Abstract
In response to stress, cellular compartments activate signaling pathways that mediate transcriptional programs to promote survival and reestablish homeostasis. Manipulation of the magnitude and duration of the activation of stress responses has been proposed as a strategy to prevent or repair the damage associated with aging or degenerative diseases. However, as these pathways likely evolved to respond specifically to transient perturbations, the unpredictability of prolonged activation should be considered.
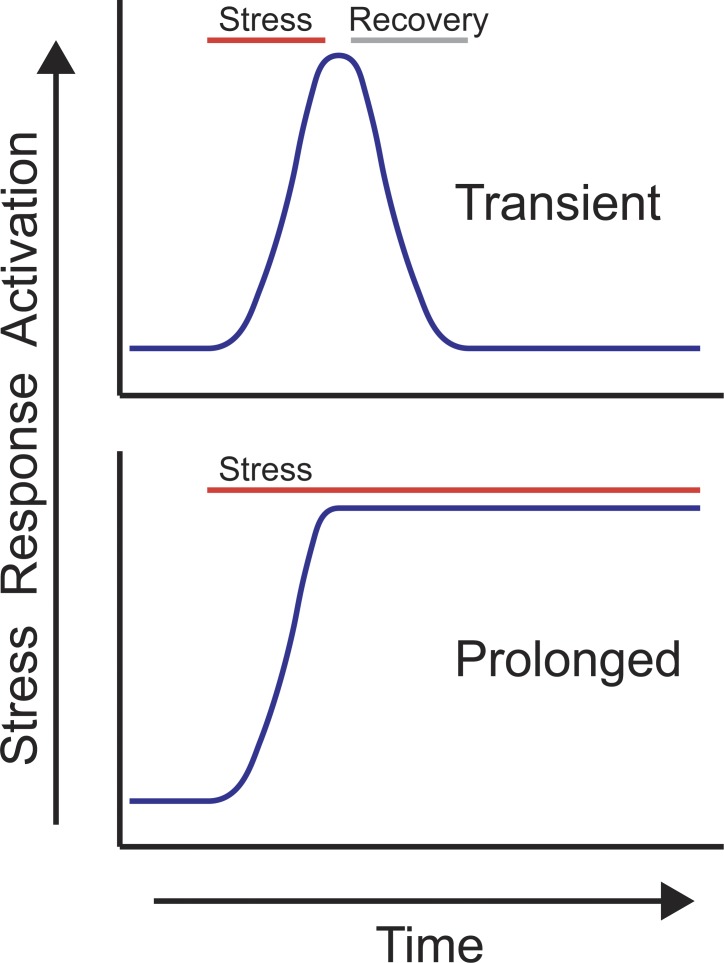
Cellular stresses, such as unfolded or misfolded protein accumulation and organelle deterioration, are associated with numerous diseases as well as the aging process. Thus, enhanced activation of pathways that have evolved to protect against these defects may protect against degenerative diseases such as Parkinson’s and Alzheimer’s or the ill effects of normal aging (Powers et al., 2009; Bratic and Larsson, 2013). Stress response pathways are typically maintained in the off state or at a baseline level. Upon organelle perturbation, they are activated to the appropriate magnitude and duration to efficiently promote cellular survival and organelle recovery. Once homeostasis is reestablished, the pathway is down-regulated so that cells can properly respond to future stress (Fig. 1 A).
Figure 1.
Transient versus perpetual or prolonged stress response activation. (A) The HSR, UPR, and the UPRmt remain in an “off” or “low-activity” state until they are activated by compartment-specific stress. As these pathways are not constitutively active, these stress responses likely evolved to promote survival during temporary stressful conditions and ultimately recover once the condition causing the cellular dysfunction is alleviated. Transient stresses include environmental effects such as temperature shifts, exposure to toxins, or altered nutritional status. (B) Genetic mutations that perturb cytosolic, ER, or mitochondrial proteostasis are typically selected against evolutionarily as they cause cellular dysfunction. However, genotoxic perturbations, or damage that has accrued over long periods of time during aging or disease, may cause prolonged or perpetual activation of the HSR, UPR, and UPRmt, as the mutation cannot be rectified. Prolonged or perpetual stress response activation is potentially very different than transient activation as there is never a recovery. As evolution did not select for prolonged stress response activation, we hypothesize that it is difficult to predict the outcome of prolonged stress response activation.
Manipulations of these pathways can mitigate the intracellular damage that occurs during aging or in degenerative diseases. However, these pathways did not likely evolve to deal with prolonged stress or to be activated for extended periods of time (Fig. 1 B). If continued activation were entirely beneficial, these pathways would likely have evolved to be hard-wired into developmental or cell-specific programs rather than to be stress inducible. We hypothesize that prolonged stress response activation has not been subject to evolution, as conditions that cause perpetual activation, such as deleterious gene mutations, result in cellular damage and would be selected against. Thus, the potential outcomes of prolonged stress response activation are difficult to predict. Here, we review the evidence suggesting that stress response pathways evolved to be transiently activated to a precise magnitude to match the level of dysfunction and allow the most efficient recovery, and consider the positive and negative effects of enhanced stress response activation. We also consider approaches to therapeutically engage stress response signaling.
