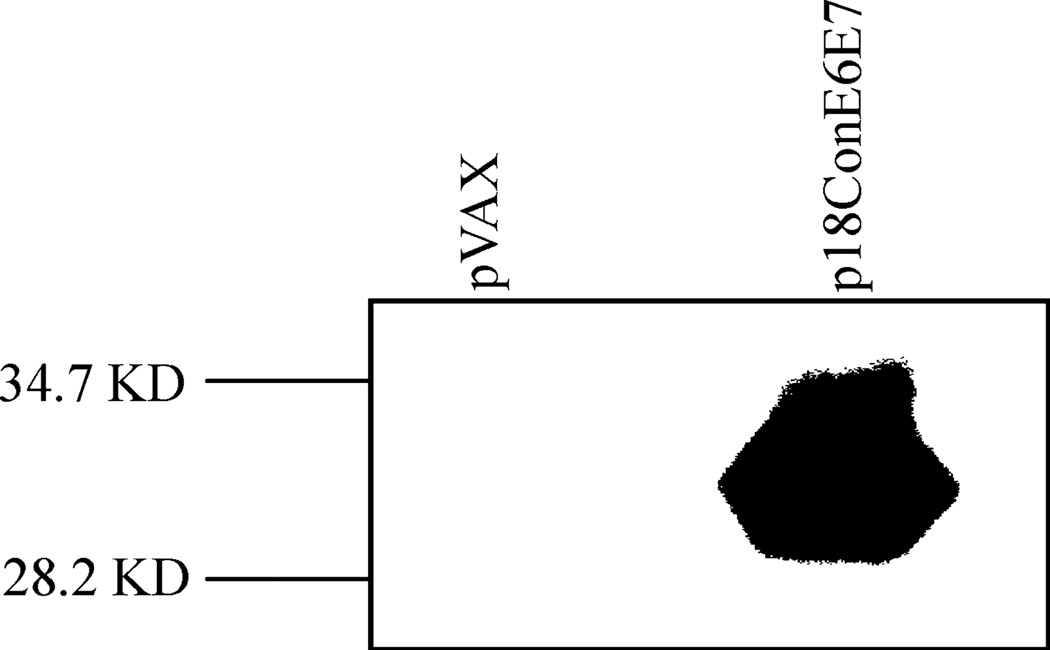
The expression of p18ConE6E7 was confirmed by T7 coupled transcription and translation reaction. After immunoprecipitation with the HPV 18 E6-specific monoclonal antibody, the expression of E6 and E7 genes was assessed by SDS-PAGE gel (15%). The E6/E7 fusion protein migrated corresponding to a molecular weight of approximately 31 kDa when immunoprecipitated with the E6-specific antibody. For the negative control, no protein band could be seen in the pVAX group (Fig. 3).
Fig. 3.
In vitro translation to detect the expression of HPV 18 E6 and E7 fusion gene. The gene product was immunoprecipitated using a HPV 18 E6-specific monoclonal antibody, ran on SDS-PAGE gel, and detected by autoradiography.
