Kar1 interacts with the C-terminal (CT) region of Sfi1 to tether the yeast centrosome’s bridge to the nuclear envelope; Sfi1-CT and C-terminal Cdc31-binding regions form the antiparallel Sfi1 overlap in the bridge.
Abstract
The yeast spindle pole body (SPB) is the functional equivalent of the mammalian centrosome. The half bridge is a SPB substructure on the nuclear envelope (NE), playing a key role in SPB duplication. Its cytoplasmic components are the membrane-anchored Kar1, the yeast centrin Cdc31, and the Cdc31-binding protein Sfi1. In G1, the half bridge expands into the bridge through Sfi1 C-terminal (Sfi1-CT) dimerization, the licensing step for SPB duplication. We exploited photo-activated localization microscopy (PALM) to show that Kar1 localizes in the bridge center. Binding assays revealed direct interaction between Kar1 and C-terminal Sfi1 fragments. kar1Δ cells whose viability was maintained by the dominant CDC31-16 showed an arched bridge, indicating Kar1’s function in tethering Sfi1 to the NE. Cdc31-16 enhanced Cdc31–Cdc31 interactions between Sfi1–Cdc31 layers, as suggested by binding free energy calculations. In our model, Kar1 binding is restricted to Sfi1-CT and Sfi1 C-terminal centrin-binding repeats, and centrin and Kar1 provide cross-links, while Sfi1-CT stabilizes the bridge and ensures timely SPB separation.
Introduction
Microtubule organizing centers (MTOCs), such as the mammalian centrosome (Bornens, 2012) and their yeast equivalent spindle pole body (SPB; Jaspersen and Winey, 2004), acquire their microtubule organizing activity by recruiting γ-tubulin complexes (Kollman et al., 2011). Both centrosomes and SPBs duplicate only once in the cell cycle and use the existing structure as the site for assembly of the daughter organelle (Nigg and Stearns, 2011).
The SPB of Saccharomyces cerevisiae consists of layered plaques and remains embedded in the nuclear envelope (NE) throughout the cell cycle. A specialized substructure called the half bridge is essential for SPB duplication. The half bridge is a one-sided extension of the central plaque that is layered on top of the cytoplasmic and nuclear sides of the NE (Byers and Goetsch, 1975). In early G1, the half bridge elongates into a bridge structure. A miniature version of the SPB called the satellite develops at the distal end of the bridge on the cytoplasmic side of the NE. After the start of the cell cycle, the satellite elongates into a duplication plaque that is subsequently inserted into the NE (Adams and Kilmartin, 2000).
Four proteins constitute the SPB half bridge/bridge and are all essential for SPB duplication. The membrane-anchored protein Kar1 is accompanied by Sfi1 on the cytoplasmic side of the half bridge/bridge (Rose and Fink, 1987; Spang et al., 1995). The yeast centrin Cdc31, a conserved Ca2+-binding protein similar to calmodulin, directly interacts with both Sfi1 and Kar1 (Spang et al., 1993; Biggins and Rose, 1994; Wiech et al., 1996; Kilmartin, 2003). The SUN domain protein Mps3 was suggested as the sole component of the nuclear half bridge side (Jaspersen et al., 2002, 2006).
Sfi1 is a long, α-helical protein that longitudinally spans the entire length of the half bridge (Kilmartin, 2003). It consists of an unstructured N-terminal region (Sfi1-NT), central Cdc31 binding sites, and a disordered C terminus (Sfi1-CT; Li et al., 2006). All Sfi1 molecules are aligned with the same orientation in the half bridge where the N terminus is embedded in the SPB’s central plaque and the C terminus marks the distal end of the half bridge. By C-tail–to–C-tail interaction of Sfi1 molecules, half bridge-into-bridge extension occurs (Kilmartin, 2003; Li et al., 2006; Elserafy et al., 2014). This arrangement exposes a raft of Sfi1 N termini, proposed to function as the satellite assembly platform (Adams and Kilmartin, 2000). In S phase, Sfi1-CT becomes phosphorylated by cyclin-dependent kinase 1 (Cdk1) to separate the bridge after SPB duplication and to restrict this event to once per cell cycle (Avena et al., 2014; Elserafy et al., 2014).
Besides its function in karyogamy where Kar1 recruits the γ-tubulin receptor Spc72 and the motor protein Kar3 to the bridge (Pereira et al., 1999; Gibeaux et al., 2013), Kar1 has an important role in SPB duplication (Rose and Fink, 1987). Region I around Kar1’s Cdc31 binding site is essential for SPB duplication, although the molecular role of this region is not understood (Vallen et al., 1992a; Spang et al., 1995). Interestingly, several single point mutations in CDC31 suppress Kar1’s function in SPB duplication by a mechanism currently not understood (Vallen et al., 1994).
Centrin binding to MTOCs is conserved. In yeast, Kar1 harbors a single Cdc31-binding site, whereas Sfi1 contains ∼20–21 binding sites in its center (Li et al., 2006). In higher eukaryotes, centrin forms complexes with multi-centrin binding proteins named hSfi1 and Poc5 in the lumen of centrioles (Kilmartin, 2003; Azimzadeh et al., 2009).
Here, we describe the interaction of Kar1 and Cdc31 with Sfi1, elucidate the mechanism for the bypassing of Kar1 by CDC31 suppressor mutants, and provide a comprehensive model for the role of Kar1 and Cdc31 in SPB duplication.
Results
Kar1 and Sfi1 are stably associated with the SPB throughout the cell cycle
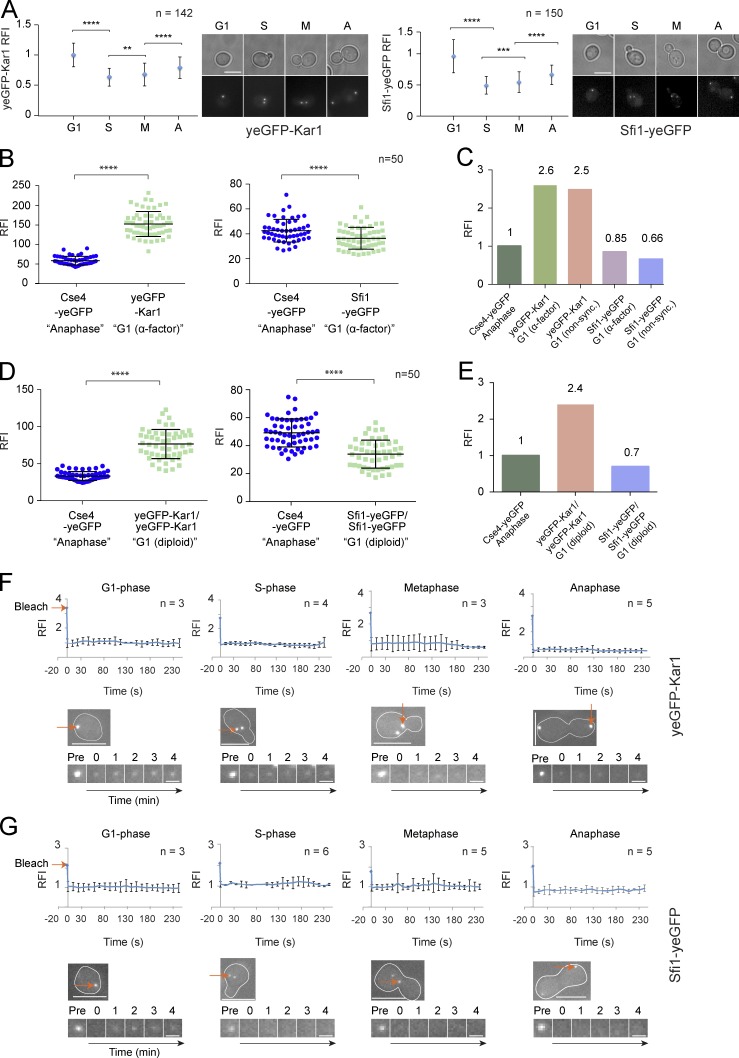
To understand how Kar1 and Sfi1 behave during the cell cycle, we generated tagged yeGFP-KAR1 and SFI1-yeGFP cells. yeGFP-Kar1 was chosen, as a tag at the C-terminal membrane anchor of Kar1 severely affects SPB duplication (Vallen et al., 1992a,b). Tagging of the proteins had no impact on cell growth, or on their recruitment to the SPB (Figs. 1 A and S1 A). Cdc31 was not included in this analysis, as N- and C-terminal fusions render the protein nonfunctional (Kilmartin, 2003).
Figure 1.
Kar1 and Sfi1 show coordinated behavior and are stably integrated into the half bridge and bridge during the cell cycle. (A) Kar1 and Sfi1 levels at the SPB. The RFI of yeGFP-Kar1 and Sfi1-yeGFP at SPBs was measured during G1 phase, S phase (S), metaphase (M), and anaphase (A). (B) RFI of yeGFP-Kar1 and Sfi1-yeGFP in α-factor–arrested cells. Anaphase CSE4-yeGFP cells were used as a fluorescence standard. (C) Quantification of yeGFP-Kar1 and Sfi1-yeGFP in α-factor–synchronized and unsynchronized cells. (D) Quantification of yeGFP-Kar1 and Sfi1-yeGFP in G1 unsynchronized diploid cells. (E) Quantification of D. (F and G) FRAP of SPB-associated yeGFP-Kar1 and Sfi1-yeGFP in G1, S, M, and A cells. Red arrows indicate the position of bleaching. n, number of analyzed SPBs. ****, P ≤ 0.0001; ***, P ≤ 0.001; **, P ≤ 0.01; ns, P > 0.05. Error bars indicate SD. Bars: (A, F, and G, top images) 5 µm; (F and G, bottom images) 1 µm.
The fluorescence intensity of a GFP signal is directly proportional to the number of GFP-tagged proteins (Wu and Pollard, 2005). We determined the relative fluorescence intensity of Sfi1-yeGFP and yeGFP-Kar1 in haploid strains and normalized it to the constant GFP signal of haploid CSE4-yeGFP cells in anaphase (Fig. 1, B and C; Erlemann et al., 2012). Polyploidization is a common phenotype that arises from half bridge gene manipulations. Using FACS analysis, we confirmed the haploidy of yeGFP-tagged strains (Fig. S1 B).
During the elongation of the half bridge, the fluorescence signal for Kar1 and Sfi1 reached maximal intensities before each signal declined by around half with bridge separation in S phase (Fig. 1 A). A slight increase in the SPB signals of each protein was observed at mitotic entry, possibly reflecting the lateral extension of the half bridge. This cycling behavior of Kar1 and Sfi1 points toward coordinated behavior of both proteins.
Kar1 was present in a threefold molar excess over Sfi1 at the SPB in α-factor–arrested G1 cells (Fig. 1, B and C). To exclude secondary effects of pheromone treatment on half bridge/bridge structure, the signals of both proteins were measured in unsynchronized, morphology-selected G1 cells. The values resembled the threefold excess of Kar1 over Sfi1 in synchronized cells (Fig. 1 C).
Because EM analysis of SPBs showed a significant size difference between the core SPB of diploid and haploid cells (Byers, 1981), we asked whether the size of the half bridge/bridge structure of diploid cells increased in a similarly coordinated manner. The yeGFP SPB signal of diploid cells was normalized to haploid CSE4-yeGFP cells (Fig. S1, B–D). The SPB signal intensities of yeGFP-KAR1/yeGFP-KAR1 and SFI1-yeGFP/SFI1-yeGFP cycling diploid G1 cells were similar to that of haploid cells (Fig. 1, C and E). This shows that the half bridge/bridge possess a similar architecture irrespective of the size of the core SPB.
We next tested whether the half bridge/bridge components are in constant exchange with soluble pools. To this end, yeGFP-Kar1 and Sfi1-yeGFP were analyzed using FRAP (Fig. 1, F and G). Neither protein displayed dynamic behavior at any cell cycle phase. To exclude the possibility that most of the Kar1 and Sfi1 pools resided at the SPB and were bleached in the FRAP experiment without the chance of recovery, we performed fluorescence loss in photobleaching (FLIP). As proof of principle, we analyzed the GFP signal of tagged polo-like kinase Cdc5 at SPBs. The SPB-associated Cdc5 signal diminished when constant laser pulses bleached the nucleus (Fig. S1 E). FLIP of SFI1-yeGFP cells did not affect the SPB-associated signal (Fig. S1 F). These data show that the half bridge/bridge of the SPB is a stable structure and that Kar1 and Sfi1 show coordinated behavior.