Stress detection and matched transcriptional activation
Several organelle or stress-specific stress responses have been identified and are described in more detail elsewhere (Åkerfelt et al., 2010; Walter and Ron, 2011; Jensen and Jasper, 2014). Here, we focus on specific responses that are activated by cytosolic, ER, or mitochondrial stress or dysfunction.
The heat shock response (HSR)
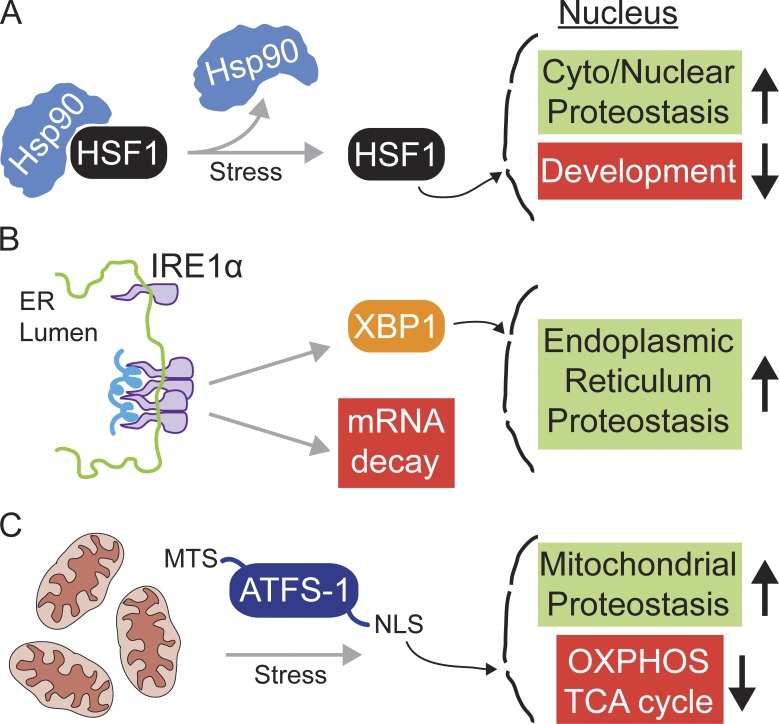
The HSR is mediated by the transcription factor HSF1 and occurs during conditions that cause an increase in unfolded or misfolded proteins primarily in the cytosol and nucleus, such as increased temperature, oxidative stress, and exposure to heavy metals (Ananthan et al., 1986; Åkerfelt et al., 2010). However, it can also be activated independently of misfolded proteins, as stalled ribosome complexes also activate the response (Brandman et al., 2012). The HSR is a transcriptional program that involves the induction of ∼500 genes, including cytosolic and nuclear-localized protein homeostasis (proteostasis) machinery such as molecular chaperones and genes involved in protein synthesis, the cell cycle, and the regulation of cell death (Mendillo et al., 2012; Ryno et al., 2014). While the induction of chaperones and proteases garner much of the attention, the HSR also includes the repression of ∼1,000 genes including developmental, immune, apoptotic (Mendillo et al., 2012; Ryno et al., 2014), and cytoskeletal maintenance components (Baird et al., 2014; Fig. 2 A).
Figure 2.
The heat shock response, the UPR, and the mitochondrial UPR. (A) The HSR is mediated by the transcription factor HSF1. Normally, HSF1 is repressed or maintained in the “off” state by interacting with the molecular chaperone Hsp90. However, when unfolded proteins accumulate in the cytosol or nucleus, HSF1 dissociates from Hsp90 and binds the promoters of HSR genes. The HSR includes the induction (green) of proteostasis machinery including molecular chaperones as well as the repression (red) of many genes required for development. Once proteostasis is recovered, HSF1 is degraded and the HSR is attenuated. (B) The UPR is mediated by at least three ER stress sensor molecules, the most conserved of which is IRE1. Upon detection of ER stress, IRE1 oligomerizes, activating its cytosolic RNase domain which results in (1) the cleavage and subsequent degradation of ER-localized mRNAs, reducing the incoming protein load on the stressed organelle; and (2) the activation of the transcription factor XBP1, which results in induction of a broad response including ER proteostasis machinery, lipid synthesis to expand ER volume, and increased secretory components. Once ER homeostasis is reestablished, IRE1 signaling is attenuated by association with ER chaperones and XBP1 is degraded. (C) The UPRmt is a mitochondrial-specific stress response mediated by ATFS-1. ATFS-1 is activated when mitochondrial protein import is impaired, which can be caused by imbalanced mitochondrial proteostasis or respiratory chain defects. Cytosolic ATFS-1 then traffics to the nucleus and activates the UPRmt, which includes an increase (green) in mitochondrial proteostasis machinery such as mitochondrial chaperones and ROS-detoxifying components. The UPRmt also involves the repression (red) or limited expression of some of the most highly expressed mitochondrial proteins including components of the TCA cycle and the oxidative phosphorylation (OXPHOS) complexes. Once mitochondrial function is recovered, ATFS-1 is degraded and the UPRmt is down-regulated.