3.3. Vaccination with p18ConE6E7 generates strong E6- and E7-specific T-cell immune responses in C57BL/6 mice
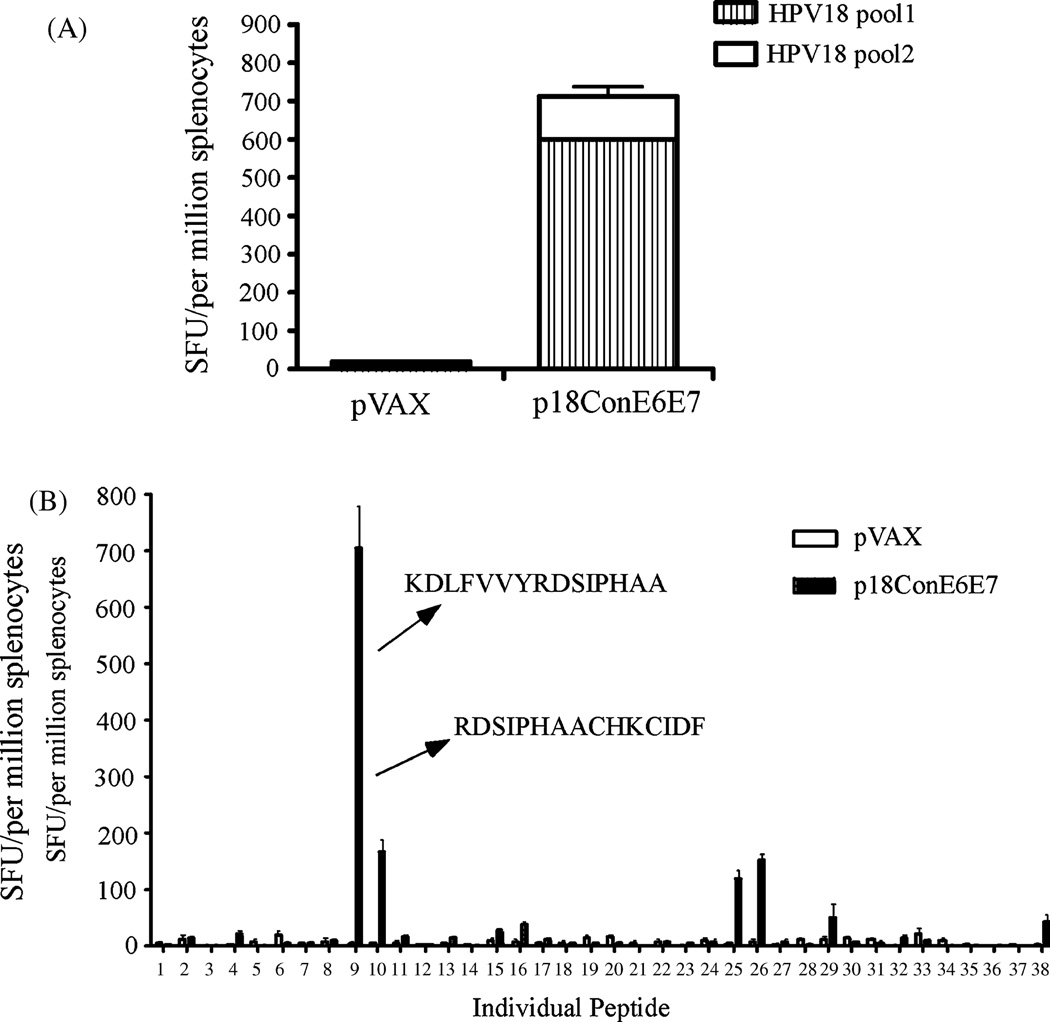
After confirmation of the expression of p18ConE6E7, we determined whether this construct could induce CTL responses. C57BL/6 mice were immunized with p18ConE6E7, and ELISpot analysis was performed to determine the IFN-γ response to stimulation with two pools of peptides from HPV18 E6 and E7 consensus fusion protein. As shown in Fig. 4A, p18ConE6E7 was able to induce strong cellular immune responses in C57BL/6 mice after three immunizations.
Fig. 4.
p18ConE6E7 induced strong cell-mediated immune responses in C57BL/6 mice. Frequencies of HPV-18 consensus E6 and E7-specific IFN-γ spot forming units (SFU) per million total splenocytes after DNA vaccination with p18ConE6E7 were determined by ELISpot assay. (A) The splenocytes were isolated from individual immunized mice (five mice per group) and stimulated in vitro with overlapping consensus HPV-18 E6/E7 peptides pools. (B) Characterization of HPV-18 E6- and E7-specific dominant epitopes. The splenocytes collected from p18ConE6E7 vaccinated C57BL/6 mice were cultured with individual HPV-18 E6 and E7 peptide for 24 hours. IFN-γ secreting cells were determined by ELISpot assay as described above. pVAX immunized mice were included as a negative control.
