Summary
Circadian clocks are oscillatory systems that schedule daily rhythms of organismal behavior. The ability of the clock to reset its phase in response to external signals is critical for proper synchronization with the environment. In the model clock from cyanobacteria, the KaiABC proteins that comprise the core oscillator [1, 2] are directly sensitive to metabolites. Reduced ATP/ADP ratio and the oxidized quinones cause clock phase shifts in vitro [3, 4]. But it is unclear what determine the metabolic response of the cell to darkness and thus the magnitude of clock resetting. We show that the cyanobacterial circadian clock generates a rhythm in metabolism that causes cells to accumulate glycogen in anticipation of nightfall. Mutation of the histidine kinase CikA creates an insensitive clock input phenotype by misregulating clock output genome-wide, leading to over-accumulation of glycogen and subsequently high ATP in the dark. Conversely, we show that disrupting glycogen metabolism results in low ATP in the dark and makes the clock hypersensitive to dark pulses. The observed changes in cellular energy are sufficient to recapitulate phase shifting phenotypes in an in vitro model of the clock. Our results show that clock input phenotypes can arise from metabolic dysregulation and illustrate a framework for circadian biology where clock outputs feed back through metabolism to control input mechanisms.
Results and Discussion
Mutants with Defective Clock Input Maintain High Energy Charge in the Dark
Multiple studies have searched for mutants with a defective clock input phenotype where the circadian clock ceases to show large phase shifts in response to a dark pulse [5–7]. A major result of these efforts was the identification of the histidine kinase cikA (circadian input kinase), a gene required for normal clock input sensitivity. However, it is currently unclear how CikA is mechanistically related to the energy and redox metabolite-based resetting mechanisms studied with the purified Kai proteins [3, 4]. One possibility is that CikA is part of an additional signal transduction mechanism that can override the direct effect of metabolites on the oscillator. An alternative possibility is that the effect of mutations on clock input is indirect: by altering the state of the cell, they could act to blunt the metabolic effect of a dark pulse and thus the signals to the clock.
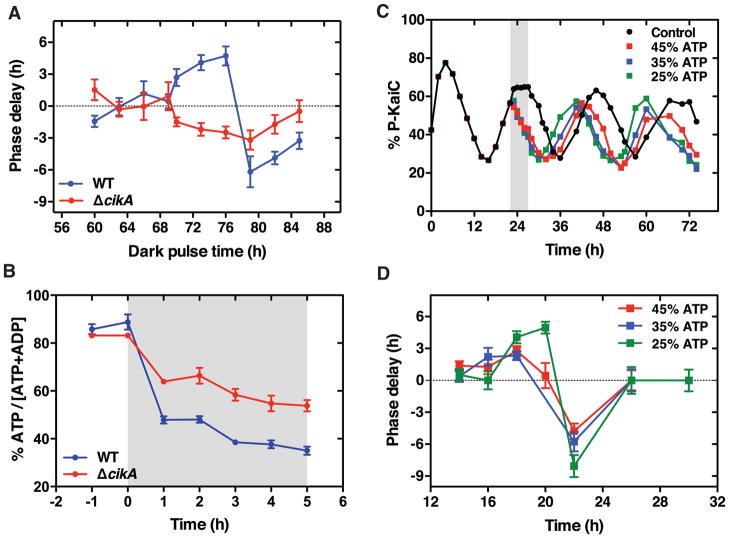
To study this latter possibility, we first measured phase resetting strength in ΔcikA and wildtype cells, confirming the resetting defect in ΔcikA (Figure 1A, Figures S1 and S2A–S2B). We then measured the ATP/ADP energy charge in wildtype and ΔcikA cells during the 5-hour dark pulses used to reset the clock (Figure 1B). Indeed, we found that while each strain had similar energy charge in the light, ΔcikA maintained a consistently higher ATP/ADP ratio in the dark (~55% vs ~40% in wild type) (Figure 1B, Figure S2C). Thus, we conclude that ΔcikA cells have an altered metabolic state that makes the impact of a light-dark transition less severe.