Normally, HSF1 is repressed by the cytosolic and nuclear-localized molecular chaperone Hsp90, which binds and maintains the transcription factor at a baseline level (Morimoto, 1998; Zou et al., 1998). As unfolded proteins increase, HSF1 is released, allowing it to bind the heat shock promoter element and regulate transcription (Fig. 2 A; Topol et al., 1985; Morgan et al., 1987). In addition to direct regulation by chaperones, HSF1 is also subject to multiple posttranslational modifications (Anckar and Sistonen, 2011). For example, the magnitude and duration of HSF1 activation are further regulated by the acetyltransferase EP300 and the deacetylase SIRT1. HSF1 acetylation by EP300 controls the quantity of HSF1 available for activation by preventing proteasome-dependent degradation (Raychaudhuri et al., 2014). Conversely, deacetylation of HSF1 by SIRT1 promotes activation of HSF1 during stress (Westerheide et al., 2009), but will eventually lead to HSF1 turnover to down-regulate the response (Raychaudhuri et al., 2014). Further regulation of the HSR occurs at the organismal level via thermosensory neurons and neuroendocrine signaling. Non–cell-autonomous HSF1 activation presumably allows for more precise matching of the HSR to the behavioral and metabolic status of the organism (Prahlad et al., 2008; Prahlad and Morimoto, 2011).
The unfolded protein response (UPR)
The ER is the site of protein synthesis and folding for the vast majority proteins that are secreted or localized within the secretory pathway. In response to increased protein flux through the ER or to conditions that perturb ER protein folding, UPR activation limits the load on the stressed organelle by reducing localized protein synthesis and activating protective ER-specific transcriptional programs to reestablish organelle homeostasis (Walter and Ron, 2011; Fig. 2 B). The most conserved branch of the UPR is regulated by the ER–membrane localized kinase IRE1 and the transcription factor XBP1 (Hac1 in yeast). The UPR is activated when accumulating unfolded proteins directly interact with the luminal domain of IRE1 (Gardner and Walter, 2011), causing it to oligomerize, activating the cytosolic RNase domain (Korennykh et al., 2009). IRE1 cleaves several ER-localized mRNAs, resulting in their degradation, and thus reducing their translation and the burden on the ER protein-folding environment (Han et al., 2009; Hollien et al., 2009). Concomitantly, IRE1 also cleaves an inhibitory intron from the transcript encoding XBP1, which upon ligation allows the translation of the active transcription factor (Cox and Walter, 1996; Yoshida et al., 2001; Calfon et al., 2002). Once translated, XBP1 activates a broad transcriptional response that includes ER-localized components that promote protein folding and quality control, compartmental expansion, and increased ER–Golgi trafficking (Travers et al., 2000; Shoulders et al., 2013; Fig. 2 B). However, if ER stress cannot be rectified, an apoptotic program is engaged to eliminate the unsalvageable cell (Tabas and Ron, 2011; Upton et al., 2012; Lu et al., 2014).
The mitochondrial UPR (UPRmt)
The UPRmt is a transcriptional response that occurs specifically during mitochondrial dysfunction to promote survival and recovery of mitochondrial activity. The UPRmt in Caenorhabditis elegans is regulated by the transcription factor ATFS-1, which is normally imported into mitochondria and degraded (Nargund et al., 2012; Haynes et al., 2013). However, during conditions that impair mitochondrial protein import such as respiratory chain dysfunction, mitochondrial unfolded protein accumulation, or high levels of reactive oxygen species (ROS), general mitochondrial import efficiency is reduced, causing mitochondrial proteins to accumulate in the cytosol (Wright et al., 2001; Nargund et al., 2012; Harbauer et al., 2014). As ATFS-1 has a nuclear localization sequence, some of the cytosolic ATFS-1 pool then traffics to the nucleus to mediate UPRmt activation (Nargund et al., 2012; Fig. 2 C). Similar to the HSR and UPR, the UPRmt receives non–cell-autonomous regulatory inputs (Durieux et al., 2011; Owusu-Ansah et al., 2013; Taylor and Dillin, 2013).
ATFS-1 activation increases mitochondrial protein folding capacity and promotes mitochondrial recovery by increasing mitochondrial chaperones, proteases, respiratory chain complex assembly factors, import, and fission components (Nargund et al., 2012, 2015). Concomitantly, ATFS-1 limits expression of the tricarboxylic acid (TCA) cycle and respiratory chain components, suggesting that ATFS-1 promotes mitochondrial recovery by increasing protein folding and complex assembly capacity while slowing the rate of respiratory complex biogenesis to match the stressed organelle’s capacity (Nargund et al., 2015; Fig. 2 C). To facilitate organelle repair, ATFS-1 increases expression of all glycolysis components in order to maintain energy levels. Thus, the UPRmt includes a shift in cellular metabolism to promote survival during mitochondrial dysfunction that is reminiscent of the metabolism in rapidly dividing cells (Vander Heiden et al., 2009), and which presumably must be down-regulated upon return to homeostasis.
