Binding of STIL activates Plk4, and the subsequent phosphorylation of STIL by Plk4 primes the binding of STIL to SAS6 to promote centriole assembly.
Abstract
Centriole duplication occurs once per cell cycle in order to maintain control of centrosome number and ensure genome integrity. Polo-like kinase 4 (Plk4) is a master regulator of centriole biogenesis, but how its activity is regulated to control centriole assembly is unclear. Here we used gene editing in human cells to create a chemical genetic system in which endogenous Plk4 can be specifically inhibited using a cell-permeable ATP analogue. Using this system, we demonstrate that STIL localization to the centriole requires continued Plk4 activity. Most importantly, we show that direct binding of STIL activates Plk4 by promoting self-phosphorylation of the activation loop of the kinase. Plk4 subsequently phosphorylates STIL to promote centriole assembly in two steps. First, Plk4 activity promotes the recruitment of STIL to the centriole. Second, Plk4 primes the direct binding of STIL to the C terminus of SAS6. Our findings uncover a molecular basis for the timing of Plk4 activation through the cell cycle–regulated accumulation of STIL.
Introduction
Centrioles are characterized by an evolutionarily conserved ninefold rotational symmetry (Gönczy, 2012). In cycling cells, a pair of centrioles forms the core of the centrosome, the cell’s major microtubule-organizing center. This centriole pair duplicates once in each cell cycle by forming one new centriole on the wall of each of the two preexisting parental centrioles (Tsou and Stearns, 2006; Nigg and Raff, 2009). This tightly coordinated process ensures that the single interphase centrosome reproduces exactly once before mitosis. The two centrosomes then separate and instruct the formation of the bipolar spindle apparatus upon which chromosomes are segregated. Abnormalities in centriole duplication can result in the production of extra copies of centrosomes, a feature commonly observed in human cancers and widely implicated in contributing to the pathogenesis of the disease (Basto et al., 2008; Castellanos et al., 2008; Ganem et al., 2009; Silkworth et al., 2009; Chan, 2011; Godinho et al., 2014).
Pioneering work in Caenorhabditis elegans has led to the identification of a conserved set of five core proteins required for centriole assembly: ZYG-1/Plk4, SPD2/CEP192, SAS6, SAS5/STIL/Ana2, and SAS4/CPAP (O’Connell et al., 2001; Kirkham et al., 2003; Leidel and Gönczy, 2003; Dammermann et al., 2004; Delattre et al., 2004; Kemp et al., 2004; Pelletier et al., 2004; Leidel et al., 2005). Of these components, ZYG-1/Plk4 has emerged as a central, upstream regulator of centriole biogenesis. The abundance of Plk4 must be carefully controlled: reducing Plk4 levels leads to a failure of centriole duplication, whereas Plk4 overexpression drives the formation of supernumary centrioles. Plk4 levels are self-regulated by a negative feedback loop in which the kinase phosphorylates itself to trigger capture by an E3 ubiquitin ligase, leading to ubiquitylation and destruction of the active kinase (Cunha-Ferreira et al., 2009, 2013; Rogers et al., 2009; Guderian et al., 2010; Holland et al., 2010, 2012; Klebba et al., 2013).
In early G1 phase, Plk4 is localized around the entire wall of the parental centriole and transitions at the beginning of S phase to an asymmetric spot on the parental centriole that marks the site of cartwheel assembly (Kim et al., 2013; Sonnen et al., 2013; Ohta et al., 2014). The cartwheel appears at the beginning of procentriole assembly and is formed by the oligomerization of the centriole protein SAS6 (Kitagawa et al., 2011; van Breugel et al., 2011, 2014). In C. elegans and Drosophila melanogaster, SAS6 interacts directly with another cartwheel protein SAS5/Ana2 (Leidel et al., 2005; Stevens et al., 2010a,b). Although initial studies failed to detect a direct interaction between STIL (the human counterpart of SAS5/Ana2) and SAS6 (Tang et al., 2011; Arquint et al., 2012), it was recently reported that phosphorylation of the conserved STAN domain of STIL/Ana2 creates a binding site for SAS6 that is required for SAS6 recruitment to the site of procentriole assembly (Dzhindzhev et al., 2014; Ohta et al., 2014). Plk4 was also shown to phosphorylate STIL in vitro and when overexpressed in cells (Dzhindzhev et al., 2014; Ohta et al., 2014; Kratz et al., 2015). Nevertheless, Plk4 is a low-abundance enzyme and it remains unclear if endogenous Plk4 phosphorylates the STIL STAN domain in vivo. Furthermore, Plk4 localizes to the centriole throughout the cell cycle in human cells (Sonnen et al., 2012), but how its activity is regulated to trigger procentriole formation remains unknown. A major limitation in addressing these questions is the lack of tools to rapidly and specifically control Plk4 kinase activity in vivo.
A recent study reported the development of CFI-400945, a potent small-molecule ATP-competitive inhibitor of Plk4 kinase activity (Laufer et al., 2013; Mason et al., 2014). However, along with inhibiting Plk4, CFI-400945 also strongly inhibited Aurora B kinase both in vitro and in vivo, complicating the use of this inhibitor for studying Plk4 targets in cells (Holland and Cleveland, 2014). We previously showed that the mutation of a single amino acid in the ATP-binding pocket of Plk4 creates an analogue-sensitive (AS) kinase that can be inhibited in a highly specific manner with cell-permeable, nonhydrolyzable, bulky ATP analogues (Holland et al., 2010). Here, we knocked-in the AS mutation into both endogenous Plk4 alleles in a human cell line. Using chemical genetics and phospho-specific antibodies, we demonstrate that STIL is a target of endogenous Plk4 in vivo. STIL phosphorylation by Plk4 is shown to be required for centriole duplication, establishing STIL as a key target of Plk4 in centriole biogenesis. Most importantly, we show that STIL binding activates Plk4 kinase activity. Given that STIL is degraded after mitosis and accumulates at the beginning of S phase (Tang et al., 2011; Arquint et al., 2012; Arquint and Nigg, 2014), our data offer a molecular basis for controlling the timing of Plk4 activation and centriole assembly.