Next, we mapped the dominant epitopes of the cellular immune responses induced by p18ConE6E7. Accordingly, ELISpot assay was performed against individual peptides spanning HPV 18 consensus E6/E7 fusion protein (Fig. 4B). The results indicated that peptides KDLFVVYRDSIPHAA (peptide 9) and RDSIPHAACHKCIDF (peptide 10) were recognized as dominant and subdominant epitopes for E6 immunogen, respectively. The dominant peptide overlaps with an epitope (FAFKDLFVV) predicted to have high binding activity for HLA-A2 molecules and the subdominant epitope overlaps with a well-characterized E6 dominant epitope KCIDFYSRI that was shared between HPV18 and HPV45 (31), supporting the normal effective processing of this artificial fusion antigen during vaccination.
3.4. Vaccination with p18ConE6E7 generates strong E6- and E7-specific T-cell immune responses in Rhesus Monkeys
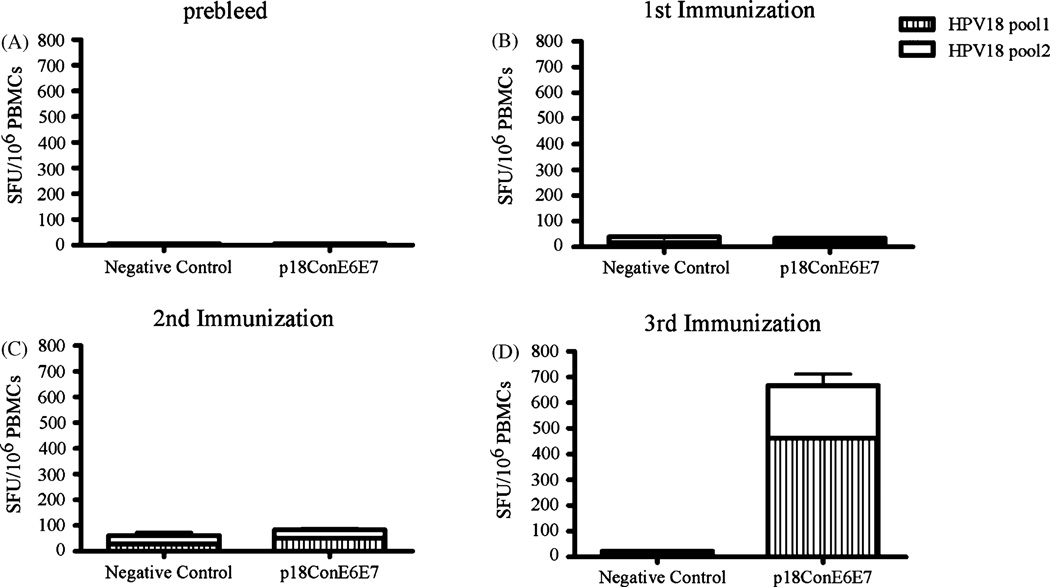
Despite the success of cancer DNA vaccines in small animal models, little has been done to test the ability of cancer DNA vaccines to elicit anti-tumor cellular immune responses in large animals. We thought the data obtained from non-human primates would be important. Therefore, we moved forward to determine the immunogenicity of p18ConE6E7 in Rhesus Monkeys after we confirmed that p18ConE6E7 induced strong cellular immunity in mice. The monkeys were immunized IM/EP three times at weeks 0, 3, 8. Blood was collected two weeks after each immunization and IFN-γ ELISpot assays were performed. The prebleed blood samples were used to show the background immune response of each monkey (Fig. 5A).
Fig. 5.
p18ConE6E7 induced strong cell-mediated immune responses in Rhesus monkeys. Frequencies of HPV-18 consensus E6 and E7-specific IFN-γ spot forming units (SFU) per million total PBMC after each DNA immunization with p18ConE6E7 were determined by ELISpot assay.
There were no detectable immune responses in the p18ConE6E7-immunized monkeys (32 ± 12 SFU/106 PBMCs) (Fig. 5B) after the first immunization and only very weak responses (86 ± 21 SFU/106 PBMCs) were able to detect after the second immunization (Fig. 5C). However, the average IFN-γ responses for the monkeys immunized with p18ConE6E7 increased significantly with the third immunization. The average number of IFN-γ producing cells in these animals went up to 669 ± 369 SFU/106 PBMCs (Fig. 5D). Hence, immunization of Rhesus macaques with p18ConE6E7 could elicit clear HPV18 E6- and E7-specific cellular responses.