Kar1 localizes to the center of the bridge
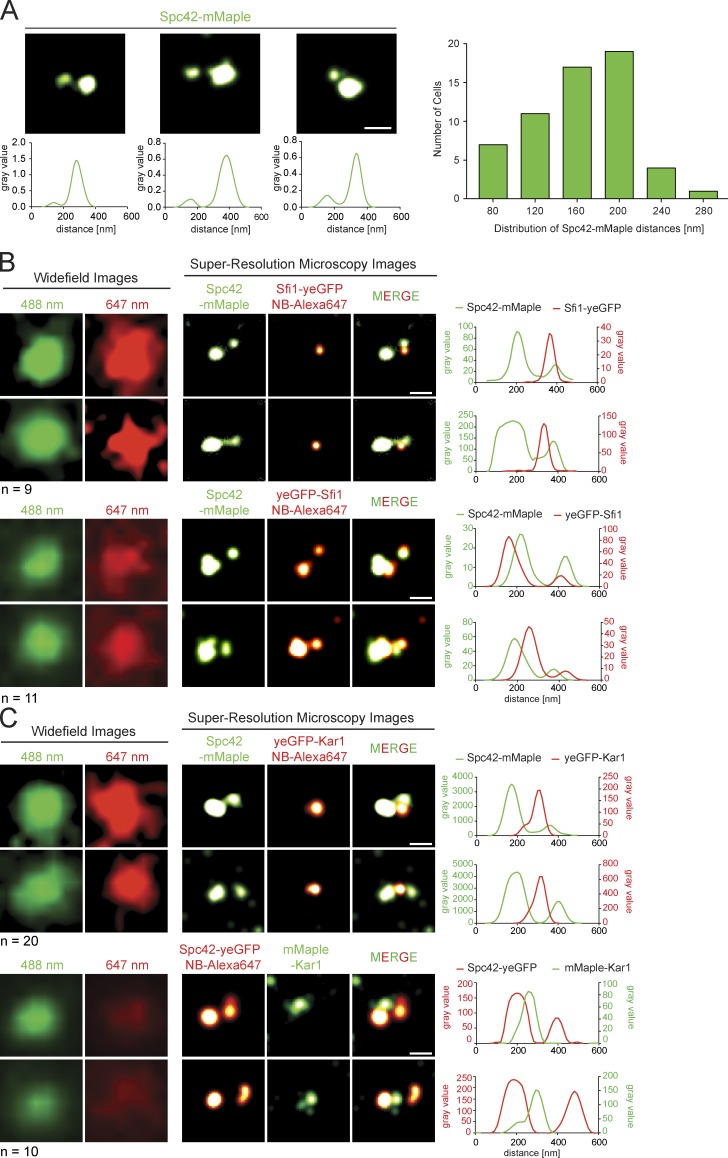
The SPB half bridge has a length of ∼60 nm, which is doubled upon bridge formation (Li et al., 2006). Therefore, determining the exact position of Kar1 along the bridge is difficult to achieve by conventional light microscopy. Here we used photo-activated localization microscopy (PALM) and direct stochastic optical reconstruction microscopy (dSTORM), both of which can reach a spatial resolution of 20 nm (Leung and Chou, 2011). As proof of principle, we analyzed α-factor cells that carried a satellite structure on their bridge (Fig. 2 A). Photo-convertible Spc42-mMaple enabled us to resolve the SPB central plaque and the smaller satellite (Donaldson and Kilmartin, 1996). From the 2D projection images, we calculated the distance between the two Spc42-mMaple signals to be 200 nm on average (Fig. 2 A), which is consistent with published bridge length measurements (Li et al., 2006). In α-factor–arrested cells, the SPB component Spc110 is only associated with the mature mother SPB but not with the satellite (Adams and Kilmartin, 1999). By using Spc110-mMaple or Spc110-yeGFP and Alexa Fluor 647–nanobody labeling, we confirmed that the weaker of the two Spc42 signals resembled the satellite (Fig. S2 A).
Figure 2.
Kar1 and Sfi1 localize to the center of the bridge. (A) PALM images of Spc42-mMaple in α-factor–arrested cells. The distribution of the signal distances is given. (B and C) Fine localization of Sfi1 and Kar1 at the SPB. Combined PALM and dSTORM dual-color images of indicated α-factor–arrested cells are shown. Corresponding line plot profiles are given. Owing to the 2D projection, the Sfi1-yeGFP signal in B was found in ∼60% of the cells slightly shifted from the bridge center. The yeGFP-Sfi1 signal at the satellite in B is probably lower compared with the signal at the mother SPB because of reduced nanobody accessibility. Data from a single representative experiment out of three repeats are shown. n, number of cells analyzed. Bars, 200 nm.
The bridge is composed of two stacks of parallel, similarly oriented Sfi1 proteins that overlap in an antiparallel arrangement at the center of the bridge (Kilmartin, 2003; Li et al., 2006). Consistent with immuno-EM data (Kilmartin, 2003; Li et al., 2006), the Sfi1-yeGFP signal was detected in between the SPB and the satellite while yeGFP-Sfi1 localized with the two Spc42-mMaple signals (Fig. 2 B). The lack of Sfi1-yeGFP resolution in the bridge indicates that the distance between overlapping Sfi1 C termini must be below the 20 nm resolution limit of our microscope system.
To study the localization of Kar1 within the half bridge/bridge, we combined either yeGFP- or mMaple-Kar1 with Spc42-mMaple or yeGFP, respectively. α-Factor–arrested cells displayed a single mMaple-Kar1 signal that resided between the two Spc42 dots (Fig. 2 C). This suggests that Kar1 localizes in the center of the bridge. Similar results were observed in yeGFP-KAR1 cells. It is noteworthy that in 40% of cells, the yeGFP-Kar1 signal was resolved into two adjacent dots that were positioned in between the Spc42 signals of the SPB and satellite (Fig. S2 B). This split yeGFP-Kar1 signal was restricted to the nanobody label and was not observed with mMaple-Kar1. We suggest that in some cells the nanobodies had limited access to yeGFP-Kar1 in the bridge center. In summary, both approaches detected Kar1 in the bridge center.
Specific binding of Kar1 to Sfi1-CT and neighboring Cdc31 binding sites
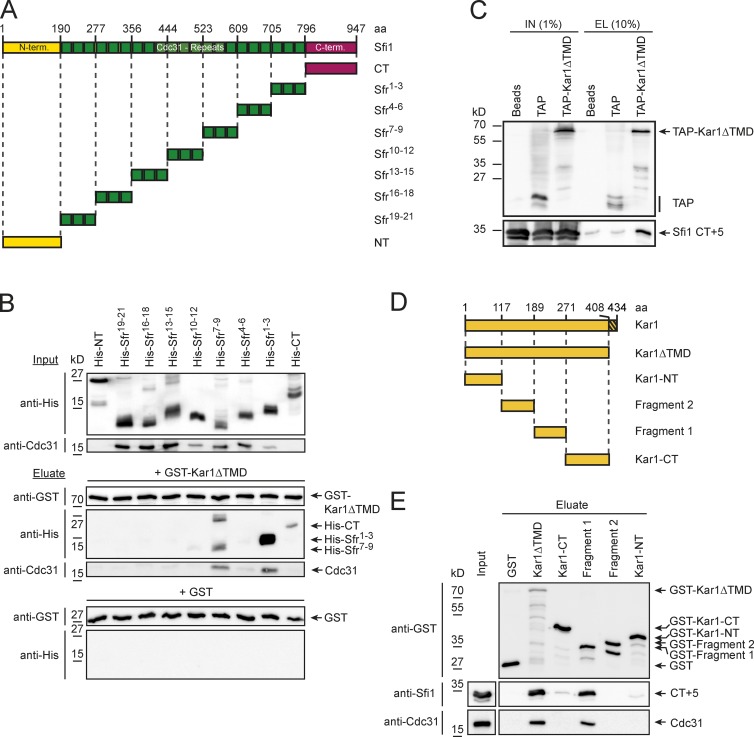
Our localization data raise the possibility that Kar1 directly interacts with the C-terminal region of Sfi1. To test this notion further, we constructed N- and C-terminal Sfi1 fragments (Sfrs) lacking Cdc31 binding sites (NT and CT) as well as several internal Sfrs plus the respective Cdc31 binding repeat number, counted from the C terminus (Fig. 3 A). Each of the Sfrs contained three predicted Cdc31 binding sites and were bacterially coexpressed with Cdc31 using dicistronic vectors (Kilmartin, 2003).
Figure 3.
Kar1 binds to the Sfi1 C-terminal region and Sfi1 binds to the Kar1 SPB duplication region. (A) Representation of Sfrs, tested for interaction with Kar1. (B) Kar1 binding to Sfrs from E. coli lysates. The fragments of A were expressed as N-terminal His tag fusions (top/input panel) and incubated with Sepharose-bound GST-Kar1ΔTMD (middle) or GST (bottom). Immunoblots with anti-His, anti-GST, and anti-Cdc31 are shown. (C) Kar1–Sfi1 interaction in S. cerevisiae cell lysates. A C-terminal construct of Sfi1 bearing the last five Cdc31 binding repeats (CT+5) was expressed alone or together with TAP-Kar1ΔTMD or TAP tag. After IgG Sepharose incubation, proteins were analyzed by immunoblotting with anti-TAP (top) or anti-Sfi1 (bottom) antibodies. (D) Cartoon of Kar1 fragments used in E. Hatched box, TMD domain. (E) Sfi1 binding to Kar1 fragments. GST-tagged Kar1 fragments from E. coli were incubated with Sfi1 CT+5 as in B. Immunoblots with indicated antibodies are shown. aa, amino acid; NT, N terminus; CT, C terminus; Sfr, Sfr.
We tested the interaction of each Sfr with Kar1 in vitro. Bacterially expressed GST or GST-Kar1ΔTMD (transmembrane domain) on Glutathione Sepharose was incubated with Escherichia coli extracts containing either His-tagged Sfi1-NT, -CT, or Sfrs-Cdc31. It is noteworthy that the expression of full-length Kar1 with the hydrophobic C-terminal TMD leads to protein aggregation (Spang et al., 1995). Analysis of the Kar1 beads revealed Kar1 interactions with Sfi1-CT and the C-terminally located Sfr1–3 and Sfr7–9 (Fig. 3 B). In contrast, Sfr4–6-Cdc31 and the more N-terminally located Sfrs did not associate with Kar1. These data support the view that Kar1 selectively binds to Sfi1-CT and its neighboring Cdc31 binding sites.
We next performed coimmunoprecipitation experiments from yeast cell lysates, in which tandem affinity purification (TAP)-tagged Kar1ΔTMD was co-overexpressed with a fragment containing Sfi1-CT and the last five centrin binding sites (CT+5) (Figs. 3 C and S3, A and B). Expression of kar1Δtmd was not toxic for cells because of the lack of the C-terminal membrane anchor (Fig. S3 A; Vallen et al., 1992b). Interaction between the C-terminal Sfr and the Kar1 protein confirmed the result of the in vitro capture data.
The half bridge extension model predicts self-interactions of Sfi1-CT (Li et al., 2006). Consequently a Sfi1 C-terminal fragment should have the ability to pull down another C-terminal fragment from cell lysates. To test this binding activity of C-terminal Sfrs, we bacterially expressed Sfrs, with a varying number of centrin binding repeats (Fig. S3 B). When these fragments were bound to Sepharose and incubated with bacterial cell lysates expressing an identical copy of these Sfrs but with a different tag, self-association was observed between C-terminal fragments (Sfi1 CT+1 and CT+5) and bare C termini. In contrast, no binding was detected between the C and the N terminus of Sfi1 (Fig. S3 C). Thus, the C-terminal Sfi1 region not only binds to Kar1 and Cdc31 but can also interact with itself. Note that our data do not discriminate between parallel or antiparallel interaction of fragments.
Genetic experiments have identified a region in KAR1 (region I; residues 191–246) that is essential for SPB duplication (Vallen et al., 1992a). We generated four Kar1 fragments to test their ability to interact with Sfi1 CT+5 (Fig. 3 D). Only Kar1ΔTMD and fragment 1 of Kar1, which resembles region I, were able to interact with Sfi1 CT+5 (Fig. 3 E). These experiments reveal an association of the C terminus of Sfi1 with the essential region I of Kar1.