Results
A chemical genetics system for controlling Plk4 activity in cells
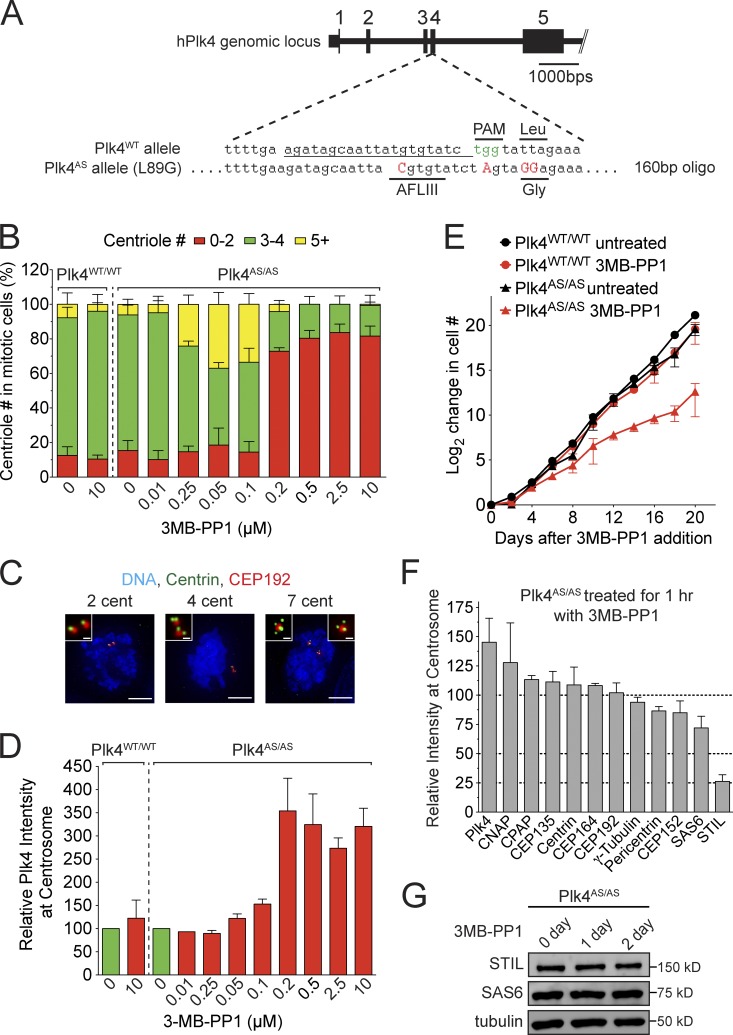
To study the function of Plk4 kinase activity in cells, we used CRISPR/Cas9 genome engineering to knock-in the Plk4 AS mutation (L89G) into both endogenous Plk4 alleles in the DLD-1 colon cancer cell line (Fig. 1 A). Two homozygous Plk4AS/AS clones were identified that behaved similarly in all assays and are hereafter presented together. Importantly, Plk4AS/AS cells underwent normal centriole duplication, demonstrating the functionality of the Plk4AS allele in vivo (Fig. 1, B and C).
Figure 1.
Plk4 kinase activity is required to maintain STIL at the centriole. (A) Schematic of the strategy used to knock-in the AS mutation into both alleles of Plk4 in human DLD-1 cells. The repair oligonucleotide introduced the AS mutation (L89G), a silent AflIII restriction site, and a mutation in the protospacer adjacent motif (PAM) to prevent recutting by SpCas9 after homology-directed repair. (B) Plk4WT/WT or Plk4AS/AS cells were treated with 3MB-PP1 for 20 h and nocodazole was added for the final 4 h of the treatment. The graph shows the fraction of mitotic cells with the indicated number of centrioles. Bars represent the mean of three independent experiments, with >20 cells counted per experiment. (C) Selected images of mitotic Plk4AS/AS cells from B stained with Centrin and CEP192. Bars: (large images) 5 µm; (inset images) 0.5 µm. (D) Quantification of the relative levels of Plk4 at the centrosome of interphase cells 20 h after addition of 3MB-PP1. Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. (E) Graph showing the increase in cell number at various times after addition of 3MB-PP1. Points show the mean of at least three independent experiments. (F) Quantification of relative protein abundance at the centrosome of S/G2 phase cells 1 h after the addition of 3MB-PP1. Bars represent the mean of three independent experiments, with >40 cells counted per experiment. (G) Immunoblot showing no change in the level of endogenous STIL and SAS6 at 1 or 2 d after Plk4 inhibition with 3MB-PP1. All error bars represent the SEM.
Inhibition of Plk4 kinase activity leads to an increase in the level of the kinase (Holland et al., 2010). We therefore used the abundance of Plk4 at the centrosome as a readout of kinase inhibition. Treatment of Plk4WT/WT cells with 10 µM of the bulky purine analogue 3MB-PP1 did not affect centriole number or Plk4 levels (Fig. 1, B and D). In contrast, treatment of Plk4AS/AS cells with increasing concentrations of 3MB-PP1 led to a dose-dependent increase in Plk4 levels at the centrosome. Maximal Plk4 stabilization was achieved at 0.2 µM 3MB-PP1, which indicates complete inhibition of Plk4 activity at this dose (Fig. 1 D). Consistently, Plk4AS/AS cells treated with 0.2 µM 3MB-PP1 for one cell cycle failed centriole duplication (>70% cells contained 0–2 centrioles in mitosis; Fig. 1, B and C). Treatment of Plk4AS/AS cells with lower doses of 3MB-PP1 partially increased Plk4 abundance and gave rise to modest centriole amplification in mitotic cells (>20% of cells treated with between 0.025 and 0.1 µM of 3MB-PP1 contained more than four centrioles in mitosis; Fig. 1, B–D). This effect may be due to the formation of heterodimers between kinase inactive and catalytically active Plk4 that leads to an increase in the abundance of the wild type (WT) kinase (Guderian et al., 2010; Mason et al., 2014).