DISCUSSION
Cellular immune responses have been reported to be effective for tumor cell destruction and control of HPV infection, as HPV-associated cervical lesions are more prevalent in immunosuppressed patients (30, 32). Studies have suggested that cytotoxic T lymphocytes may mediate tumor regression in animal models and protect against persistant viruses (33–35). The E6 and E7 proteins are intracellular and represent genuine tumor-specific antigens that could act as targets for destruction of tumor cells without damage of healthy host cells, thus providing a clear opportunity for cervical cancer immunotherapy. Here, we designed an engineered HPV 18 E6 and E7 vaccine for use in the treatment of human cervical cancer. We showed that this vaccine could elicit CTLs in a murine model. Importantly, although there is no challenge animal model available for HPV 18, the fact that this vaccine could also elicit CTLs in a non-human primate model is promising.
DNA vaccines have emerged as an attractive approach for antigen-specific immunotherapy. This technology has significant potential, compared to traditional protein vaccines, in terms of priming CTL responses and generating memory CD8 T-cell responses (36, 37). Other major theoretical advantages of DNA vaccination are that they can be used for repeat administration as the efficacy of plasmid vectors are not influenced by pre-existing neutralizing antibodies, which is very important in cancer therapy. In the Rhesus monkey study, the immune responses increased significantly when the third immunization was conducted, suggesting that enhanced CTLs may be obtained by multiple vaccinations, further supporting this hypothesis.
The phylogenetic analysis of E6 gene sequences conducted by Chan et al indicated that the ongenic HPV virus types largely associated with two classes of HPV, class A and C. Class A viruses include HPV types 16, 31, 33, 35, 52, 58 and 67, whereas class C comprise HPV types 18, 39, 45, 59 and 68 (38). The homology of E6 and E7 proteins within each class suggest the possibility that a broad coverage may be achieved by using a HPV 16 or HPV18 vaccine. In fact, a shared T cell epitope (KCIDFYSRI) that bound to HLA-A*0201 but not Db or Kb molecules between HPV 18 and 45 was identified (31). In order to maximize potential immune cross-reactivity, especially at the T-cell level, we generated a HPV 18 consensus E6 and E7 DNA immunogen. Based on previous research in our lab (21), we hypothesized that this consensus-based vaccine could enhance cross-reactivity. We found that the subdominant peptide indeed overlapped with the cross-reactive human T cell epitope shared between 18 and 45. While beyond the scope of the present study, it will be important to perform additional studies to determine the ability of p18ConE6E7 to induce cross-reactive responses against other phylogenetically related HPV types in class C.
Taken together, we have studied a novel HPV18 DNA vaccine encoding an E6/E7 fusion consensus protein. We observe that vaccination of p18ConE6E7 can generate clear and important cellular immunity in both mice and rhesus monkeys. Further study of this vaccine strategy is in progress.