Figure 1. Weak phase resetting in ΔcikA cells can be explained by high energy charge in the dark.
(A) Phase delay of the bioluminescence rhythm (PkaiBC::luxAB) in wild-type (WT) and ΔcikA cells in response to 5-hour dark pulses at the indicated time. Cells were entrained by one light-dark cycle (12 h:12 h) and then released into constant light (LL). Phase delays relative to an untreated control were estimated by fitting the time series to a sinusoid. Error bars represent standard deviations (n = 12).
(B) Drop in the ATP/(ATP + ADP) energy charge during a 5-hour dark pulse (begins at shaded region) in WT (blue symbols) and ΔcikA (red symbols) cells. Error bars represent standard errors (n = 4).
(C) Phase shifts induced in the KaiC phosphorylation rhythm in purified KaiABC reactions. ATP/(ATP + ADP) was lowered from 80% (control) to various levels by addition of ADP during a 5-hour pulse (shaded bar), then reactions were returned to the initial buffer via desalting column.
(D) In vitro phase response curves. Reactions were transferred from 80% ATP/(ATP + ADP) to various levels for 5 hours, as in (C). Phase delays relative to an untreated control were determined by fitting the time series to a sinusoid. Error bars show the uncertainty estimates from the fit.
High Energy Charge Weakens Phase Resetting in vitro
We then asked whether the differences in dark ATP/ADP we observed in ΔcikA relative to wildtype were sufficient to explain the weak phase shift phenotype. To address this question, we used an in vitro model of clock input where manipulating ATP and ADP concentrations cause phase shifts in the rhythm of KaiC phosphorylation [3]. We initiated reactions using an ATP/ADP ratio that mimics growth in light (80%), then used a buffer exchange technique to deliver 5-hour pulses of nucleotides at a range of ATP/ADP ratios. We found that elevated ATP/ADP conditions (as seen in ΔcikA cells in the dark) caused phase shifts that were substantially lower in magnitude than the wildtype dark-like conditions (Figures 1C–1D, Figure S2D). These smaller phase shifts in vitro are similar to the weakened phase resetting seen in vivo, and we therefore conclude that the changes in energy metabolism in the ΔcikA mutant can explain much of its clock input phenotype.
The Clock Generates Rhythms in Energy Storage Metabolism
CikA is known to interact with clock components [8], and the metabolic phenotype of ΔcikA led us to ask whether the clock normally generates rhythms in the metabolic state of the cell that help prepare cells for darkness. Dark carbon metabolism in autotrophic cyanobacteria is thought to be largely supported by catabolism of glycogen reserves accumulated during the day [9], and genes involved in glycolysis and glycogen metabolism are some of the highest amplitude oscillating transcripts in constant light [10, 11]. We therefore investigated whether the clock output regulates glycogen storage. We found that the circadian clock generates high amplitude rhythms in energy storage metabolism, forcing cells to accumulate glycogen during the subjective day, and break it down during the subjective night, even when the lights are on (Figure 2A, Figure S3A). In cells without a functional circadian clock (ΔkaiBC) the rhythmicity in glycogen oscillation is lost (Figure 2B). These results are analogous to a known circadian function in other organisms, directing carbohydrate reserve storage during the day, and assuring that its consumption is appropriately regulated during the night [12–16].