Human DLD-1 cells continue to proliferate in the absence of centrioles
Recent work has shown that loss of the p53 tumor suppressor is necessary for the proliferation of cells lacking centrioles (Bazzi and Anderson, 2014; Izquierdo et al., 2014). DLD-1 cell lines express a mutant form of p53 with compromised function (Sur et al., 2009). Consequently, chronic treatment of DLD-1 Plk4AS/AS cells with 3MB-PP1 resulted in the step-wise reduction in centriole number as cells failed centriole duplication but continued to divide (Fig. S1, A and B). The cell cycle profile of Plk4AS/AS cells was unaltered at day 5 after 3MB-PP1 addition, by which point >90% of cells lacked centrioles (Fig. S1, A–C). DLD-1 Plk4AS/AS cells lacking centrioles exhibited a significant increase in the level of aneuploidy and a modestly reduced proliferation rate (Figs. 1 E and S1 D), but had no reduction in long-term clonogenic survival capability (Fig. S1, E and F). These data are consistent with the view that centrosomes are not essential for cell growth, but increase the fidelity of chromosome segregation (Khodjakov and Rieder, 2001; Debec et al., 2010; Sir et al., 2013). Strikingly, washout of 3MB-PP1 in acentriolar DLD-1 cells led to the reactivation of Plk4 and the formation of de novo centrioles (Fig. S1 G). Plk4 activity is therefore dose-limiting for canonical and de novo centriole biogenesis.
The centriole localization of STIL requires Plk4 kinase activity
To determine which proteins require Plk4 kinase activity for centriole recruitment, we inhibited Plk4 for 1 h and measured the abundance of 12 proteins at the centrosome of S/G2 cells (Fig. 1 F). As expected, 1 h after inhibiting Plk4, the abundance of the kinase at the centriole increased. RNAi depletion of STIL has been reported to dramatically decrease the centriole-localized pool of SAS6 (Tang et al., 2011; Arquint et al., 2012; Vulprecht et al., 2012; see Fig. 5 E). Surprisingly, while the abundance of STIL at the centriole declined to 26% of control levels 1 h after inhibition of Plk4, the abundance of SAS6 declined to only 72% in the same time period (Fig. 1 F). Prolonged treatment with 3MB-PP1 led to a progressive decline in the level of SAS6 at the centriole, reaching 31% of control levels by 2 d after 3MB-PP1 addition (Fig. S1 H). Importantly, STIL and SAS6 cellular protein levels were not altered after chronic Plk4 inhibition (Fig. 1 G). We conclude that unlike SAS6, STIL dissociates from the centriole with extremely rapid kinetics after Plk4 inhibition.
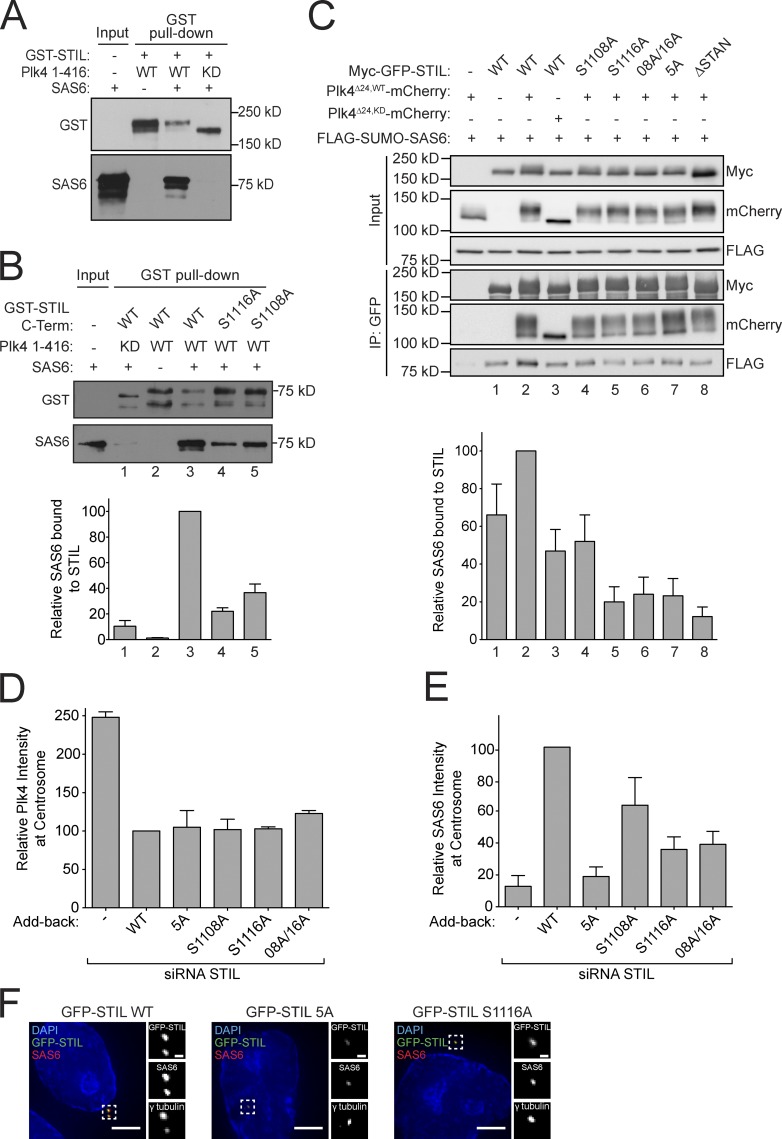
Figure 5.