ACKNOWLEDGMENT
This work was supported by NIH grants awarded to Dr. David B. Weiner and NCI grant K08 CA097218 awarded to Dr. Duane Sewell.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Franceschi S, Rajkumar T, Vaccarella S, et al. Human papillomavirus and risk factors for cervical cancer in Chennai, India: a case-control study. Int J Cancer. 2003;107:127–133. doi: 10.1002/ijc.11350. [DOI] [PubMed] [Google Scholar]
- 2.de Villiers EM. Taxonomic classification of papillomaviruses. Papillomavirus Rep. 2001;12:57–63. [Google Scholar]
- 3.Munoz N, Bosch FX, Castellsague X, et al. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111:278–285. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 4.Bosch FX, Manos MM, Munoz N, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 5.Hildesheim A, Hadjimichael O, Schwartz PE, et al. Rish factors for rapid-onset cervical cancer. Am J Obstet Gynecol. 1999;180:517–577. doi: 10.1016/s0002-9378(99)70256-5. [DOI] [PubMed] [Google Scholar]
- 6.Munger K, Howley PM. Human papillomavirus immortalization and transformation functions. Virus Res. 2002;89:213–228. doi: 10.1016/s0168-1702(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 7.Eiben GL, Velders MP, Kast WM. The cell-mediated immune response to human papillomavirus-induced cervical cancer: implications for immunotherapy. Adv Cancer Res. 2002;86:113–148. doi: 10.1016/s0065-230x(02)86004-3. [DOI] [PubMed] [Google Scholar]
- 8.Peng S, Trimble C, He L, et al. Characterization of HLA-A2-restricted HPV-16 E7-specific CD8(+) T-cell immune responses induced by DNA vaccines in HLA-A2 transgenic mice. Gene Ther. 2006;13:67–77. doi: 10.1038/sj.gt.3302607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CT, Tsai YC, He L, et al. A DNA vaccine encoding a codon-optimized human papillomavirus type 16 E6 gene enhances CTL response and anti-tumor activity. J Biomed Sci. 2006;13:481–488. doi: 10.1007/s11373-006-9086-6. [DOI] [PubMed] [Google Scholar]
- 10.Wu TC, Guarnieri FG, Staveley-O'Carroll KF, et al. Engineering an intracellular pathway for MHC class II presentation of HPV-16 E7. Proc Natl Acad Sci U S A. 1995;92:11671–11675. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji H, Wang TL, Chen CH, et al. Targeting human papillomavirus type 16 E7 to the endosomal/lysosomal compartment enhances the antitumor immunity of DNA vaccines against murine human papillomavirus type 16 E7-expressing tumors. Hum Gene Ther. 1999;10:2727–2740. doi: 10.1089/10430349950016474. [DOI] [PubMed] [Google Scholar]
- 12.Chen CH, Wang TL, Hung CF, Pardoll DM, Wu TC. Boosting with recombinant vaccinia increases HPV-16 E7-specific T cell precursor frequencies of HPV-16 E7-expressing DNA vaccines. Vaccine. 2000;18:2015–2022. doi: 10.1016/s0264-410x(99)00528-9. [DOI] [PubMed] [Google Scholar]
- 13.Martin T, Parker SE, Hedstrom R, et al. Plasmid DNA malaria vaccine: the potential for genomic integration after intramuscular injection. Hum. Gene Ther. 1999;10:759–768. doi: 10.1089/10430349950018517. [DOI] [PubMed] [Google Scholar]
- 14.Nichols WW, Ledwith BJ, Manam SV, Triolo PJ. Potential DNA vaccine integration into host cell genome. Ann. N. Y. Acad. Sci. 1995;772:30–39. doi: 10.1111/j.1749-6632.1995.tb44729.x. [DOI] [PubMed] [Google Scholar]
- 15.Chattergoon M, Boyer J, Weiner DB. Genetic immunization: A new era in vaccines and immune therapies. FASEB J. 1997;11:753–763. doi: 10.1096/fasebj.11.10.9271360. [DOI] [PubMed] [Google Scholar]
- 16.Andre S, Seed B, Eberle J, et al. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deml L, Bojak A, Steck S, et al. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 gag protein. J. Virol. 2001;75:10991–11001. doi: 10.1128/JVI.75.22.10991-11001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muthumani K, Zhang D, Dayes NS, et al. Novel engineered HIV-1 East African Clade-A gp160 plasmid construct induces strong humoral and cell-mediated immune responses in vivo. Virology. 2003;314:134–146. doi: 10.1016/s0042-6822(03)00459-8. [DOI] [PubMed] [Google Scholar]