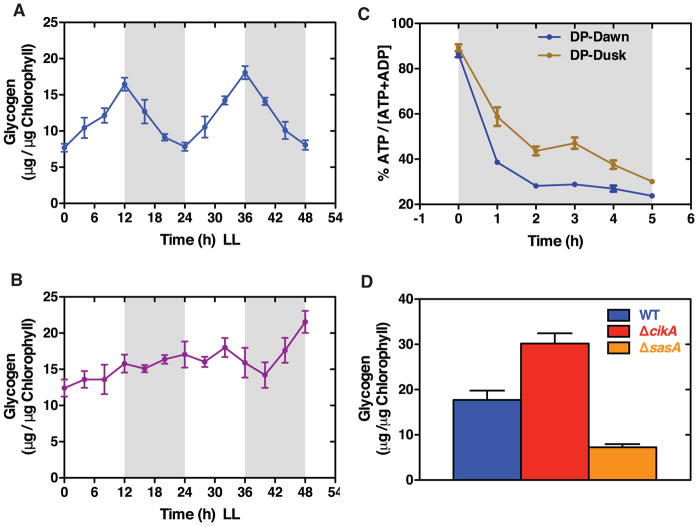
Figure 2. A circadian rhythm in energy storage metabolism protects cells against light-dark transitions.
(A–B) Glycogen time course in WT (A) and ΔkaiBC cells (B) in LL. Cultures were entrained by a 12 h:12 h light-dark cycle and subsequently released into LL (t = 0). Shaded regions represent the subjective nights. Error bars represent standard deviations (n = 4).
(C) Drop in the ATP/(ATP + ADP) energy charge in WT cells during a 5-hour dark pulse (DP) given at subjective dawn (t =25 h, blue symbols) or subjective dusk (t = 37 h, brown symbols). Cultures were grown as in (A). Error bars represent standard errors (n = 4–8).
(D) Glycogen phenotypes of clock-related histidine kinases (WT, blue bar; ΔcikA, red bar; ΔsasA, orange bar). Cultures were grown as in (A), samples collected at t = 12 h. Error bars represent standard deviations (n = 7).
We hypothesized that the metabolic rhythms produced by the clock serve to optimize growth during the day while anticipating dusk thus allowing the cell to prepare for loss of photosynthetic energy production during the night. Indeed, we found that cellular energy (ATP/ADP ratio) falls faster and reaches lower levels when a dark pulse occurs near subjective dawn (when darkness is not anticipated) compared to subjective dusk (Figure 2C). These differences in the cellular energy mirror the rhythmic accumulation and degradation of glycogen, suggesting that glycogen reserves are important to support metabolism in the dark.
Genetic analysis has indicated a role for CikA in modulating clock output [17], and CikA has recently been shown to be a cognate histidine kinase for the clock output transcription factor RpaA [18]. We therefore asked if the clock-generated glycogen rhythms would be distorted in a ΔcikA strain. Indeed, we found that ΔcikA cells over-accumulate glycogen in constant light, and glycogen content is not strongly rhythmic in this strain (Figure 2D, Figures S3B–S3C). The histidine kinase SasA opposes the action of CikA on RpaA [18]. Consistent with this picture of opposed output signaling enzymes, we find that ΔsasA cells store little glycogen, a dawn-like metabolic state in contrast to the dusk-like state of ΔcikA (Figure 2D).
ΔcikA Creates a Dusk-like Transcriptional State Genome-wide
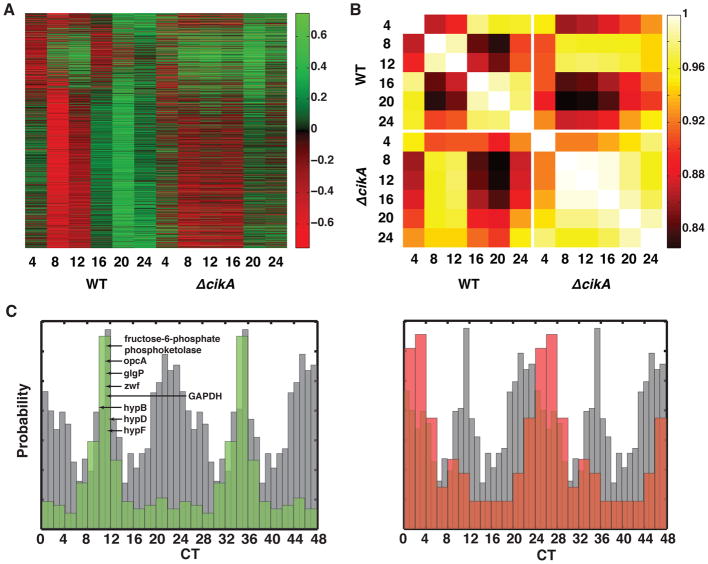
Given the participation of CikA in the clock output, we hypothesized that the metabolic phenotypes of ΔcikA might be caused by misregulation of gene expression, and that ΔcikA would cause genome-wide changes in the circadian transcriptional program. We performed a comparative microarray timecourse analysis between wildtype and ΔcikA cells (Figure 3A). To compare the profiles of transcription between these strains in absolute terms, we computed a genome-wide rank-order (Spearman) correlation between all pairs of data sets. Visualized as a matrix of correlation coefficients, these data show an expected block of correlations between subjective day and subjective night time points in the wildtype, reflecting the predominance of two distinct phases of clock-driven gene expression [10, 11]. In contrast, genome-wide rhythms in the ΔcikA cells are much less prominent, and the ΔcikA time points are generally similar to the CT 8 and CT 12 (subjective dusk) timepoints in the wild-type cells (Figure 3B).