The SPB localization of Kar1 is dependent on Sfi1
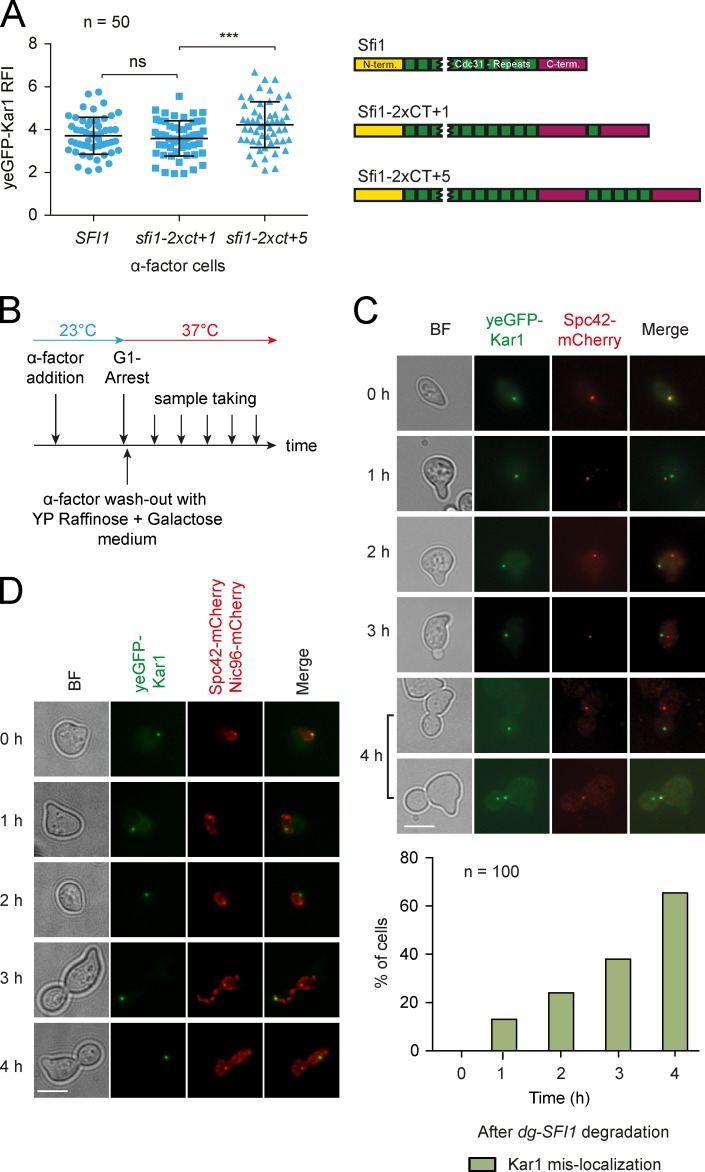
Next, we sought evidence for an interaction between Sfi1 and Kar1 in vivo. Since Kar1 binds to the Sfi1 C-terminal region in vitro, we asked whether we could detect an increase in Kar1 molecules when we artificially increased the size of the C-terminal region of Sfi1. Two SFI1 constructs were generated. The first doubled the size of the C terminus and included the last Cdc31 binding site (sfi1-2xct+1), whereas the second doubled the C terminus and the last five binding repeats (sfi1-2xct+5; Fig. 4 A). The gene fusions replaced the wild-type SFI1 through the use of a SFI1 shuffle strain. As both constructs led to artificial diploidization, the strains were compared with diploid SFI1 cells (Fig. S4 A). Only sfi1-2xct+5 cells, but not sfi1-2xct+1, showed a significant increase in yeGFP-Kar1 signal at SPBs (Fig. 4 A). Immunoblotting revealed similar expression levels of the two elongated Sfi1 versions (Fig. S4 B). This finding emphasized the importance of the C-terminal Cdc31 binding sites in Sfi1 for Kar1 binding.
Figure 4.
In vivo SPB localization of Kar1 depends on Sfi1. (A) Kar1 levels at the SPB in C-terminally elongated Sfi1 cells. The RFI of yeGFP-Kar1 at the SPBs was measured in the indicated α-factor–arrested cells. (B) Timeline of experiments performed in C and D. (C) Localization of yeGFP-Kar1 upon degradation of dg-SFI1. The percentage of cells showing Kar1 mislocalization at 37°C is provided. (D) As in C, but with Nic96-mCherry as NE marker. n, number of analyzed SPBs per time point. ***, P ≤ 0.001; ns, P > 0.05. Error bars indicate SD. Bars, 5 µm.
Next, we asked whether SPB localization of Kar1 requires Sfi1. The Sfi1 degron fusion (dg-SFI1) in Gal1-UBR1 cells was depleted to address the interdependency of the Kar1–Sfi1 interaction (Kanemaki et al., 2003; Elserafy et al., 2014; Fig. 4 B). Kar1 did not colocalize with the SPB in >60% of dg-SFI1 cells, which indicates that Sfi1 recruits Kar1 to the half bridge/bridge (Fig. 4 C). For some cells, the deficiency in Kar1 localization was not fully penetrant (Fig. 4 C, 4 h, lower panel), most probably due to the slow turnover rate of Sfi1 (Fig. 1 G). Remarkably, the SPB mislocalized Kar1 did not result in a diffuse signal but rather formed puncta on the NE, as indicated by the colocalization with the nuclear pore complex marker Nic96-mCherry (Fig. 4 D). Complementation of dg-SFI1 cells with an SFI1-encoding plasmid rescued Kar1 mislocalization (Fig. S4 C).
To evaluate the graded importance of the C-terminal Cdc31 repeats in Sfi1 for Kar1 binding, we performed SFI1 overexpression experiments (Fig. S4 D). All SFI1 constructs were coexpressed together with CDC31, as the overexpression of only Sfi1 CT+1 caused cell death by depletion of Cdc31 (Fig. S4 E). Overexpression of Sfi1 CT+5 mildly affected the growth of yeast cells. Longer C-terminal constructs, especially CT+7 and +8, severely compromised cell growth, and CT+9 completely abrogated it. These findings are consistent with the data from the in vitro capturing assay and emphasize the importance of Sfi1 C-terminal Cdc31 binding repeats for Kar1 binding and Sfi1 self-interaction (Fig. 3 B).
Bridge arching in kar1Δ CDC31-16 cells
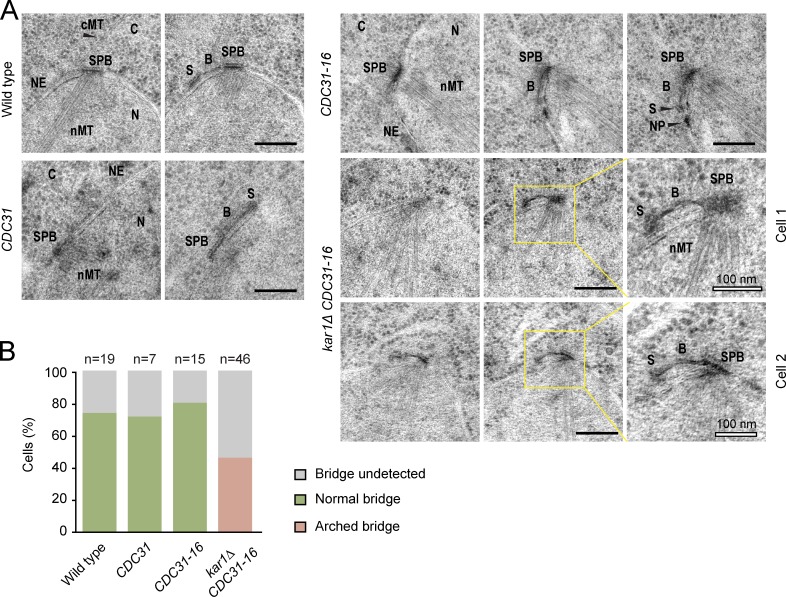
Several CDC31 mutations have been reported to bypass the essential need for KAR1 (Vallen et al., 1994). We analyzed the bridge morphology in kar1Δ CDC31-16 cells by EM to better understand Kar1 function in SPB duplication. Strikingly, the bridge was no longer running parallel along the top of the NE as was the case for wild-type, 2 µm-CDC31, and 2 µm-CDC31-16 cells (Figs. 5 A and S5 A). Instead, the bridge arched away from the NE yet still retained the connection between the mother SPB and the satellite. This arched bridge was observed in all kar1Δ CDC31-16 cells with a detectable bridge but never in control cells (Fig. 5 B). These data strongly support a function of Kar1 as an anchor that tethers the C-terminal region of Sfi1 to the NE.
Figure 5.
Deletion of Kar1 leads to an arched bridge. (A) Shown is the bridge morphology of ESM356 cells with the 2 µm-based plasmids pRS425 (wild type), pRS425-CDC31 (CDC31), and pRS425-CDC31-16 (CDC31-16) on top of endogenous CDC31 or of a KAR1 shuffle strain (kar1Δ CEN-URA3-KAR1) expressing pRS425-CDC31-16 (kar1Δ CDC31-16) after 5-FOA. Serial sections of α-factor–arrested cells are shown. NE, nuclear envelope; cMT, cytoplasmic microtubules; nMT, nuclear microtubules; C, cytoplasm; N, nucleus; B, bridge; S, satellite; NP, nucleopore. Panels on the right show enlarged views of the boxed regions. Bars, 200 nm (where another value is not given). (B) Quantification of A. n, number of cells analyzed.
Artificial cross-linking of Kar1 and Sfi1 in vivo bypasses Kar1’s essential region I
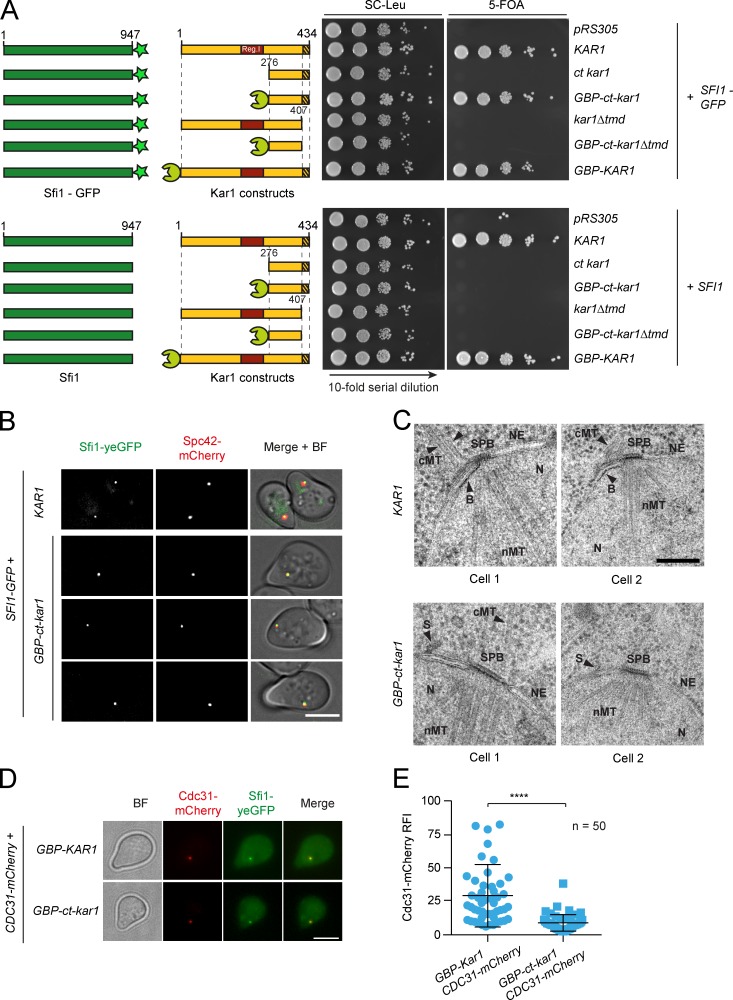
We considered the possibility of bypassing the essential function of Kar1’s region I by in vivo cross-linking of Sfi1-GFP to the TMD of Kar1 with the GFP-binding protein (GBP; Rothbauer et al., 2008). Such a bypass might be expected if this Kar1 region tethers Sfi1 to the NE. A series of constructs was generated in which different N-terminal deletions of Kar1 were fused to GBP (Fig. 6 A). In the most extreme case, residues 2–276, comprising essential region I (Vallen et al., 1992a), were substituted by GBP (GBP-ct-kar1). The function of the Kar1 truncations was tested in KAR1 shuffle strains expressing SFI1 or SFI1-yeGFP (Fig. 6 A). Strikingly, cells only survived the replacement of Kar1’s region I in the presence of SFI1-yeGFP. As expected, Sfi1-yeGFP still associated with the SPB in GBP-ct-kar1 cells (Fig. 6 B). Cells were inviable when Kar1’s TMD was deleted (Fig. 6 A). Immunoblotting confirmed the absence of wild-type Kar1 protein from the GBP-containing strains after passaging on 5-FOA–containing media (Fig. S5 B). We analyzed the temperature growth profile of the 5-FOA surviving strains (Fig. S5 C). Mild growth defects were only detected at 37°C. FACS analysis revealed diploidization of these mutant cells (Fig. S5 D). Thus, cross-linking of Sfi1-CT to a membrane anchor is sufficient to reconstitute Kar1’s essential function for SPB duplication and restores cell viability.