Regulation of response duration and recovery
In addition to the magnitude of a specific stress response, which is largely governed by activating mechanisms, the duration of the response must be tightly regulated to match cell physiology and promote efficient recovery. Consistent with the idea that prolonged activation of each pathway is potentially detrimental, multiple mechanisms exist to limit and down-regulate stress response activation. Included in the HSR, UPR and UPRmt are components that down-regulate HSF1, XBP1, and ATFS-1, respectively, via negative feedback loops. For example, HSF1 induces expression of Hsp70 and Hsp90, which in addition to promoting efficient protein folding also associates with active HSF1 to dampen the response (Shi et al., 1998). Similarly, XBP1 induces expression of ER-localized chaperones, which associate with IRE1 to attenuate signaling upon proteostasis recovery (Todd-Corlett et al., 2007; Eletto et al., 2014). ATFS-1 also induces multiple components that promote mitochondrial protein import efficiency, which serves to reduce cytosolic and ultimately nuclear accumulation of ATFS-1 (Nargund et al., 2012). Furthermore, all three pathways increase components that target the active transcription factor for degradation, including ubiquitin ligases. In addition to negative regulation of the response regulators, the outputs of the responses are also down-regulated once proteostasis is reestablished. For example, as misfolded or unfolded proteins are depleted, the HSF1-induced chaperone Hsp70 is ubiquitinated by the HSF1-induced ubiquitin ligase CHIP and is degraded by proteasomes, returning the chaperone capacity to baseline levels (Qian et al., 2006).
Effects of prolonged activation
HSF1, XBP1, and ATFS-1 have all been shown to be protective during organelle-specific stress, promoting survival and cellular proliferation during conditions that perturb cytosolic and nuclear (Morano et al., 1999; Xiao et al., 1999; Hsu et al., 2003; Morley and Morimoto, 2004), ER (Cox et al., 1993; Shen et al., 2001; Lin et al., 2009; Richardson et al., 2010), or mitochondrial proteostasis (Baker et al., 2012; Nargund et al., 2012), respectively. Interestingly, cellular damage that accrues in aging animals activates each pathway when it occurs in young animals. However, all three pathways (Yoneda et al., 2004; Ben-Zvi et al., 2009; David et al., 2010; Durieux et al., 2011) are attenuated and less effective in aging animals, which coincides with a proteostatic collapse (Ben-Zvi et al., 2009), further suggesting that enhanced activation may be beneficial.
Several interesting observations reveal their protective effects against age-associated cellular damage. Impaired insulin signaling, which extends worm lifespan considerably, requires multiple transcription factors including HSF1 and XBP1 (Kimura et al., 1997; Lin et al., 1997; Hsu et al., 2003; Henis-Korenblit et al., 2010). Similarly, modest mitochondrial dysfunction that activates the UPRmt also extends lifespan in multiple species including mice, flies, and worms (Dillin et al., 2002b; Durieux et al., 2011; Houtkooper et al., 2013; Owusu-Ansah et al., 2013; Schieber and Chandel, 2014). Thus, pathway activation can mitigate age-associated damage; however, it should be noted that this often comes at the expense of fecundity and normal development (Dillin et al., 2002a).
HSR
Impressively, HSF1 activation is sufficient to extend the lifespan of normal worms, indicating that the HSR can be protective over longer periods of time (Hsu et al., 2003; Westerheide et al., 2009). HSF1 activity positively affects proteostasis and reduces aggregation of disease-associated proteins in multiple organisms such as those containing polyglutamine stretches (Calamini et al., 2012; Brunquell et al., 2014), α-synuclein (Hamamichi et al., 2008), prion protein (PrP; Steele et al., 2008), and Aβ1–42 (Cohen et al., 2006; Calamini et al., 2012).
In addition to these protective effects, accumulating evidence indicates that HSF1 activation can also negatively affect proteostasis. Defects in folding and trafficking of the CFTR protein caused by an amino acid deletion result in cystic fibrosis. While expression of mutated CFTR activates HSF1, it was recently shown that HSF1 inhibition increases CFTR trafficking and function, suggesting that prolonged HSF1 activation creates a maladaptive state (Wang et al., 2006; Roth et al., 2014). Similarly, HSF1 overexpression has been shown to exacerbate aggregation of the polyglutamine protein Huntingtin (Bersuker et al., 2013). Lastly, the HSF1 expression level is associated with poor prognoses in breast cancers, which is consistent with many cancer types requiring HSF1 activity to promote proliferation (Dai et al., 2007; Santagata et al., 2011), highlighting the importance of appropriate HSF1 activation.
UPR
Similar to HSF1, expression of XBP1 is sufficient to counteract the secretory pathway dysfunction that occurs during worm aging and results in lifespan extension (Taylor and Dillin, 2013). This suggests that approaches to promote UPR activation may be effective against diseases associated with ER stress, which include neurodegenerative and metabolic diseases as well as those associated with mutations causing expression of terminally misfolded secretory proteins (Ryno et al., 2013). Enhanced UPR activation has been demonstrated to reduce the secretion of a misfolded and dysfunctional variant of rhodopsin that results in photoreceptor cell death (Chiang et al., 2012), promote proper folding and function of mutant lysosomal proteins associated with lysosomal storage disease (Mu et al., 2008), reduce the secretion of amyloidogenic aggregation-prone proteins (Cooley et al., 2014), and limit the neurodegeneration in mouse models of Charcot-Marie Tooth disease and amyotrophic lateral sclerosis (Das et al., 2015).