Figure 3. The ΔcikA phase resetting mutant has a dusk-like transcriptional state.
(A) Heat map showing circadian gene expression time courses in WT and ΔcikA strains. Transcript abundance, as measured by microarray, was log-transformed and normalized across both data sets to give zero mean and unit amplitude in the WT rhythm. The data were then sorted by WT circadian phase. Green indicates high relative expression, and red indicates low relative expression. Cultures were entrained with two light-dark cycles; numbers are time in hours after release into LL.
(B) Cross-correlation matrix showing genome-wide Spearman (rank-order) correlations between expression of circadian genes in each data set. Numbers are times in hours after release into LL.
(C) Normalized histograms showing WT phasing of circadian genes called as over-expressed (green) or under-expressed (red) in ΔcikA relative to the WT time series (p < 0.01, two-sided t test). Selected metabolic genes are called out with arrows. The phasing histogram of all circadian genes is superimposed in gray. Data are double plotted to show periodicity. Circadian phasing data are taken from [11].
To characterize how transcriptional output is altered in this mutant, we looked for genes whose average expression level was significantly higher or lower in ΔcikA relative to the expression in wildtype during the circadian cycle. We then plotted an estimate of the peak expression time of these genes in the normal circadian cycle [11]. Highly upregulated genes in ΔcikA were primarily those whose wildtype expression peaks near subjective dusk, while the downregulated genes tended to have wildtype peak expression near subjective dawn. Notably, the upregulated genes included glycolytic enzymes and pentose-phosphate pathway enzymes, indicating the impact of clock-driven transcription on metabolic regulation (Figure 3C, Table S1). Similar transcriptional effects have been observed as a result of overexpressing core clock genes [19]. These results indicate that ΔcikA severely distorts the genome-wide output of the circadian clock, leaving cells in an exaggerated dusk-like state both transcriptionally and metabolically.