- 19.Schneider R, Campbell M, Nasioulas G, Felber BK, Pavlakis GN. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JS, Kim JJ, Hwang DS, et al. Induction of potent Th1-Type immune responses from a novel DNA vaccine for West Nile Virus New York Isolate (WNV-NY1999) J. Infect Diseases. 2001;184:809–816. doi: 10.1086/323395. [DOI] [PubMed] [Google Scholar]
- 21.Yan J, Yoon H, Kumar K, et al. Enhanced diversity and magnitude of cellular immune responses elicited by a novel engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Molecular Therapy. 2007:411–421. doi: 10.1038/sj.mt.6300036. [DOI] [PubMed] [Google Scholar]
- 22.Laddy DJ, Yan J, Corbitt N, et al. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25:2984–2989. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirao LA, Wu L, Khan AS, et al. Intradermal/subcutaneous immunization by electroporation improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine. 2008;26:440–448. doi: 10.1016/j.vaccine.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 24.Luckay A, Sidhu M, Kjeken R, et al. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in Rhesus macaques. J. Virol. 2007;81:5257–5269. doi: 10.1128/JVI.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolland M, Jensen MA, Nickle DC, et al. Reconstruction and function of ancestral central-of-tree (COT) HIV-1 proteins. J. Virol. 2007 doi: 10.1128/JVI.02683-06. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Weaver EA, Lu Z, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J. Virol. 2005;79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyer J, Robinson T, Kutzler M, et al. SIV DNA vaccine co-administered with IL-12 expression plasmid enhances CD8 SIV cellular immune responses in cynomolgus macaques. J. Med. Pramatol. 2005;34:262–270. doi: 10.1111/j.1600-0684.2005.00124.x. [DOI] [PubMed] [Google Scholar]
- 28.Pim D, Storey A, Thomas M, Massimi P, Banks L. Mutational analysis of HPV-18 E6 identifies domains required for p53 degradation in vitro, abolition of p53 transactivation in vivo and immortalisation of primary BMK cells. Oncogene. 1994;9:1869–1876. [PubMed] [Google Scholar]
- 29.Crook T, Tidy JA, Vousden K. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 30.Boursnell ME, Rutherford E, Hickling JK, et al. Construction and characterisation of a recombinant vaccinia virus expressing human papillomavirus proteins for immunotherapy of cervical cancer. Vaccine. 1996;14:1485–1494. doi: 10.1016/S0264-410X(96)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy C, Youde SJ, Man S. Definition of an HPV18/45 cross-reactive human T-cell epitope after DNA immunisation of HLA-A2/KB transgenic mice. Int J Cancer. 2006;118:2514–2521. doi: 10.1002/ijc.21643. [DOI] [PubMed] [Google Scholar]
- 32.Petry KU, Scheffel D, Bode U, et al. Cellular immunodeficiency enhances the progression of human papillomavirus-associated cervical lesions. Int J Cancer. 1994;57:836–840. doi: 10.1002/ijc.2910570612. [DOI] [PubMed] [Google Scholar]
- 33.Lamikanra A, Pan ZK, Isaacs SN, Wu TC, Paterson Y. Regression of established human papillomavirus type 16 (HPV-16) immortalized tumors in vivo by vaccinia viruses expressing different forms of HPV-16 E7 correlates with enhanced CD8(+) T-cell responses that home to the tumor site. J Virol. 2001;75:9654–9664. doi: 10.1128/JVI.75.20.9654-9664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng WF, Hung CF, Pai SI, et al. Repeated DNA vaccinations elicited qualitatively different cytotoxic T lymphocytes and improved protective antitumor effects. J Biomed Sci. 2002;9:675–687. doi: 10.1159/000067285. [DOI] [PubMed] [Google Scholar]
- 35.Mayordomo JI, Zorina T, Storkus WJ, et al. Bone marrow derived dendritic cells pulsed with synthetic tumor peptides elicit protective and therapeutic antitumor immunity. Nat Med. 1995;1:1297–1302. doi: 10.1038/nm1295-1297. [DOI] [PubMed] [Google Scholar]
- 36.Van Herck K, Van Damme P, Lievens M, Stoffel M. Hepatitis A vaccine: indirect evidence of immune memory 12 years after the primary course. J Med Virol. 2004;72:194–196. doi: 10.1002/jmv.10574. [DOI] [PubMed] [Google Scholar]
- 37.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization*. Annu Rev Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 38.Chan SY, Delius H, Halpern AL, Bernard HU. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]