However, numerous studies indicate that prolonged or inappropriate UPR signaling can be toxic, even if apoptotic induction is avoided (Tabas and Ron, 2011). Phospho-transfer by IRE1’s cytosolic kinase domain is not required for activation of the RNase domain; rather, it is required to down-regulate signaling as ER stress is alleviated (Chawla et al., 2011; Rubio et al., 2011). Cells expressing IRE1 with impaired phosphor-transfer activity efficiently activate the UPR but are unable to attenuate IRE1 activity. The prolonged UPR activation in these cells fails to return the organelle to proteostasis and is at least partially due to the sustained production of ER-targeted proteins (Rubio et al., 2011), which can lead to developmental arrest or apoptosis (Eletto et al., 2014). Additionally, prolonged turnover of ER-localized mRNAs by IRE1 likely has negative consequences for secretory pathway activity.
UPRmt
While ATFS-1 is necessary for longevity associated with modest mitochondrial dysfunction (Rea et al., 2007; Schieber and Chandel, 2014), ATFS-1 is not sufficient to promote longevity independent of mitochondrial stress. Mutations in ATFS-1’s mitochondrial targeting sequence that cause it to redistribute to the nucleus under normal conditions are quite toxic, impeding development and reducing lifespan (Rauthan et al., 2013). These results may be explained by ATFS-1 functioning as a single component within a broader signaling network that must be integrated to exude protective effects during stress. For example, autophagy (Lapierre et al., 2013), altered protein synthesis (Baker et al., 2012), and additional transcription programs (Lee et al., 2010; Walter et al., 2011) are also required to promote longevity during mitochondrial dysfunction. Highlighting the protective effects of activating the UPRmt to appropriately match the level of mitochondrial dysfunction, worms expressing activated ATFS-1 are resistant when chronically exposed to mitochondrial toxins, statins (Rauthan et al., 2013), or the pathogenic bacteria Pseudomonas aeruginosa (Pellegrino et al., 2014), which perturbs mitochondrial function. These results indicate that enhanced UPRmt activation can be protective, but the magnitude and duration of the response should be considered as well as other factors that potentially coordinate with the UPRmt.
Conclusions
Manipulations to enhance stress response activation hold promise therapeutically to mitigate the cellular damage that accrues during aging and disease (Calamini et al., 2012; Mouchiroud et al., 2013). Response activation in principle can be achieved by (1) perturbing the protein folding environment, (2) activating the transcription factor directly, or (3) impairing the turnover of the active transcription factors. But the evidence reviewed here suggests that it is difficult to predict the outcome of prolonged activation, as these responses likely evolved to resolve transient proteotoxic stress (Fig. 1 A). Therefore, manipulation of these stress response pathways as a therapeutic measure will require careful consideration of the effects of prolonged activation (Fig. 1 B). Considering the number of transcripts XBP1, ATFS-1, and HSF1 affect in addition to those promoting proteostasis, prolonged activation may alter fundamental aspects of a particular cell. For example, to promote mitochondrial recovery, ATFS-1 activation shifts metabolism to that typically observed in rapidly proliferating cells, which may be detrimental to postmitotic cells if activation is prolonged (Fig. 2 C).
Despite the challenges in manipulating these pathways to promote organelle recovery and cell survival, we are optimistic that as more knowledge is gained regarding pathway regulation and outputs, therapeutic manipulations can be tailored to limit cellular damage so as to avoid unintended effects of prolonged alterations. A particularly exciting example has been the development of phosphatase inhibitors that partially prolong the effects of the UPR branch that attenuates translation during ER stress (Novoa et al., 2001; Boyce et al., 2005). Impaired activation, or extreme prolonged activation, of the translational control branch of the UPR results in cell death and developmental arrest (Harding et al., 2000, 2009). However, guanabenz inhibits only one of the two phosphatases that attenuate the pathway (Tsaytler et al., 2011). Impressively, guanabenz and, more recently, a related compound have been shown to be protective in a variety of cultured cell lines as well as in mouse models of neurodegeneration (Das et al., 2015; Way et al., 2015).
Acknowledgements
We apologize to those colleagues whose work on the HSR, UPR, and UPRmt we could not include due to space limitations.
This work was supported by the National Institutes of Health (grants R01AG040061 and R01AG047182 to C.M. Haynes).
The authors declare no competing financial interests.