Figure 6.
Cross-linking of Kar1 and Sfi1 rescues the deletion of Kar1 SPB duplication region I. (A) Viability of kar1 mutants fused to the GBP. Indicated alleles were genomically integrated into KAR1 shuffle strains (kar1Δ CEN-URA3-KAR1 SFI1-yeGFP [top] or kar1Δ CEN-URA3-KAR1 SFI1 [bottom]) and tested for growth at 23°C on selection plates. Bright green star, GFP; red box, region I; hatched box, TMD; lime green sphere, GBP. (B) SPB localization of Sfi1-yeGFP was analyzed in GBP-ct-kar1 SFI1-yeGFP SPC42-mCherry cells. (C) SPB morphology of GBP-ct-kar1 SFI1-yeGFP cells. Representative electron micrographs of indicated cells are shown. NE, nuclear envelope; cMT, cytoplasmic microtubules; nMT, nuclear microtubules; N, nucleus; B, bridge; S, satellite. (D) Localization of CEN plasmid–expressed Cdc31-mCherry in GBP-ct-kar1 cells. (E) Quantification of D. n, number of cells analyzed. ****, P ≤ 0.0001. Error bars indicate SD. Bars: (B and D) 5 µm; (C) 200 nm.
We next analyzed the bridge morphology of GBP-ct-kar1 cells by EM. In wild-type KAR1 SFI1-yeGFP cells, the bridge extended parallel to the NE (Fig. 6 C). The cytoplasmic side of the bridge organizes cytoplasmic MTs since the Kar1 N terminus anchors the γ-tubulin complex receptor protein Spc72 to the bridge (Pereira et al., 1999). In GBP-ct-kar1 SFI1-yeGFP cells, the bridge was also appropriately aligned along the NE. However, the absence of the N terminus of Kar1 meant that no cytoplasmic MTs were organized from the bridge in these cells.
Previous work suggested that Kar1’s main function is to bind Cdc31 to the SPB (Biggins and Rose, 1994). To test this possibility, we made use of the GBP-ct-kar1 SFI1-yeGFP strain, in which we analyzed Cdc31 abundance at SPBs. Although not functional, Cdc31-mCherry has been used as reporter for Cdc31 localization (Fischer et al., 2004). Cdc31-mCherry localized to the SPB in GBP-KAR1 cells (Fig. 6 D). The replacement of GBP-KAR1 by GBP-ct-kar1, which lacks the Cdc31 binding region of Kar1, reduced Cdc31 binding to SPBs by a factor of 3 (Fig. 6 E). However, a portion of Cdc31 was still SPB-associated, probably through Sfi1 binding.
Collectively, these results demonstrate that the main function of Kar1 in SPB duplication is the anchoring of Sfi1’s C-terminal region onto the NE.
Sfi1-CT is important for SPB duplication and bridge stability
To understand the contributions of Sfi1-CT and the adjacent Cdc31 binding sites for Kar1 binding and bridge function, we analyzed the viability of C-terminal SFI1 truncation mutants. Viability tests using a shuffle approach showed that the C terminus and the last three Cdc31 binding repeats of Sfi1 (CT+3) were dispensable for growth at 23°C but not at 37°C (Fig. S5 E). Yeast strains expressing a truncated SFI1 version greater than ΔCT+3 were no longer viable. FACS analysis revealed diploidization in the viable SFI1 truncations (Fig. S5 F). Thus, surprisingly, the CT of Sfi1 is not essential for viability.
sfi1Δct cells may be viable because Kar1 also binds to the Cdc31 binding sites at the C terminus of Sfi1 (Fig. 3). Indeed, analysis of yeGFP-Kar1 localization in sfi1Δct cells revealed localization in the bridge center (Fig. S2 C). Attempts to localize yeGFP-Kar1 in sfi1Δ(ct+1) cells failed because of the inviability of such cells.
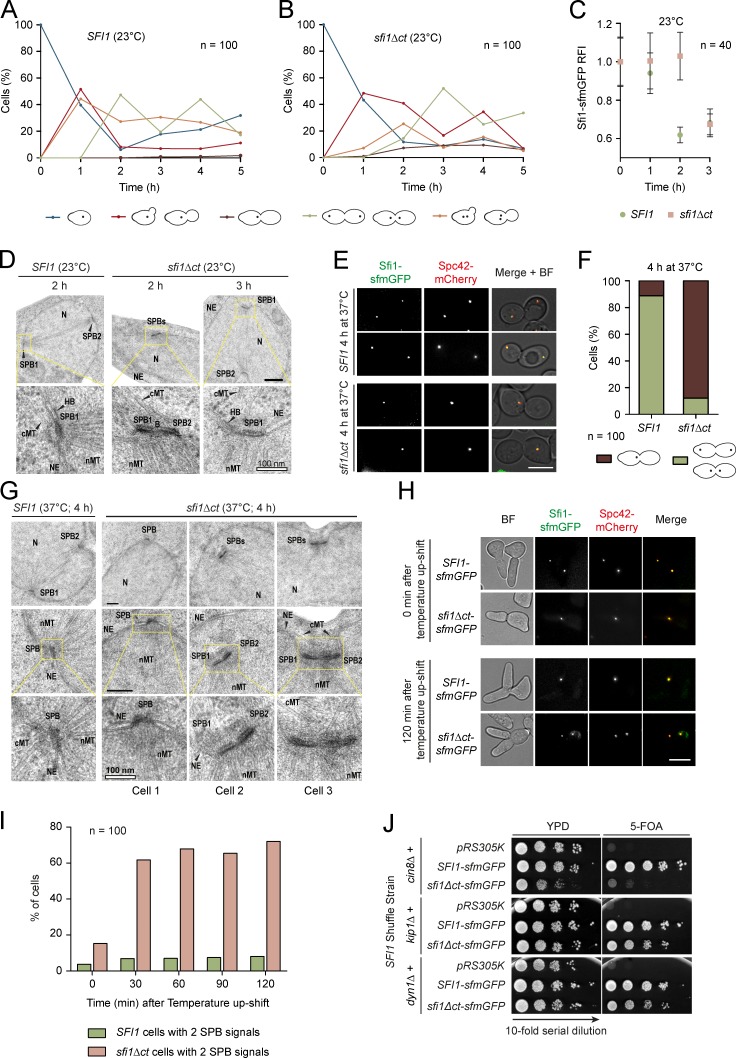
We next analyzed SPB duplication and bridge stability in sfi1Δct cells. At the permissive temperature, sfi1Δct cells required 2–3 h to separate the SPBs. In SFI1 cells, SPB separation happened 1 h faster (Fig. 7, A–C). EM analysis indicated 50% side-by-side SPBs in sfi1Δct and SFI1 cells 1 h after the release of the cell cycle block (n = 10 cells). After 2 h, all analyzed SFI1 cells had separated SPBs, whereas in 40% of sfi1Δct cells, SPBs were still side-by-side. After 3 h, most sfi1Δct cells had separated SPBs (Fig. 7 D). Thus, sfi1Δct cells at 23°C separate their SPBs with a delay.
Figure 7.
Sfi1 C terminus is important for bridge integrity. (A and B) The effect of sfi1Δct on cellular fitness at 23°C. The Sfi1-sfmGFP signal was followed in α-factor–synchronized cells. The different cell morphologies observed are indicated. (C) Quantification of the Sfi1-sfmGFP signal of the major morphologies observed at time points 0–3 h in A and B. (D) SPB morphology of sfi1Δct cells at 23°C. Representative EM pictures are shown. 10 cells were analyzed per time point. NE, nuclear envelope; cMT, cytoplasmic microtubules; nMT, nuclear microtubules; N, nucleus; B, bridge. (E) The effect of sfi1Δct on SPB duplication at 37°C. Synchronized cells were shifted to 37°C upon α-factor washout and cell fate was followed over 4 h. Two representative large-budded cells of the indicated strains are shown. (F) Quantification of metaphase cells from E. (G) SPB morphology of SFI1 and sfi1Δct cells from E. In D and G, panels boxed in yellow are enlarged below. (H) Importance of Sfi1-CT for bridge stability. SFI1 and sfi1Δct cells were arrested in G1/S by SIC1 overexpression at 23°C. Cells were shifted to 37°C and the SPB signal was monitored over time. (I) Quantification of H. (J) Synthetic lethality test between sfi1Δct and cin8Δ, kip1Δ, and dyn1Δ. Serial dilutions of cells were tested for growth at 23°C. n, number of cells analyzed per time point. Bars: (D and G) 200 nm, where not otherwise indicated; (E and H) 5 µm.
Shifting α-factor–synchronized sfi1Δct cells to 37°C led to an arrest of large-budded cells with one SPB signal (Fig. 7, E–F). EM analysis of these cells showed mostly two side-by-side SPBs, closely connected by an ill-defined structure (Fig. 7 G). The side-by-side SPBs of sfi1Δct cells were found in invaginations of the NE (Fig. S5 G). In ∼20% of the arrested cells, only one SPB was detected in thin serial sections of yeast nuclei (Fig. 7 G). Thus, sfi1Δct cells that started SPB duplication with a bridge and a satellite upon shifting cells to the restrictive temperature duplicated the SPB but then failed to separate the SPBs.
We next asked whether the Sfi1-CT is needed for bridge stability when cells first duplicated the SPBs at the permissive temperature and then were shifted to 37°C. We arrested SFI1 and sfi1Δct cells in G1/S by the overexpression of the Cdk1-Clb inhibitor SIC1. Cells arrested in this way carry two side-by-side SPBs that were still connected by the bridge (Haase et al., 2001). The side-by-side configuration of SFI1 cells was unaffected by the impact of shifting the cells to 37°C (Fig. 7, H and I). In contrast, sfi1Δct SIC1 cells shifted to 37°C showed two separated SPB signals, probably because of bridge severing between the two Sfi1 rafts that extend from each SPB, as indicated by the equal SPB distribution of Sfi1-sfmGFP (Fig. 7, H and I; and Fig. S5 H). Together, these data suggest that, depending on the growth condition, Sfi1-CT is important for either SPB separation or bridge stability but less important for Kar1 recruitment.
The missing phosphorylation platform raises the question as to how bridge severing can be promoted in sfi1Δct cells at the permissive temperature. The kinesin motor protein Cin8 has been implicated in bridge fission (Crasta et al., 2008). Interestingly, sfi1Δct genetically interacted with the deletion of CIN8 but not KIP1 or DYN1 (Fig. 7 J). Thus, in the absence of the regulatory Sfi1-CT platform, motor forces provided by Cin8 facilitate bridge severing and spindle assembly.