Disrupting Energy Storage Metabolism Makes the Clock Hypersensitive to Darkness
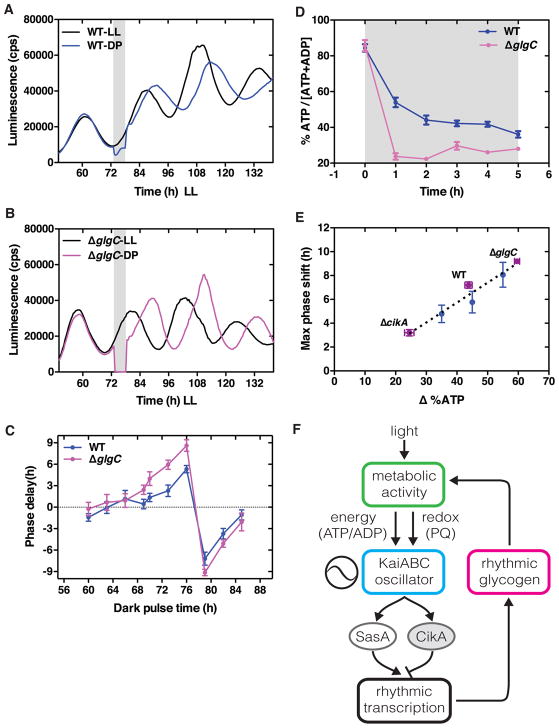
Having found that the clock output modulates clock input strength through rhythmic control of metabolism, we reasoned that it should be possible to make mutations that disrupt dark metabolism and thereby create cells that have a hypersensitive clock response to dark pulses. We deleted glgC, the first committed enzyme in glycogen biosynthesis (glucose-1-phosphate adenylyl transferase), resulting in a strain that does not accumulate glycogen (Figure S3D). We measured circadian rhythms and phase shifts in response to 5-hour dark pulses in both wildtype and ΔglgC cells, and used these data to construct phase response curves (Figures 4A–4C). Though both strains remain unresponsive to dark pulses during the subjective night, a time when the core oscillator machinery is intrinsically insensitive [3], ΔglgC cells respond more strongly to dark pulses during the subjective day. ΔglgC cells also have a shorter free-running period (23.6 h) than the wildtype (24.8 h). These results indicate that metabolic defects can render the clock input more sensitive to environmental changes.
Figure 4. Disrupting glycogen synthesis sensitizes the circadian clock to dark pulses.
(A–B) Bioluminescence rhythms (PkaiBC::luxAB) in WT (A) and ΔglgC (B). Control samples (black) kept in LL while experimental samples (red or blue) subjected to a 5-hour dark pulse (shaded bar). Cells were entrained by one light-dark cycle (12 h:12 h) and then released into LL.
(C) Phase delay of the bioluminescence rhythm (PkaiBC::luxAB) in WT (blue) and ΔglgC (magenta) cells in response to 5-hour dark pulses at the indicated time. Cells were entrained by one light-dark cycle (12 h:12 h) and then released into LL. Phase delays relative to an untreated control were estimated as in Figure 1A. Error bars represent standard deviations (n = 12).
(D) Drop in the ATP/(ATP + ADP) energy charge during a 5-hour dark pulse (begins at shaded region) in WT (blue symbols) and ΔglgC (magenta symbols) cells. Error bars represent standard errors (n = 6).
(E) Correlation between maximum observed phase shift and magnitude of drop in % ATP/(ATP + ADP) during the perturbation across all in vivo (purple symbols) and in vitro (blue symbols) conditions studied here. Trend line shows linear regression.
(F) Schematic for metabolic feedback loop in the circadian clock. Day-night cycles cause metabolic fluctuations that entrain the core KaiABC oscillator. The opposing histidine kinases SasA and CikA transduce the signal from the core oscillator into a transcriptional rhythm. The clock output creates rhythms in energy storage metabolism. The rhythmic availability and regulation of these energy stores control the ability of the clock to reset, closing a metabolic feedback loop from output back to input.
Consistent with our hypothesis that accumulated energy storage metabolites are used to support metabolic flux and thus the ATP/ADP energy charge in the dark, we found that ATP/ADP fell to lower levels in the ΔglgC mutant relative to wildtype (Figure 4D). Conversely, when wildtype cells are grown under dim light, the magnitude of the drop in ATP/ADP following a light-dark transition is less severe, and the ability of the clock to phase shift is analogously reduced (Figure S4) [20]. Thus we can summarize our in vivo and in vitro results as a trend across data sets: the extent of metabolic change (mean drop in %ATP) is predictive of the average magnitude of the clock phase shift (Figure 4E). These results strongly suggest that the sensitivity of the Kai system to the metabolic state of the cells is the primary mechanism of circadian clock input.