STIL STAN domain phosphorylation is required for direct binding to SAS6. (A and B) GST-STIL or GST-STIL C-term (aa 898–1287) were phosphorylated in vitro with kinase-active or inactive His-Plk4 and incubated with SAS6. GST pull-downs were analyzed by immunoblotting with the indicated antibodies. The graph shows the quantification from B of the relative amount of SAS6 bound to GST-STIL C-term. Bars represent the mean of three independent experiments. (C, top) Cells were cotransfected and subject to coimmunoprecipitation analysis with the indicated antibodies. (C, bottom) Quantification of the relative amount of SAS6 bound to Myc-GFP-STIL. Bars represent the mean of three independent experiments. (D and E) Quantification showing the relative level of Plk4 or SAS6 at the centrosome of cells in which endogenous STIL had been depleted and replaced with the indicated Myc-GFP-STIL transgene. Bars represent the mean of at least three independent experiments, with >50 cells counted per experiment. (F) Selected images of cells showing Myc-GFP-STIL and SAS6 staining. Bars: (left) 5 µm; (right) 0.5 µm. All error bars represent the SEM.
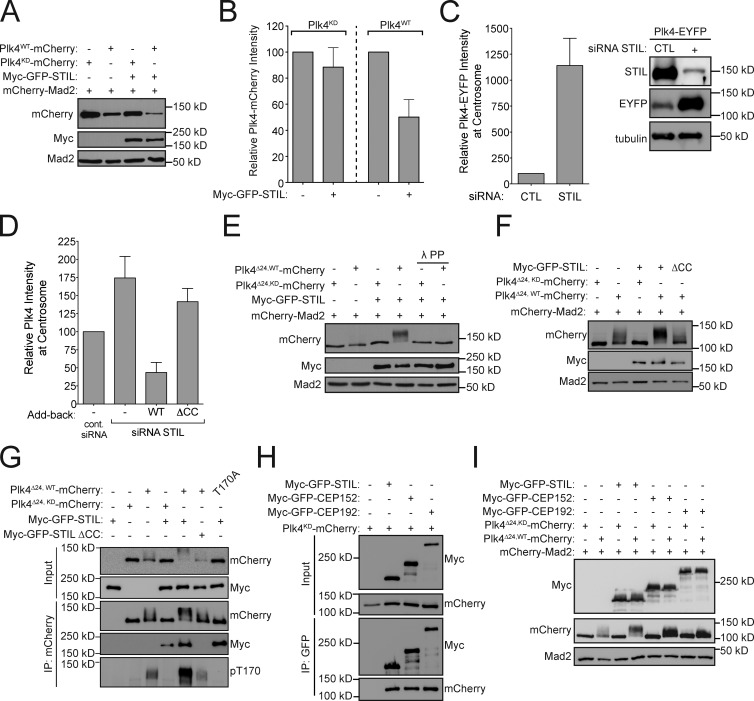
Plk4 directly binds and facilitates the recruitment of STIL to the centriole
Because Plk4 kinase activity is required for the recruitment of STIL to the centriole, we investigated whether STIL and Plk4 form a complex in cells. Cells were transfected with full-length Myc-GFP-STIL and either kinase-active (Plk4WT) or kinase-dead (Plk4KD) Plk4-mCherry. Both active and inactive Plk4-mCherry copurified with Myc-GFP-STIL (Fig. S2 A). Plk4 is a suicide kinase that promotes its own destruction through self-phosphorylation of a 24-aa multi-phosphodegron (MPD; Holland et al., 2010). As expected, deletion of the MPD (aa S282–S305) stabilized kinase active Plk4 (Plk4Δ24,WT) and increased the amount of active Plk4 that coimmunoprecipitated with Myc-GFP-STIL (Fig. S2 A). To examine whether Plk4 directly associates with STIL, we performed GST pull-down assays in the absence of ATP using purified recombinant STIL and GST-Plk4. GST-Plk4 could specifically pull down STIL, demonstrating a direct, kinase-independent association (Fig. S2 B). Together, these data show that Plk4 and STIL form a complex independent of Plk4 kinase activity. This contrasts with a recent study that proposed that Plk4 interacted with STIL in a kinase activity–dependent manner (Ohta et al., 2014).
To map the domain of STIL that interacts with Plk4, cells were transfected with a series of Myc-GFP-STIL truncation constructs, and the interaction with Plk4KD-mCherry was examined. Using this approach, we mapped amino acids 715–850 as a region of STIL sufficient for binding to Plk4 (Fig. S2, C and D). This region contains a highly conserved sequence (aa 721–746) that forms a predicted coiled-coil (Fig. S3 A). Deletion of aa 721–746 (ΔCC) from full-length Myc-GFP-STIL decreased binding to both kinase-active and inactive Plk4Δ24-mCherry (Fig. S3, B and C). We next analyzed whether Plk4 binding was required for the localization of STIL to the centriole. Myc-GFP-STIL WT and ΔCC were expressed in cells depleted of endogenous STIL by siRNA. Deletion of the predicted coiled-coil domain reduced the abundance of STIL at the centriole to 6% of that observed in control cells and failed to rescue centrosome duplication in cells depleted of endogenous STIL (Fig. S3, D and E). We conclude that binding to Plk4 is necessary for STIL to target to the centriole and function in centriole duplication.
STIL binding to Plk4 promotes kinase activity
Cotransfection of Myc-GFP-STIL reduced the abundance of Plk4WT by 50%, but had little effect on the level of Plk4KD (Fig. 2, A and B). Since Plk4 promotes its own destruction, we hypothesized that STIL expression stimulated Plk4 kinase activity and thus destruction. To test this hypothesis, we first examined the abundance of stably overexpressed Plk4-EYFP in the presence and absence of STIL. The total cellular pool, and levels of centrosome-localized Plk4-EYFP, increased dramatically after STIL depletion (Fig. 2 C). To test whether STIL regulates the abundance of endogenous Plk4, Myc-GFP-STIL WT and ΔCC were expressed in cells depleted of STIL. STIL knockdown increased the level of endogenous Plk4 at the centrosome (Fig. 2 D), whereas overexpression of Myc-GFP-STIL WT decreased the abundance of centrosomal Plk4. This decrease required STIL binding to Plk4, as overexpression of Myc-GFP-STILΔCC had little effect on Plk4 levels (Fig. 2 D).