Cdc31 stabilizes Sfi1 layers at SPBs
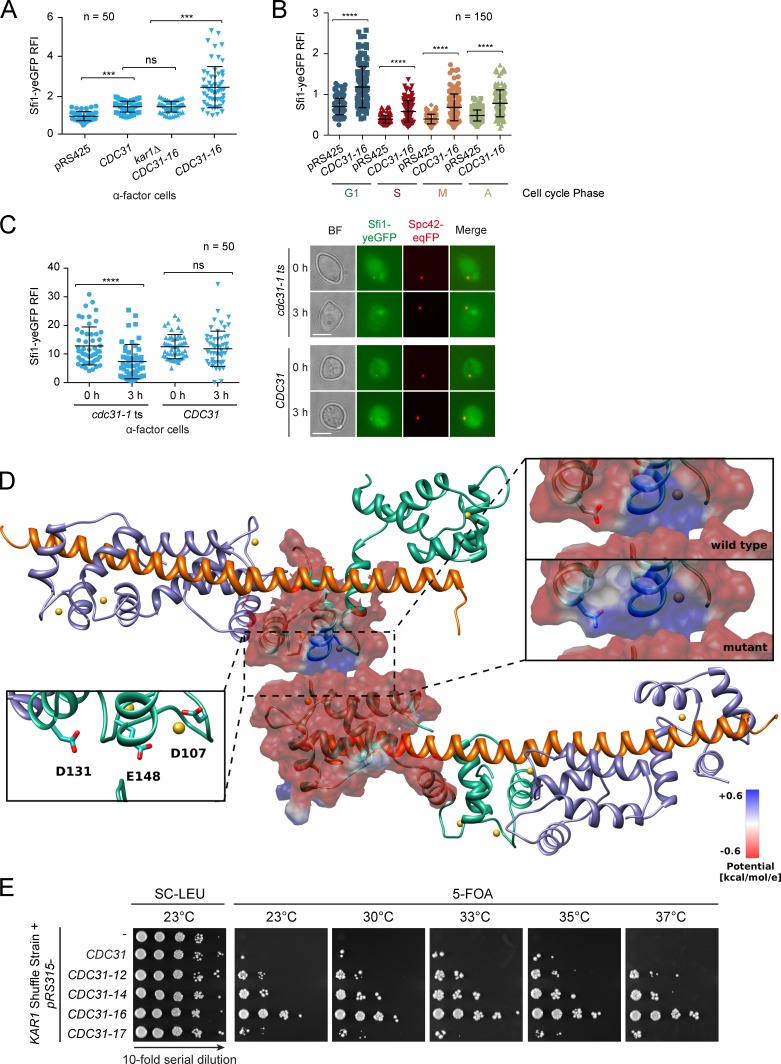
CDC31-16 is a suppressor of kar1Δ lethality, and in kar1Δ CDC31-16 cells, the bridge is able to withstand mechanical stress applied by bridge arching. This suggests an important role for Cdc31 in mediating interactions between Sfi1 layers of the half bridge/bridge. Biochemical analysis of Cdc31 function in the Sfi1–Cdc31 complex was complicated by the insolubility of Sfrs expressed without Cdc31. Therefore, we quantified the Sfi1 content in G1 arrested cells expressing different CDC31 variants (Fig. 8 A). When CDC31 or CDC31-16 were expressed alongside the endogenous CDC31, the Sfi1-yeGFP signal at the SPB increased. However, the intensity of the Sfi1-yeGFP signal in kar1Δ CDC31-16 cells was lower than that of cells expressing CDC31-16, indicating that Kar1 and Cdc31 may have overlapping functions in recruiting Sfi1 to the SPB. The Sfi1 signal also increased to the same extent in all cell cycle phases when CDC31-16 was expressed (Fig. 8 B). Kar1 levels were unaffected by CDC31 or CDC31-16 expression (Fig. S5 I). All yeast strains were haploid according to FACS analysis (Fig. S5, J–K). In short, both Cdc31 and Cdc31-16 recruit additional Sfi1 to the SPB, yet the Cdc31-16 mutant is more potent than its wild-type counterpart.
Figure 8.
Dominant mutations in CDC31 lead to an increased Sfi1 recruitment to the bridge by favoring Cdc31–Cdc31 contacts. (A and B) Sfi1 recruitment to the SPB in different genetic backgrounds (see Fig. 5 A). The RFI at the SPB was measured in α-factor–arrested (A) or unsynchronized cells (B) at 23°C. (C) Sfi1 SPB localization in cdc31-1 cells. Cells were shifted to 37°C for 3 h. n, number of analyzed SPBs per time point. ****, P ≤ 0.0001; ***, P ≤ 0.001; ns, P > 0.05. Error bars indicate SD. Bar, 5 µm. (D) Model of the dimeric structure of two Sfi1–Cdc31 complexes. The interface between the interacting Cdc31 molecules is given by the crystal contacts in PDB accession no. 2GV5 (Li et al., 2006). The proteins are shown with an illustrated representation (Sfi1, orange; Cdc31, green and lilac); the calcium ions are shown in yellow. The residues, mutated in the suppressing mutants, are shown as sticks (left inset). The electrostatic potential of CDC31 (top right inset) and CDC31-16 (bottom right inset) is shown mapped onto the molecular surface. (E) The KAR1 shuffle strain bearing a CEN-based plasmid encoding for one of the indicated CDC31 variants was tested for growth.
Our notion of Cdc31 as a half bridge/bridge stabilizer was further supported by the analysis of α-factor–arrested cells that harbored the conditional lethal cdc31-1 allele. After 3 h of shifting cells to restrictive temperature, the signal of Sfi1 at the SPB decreased significantly in comparison to CDC31 cells (Fig. 8 C). Thus, Cdc31 is required to stabilize Sfi1 at the half bridge/bridge.
Modeling of Sfi1–Cdc31 complexes and computation of the binding free energy suggest that the KAR1-suppressing CDC31 mutations enhance inter-Sfi1 strand interactions
We analyzed the interaction interfaces of a Sfi1–Cdc31 complex in a crystal structure (PDB accession no. 2GV5; Li et al., 2006) and investigated the positions of the mutated residues D131, E148, and D107 in the Cdc31-12, -14, -16, and -17 mutants, which suppress the need for Kar1. The critical residues localized on the same side of one Cdc31 molecule in the vicinity of a crystal contact between two Cdc31 molecules attached to antiparallel Sfi1 helices (Fig. 8 D). This crystal structure was used as a model for computing the binding free energies between two Sfi1–Cdc31 complexes interacting via their centrins. The energies computed with the Amber99 force field were −9.6 kcal/mol for Cdc31 and −11.1, −12.0, −12.1, and −9.9 kcal/mol for Cdc31-12, 14, -16, and -17, respectively (Table S1). Thus, all of these suppressor mutants favor Sfi1–Cdc31–Cdc31–Sfi1 complex formation more than wild-type Cdc31 does. The relative binding constants of the Cdc31-12, -14, -16, and -17 mutant complexes were 11.9-, 56.3-, 64.4-, and 1.5-fold that of wild-type Cdc31. Calculations performed with two other force fields gave a similar ranking of the stability of the mutant complexes, with the exception of a prediction that Cdc31-17 binding may be less energetically favorable than wild type (Table S1). The differences in binding free energy arise primarily from differences in the electrostatic component. The electrostatic potentials of Cdc31 and Cdc31-16 clearly demonstrate how the electrostatic complementarity is improved by the KAR1-suppressing CDC31 mutations (Fig. 8 D). Remarkably, we found that the ability of the dominant CDC31 mutants to suppress kar1Δ lethality follows the free energy calculations (Fig. 8 E). Together, these data support the contribution of Cdc31 to half bridge/bridge stability by cross-linking Sfi1 molecules.
Elevated Kar1 levels lead to an extraordinary elongated bridge
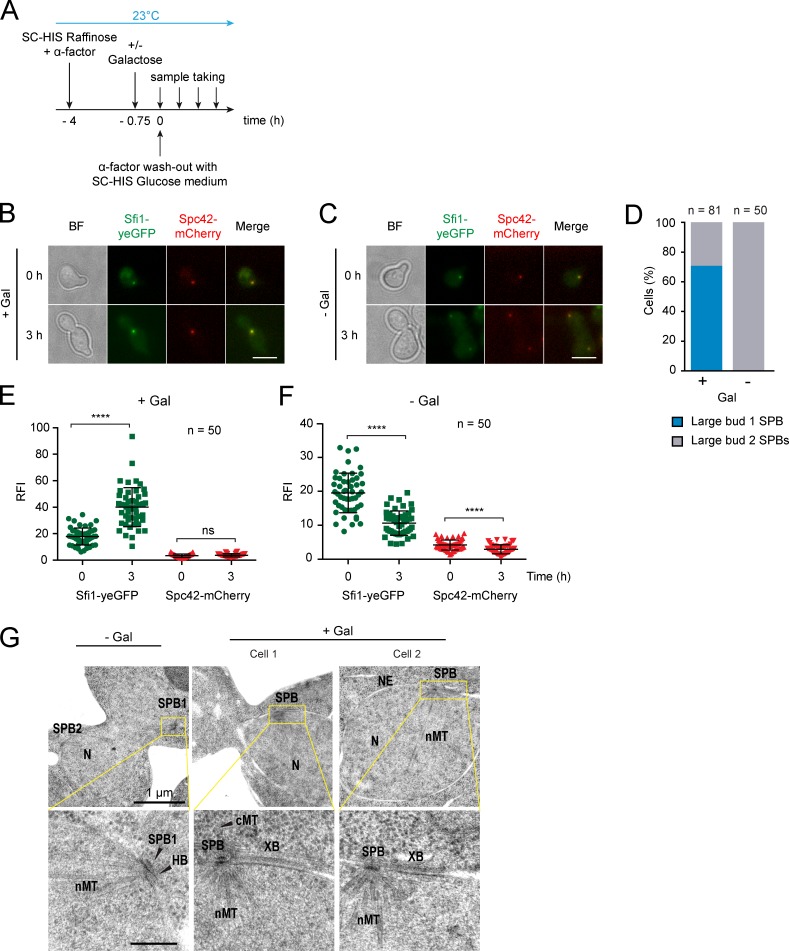
Our data suggest that Cdc31 and Kar1 cooperate in stabilizing the bridge structure. Early studies on KAR1 revealed its toxic character when overexpressed (Rose and Fink, 1987; Vallen et al., 1994). Considering our findings on Kar1 being an anchor for the SPB half bridge/bridge and possibly also a Sfi1 cross-linker, we assumed that Kar1 overexpression would lead to stabilization of the Sfi1 layers in the bridge. To test this, a pulse of KAR1 expression was set and cell fate was followed upon α-factor washout (Fig. 9 A). 70% of cells arrested with a large bud and only one SPB signal (Fig. 9, B and D). Quantification of Kar1, Sfi1, and Spc42 contents at these evoked SPBs indicated a strong increase in Kar1 and Sfi1 without affecting the Spc42 signal (Figs. S5 L and 9 E). Without Gal1-KAR1 overexpression, the SPB duplicated and subsequently separated (Fig. 9, C and F).
Figure 9.
Elevated Kar1 dosage leads to an extraordinarily elongated bridge. (A) Timeline of performed experiments. (B and C) The effect of KAR1 overexpression on SPB separation. Sfi1-yeGFP and Spc42-mCherry signals were monitored. (D) Quantification of SPB number of large-budded cells from B and C at 3 h. (E and F) Quantification of the Sfi1-yeGFP and Spc42-mCherry signals at the SPB of cells from B and C. Note: at 0 h, the SPB signal consists of the mother SPB and the satellite. In cases where two SPB signals were visible at 3 h (F), each SPB was quantified separately. ****, P ≤ 0.0001; ns, P > 0.05. Error bars indicate SD. (G) EM images of cells from B and C. NE, nuclear envelope; cMT, cytoplasmic microtubules; nMT, nuclear microtubules; N, nucleus; XB, extraordinary elongated bridge. The boxed regions are enlarged in the panels below. n, number of cells analyzed. Bars: (B and C) 5 µm; (G) 200 nm unless otherwise indicated.
EM analysis of cells with elevated Kar1 numbers depicted an extraordinary elongated bridge (XB), with no satellite attached to its distal end (Fig. 9 G). The length of this XB was 221 ± 55 nm (n = 10), approximately twofold longer than the wild-type bridge (Li et al., 2006). Thus, elevated Kar1 leads to an extraordinarily elongated bridge, which cannot organize a satellite and therefore fails in SPB duplication.