References
- Åkerfelt M., Morimoto R.I., and Sistonen L.. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11:545–555. 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthan J., Goldberg A.L., and Voellmy R.. 1986. Abnormal proteins serve as eukaryotic stress signals and trigger the activation of heat shock genes. Science. 232:522–524. 10.1126/science.3083508 [DOI] [PubMed] [Google Scholar]
- Anckar J., and Sistonen L.. 2011. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80:1089–1115. 10.1146/annurev-biochem-060809-095203 [DOI] [PubMed] [Google Scholar]
- Baird N.A., Douglas P.M., Simic M.S., Grant A.R., Moresco J.J., Wolff S.C., Yates J.R. III, Manning G., and Dillin A.. 2014. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 346:360–363. 10.1126/science.1253168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.M., Nargund A.M., Sun T., and Haynes C.M.. 2012. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 8:e1002760 10.1371/journal.pgen.1002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A., Miller E.A., and Morimoto R.I.. 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. USA. 106:14914–14919. 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersuker K., Hipp M.S., Calamini B., Morimoto R.I., and Kopito R.R.. 2013. Heat shock response activation exacerbates inclusion body formation in a cellular model of Huntington disease. J. Biol. Chem. 288:23633–23638. 10.1074/jbc.C113.481945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M., Bryant K.F., Jousse C., Long K., Harding H.P., Scheuner D., Kaufman R.J., Ma D., Coen D.M., Ron D., and Yuan J.. 2005. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science. 307:935–939. 10.1126/science.1101902 [DOI] [PubMed] [Google Scholar]
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.W., Zhou S., King D., Shen P.S., Weibezahn J., et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 151:1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic A., and Larsson N.G.. 2013. The role of mitochondria in aging. J. Clin. Invest. 123:951–957. 10.1172/JCI64125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunquell J., Bowers P., and Westerheide S.D.. 2014. Fluorodeoxyuridine enhances the heat shock response and decreases polyglutamine aggregation in an HSF-1-dependent manner in Caenorhabditis elegans. Mech. Ageing Dev. 141-142:1–4. 10.1016/j.mad.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Calamini B., Silva M.C., Madoux F., Hutt D.M., Khanna S., Chalfant M.A., Saldanha S.A., Hodder P., Tait B.D., Garza D., et al. 2012. Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8:185–196. 10.1038/nchembio.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., and Ron D.. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Chawla A., Chakrabarti S., Ghosh G., and Niwa M.. 2011. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J. Cell Biol. 193:41–50. 10.1083/jcb.201008071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W.C., Messah C., and Lin J.H.. 2012. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol. Biol. Cell. 23:758–770. 10.1091/mbc.E11-08-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E., Bieschke J., Perciavalle R.M., Kelly J.W., and Dillin A.. 2006. Opposing activities protect against age-onset proteotoxicity. Science. 313:1604–1610. 10.1126/science.1124646 [DOI] [PubMed] [Google Scholar]
- Cooley C.B., Ryno L.M., Plate L., Morgan G.J., Hulleman J.D., Kelly J.W., and Wiseman R.L.. 2014. Unfolded protein response activation reduces secretion and extracellular aggregation of amyloidogenic immunoglobulin light chain. Proc. Natl. Acad. Sci. USA. 111:13046–13051. 10.1073/pnas.1406050111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.S., and Walter P.. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 87:391–404. 10.1016/S0092-8674(00)81360-4 [DOI] [PubMed] [Google Scholar]
- Cox J.S., Shamu C.E., and Walter P.. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73:1197–1206. 10.1016/0092-8674(93)90648-A [DOI] [PubMed] [Google Scholar]
- Dai C., Whitesell L., Rogers A.B., and Lindquist S.. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 130:1005–1018. 10.1016/j.cell.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I., Krzyzosiak A., Schneider K., Wrabetz L., D’Antonio M., Barry N., Sigurdardottir A., and Bertolotti A.. 2015. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science. 348:239–242. 10.1126/science.aaa4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D.C., Ollikainen N., Trinidad J.C., Cary M.P., Burlingame A.L., and Kenyon C.. 2010. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8:e1000450 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Crawford D.K., and Kenyon C.. 2002a Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 298:830–834. 10.1126/science.1074240 [DOI] [PubMed] [Google Scholar]
- Dillin A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., and Kenyon C.. 2002b Rates of behavior and aging specified by mitochondrial function during development. Science. 298:2398–2401. 10.1126/science.1077780 [DOI] [PubMed] [Google Scholar]
- Durieux J., Wolff S., and Dillin A.. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144:79–91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D., Eletto D., Dersh D., Gidalevitz T., and Argon Y.. 2014. Protein disulfide isomerase A6 controls the decay of IRE1α signaling via disulfide-dependent association. Mol. Cell. 53:562–576. 10.1016/j.molcel.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner B.M., and Walter P.. 2011. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 333:1891–1894. 10.1126/science.1209126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamichi S., Rivas R.N., Knight A.L., Cao S., Caldwell K.A., and Caldwell G.A.. 2008. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc. Natl. Acad. Sci. USA. 105:728–733. 10.1073/pnas.0711018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Lerner A.G., Vande Walle L., Upton J.P., Xu W., Hagen A., Backes B.J., Oakes S.A., and Papa F.R.. 2009. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 138:562–575. 10.1016/j.cell.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer A.B., Zahedi R.P., Sickmann A., Pfanner N., and Meisinger C.. 2014. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 19:357–372. 10.1016/j.cmet.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., and Ron D.. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904. 