Figure 2.
STIL binding stimulates Plk4 activity. (A) Cells were cotransfected with the indicated constructs and protein levels were analyzed by immunoblotting. mCherry-Mad2 serves as a transfection control. (B) Quantification of the protein levels shown in A. Bars represent the mean of three independent experiments. (C) STIL was depleted by siRNA and 24 h later doxycycline was added to induce expression of Plk4-EYFP. The immunoblot shows the relative levels of STIL and Plk4-EYFP in control or STIL siRNA–depleted cells. The graph shows quantification of the relative level of Plk4-EYFP at the centrosome of S/G2 phase cells. Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. (D) Endogenous STIL was depleted by siRNA and replaced with either Myc-GFP-STIL WT or ΔCC using the scheme outlined in Fig. 4 A. The graph shows quantification of the relative levels of Plk4 at the centrosome of S/G2 phase cells. Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. (E and F) Cells were cotransfected with the indicated constructs, and protein levels were analyzed by immunoblotting. Where indicated, lambda protein phosphatase (λ PP) was incubated with the cell lysate for 60 min before immunoblotting. (G and H) Cells were cotransfected with the indicated constructs and subjected to coimmunoprecipitation analysis with the indicated antibodies. (I) Cells were cotransfected with the indicated constructs and protein levels were analyzed by immunoblotting. All error bars represent the SEM.
To test if STIL was capable of promoting Plk4 self-phosphorylation, we stabilized kinase-active Plk4 by deletion of the Plk4 MPD (Plk4Δ24,WT). Strikingly, expression of Myc-GFP-STIL dramatically reduced the mobility of kinase-active Plk4Δ24-mCherry in a SDS-PAGE gel, but had no effect on the mobility of kinase-dead Plk4Δ24-mCherry (Fig. 2 E). Treatment with λ-phosphatase abolished the electrophoretic mobility shift, demonstrating that the slower migration of Plk4Δ24-mCherry was a result of increased kinase self-phosphorylation. To establish whether STIL binding stimulates Plk4 activity, we coexpressed Plk4Δ24,WT-mCherry with Myc-GFP-STIL WT or ΔCC in cells. While Myc-GFP-STIL increased Plk4Δ24,WT-mCherry self-phosphorylation, the Myc-GFP-STILΔCC mutant that was defective in Plk4 binding did not (Fig. 2 F). Importantly, a fragment of Myc-GFP-STIL (aa 715–988) that contains the coiled-coil domain and interacts with Plk4 was unable to activate the kinase, suggesting that binding to the STILis not, in itself, sufficient to activate Plk4 (Fig. S3 F). We conclude that STIL binding to Plk4 stimulates kinase activity and subsequent destruction of the kinase.
Plk4 kinase activity requires phosphorylation of threonine 170 in the activation loop (T-loop) of the kinase domain (Swallow et al., 2005; Nakamura et al., 2013). We therefore tested whether STIL binding promotes Plk4 T170 phosphorylation. Plk4Δ24-mCherry was cotransfected with or without Myc-GFP-STIL, and immunoprecipitated Plk4 was probed with an antibody that recognizes phosphorylated T170 (pT170; Nakamura et al., 2013). Importantly, expression of Myc-GFP-STIL WT dramatically increased phosphorylation of T170 on kinase-active, but not inactive, Plk4Δ24 (Fig. 2 G). In contrast, expression of the Myc-GFP-STILΔCC mutant that was defective in Plk4 binding was unable to promote Plk4-mediated T170 phosphorylation. These data suggest that STIL binding stimulates Plk4 T170 self-phosphorylation, leading to increased Plk4 activity.
Human Plk4 is targeted to the centriole through a direct interaction with the acidic N-terminal region of CEP152 and CEP192 (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010; Kim et al., 2013; Sonnen et al., 2013). However, while Myc-GFP-STIL, Myc-GFP-CEP152, and Myc-GFP-CEP192 all formed a complex with Plk4Δ24,KD-mCherry in cells, STIL was the only Plk4 binding partner that significantly stimulated self-phosphorylation of Plk4Δ24 (Fig. 2, H and I).