Discussion
Kar1 interacts with the C-terminal half of Sfi1 to tether the bridge to the NE
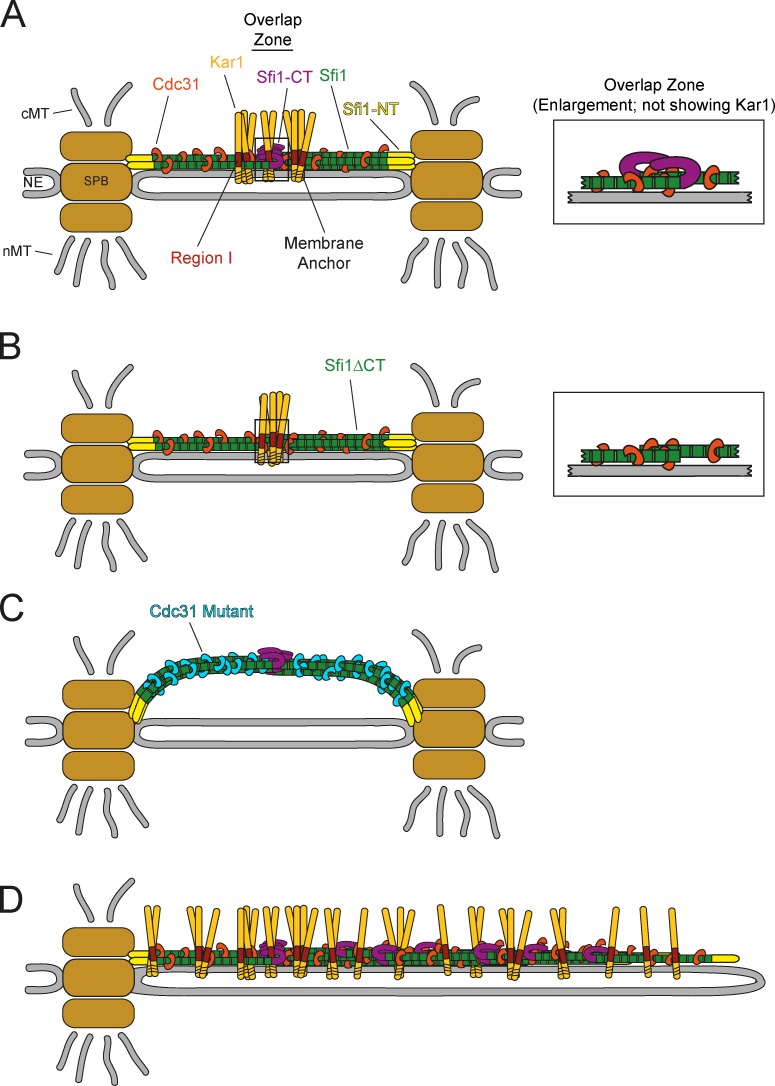
It has been shown that Kar1 associates with the SPB bridge and that it is tethered to the NE via its hydrophobic C terminus (Vallen et al., 1992a; Spang et al., 1995). Here, we define the position of Kar1 in the central region of the bridge between the SPB and its satellite by super-resolution microscopy. This localization is consistent with the direct interaction of Kar1 with the C terminus and the adjacent Cdc31 binding sites of Sfi1 (illustrated in Fig. 10 A). Further, we show a dependency of the SPB localization of Kar1 on Sfi1.
Figure 10.
Model for the SPB bridge. (A) Model of paired SPBs showing the bridge components in a wild-type situation. (B) The bridge of sfi1Δct cells. Note that Kar1 was omitted from the enlargements. (C) The arched bridge phenotype observed in kar1Δ CDC31-16 cells. (D) Model for the extraordinary elongated bridge structure of Gal1-KAR1 cells.
In sfi1Δct cells, Kar1 is still localized to the bridge center (Fig. 10 B). Thus, the interaction of Kar1 with the C-terminal Cdc31-binding repeats of Sfi1 is sufficient for correct Kar1 localization. In contrast, the localization and interaction data do not support the notion that Kar1 can interact with Sfi1-NT or the Sfi1 N-terminal Cdc31 binding repeats. How does Kar1 discriminate between the different Cdc31 binding regions in Sfi1? Crystal structures of yeast Sfrs suggest that the center of Sfi1 exists as a continuous, long-stretched α-helix, bearing ∼20 binding repeats for Cdc31 (Li et al., 2006). This predicts that the central region of Sfi1 is continuously decorated with Cdc31 molecules. Pull-down experiments, however, showed that not all of the Cdc31 binding sites are occupied (Kilmartin, 2003). Moreover, recent work on fission yeast Sfi1 demonstrated that, in fact, not all of the Cdc31 repeats are equal, and that they show differences in function and importance (Lee et al., 2014). As one possibility, the Cdc31 density along Sfi1 may determine the interaction withKar1. Alternatively, small changes in the structure of the Sfi1–Cdc31 binding regions may determine the preference for Kar1 binding. Further structural studies are required to understand the specificity of Kar1 binding to the C-terminal Cdc31 binding sites of Sfi1.
The function of Kar1 is to align Sfi1 filaments along the outer face of the NE. This conclusion is based on the arched bridge phenotype of kar1Δ CDC31-16 cells (Fig. 10 C). It is further consistent with the suppression of the essential Kar1 region I that mediates Sfi1 binding by in vivo cross-linking of Sfi1-yeGFP with GBP-ct-kar1. SFI1-yeGFP GBP-ct-kar1 cells show a remarkably normal SPB bridge morphology and therefore prove that the main function of Kar1 is to be the tether of Sfi1 to the NE.
Sfi1-CT and C-terminal Cdc31-binding regions form the antiparallel Sfi1 overlap in the bridge
A reasonable model is that the Sfi1 C termini are interlocked like the fingers of a newly married couple (Fig. 10 A). Importantly, a small number of Cdc31-binding sites are most likely part of the antiparallel Sfi1 overlap. Binding of Kar1 to the Sfi1-CT and the adjacent Sfi1 centrin binding sites probably promotes the Sfi1-C-C-Sfi1 bridge interactions. This model is in agreement with the finding that deletion of the Sfi1-CT does not lead to the collapse of the bridge at 23°C and that Kar1 still localizes in the bridge center in sfi1Δct cells (Fig. 10 B).
What is the function of the C terminus of Sfi1? If the C terminus of Sfi1 is present, phosphorylation of Sfi1-CT by Cdk1, as indicated by the SPB separation defect of the phospho-inhibitory sfi1-6A allele, is essential for SPB separation and cell viability (Avena et al., 2014; Elserafy et al., 2014). Here we show that at 23°C the bridge of sfi1Δct cells is robust enough to ensure SPB duplication and separation. However, because of the lack of the C-terminal Cdk1 regulation platform, SPB separation was delayed in sfi1Δct cells. At the restrictive temperature, most sfi1Δct cells duplicate the SPB. However, the side-by-side SPBs did not separate. The precise SPB defect of sfi1Δct cells remains unclear. Similar bilobed SPBs have been reported for spc29(ts) or spc42(ts) conditional lethal mutant cells (Donaldson and Kilmartin, 1996; Elliott et al., 1999).
Model for suppression of KAR1 deletion by dominant CDC31 alleles
Mutations in CDC31 that could either suppress the temperature-sensitive growth defect of kar1(ts) or even the complete deletion of KAR1 were previously identified (Vallen et al., 1994). Here, we propose a simple model to explain how suppressor mutations in the yeast centrin can bypass the requirement for KAR1. The kar1Δ suppressors CDC31-12, -14, -16, and -17 lead to higher affinity binding between Sfi1–Cdc31 filaments. This is the case for antiparallel Sfi1 interactions in the bridge overlap, as indicated by the binding free energy calculations but probably also for parallel Sfi1 interactions, as a high gene dosage of CDC31-16 recruited additional Sfi1 to SPBs during all cell cycle phases. Although the gain in binding free energy upon mutation is small for one binding site, the stabilizing impact of each mutation would be additive due to the alignment of several Sfi1 molecules in the half bridge or bridge. This generates a strong, cohesive, “mass action” association (Li et al., 2006). We therefore propose that the suppressing activity of Cdc31-16 arises from an increase in lateral Sfi1–Cdc31 interactions, through which the overlap region in the center of bridge becomes more rigid, and so is able to bypass the original task of Kar1, the interconnection of the single Sfi1 filaments. Although they rescue viability, the dominant CDC31-16 suppressor mutation cannot tether the stabilized Sfi1 filaments to the membrane (Fig. 10 C). Data from fission yeast and green algae further support a cross-linking function of centrin. Sfi1–Cdc31 filaments found at the SPB of Schizosaccharomyces pombe or in the basal body contractile fibers in Chlamydomonas reinhardtii are stabilized by interconnecting centrin–centrin interactions (Martinez-Sanz et al., 2006; Bouhlel et al., 2015).
When we increased Kar1 concentration, SPB duplication was blocked and cells arrested in the cell cycle with a large bud (Vallen et al., 1994). Elevated Kar1 at SPBs led to an extraordinarily elongated bridge without a satellite. We suggest that supernumerary Kar1 binds to the bridge via Sfi1. This Kar1 then recruits an excess of Sfi1 in a random fashion accompanied by bridge elongation (Fig. 10 D). In our model, the absence of a coherent Sfi1 N-terminal border at the distal end of the bridge interferes with satellite binding to the bridge and SPB duplication.
In vertebrate cells the role of the different centrins and their function in centriole duplication is a matter of debate (Middendorp et al., 1997; Laoukili et al., 2000; Salisbury et al., 2002; Strnad et al., 2007; Dantas et al., 2011). Recent findings now indicated centrin2 as a regulator of primary cilia formation in human retinal pigmented epithelial cells by controlling CP110 levels at basal bodies (Prosser and Morrison, 2015).
Besides Sfi1, whose well-conserved human homologue associates with centrioles, the centriolar proteins hPoc5 and CP110 also bind to centrin (Kilmartin, 2003; Tsang et al., 2006; Azimzadeh et al., 2009). Studies have revealed the importance of hPoc5 and CP110 for correct centriole maturation (Chen et al., 2002; Azimzadeh et al., 2009; Schmidt et al., 2009), but detailed in vivo studies on Sfi1’s function in human cells have yet to be performed. Our findings on the organization of the yeast bridge by centrin and Sfi1 provide an important paradigm to inform models for the interrogation of hSfi1/hPoc5-centrin function in human centrioles.
Materials and methods
Yeast strains, DNA manipulations, and culture conditions
Yeast cells were grown in SC (synthetic complete) medium, SC-selection, or YPRaf (yeast extract, peptone, and raffinose) at 23°C. 20% galactose was added to cells in YPRaf medium at a dilution of 1:10 to induce expression of genes under a pGAL1 promoter. To arrest cells in G1, α-factor (final concentration 10 µg/ml) was added to the cell culture. To release cells from the arrest, cells were washed twice with α-factor–free medium. Alkaline lysis and TCA precipitation were used to prepare yeast extracts (Janke et al., 2004). The QuikChange method (Agilent Technologies) was used to perform site-directed mutagenesis. The various KAR1 constructs used in Fig. 6 were inserted into the genome using the pRS305 vector. The single integration vector (pRS305K, LEU2-based integration vector, with a KanMX4 selection marker) was used to genomically integrate different alleles of SFI1 (Taxis and Knop, 2006). Single integration was confirmed by colony PCR. Endogenous gene tagging and deletion was done using cassette PCR-based methods. In short, the sequence of fluorescent proteins or markers was PCR amplified from a plasmid, using appropriate primer pairs, bearing homology sequences for the amplicon as well as the genomic integration locus (Knop et al., 1999; Janke et al., 2004). To test cellular fitness, yeast cells were grown in the indicated selection medium at 23°C overnight before cell density was adjusted to OD600 = 1 using the same medium. Starting from this suspension, 10-fold serial dilutions were spotted on the desired plates and incubated at different temperatures as indicated. All yeast strains and plasmids used in the study are listed in Tables S2 and S3.
For immuno-affinity isolation, Kar1 was PCR amplified, deleting its C-terminal TMD, and fused to the TAP tag (Rigaut et al., 1999). Subsequently, the Kar1 fusion construct was transferred into a uracil-based, galactose-inducible yeast expression plasmid (pCS180-1, p426-pGal1-TAP-kar1Δtm). It was cotransformed in ESM356 together with Sfi1’s C terminus and the last five C-terminal Cdc31 binding repeats (pCS177-1, p423-pGal1-sfi-ct+5). The genes within these plasmids were expressed by galactose induction for 8 h at 30°C. After harvesting, the cell pellet was lysed in TAP-A buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1× protease inhibitor cocktail [Roche], and 1 mM PMSF) and coupled to IgG Sepharose 6 Fast Flow beads (GE Healthcare). After incubation for 1.5 h, the beads were washed with TAP-A buffer and finally eluted with 0.5 M ammonium hydroxide. The Kar1 and Sfi1 interaction was tested by Western blot analysis. For TAP-Kar1ΔTMD detection, an anti-TAP primary antibody (from rabbit; Open Biosystems) was used. To detect untagged Sfi1, we generated an antibody in rabbits, directed against the C terminus (residues 796–947) of the protein.