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Scheuner D., Chen J.J., Kaufman R.J., and Ron D.. 2009. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2α) dephosphorylation in mammalian development. Proc. Natl. Acad. Sci. USA. 106:1832–1837. 10.1073/pnas.0809632106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C.M., Fiorese C.J., and Lin Y.F.. 2013. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 23:311–318. 10.1016/j.tcb.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S., Zhang P., Hansen M., McCormick M., Lee S.J., Cary M., and Kenyon C.. 2010. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl. Acad. Sci. USA. 107:9730–9735. 10.1073/pnas.1002575107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J., Lin J.H., Li H., Stevens N., Walter P., and Weissman J.S.. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186:323–331. 10.1083/jcb.200903014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., and Auwerx J.. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497:451–457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A.L., Murphy C.T., and Kenyon C.. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 300:1142–1145. 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]
- Jensen M.B., and Jasper H.. 2014. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 20:214–225. 10.1016/j.cmet.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K.D., Tissenbaum H.A., Liu Y., and Ruvkun G.. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 277:942–946. 10.1126/science.277.5328.942 [DOI] [PubMed] [Google Scholar]
- Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Zhang C., Shokat K.M., Stroud R.M., and Walter P.. 2009. The unfolded protein response signals through high-order assembly of Ire1. Nature. 457:687–693. 10.1038/nature07661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L.R., De Magalhaes Filho C.D., McQuary P.R., Chu C.C., Visvikis O., Chang J.T., Gelino S., Ong B., Davis A.E., Irazoqui J.E., et al. 2013. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4:2267 10.1038/ncomms3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Hwang A.B., and Kenyon C.. 2010. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20:2131–2136. 10.1016/j.cub.2010.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Dorman J.B., Rodan A., and Kenyon C.. 1997. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 278:1319–1322. 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- Lin J.H., Li H., Zhang Y., Ron D., and Walter P.. 2009. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS ONE. 4:e4170 10.1371/journal.pone.0004170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Lawrence D.A., Marsters S., Acosta-Alvear D., Kimmig P., Mendez A.S., Paton A.W., Paton J.C., Walter P., and Ashkenazi A.. 2014. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 345:98–101. 10.1126/science.1254312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo M.L., Santagata S., Koeva M., Bell G.W., Hu R., Tamimi R.M., Fraenkel E., Ince T.A., Whitesell L., and Lindquist S.. 2012. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 150:549–562. 10.1016/j.cell.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano K.A., Santoro N., Koch K.A., and Thiele D.J.. 1999. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 19:402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan W.D., Williams G.T., Morimoto R.I., Greene J., Kingston R.E., and Tjian R.. 1987. Two transcriptional activators, CCAAT-box-binding transcription factor and heat shock transcription factor, interact with a human hsp70 gene promoter. Mol. Cell. Biol. 7:1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R.I. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12:3788–3796. 10.1101/gad.12.24.3788 [DOI] [PubMed] [Google Scholar]
- Morley J.F., and Morimoto R.I.. 2004. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 15:657–664. 10.1091/mbc.E03-07-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y.S., Viswanathan M., Schoonjans K., et al. 2013. The NAD+/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 154:430–441. 10.1016/j.cell.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu T.W., Ong D.S., Wang Y.J., Balch W.E., Yates J.R. III, Segatori L., and Kelly J.W.. 2008. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 134:769–781. 10.1016/j.cell.2008.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund A.M., Pellegrino M.W., Fiorese C.J., Baker B.M., and Haynes C.M.. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 337:587–590. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund A.M., Fiorese C.J., Pellegrino M.W., Deng P., and Haynes C.M.. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell. 58:123–133. 10.1016/j.molcel.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I., Zeng H., Harding H.P., and Ron D.. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Song W., and Perrimon N.. 2013. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 155:699–712. 10.1016/j.cell.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M.W., Nargund A.M., Kirienko N.V., Gillis R., Fiorese C.J., and Haynes C.M.. 2014. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 516:414–417. 10.1038/nature13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., and Balch W.E.. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78:959–991. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Prahlad V., and Morimoto R.I.. 2011. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc. Natl. Acad. Sci. USA. 108:14204–14209. 10.1073/pnas.1106557108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T., and Morimoto R.I.. 2008. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 320:811–814. 10.1126/science.1156093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S.B., McDonough H., Boellmann F., Cyr D.M., and Patterson C.. 2006. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 440:551–555. 10.1038/nature04600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M., Ranji P., Aguilera Pradenas N., Pitot C., and Pilon M.. 2013. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc. Natl. Acad. Sci. USA. 110:5981–5986. 10.1073/pnas.1218778110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Loew C., Körner R., Pinkert S., Theis M., Hayer-Hartl M., Buchholz F., and Hartl F.U.. 2014. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell. 156:975–985. 10.1016/j.cell.2014.01.055 [DOI] [PubMed] [Google Scholar]