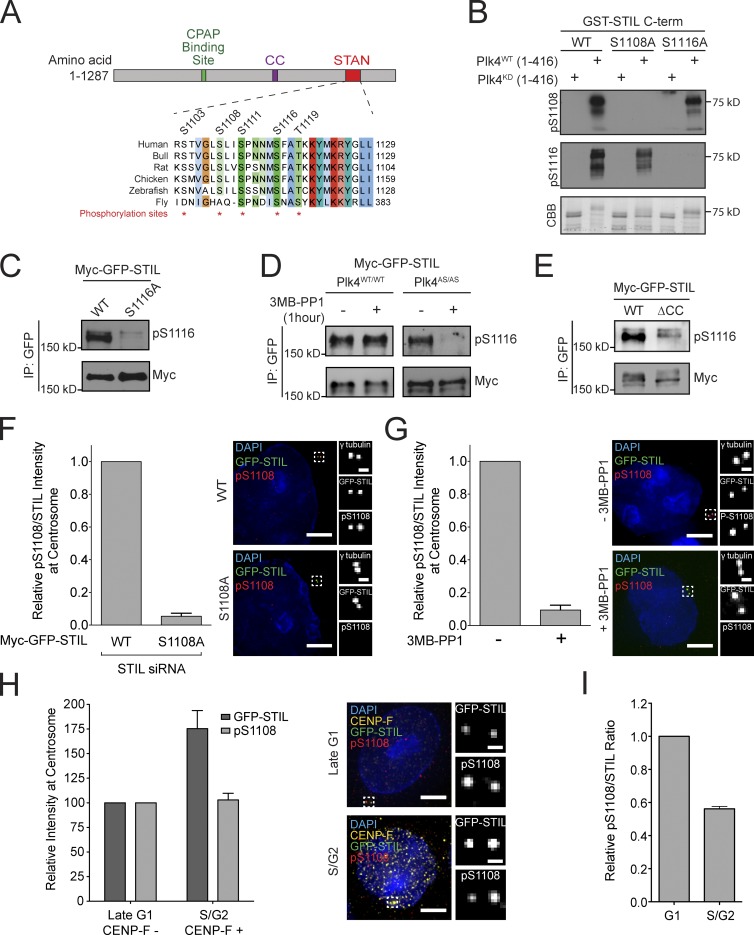
Plk4 phosphorylates the STIL STAN domain in vivo
Given that STIL directly binds and stimulates Plk4 activity, we investigated whether Plk4 phosphorylates STIL to control centriole assembly. We mapped in vitro Plk4 phosphorylation sites on STIL using mass spectrometry (Fig. S4 A). STIL-related proteins show high sequence homology in a short ∼90-aa region known as the STAN (STIL/Ana2) motif (Stevens et al., 2010a). This region contains five conserved residues that are phosphorylated in cells: S1103, S1108, S1111, S1116, and T1119 (Hoffert et al., 2006; Huttlin et al., 2010; Fig. 3 A). Of these five sites, S1108 and S1116 were phosphorylated by Plk4 in vitro and closely matched the Plk4 consensus phosphorylation sequence (Johnson et al., 2007; Kettenbach et al., 2012). To facilitate analysis of these phosphorylation sites, we generated phospho-specific antibodies. The affinity-purified pS1108 and pS1116 antibodies recognized recombinant GST-STIL and GST-STIL C terminus (C-term) only in the presence of kinase-active Plk4 (Figs. 3 B and S4 B). Moreover, recognition of phosphorylated GST-STIL C-term by the pS1108 and pS1116 antibody was abolished by mutation of S1108A and S1116A, respectively (Fig. 3 B). These observations confirm that Plk4 phosphorylates STIL at S1108 and S1116 in vitro, and demonstrate the specificity of the pS1108 and pS1116 antibodies for revealing the phosphorylation status of STIL.
Figure 3.
Plk4 phosphorylates the STIL SAN domain in vivo. (A) Schematic of STIL showing the CPAP binding domain, coiled-coil domain (CC), and the conserved STAN domain. Alignment shows the position of five amino acids in the STAN domain that are phosphorylated in vivo. (B) GST-STIL C-term (aa 898–1287) was phosphorylated in vitro with kinase-active or inactive His-Plk4 and analyzed by immunoblotting with the indicated antibodies. The Coomassie (CBB)-stained gel shows the purified protein. (C) Myc-GFP-STIL WT or S1116A were immunopurified from cells and analyzed by immunoblotting with the indicated antibodies. (D) Plk4WT/WT or Plk4AS/AS cells were treated with 3MB-PP1 for 1 h. Myc-GFP-STIL was then immunopurified from cells and analyzed by immunoblotting with the indicated antibodies. (E) Myc-GFP-STIL WT or ΔCC were immunopurified from cells and analyzed by immunoblotting with the indicated antibodies. (F, left) Endogenous STIL was replaced with either Myc-GFP-STIL WT or S1108A. The graph shows quantification of the relative levels of pS1108/STIL at the centrosome of S/G2 phase cells. Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. (F, right) Selected images of cells showing Myc-GFP-STIL and pS1108 staining. Bars: (left) 5 µm; (right) 0.5 µm. (G, left) Plk4AS/AS cells were treated with or without 3MB-PP1 for 1 h. The graph shows quantification of the relative levels of pS1108/STIL at the centrosome of S/G2 phase cells. Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. (G, right) Selected images of cells showing Myc-GFP-STIL and pS1108 staining. Bars: (left) 5 µm; (right) 0.5 µm. (H, left) Quantification showing the relative levels of Myc-GFP-STIL and pS1108 at the centrosome of G1 (CENP-F negative) and S/G2 (CENP-F positive) phase cells. Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. (H, right) Selected images of cells showing Myc-GFP-STIL and pS1108 staining. Bars: (left) 5 µm; (right) 0.5 µm. (I) Quantification showing the relative levels of pS1108/STIL at the centrosome of G1 or S/G2 phase cells. Ratio is calculated from the data shown in H. All error bars represent the SEM.
The pS1116 antibody recognized Myc-GFP-STIL WT purified from cells, but not Myc-GFP-STIL containing a S1116A mutation (Fig. 3 C). To establish whether Plk4 was responsible for phosphorylating STIL S1116 in cells, we treated Plk4WT/WT and Plk4AS/AS cells with 3MB-PP1 for 1 h and examined phosphorylation of STIL S1116. Treatment with 3MB-PP1 abolished phosphorylation of Myc-GFP-STIL S1116 in Plk4AS/AS cells, but did not affect phosphorylation of this site in Plk4WT/WT cells, demonstrating that Plk4 phosphorylates STIL S1116 in cells (Fig. 3 D). Deletion of the coiled-coil region of STIL dramatically reduced phosphorylation of STIL S1116, suggesting that phosphorylation of this site requires Plk4 binding to STIL and/or the recruitment of STIL to the centriole (Fig. 3 E).