EM
For EM analysis, cells were high pressure frozen, freeze-substituted, sectioned, and stained as described previously (Giddings et al., 2001). First, vacuum filtration was performed to collect cells onto a 0.45 µm polycarbonate filter (EMD Millipore). Cells were transferred and cryo-immobilized by using the EM PACT2 (Leica), a high-pressure freezing machine operating at a pressure of ∼2,045 bar. For freeze-substitution, an EM-AFS2 device (Leica) together with a freeze-substitution solution of 0.1% glutaraldehyde, 0.2% uranyl acetate, and 1% water dissolved in anhydrous acetone was used, followed by stepwise infiltration in Lowicryl HM20 (Polysciences, Inc.), starting at a low temperature of −90°C. The samples were then exposed to UV light at −45°C for 48 h and gradually warmed up to 20°C to allow polymerization. Serial sectioning of embedded cells, to generate 60–70-nm-thick sections, was done using a Reichert Ultracut S Microtome (Leica). Sections were poststained with uranyl acetate and lead citrate. Finally, images were captured with an electron microscope (CM120; Philips Electronics) operated at 120 kV.
Fluorescence light microscopy
A DeltaVision RT system (Olympus IX71 based; Applied Precision) equipped with a Photometrics CoolSnap HQ camera (Roper Scientific), a 100×/1.4 NA UPlan-SApochromat objective lens (Olympus), a mercury arc light source, and softWoRx software (Applied Precision) was used for cell imaging and fluorescence signals recording. Imaging was done at 23°C, using the GFP or the mCherry channels with different exposure times according to the fluorescence intensity of each protein. For quantification of Sfi1-yeGFP, yeGFP-Kar1, and Cse4-yeGFP signals in different strains, cells were analyzed without fixation. yeGFP-KAR1 or SFI1-yeGFP cells were arrested in α-factor and then mixed with unsynchronized CSE4-yeGFP SPC42-mCherry cells. The additional mCherry signal in the CSE4-yeGFP SPC42-mCherry cells allowed the differentiation between the cell types. The relative fluorescence intensity (RFI) of the fluorescence Cse4-yeGFP standard was set to 1 in cases when different yeast strains were imaged independently to allow comparison of the RFI of Sfi1-yeGFP or yeGFP-Kar1 from these cells. Each quantification experiment was repeated at least three times and a representative example was shown. Image processing used the ImageJ software package (National Institutes of Health).
FRAP
Yeast strains were grown to log phase in sterile-filtered SC medium at 23°C. Cells in the respective cell cycle phases were selected according to their morphology: G1 (one SPB, no bud), S (small bud, two separated, nearby SPBs), M (metaphase; large bud, two separated SPBs), and A (anaphase; large bud, one SPB in the mother cell and one in the bud). For imaging, cells were immobilized onto glass-bottom dishes. Dishes were incubated with 100 µl Concanavalin A solution (6% Concanavalin A, 100 mM Tris-Cl, pH 7.0, and 100 mM MnCl2) for 5 min and subsequently washed with 300 µl of distilled water. Yeast cells were attached to the dish for 5–15 min at 30°C and subsequently washed and overlaid with prewarmed medium. Fluorescent images were acquired with the DeltaVision RT system. The first five prebleach images were acquired before the target SPB (region of interest [ROI]) was bleached with a 50-ms laser pulse with 50% laser power. 27 post-bleach images were then captured over a total experiment duration of 4 min. To obtain a normalized FRAP curve and correct for acquisition bleaching, the double normalization method from Phair et al. (2004) was performed. The 50 mW, 488 nm laser system (DeltaVision QLM; Applied Precision) was used for photobleaching experiments. Data analysis was performed with the ImageJ software package and values obtained from measurements were further analyzed with Microsoft Excel.
FLIP
Before bleaching, one image was acquired with a 0.6 s exposure in the GFP channel, and a reference image was taken in the bright field channel (0.05 s exposure). Then an area outside the SPB was continuously bleached with 20-ms laser pulses with 50% laser power, and images were acquired before and after bleaching every 1 s with a 0.5-s exposure time in the GFP channel. Fluorescence intensities of a 5 × 5 pixel ROI of the SPB were measured using the ImageJ software package. A region outside the cell and a SPB signal of another cell were used for background subtraction from the ROI.
FACS analysis
To measure the DNA content of the cells, FACS analysis was performed. Yeast cultures were grown at 23°C to log phase. From this culture, 1 ml was harvested. The pellet was resuspended in 1 ml 70% cold ethanol and incubated for 2 h at 4°C. The cells were centrifuged at 14,000 rpm for 2 min and washed with 1 ml sodium citrate buffer, pH 7.4. Subsequently, the buffer was removed and the cells were incubated at 37°C overnight in 1 ml 0.25 mg/ml RNase A. The cells were pelleted and resuspended in 500 µl of 5 mg/ml pepsin and incubated at 37°C for 2 h. Cells were transferred into sodium citrate buffer of pH 7.4 and stained with propidium iodide. FACS analysis was done using a benchtop analyzer (FACSCanto II; BD). 10,000 events were read per sample and files were processed using FACSDiva software.
Super-resolution microscopy
Yeast cells were prepared for microscopy in a similar way to an earlier described protocol (Ries et al., 2012). In short, cells were grown in SC medium at 23°C and arrested in α-factor. Cell suspension was placed in Concanavalin A (Sigma-Aldrich)-coated Nunc Lab-Tek #1 (Thermo Scientific) to allow the cells to settle. Subsequently, cells were fixed with 4% PFA, treated with 0.15% Triton X-100, 5% BSA in PBS, and stained with Alexa Fluor 647 custom-labeled GFP-Traps (ChromoTek). Super-resolution microscopy was performed as described previously (Heilemann et al., 2008; Köthe et al., 2014) on a customized microscope setup. In brief, 405 nm, 561 nm, and 647 nm diode lasers (Cube; Coherent) were coupled into the cellTIRF of an inverted microscope (IX81; Olympus) equipped with a 150× oil immersion objective lens (UApo; Olympus). Excitation and fluorescence emission were separated using appropriate filters (AHF), and single-molecule fluorescence was recorded with an EM-CCD camera (Ixon; Andor). Combined PALM and dSTORM imaging was realized by imaging sequentially, starting with Alexa Fluor 647, in 50 mM MEA supplemented oxygen-free PBS based buffer. Typically, 6,000 frames were recorded with an integration time of 20 Hz, for both channels. SimpleSTORM was used for single-molecule localization and image reconstruction (Köthe et al., 2014). The images were further processed with ImageJ and the dual-color images were post-aligned with the aid of multi-spectral beads (Invitrogen) added to the samples.
Protein expression and capture assays
For in vitro capture assays from E. coli cell lysates, KAR1 and SFI1 fragments of indicated lengths (see illustrations within the figures) were cloned as GST fusions in the dicistronic vector pGEX-6p-2rbs (a gift from A. Musacchio, Max Planck Institute, Dortmund, Germany) and as His-tag fusions in the dicistronic vector pETDuet-1 (EMD Millipore). All SFI1 fragments containing a binding site for Cdc31 were coexpressed with Cdc31. All proteins were expressed at 25°C with 0.3–0.5 mM of IPTG. As the first step, the bacterial cells expressing the GST or GST-tagged fragments were lysed in buffer containing 1× PBS, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, and protease inhibitors (Roche). After an ultracentrifugation step, the proteins were coupled to GST Sepharose beads (Macherey-Nagel). A quantitative portion of the beads was analyzed by SDS-PAGE to retain the same amount of bait for each fragment tested in the assay. After washing steps, the coupled beads were incubated with His-tagged constructs, prepared from E. coli cells lysed in binding buffer (1× PBS, 250 mM NaCl, and 0.25% Triton X-100). Because the expression rates of the His-tagged fragments were different, the amount of cell lysate used for the assay was adjusted. After incubation and extensive washing steps with ice-cold binding buffer, the beads were eluted with Laemmli buffer and analyzed by immunoblotting with the indicated antibodies.
Modeling of the dimer of Sfi1–CdC31 complexes for wild-type and mutant Cdc31
The crystal structure of the Sfi1–Cdc31 complex (PDB accession no. 2GV5; Li et al., 2006) at a 3.0-Å resolution consisting of one Sfr bound to two Cdc31 molecules was used for modeling and for the binding free energy calculations. To obtain a model of the structure of two Sfi1–Cdc31 complexes interacting at their Cdc31 binding sites, symmetry mates of the initial structure were generated using PyMol (PyMOL Molecular Graphics System, Version 1.7.4; Schrödinger, LLC). The Cdc31–Cdc31 binding regions of the complex structures formed by the original Sfi1–Cdc31 structure and its symmetry mates were investigated using the PDBePISA webserver (Krissinel and Henrick, 2007). Among the possible complexes, the one with all the mutated residues (D107, D131, and E148) in the binding region and the Sfi1 α-helices approximately antiparallel was chosen for calculations of binding free energy.
MSE (selenomethionine residues) entries in the PDB file were replaced with MET (methionine residues) without modifying atomic positions. Water molecules in the crystal structure were removed and the missing Ca2+ atoms were placed in the corresponding Ca2+-binding sites. All of the cysteines were treated as free residues since no disulfide bridge was observed in the crystal structure.
Single point mutations for each of the four mutants (Cdc31-12, -14, 16, and -17) were introduced using the mutagenesis tool implemented in PyMol. Among the rotamers available for each of the mutations, those that had the smallest strain without steric clashes with neighboring atoms were chosen.
Before the energy calculations, the protonation states of the ionizable residues at pH 7.0 were predicted using PropKa (Li et al., 2005; Bas et al., 2008). The PDB2PQR program (Dolinsky et al., 2004, 2007; Olsson et al., 2011) was used to add hydrogen atoms by optimizing the hydrogen bonding network and repositioning some of the atoms to prevent further steric clashes. Coordinate files were generated with partial atomic charges and atomic radii were assigned from the CHARMM22 (Brooks et al., 1983), Amber99 (Wang et al., 2000), and PARSE (Sitkoff et al., 1994) force fields for the wild-type and mutant structures.
Binding free energy calculations
The binding free energies for the wild-type and mutant complexes were computed as the sum of electrostatic and nonpolar energies using an implicit solvent model. The electrostatic binding free energy was defined as:
The binding free energy consists of the electrostatic interaction energy () between the binding partners P1 and P2, and the desolvation terms ( and ) arising from the loss of the electrostatic interactions with the solvent upon complexation. A two-step procedure was used for each of the electrostatic desolvation terms. In the first step, the electrostatic energies of each of the binding partners alone in solvent were computed. In the second, the energies for each of the partners were computed in the presence of the other partner without the other partner’s partial charges. The difference in energy obtained from the two steps resulted in the desolvation energy term for the corresponding binding partner.
The electrostatic interaction energy between the binding partners is defined as: , where Φi is the electrostatic potential of one of the binding partners at the position of the atomic charge qi of the second partner and the sum is over all charges in the second partner.
The electrostatic interaction potentials and desolvation energies were calculated by numerical solution of the nonlinear Poisson-Boltzmann equation using the UHBD (Madura et al., 1995) software and three different force fields: Amber99, CHARMM22, and PARSE. The temperature was set to 298.15 K and the relative dielectric constants of the solute and the solvent were chosen to be 2.00 and 78.54, respectively. The van der Waals surface was used for the dielectric and ionic boundaries (Pang and Zhou, 2013). The ionic strength of the solvent and the ionic radius were set to 0.05 M and 1.8 Å, respectively. Electrostatic focusing was used with grid spacing values of 0.6 Å for the coarse and 0.15 Å for the fine grids. The dimensions were set as 417 × 385 × 737 points for both grids.
The nonpolar desolvation contribution to the binding free energy was calculated using the formula:
where SASA, γ, and b stand for the solvent accessible surface area, surface tension energy, and repulsive offset, respectively. The γ and b parameters were set to 0.00542 kcal/mol Å−2 and 0.92 kcal/mol, respectively (Kuhn and Kollman, 2000). The SASA of the molecules were calculated using a probe of radius 1.4 Å with the Shrake-Rupley method (Shrake and Rupley, 1973) implemented in the APBS software (Baker et al., 2001).