- Rea S.L., Ventura N., and Johnson T.E.. 2007. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5:e259 10.1371/journal.pbio.0050259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C.E., Kooistra T., and Kim D.H.. 2010. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 463:1092–1095. 10.1038/nature08762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.M., Hutt D.M., Tong J., Bouchecareilh M., Wang N., Seeley T., Dekkers J.F., Beekman J.M., Garza D., Drew L., et al. 2014. Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol. 12:e1001998 10.1371/journal.pbio.1001998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio C., Pincus D., Korennykh A., Schuck S., El-Samad H., and Walter P.. 2011. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J. Cell Biol. 193:171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryno L.M., Wiseman R.L., and Kelly J.W.. 2013. Targeting unfolded protein response signaling pathways to ameliorate protein misfolding diseases. Curr. Opin. Chem. Biol. 17:346–352. 10.1016/j.cbpa.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryno L.M., Genereux J.C., Naito T., Morimoto R.I., Powers E.T., Shoulders M.D., and Wiseman R.L.. 2014. Characterizing the altered cellular proteome induced by the stress-independent activation of heat shock factor 1. ACS Chem. Biol. 9:1273–1283. 10.1021/cb500062n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S., Hu R., Lin N.U., Mendillo M.L., Collins L.C., Hankinson S.E., Schnitt S.J., Whitesell L., Tamimi R.M., Lindquist S., and Ince T.A.. 2011. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. USA. 108:18378–18383. 10.1073/pnas.1115031108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M., and Chandel N.S.. 2014. TOR signaling couples oxygen sensing to lifespan in C. elegans. Cell Reports. 9:9–15. 10.1016/j.celrep.2014.08.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Ellis R.E., Lee K., Liu C.Y., Yang K., Solomon A., Yoshida H., Morimoto R., Kurnit D.M., Mori K., and Kaufman R.J.. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 107:893–903. 10.1016/S0092-8674(01)00612-2 [DOI] [PubMed] [Google Scholar]
- Shi Y., Mosser D.D., and Morimoto R.I.. 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12:654–666. 10.1101/gad.12.5.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C., Yates J.R. III, Su A.I., Kelly J.W., and Wiseman R.L.. 2013. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Reports. 3:1279–1292. 10.1016/j.celrep.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A.D., Hutter G., Jackson W.S., Heppner F.L., Borkowski A.W., King O.D., Raymond G.J., Aguzzi A., and Lindquist S.. 2008. Heat shock factor 1 regulates lifespan as distinct from disease onset in prion disease. Proc. Natl. Acad. Sci. USA. 105:13626–13631. 10.1073/pnas.0806319105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., and Ron D.. 2011. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13:184–190. 10.1038/ncb0311-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.C., and Dillin A.. 2013. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 153:1435–1447. 10.1016/j.cell.2013.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd-Corlett A., Jones E., Seghers C., and Gething M.J.. 2007. Lobe IB of the ATPase domain of Kar2p/BiP interacts with Ire1p to negatively regulate the unfolded protein response in Saccharomyces cerevisiae. J. Mol. Biol. 367:770–787. 10.1016/j.jmb.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Topol J., Ruden D.M., and Parker C.S.. 1985. Sequences required for in vitro transcriptional activation of a Drosophila hsp 70 gene. Cell. 42:527–537. 10.1016/0092-8674(85)90110-2 [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., and Walter P.. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258. 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Tsaytler P., Harding H.P., Ron D., and Bertolotti A.. 2011. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 332:91–94. 10.1126/science.1201396 [DOI] [PubMed] [Google Scholar]
- Upton J.P., Wang L., Han D., Wang E.S., Huskey N.E., Lim L., Truitt M., McManus M.T., Ruggero D., Goga A., et al. 2012. IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 338:818–822. 10.1126/science.1226191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., and Thompson C.B.. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 324:1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Walter L., Baruah A., Chang H.W., Pace H.M., and Lee S.S.. 2011. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 9:e1001084 10.1371/journal.pbio.1001084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Venable J., LaPointe P., Hutt D.M., Koulov A.V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., et al. 2006. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 127:803–815. 10.1016/j.cell.2006.09.043 [DOI] [PubMed] [Google Scholar]
- Way S.W., Podojil J.R., Clayton B.L., Zaremba A., Collins T.L., Kunjamma R.B., Robinson A.P., Brugarolas P., Miller R.H., Miller S.D., and Popko B.. 2015. Pharmaceutical integrated stress response enhancement protects oligodendrocytes and provides a potential multiple sclerosis therapeutic. Nat. Commun. 6:6532 10.1038/ncomms7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide S.D., Anckar J., Stevens S.M. Jr., Sistonen L., and Morimoto R.I.. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 323:1063–1066. 10.1126/science.1165946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G., Terada K., Yano M., Sergeev I., and Mori M.. 2001. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp. Cell Res. 263:107–117. 10.1006/excr.2000.5096 [DOI] [PubMed] [Google Scholar]
- Xiao X., Zuo X., Davis A.A., McMillan D.R., Curry B.B., Richardson J.A., and Benjamin I.J.. 1999. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18:5943–5952. 10.1093/emboj/18.21.5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T., Benedetti C., Urano F., Clark S.G., Harding H.P., and Ron D.. 2004. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 117:4055–4066. 10.1242/jcs.01275 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K.. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zou J., Guo Y., Guettouche T., Smith D.F., and Voellmy R.. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 94:471–480. 10.1016/S0092-8674(00)81588-3 [DOI] [PubMed] [Google Scholar]