While the pS1116 antibody detected phosphorylated STIL via immunoblotting, it cross-reacted with an additional phosphorylated centriole protein and consequently was not useful for immunofluorescence analysis. We therefore tested whether the pS1108 antibody could detect phosphorylation of STIL S1108 by immunofluorescence staining. The pS1108 antibody stained a centriole-localized signal that colocalized with Myc-GFP-STIL (Fig. 3 F). To determine the specificity of this staining, we replaced endogenous STIL with a WT or S1108A Myc-GFP-STIL transgene. When normalized to the total level of STIL at the centriole, the centriole-localized pS1108 signal was reduced by >90% in cells expressing the S1108A mutant of Myc-GFP-STIL (Fig. 3 F). Moreover, treatment of Plk4AS/AS cells with 3MB-PP1 for 1 h resulted in a >90% reduction in STIL S1108 phosphorylation, demonstrating that STIL S1108 is a substrate for Plk4 in vivo (Fig. 3 G). To investigate the cell cycle–dependent phosphorylation of STIL S1108, we performed fluorescence intensity measurements to determine the level of Myc-GFP-STIL and pS1108 at the centriole in late G1 (CENP-F–negative) and S/G2 (CENP-F–positive) cells (Hussein and Taylor, 2002). While levels of Myc-GFP-STIL were higher in S/G2 compared with late G1 cells, the level of pS1108 staining remained unchanged (Fig. 3, H and I). Importantly, we never observed centriole-localized Myc-GFP-STIL in the absence of pS1108 staining. These data suggest that Plk4 is active from late G1 through G2 phase.
Plk4 phosphorylates the STIL STAN domain to promote centriole duplication
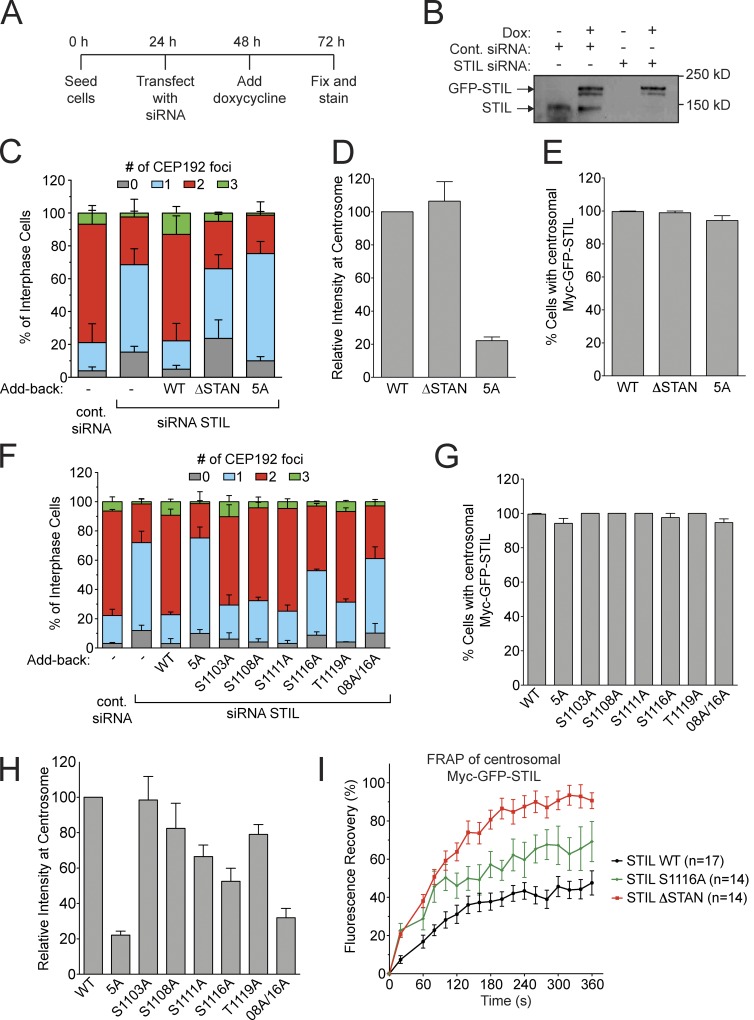
We investigated how Plk4-mediated STIL phosphorylation affects centriole biogenesis. WT or phosphorylation-defective Myc-GFP-STIL transgenes were integrated at a predefined genomic locus in a DLD-1 host cell line and expression was induced by the addition of doxycycline. All of the Myc-GFP-STIL transgenes were expressed at identical, near-endogenous levels (Fig. S4 C). Expression of Myc-GFP-STIL WT in the presence of endogenous STIL drove excessive centrosome formation in 45% of cells (Fig. S4 D). In contrast, expression of a Myc-GFP-STIL variant (5A) with all five phosphorylation sites in the STAN domain substituted to alanine had no effect on centrosome number (Fig. S4 D). Alanine substitutions at the S1108 or S1116 Plk4 phosphorylation sites substantially reduced the ability of overexpressed Myc-GFP-STIL to promote centrosome amplification (reduced to 19% and 15%, respectively), indicating that phosphorylation of these sites is important for the function of STIL in centriole biogenesis. To further characterize STIL S1108 and S1116 phosphorylation sites, we introduced phospho-mimicking mutations at these positions and assayed the ability of the Myc-GFP-STIL constructs to promote centrosome overduplication. The S1108D mutation promoted centrosome amplification as efficiently as Myc-GFP-STIL WT (Fig. S4 D). However, the S1116D mutation was indistinguishable from an alanine substitution at this site, suggesting that either the S1116D substitution failed to mimic the phosphorylated state or that centriole biogenesis requires dynamic regulation of S1116 phosphorylation.