Conclusions
We have shown here that the cyanobacterial clock is coupled to metabolism through clock output, and that there are circadian rhythms in metabolism that condition the cell to anticipate nightfall. Thus, the ability of the clock to response to resetting cues is contingent on clock output through a feedback loop that regulates energy storage (Fig. 4F). In effect, clock resetting occurs not because of darkness per se but in response to a mismatch between the clock’s metabolic predictions and the actual state of the cell. In other words, the clock functions as a feedback controller on metabolism that operates on the timescale of a day. This framework has allowed us to understand clock resetting phenotypes caused either by direct perturbation of a metabolic pathway or indirectly by misregulating clock output. Here, metabolism cannot be simply classified as either upstream or downstream of the clock because the causal connection between the two is bidirectional, a general phenomenon also observed with signaling networks in mammalian clocks [21]. We anticipate that the quantitative study of cyanobacterial clock resetting and the pathways used to rhythmically condition the metabolic state of the cell for nightfall will reveal principles not only of circadian regulation, but of the general mechanisms that cells use to regulate metabolic flux and growth rate in fluctuating environments.
Experimental Procedures
Cyanobacterial Strains
All the bioluminescent reporter strains used in this study are based on bacterial (Vibrio harveyi) luciferase system and are derivatives of Synechococcus elongatus PCC 7942 [22]. Description of reporter strains and molecular cloning and construction of mutant strains are detailed in Supplemental Experimental Procedures.
Culture Conditions
S. elongatus cells for metabolite measurements were grown in modified BG-11M liquid medium [23] supplemented with 20 mM HEPES pH 8.0 with appropriate combinations of antibiotics at 30°C under continuous illumination (LL) ~60 μmol photons m−2 s−1; cool white fluorescent light (Phillips)] with shaking at 200 rpm, except as noted. Cell densities were monitored by measuring the optical density at 750 nm (OD750).
Circadian Bioluminescence Measurements
Bioluminescence rhythms were assayed from luciferase reporter strains illuminated by a custom-built LED array on a PerkinElmer TopCount luminometer as detailed in Supplemental Experimental Procedures.
Nucleotide Analysis
We extracted nucleotides from cyanobacterial cultures as described previously [20] with modification. Luciferase assay for ATP and ADP was done using a luminometer (Glomax, Promega). Detailed methods are described in Supplemental Experimental Procedures.
KaiABC in vitro Oscillator Reactions
Expression and purification of recombinant Kai proteins are detailed in Supplemental Experimental Procedures. The Kai proteins were mixed into master reactions in an 80% ATP reaction buffer (20 mM Tris pH 8, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 2 mM ATP, 0.5 mM ADP, 0.5 mM EDTA) and incubated at 30°C. Prior to the ADP pulses, aliquots were sampled into 3x SDS-PAGE sample buffer in open 96 well plates directly from the master reaction., The master reaction was then split into separate reactions for each pulse start time. ADP pulses were initiated by the addition of an appropriate volume of a 400 mM ADP pH 7.5 stock solution to lower the ATP/ADP ratio in the reaction mixtures. ADP pulses were then terminated 5 hours later by passing the reactions through P-6 polyacrylamide spin columns (BioRad) pre-equilibrated with the 80% ATP reaction buffer, thus returning the oscillatory reaction to pre-pulse nucleotide conditions. To prevent dilution of proteins below the functional concentration range by this buffer exchange step, the Kai proteins were initially at twice our standard in vitro reaction concentration ([KaiA] = 3 μM, [KaiB] = [KaiC] = 7 μM). A control, no ADP pulse reaction was subjected to the same buffer exchange procedure. KaiC phosphorylation was assessed by SDS-PAGE analysis as described previously [24].
Glycogen Measurement
Glycogen content from the palettes was determined using a glucose hexokinase assay (Sigma) as described [25] and detailed in Supplemental Experimental Procedures.