Online supplemental material
Fig. S1 shows the functionality of yeGFP-KAR1 and SFI1-yeGFP, and it provides FLIP experiments on Cdc5 and Sfi1. Fig. S2 presents controls for the super-resolution microscopy experiments of Fig. 2 and shows Kar1 localization in sfi1Δct cells. Fig. S3 shows the overexpression of KAR1 and kar1Δtmd and depicts the Sfi1 C-terminal self-interaction. Fig. S4 provides controls for the experiments shown in Fig. 4. It emphasizes the role of Sfi1’s C-terminally located Cdc31 repeats for Kar1 binding and SPB duplication. Fig. S5 presents additional electron micrographs of kar1Δ CDC31-16 cells and provides further analysis of GBP-kar1 truncated strains. Fig. S5 further shows the viability of C-terminally truncated Sfi1 cells, provides the Kar1 content in cells overexpressing Cdc31 variants, and confirms the Kar1 overexpression as anticipated in Fig. 9. Table S1 shows the computed binding free energies of the dimer of Sfi1–Cdc31 complexes for CDC31 and its dominant mutants. Tables S2 and S3 show the plasmids and yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412050/DC1.
Supplementary Material
Acknowledgements
We thank all members of the Schiebel Laboratory as well as Dr. G. Pereira for stimulating discussions and suggestions. We acknowledge the European Molecular Biology Laboratory EM facility for providing technical support and expertise. We thank Drs. A. Musacchio, E. Schwob and K. Labibfor strains and plasmids.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) Schi295/5-1 and by the Klaus Tschira Foundation (to M. Ozboyaci and R.C. Wade).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- dSTORM
- direct stochastic optical reconstruction microscopy
- FLIP
- fluorescence loss in photobleaching
- GBP
- GFP-binding protein
- NE
- nuclear envelope
- PALM
- photo-activated localization microscopy
- RFI
- relative fluorescence intensity
- ROI
- region of interest
- Sfr
- Sfi1 fragment
- SPB
- spindle pole body
- TAP
- tandem affinity purification
- TMD
- transmembrane domain
References
- Adams I.R., and Kilmartin J.V.. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145:809–823. 10.1083/jcb.145.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams I.R., and Kilmartin J.V.. 2000. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10:329–335. 10.1016/S0962-8924(00)01798-0 [DOI] [PubMed] [Google Scholar]
- Avena J.S., Burns S., Yu Z., Ebmeier C.C., Old W.M., Jaspersen S.L., and Winey M.. 2014. Licensing of yeast centrosome duplication requires phosphoregulation of sfi1. PLoS Genet. 10:e1004666 10.1371/journal.pgen.1004666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Hergert P., Delouvée A., Euteneuer U., Formstecher E., Khodjakov A., and Bornens M.. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185:101–114. 10.1083/jcb.200808082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., and McCammon J.A.. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 98:10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas D.C., Rogers D.M., and Jensen J.H.. 2008. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 73:765–783. 10.1002/prot.22102 [DOI] [PubMed] [Google Scholar]
- Biggins S., and Rose M.D.. 1994. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 125:843–852. 10.1083/jcb.125.4.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. 2012. The centrosome in cells and organisms. Science. 335:422–426. 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Bouhlel I.B., Ohta M., Mayeux A., Bordes N., Dingli F., Boulanger J., Velve Casquillas G., Loew D., Tran P.T., Sato M., and Paoletti A.. 2015. Cell cycle control of spindle pole body duplication and splitting by Sfi1 and Cdc31 in fission yeast. J. Cell Sci. 128:1481–1493. 10.1242/jcs.159657 [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., and Karplus M.. 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- Byers B. 1981. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae In Molecular Genetics in Yeast. Vol. 16. D. Wettstein, A. Stenderup, M. Kielland-Brandt, and J. Friis, editors. Alfred Benzon Symp. 16, Munksgaard, Copenhagen. 119–133. [Google Scholar]
- Byers B., and Goetsch L.. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Indjeian V.B., McManus M., Wang L., and Dynlacht B.D.. 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 3:339–350. 10.1016/S1534-5807(02)00258-7 [DOI] [PubMed] [Google Scholar]
- Crasta K., Lim H.H., Giddings T.H. Jr., Winey M., and Surana U.. 2008. Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat. Cell Biol. 10:665–675. 10.1038/ncb1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas T.J., Wang Y., Lalor P., Dockery P., and Morrison C.G.. 2011. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J. Cell Biol. 193:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., McCammon J.A., and Baker N.A.. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32:W665–W667. 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., and Baker N.A.. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35:W522–W525. 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A.D., and Kilmartin J.V.. 1996. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) with an essential function during SPB duplication. J. Cell Biol. 132:887–901. 10.1083/jcb.132.5.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S., Knop M., Schlenstedt G., and Schiebel E.. 1999. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA. 96:6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elserafy M., Šarić M., Neuner A., Lin T.C., Zhang W., Seybold C., Sivashanmugam L., and Schiebel E.. 2014. Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle. Curr. Biol. 24:1456–1466. 10.1016/j.cub.2014.05.032 [DOI] [PubMed] [Google Scholar]
- Erlemann S., Neuner A., Gombos L., Gibeaux R., Antony C., and Schiebel E.. 2012. An extended γ-tubulin ring functions as a stable platform in microtubule nucleation. J. Cell Biol. 197:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Rodríguez-Navarro S., Pereira G., Rácz A., Schiebel E., and Hurt E.. 2004. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 6:840–848. [DOI] [PubMed] [Google Scholar]
- Gibeaux R., Politi A.Z., Nédélec F., Antony C., and Knop M.. 2013. Spindle pole body-anchored Kar3 drives the nucleus along microtubules from another nucleus in preparation for nuclear fusion during yeast karyogamy. Genes Dev. 27:335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings T.H. Jr., O’Toole E.T., Morphew M., Mastronarde D.N., McIntosh J.R., and Winey M.. 2001. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 67:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase S.B., Winey M., and Reed S.I.. 2001. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat. Cell Biol. 3:38–42. 10.1038/35050543 [DOI] [PubMed] [Google Scholar]
- Heilemann M., van de Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., and Sauer M.. 2008. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47:6172–6176. 10.1002/anie.200802376 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., and Winey M.. 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20:1–28. [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Giddings T.H.J. Jr., and Winey M.. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956. 10.1083/jcb.200208169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Martin A.E., Glazko G., Giddings T.H. Jr., Morgan G., Mushegian A., and Winey M.. 2006. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174:665–675. 10.1083/jcb.200601062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M., Sanchez-Diaz A., Gambus A., and Labib K.. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 423:720–724. 10.1038/nature01692 [DOI] [PubMed] [Google Scholar]
- Kilmartin J.V. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162:1211–1221. 10.1083/jcb.200307064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., and Schiebel E.. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 15:963–972. [DOI] [PubMed] [Google Scholar]
- Kollman J.M., Merdes A., Mourey L., and Agard D.A.. 2011. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12:709–721. 10.1038/nrm3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köthe U., Herrmannsdörfer F., Kats I., and Hamprecht F.A.. 2014. SimpleSTORM: a fast, self-calibrating reconstruction algorithm for localization microscopy. Histochem. Cell Biol. 141:613–627. 10.1007/s00418-014-1211-4 [DOI] [PubMed] [Google Scholar]
- Krissinel E., and Henrick K.. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Kuhn B., and Kollman P.A.. 2000. Binding of a diverse set of ligands to avidin and streptavidin: an accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J. Med. Chem. 43:3786–3791. 10.1021/jm000241h [DOI] [PubMed] [Google Scholar]
- Laoukili J., Perret E., Middendorp S., Houcine O., Guennou C., Marano F., Bornens M., and Tournier F.. 2000. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 113:1355–1364. [DOI] [PubMed] [Google Scholar]
- Lee I.J., Wang N., Hu W., Schott K., Bähler J., Giddings T.H. Jr., Pringle J.R., Du L.-L., and Wu J.-Q.. 2014. Regulation of spindle pole body assembly and cytokinesis by the centrin-binding protein Sfi1 in fission yeast. Mol. Biol. Cell. 25:2735–2749. 10.1091/mbc.E13-11-0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B.O., and Chou K.C.. 2011. Review of super-resolution fluorescence microscopy for biology. Appl. Spectrosc. 65:967–980. 10.1366/11-06398 [DOI] [PubMed] [Google Scholar]
- Li H., Robertson A.D., and Jensen J.H.. 2005. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 61:704–721. [DOI] [PubMed] [Google Scholar]
- Li S., Sandercock A.M., Conduit P., Robinson C.V., Williams R.L., and Kilmartin J.V.. 2006. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 173:867–877. 10.1083/jcb.200603153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura J.D., Briggs J.M., Wade R.C., Davis M.E., Luty B.A., Ilin A., Antosiewicz J., Gilson M.K., Bagheri B., Scott L.R., and McCammon J.A.. 1995. Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics program. Comput. Phys. Commun. 91:57–95. [Google Scholar]
- Martinez-Sanz J., Yang A., Blouquit Y., Duchambon P., Assairi L., and Craescu C.T.. 2006. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 273:4504–4515. [DOI] [PubMed] [Google Scholar]
- Middendorp S., Paoletti A., Schiebel E., and Bornens M.. 1997. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA. 94:9141–9146. 10.1073/pnas.94.17.9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Stearns T.. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13:1154–1160. 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M.H.M., Søndergaard C.R., Rostkowski M., and Jensen J.H.. 2011. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7:525–537. 10.1021/ct100578z [DOI] [PubMed] [Google Scholar]
- Pang X., and Zhou H.-X.. 2013. Poisson-Boltzmann calculations: van der Waals or molecular surface? Commun. Comput. Phys. 13:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Grueneberg U., Knop M., and Schiebel E.. 1999. Interaction of the yeast γ-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J. 18:4180–4195. 10.1093/emboj/18.15.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R.D., Gorski S.A., and Misteli T.. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375:393–414. [DOI] [PubMed] [Google Scholar]
- Prosser S.L., and Morrison C.G.. 2015. Centrin2 regulates CP110 removal in primary cilium formation. J. Cell Biol. 208:693–701. 10.1083/jcb.201411070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries J., Kaplan C., Platonova E., Eghlidi H., and Ewers H.. 2012. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods. 9:582–584. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., and Séraphin B.. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Rose M.D., and Fink G.R.. 1987. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 48:1047–1060. 10.1016/0092-8674(87)90712-4 [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Salisbury J.L., Suino K.M., Busby R., and Springett M.. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12:1287–1292. 10.1016/S0960-9822(02)01019-9 [DOI] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.-D., and Nigg E.A.. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Shrake A., and Rupley J.A.. 1973. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 79:351–371. 10.1016/0022-2836(73)90011-9 [DOI] [PubMed] [Google Scholar]
- Sitkoff D., Sharp K.A., and Honig B.. 1994. Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 98:1978–1988. 10.1021/j100058a043 [DOI] [Google Scholar]
- Spang A., Courtney I., Fackler U., Matzner M., and Schiebel E.. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123:405–416. 10.1083/jcb.123.2.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Courtney I., Grein K., Matzner M., and Schiebel E.. 1995. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128:863–877. 10.1083/jcb.128.5.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., and Gönczy P.. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213. 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., and Knop M.. 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques. 40:73–78. 10.2144/000112040 [DOI] [PubMed] [Google Scholar]
- Tsang W.Y., Spektor A., Luciano D.J., Indjeian V.B., Chen Z., Salisbury J.L., Sánchez I., and Dynlacht B.D.. 2006. CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol. Biol. Cell. 17:3423–3434. 10.1091/mbc.E06-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Hiller M.A., Scherson T.Y., and Rose M.D.. 1992a Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J. Cell Biol. 117:1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Scherson T.Y., Roberts T., van Zee K., and Rose M.D.. 1992b Asymmetric mitotic segregation of the yeast spindle pole body. Cell. 69:505–515. 10.1016/0092-8674(92)90451-H [DOI] [PubMed] [Google Scholar]
- Vallen E.A., Ho W., Winey M., and Rose M.D.. 1994. Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae. Genetics. 137:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cieplak P., and Kollman P.A.. 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21:1049–1074. [DOI] [Google Scholar]
- Wiech H., Geier B.M., Paschke T., Spang A., Grein K., Steinkötter J., Melkonian M., and Schiebel E.. 1996. Characterization of green alga, yeast, and human centrins. Specific subdomain features determine functional diversity. J. Biol. Chem. 271:22453–22461. [DOI] [PubMed] [Google Scholar]
- Wu J.Q., and Pollard T.D.. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314. 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.