RNA Preparation and Microarray Analysis
For RNA isolation, cultures (120 mL, OD750 ~0.2) were synchronized with two light-dark (12 h:12h) cycles before release into LL. Cultures were kept semi-turbidostatically by manual dilution and collected at 4 h intervals for the next 24 h and snap frozen in liquid nitrogen, and stored at −80°. Total RNA was extracted from frozen cells and subjected to DNase I (Promega) treatment as described [11]. The quality and quantity of the RNA samples were checked using a 2100 Bioanalyzer (Agilent) and Nanodrop 1000 (Thermo Scientific), respectively. Gene expression was measured using custom-designed 8 × 15 k microarrays (Agilent, Array ID 020846) as described [11]. Complete methods for the microarray experiment and the data analysis are detailed in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The circadian clock generates a rhythm in energy storage metabolism
A mutant with insensitive clock input has a high energy metabolic state
Disrupting glycogen metabolism makes phase shifts hypersensitive
A quantitative correspondence between dark metabolism and phase shifts
Acknowledgments
We thank Susan S. Golden (University of California, San Diego) and Carl Johnson (Vanderbilt University) for the generous gifts of strains and plasmids. We thank Kristen E. Cook, Quincey Justman, Joseph S. Markson, Vikram Vijayan, and members of the Rust lab for useful discussions and comments on the manuscript. We thank Guillaume Lambert for assistance with electronics. Microarray time course data is available through GEO (Accession Number: GSE59112). The work was supported by a Burroughs-Wellcome Career Award at the Scientific Interface (to M.J.R.), and the National Institutes of Health (GM107369-01).
References
- 1.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 2.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 3.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331:220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YI, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 6.Katayama M, Kondo T, Xiong J, Golden SS. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol. 2003;185:1415–1422. doi: 10.1128/JB.185.4.1415-1422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiyohara YB, Katayama M, Kondo T. A novel mutation in kaiC affects resetting of the cyanobacterial circadian clock. J Bacteriol. 2005;187:2559–2564. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman J, Karakaya H, Scanlan DJ, Mann NH. A comparison of gene organization in the zwf region of the genomes of the cyanobacteria Synechococcus sp. PCC 7942 and Anabaena sp. PCC 7120. Fems Microbiol Lett. 1995;133:187–193. doi: 10.1111/j.1574-6968.1995.tb07882.x. [DOI] [PubMed] [Google Scholar]
- 10.Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci U S A. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerveny J, Sinetova MA, Valledor L, Sherman LA, Nedbal L. Ultradian metabolic rhythm in the diazotrophic cyanobacterium Cyanothece sp. ATCC 51142. Proc Natl Acad Sci U S A. 2013;110:13210–13215. doi: 10.1073/pnas.1301171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci U S A. 2010;107:9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scialdone A, Mugford ST, Feike D, Skeffington A, Borrill P, Graf A, Smith AM, Howard M. Arabidopsis plants perform arithmetic division to prevent starvation at night. eLife. 2013;2:e00669. doi: 10.7554/eLife.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci U S A. 2010;107:3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50:288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Weyman PD, Umetani M, Xiong J, Qin X, Xu Q, Iwasaki H, Johnson CH. Circadian yin-yang regulation and its manipulation to globally reprogram gene expression. Current biology: CB. 2013;23:2365–2374. doi: 10.1016/j.cub.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallas T, Castenholz RW. Internal pH and ATP-ADP pools in the cyanobacterium Synechococcus sp. during exposure to growth-inhibiting low pH. J Bacteriol. 1982;149:229–236. doi: 10.1128/jb.149.1.229-236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Current biology: CB. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 22.Andersson CR, Tsinoremas NF, Shelton J, Lebedeva NV, Yarrow J, Min H, Golden SS. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 23.Bustos SA, Golden SS. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J Bacteriol. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phong C, Markson JS, Wilhoite CM, Rust MJ. Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc Natl Acad Sci U S A. 2013;110:1124–1129. doi: 10.1073/pnas.1212113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandyopadhyay A, Stockel J, Min H, Sherman LA, Pakrasi HB. High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nature communications. 2010;1:139. doi: 10.1038/ncomms1139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.