To address the role of Plk4-mediated STIL STAN domain phosphorylation in canonical centriole duplication, we replaced endogenous STIL with near physiological levels of Myc-GFP-STIL transgenes (Fig. 4, A and B). Depletion of STIL led to a 47% increase in the number of cells with ≤1 centrosome, and this effect was completely rescued by expression of an RNAi-resistant Myc-GFP-STIL WT transgene (Fig. 4 C). In contrast, expression of either Myc-GFP-STIL lacking the STAN domain (ΔSTAN, deletion of aa 1061–1147) or the Myc-GFP-STIL 5A mutant lacking five phosphorylation sites in this region failed to rescue centriole duplication (Fig. 4 C). Single alanine substitutions at each of the five-phosphorylation sites in the STIL STAN domain revealed that S1116 was the most important phosphorylation site for controlling centriole duplication (53% of S1116A cells contain ≤1 centrosome; Fig. 4 F). The Myc-GFP-STIL S1108A mutant was also partially defective in centriole duplication (32% S1108A cells contain ≤1 centrosome) and this defect was further exacerbated when combined with the S1116A mutation (61% of S1108A/S1116A cells contain ≤1 centrosome; Fig. 4 F). We conclude that Plk4-mediated phosphorylation of STIL S1116, and to a lesser extent STIL S1108, is required for centriole duplication.
Figure 4.
Phosphorylation of the STIL STAN domain is required for centriole duplication. (A) Outline of the experimental timeline for the STIL siRNA and add-back experiments. (B) Immunoblot showing the relative STIL expression level after replacement of endogenous STIL with a Myc-GFP-STIL WT transgene. (C) Quantification showing the number of CEP192 foci in cells in which endogenous STIL had been depleted and replaced with the indicated Myc-GFP-STIL transgene. Bars represent the mean of at least three independent experiments, with >100 cells counted per experiment. (D and E) Quantification from C showing the relative level of Myc-GFP-STIL at the centrosome of S/G2 phase cells (D) and the fraction of S/G2 phase cells with detectable Myc-GFP-STIL at the centrosome (E). Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. (F) Quantification showing the number of CEP192 foci in cells in which endogenous STIL had been depleted and replaced with the indicated Myc-GFP-STIL transgene. 08A/16A refers to a Myc-GFP-STIL S1108A/S1116A double mutant. Bars represent the mean of at least three independent experiments, with >100 cells counted per experiment. The Myc-GFP-STIL 5A mutant from C is shown alongside as a comparison. (G and H) Quantification from F showing the relative level of Myc-GFP-STIL at the centrosome of S/G2 phase cells (G) and the fraction of S/G2 phase cells with detectable cells Myc-GFP-STIL at the centrosome (H). Bars represent the mean of at least three independent experiments, with >40 cells counted per experiment. The Myc-GFP-STIL 5A mutant from D and E is shown alongside as a comparison. (I) Endogenous STIL was replaced with Myc-GFP-STIL WT, ΔSTAN, or S1116A. The graph shows the fluorescence recovery of centrosomal Myc-GFP-STIL after photobleaching. Points represent the mean of >10 cells from two independent experiments. All error bars represent the SEM.
Plk4 phosphorylation of the STIL STAN domain is required for centriole recruitment of STIL
We next analyzed if STAN phosphorylation contributes to STIL centriole targeting. Because STIL is degraded after mitosis (Tang et al., 2011; Arquint et al., 2012; Arquint and Nigg, 2014), we measured Myc-GFP-STIL levels in S/G2 cells marked by the presence of CENP-F (Hussein and Taylor, 2002). While all WT and mutant Myc-GFP-STIL transgenes localized to the centriole in the absence of endogenous STIL, the relative abundance of each phosphorylation site mutant at the centriole varied considerably. Surprisingly, although deletion of the STAN domain did not alter the centriole abundance of STIL, mutation of five phosphorylation sites in this region reduced the abundance of centriole STIL to <25% of that of WT STIL (Fig. 4, D and E). The difference in centriole abundance of STIL 5A and STIL ΔSTAN suggests that the STAN domain acts to inhibit STIL centriole localization and that phosphorylation of the STAN domain is able to overcome this inhibition to promote localization. Preventing phosphorylation of both the S1108 and S1116 Plk4 phosphorylation sites reduced the abundance of STIL at the centriole to 32% of control cells (Fig. 4, G and H). This suggests that phosphorylation by Plk4 of S1108 and S1116 overcomes the STAN domain–mediated inhibition of STIL centriole localization. Collectively, these observations offer an explanation for why the centriole levels of STIL are reduced after Plk4 inhibition (Fig. 1 F).
Mutating STIL S1108 to aspartic acid increased the centriolar abundance of Myc-GFP-STIL in excess of the WT protein (Fig. S4 E). Nevertheless, Myc-GFP-STIL S1108D was as defective in centriole duplication as the S1108A mutant protein, which suggests that STAN phosphorylation performs functions in addition to centriole recruitment (Fig. S4 F). Because the Myc-GFP-STIL S1108D promoted centrosome amplification as effectively as Myc-GFP-STIL WT when overexpressed in the presence of endogenous STIL (Fig. S4D), our data highlight differences in assays that use STIL overexpression versus functional replacement.
To examine how STAN domain phosphorylation affects the binding dynamics of centriolar STIL, we performed fluorescence recovery after photobleaching in cells expressing Myc-GFP-STIL transgenes. Myc-GFP-STIL WT and S1108A only partially recovered after bleaching, showing that STIL exists in both a mobile and immobile pool at the centriole (Fig. 4 I, Myc-GFP-STIL WT R% = 48%, t1/2 = 127 s). Deletion of the STAN domain or mutation of the S1116 phosphorylation site both increased the mobile fraction of centriolar STIL (Fig. 4 I; Myc-GFP-STIL ΔSTAN, R% = 93%, t1/2 = 110 s; Myc-GFP-STIL S1116A, R% = 69%, t1/2 = 80 s). Together, these data suggest that phosphorylation of the STAN domain is required for stable interaction of STIL with the centriole. The increased turnover of Myc-GFP-STIL ΔSTAN is likely to limit the centriolar accumulation of this mutant protein. This could explain why Myc-GFP-STIL ΔSTAN that lacks a domain inhibitory to centriole recuiment localizes to the centriole at a level similar to Myc-GFP-STIL WT.
Stable centriole recruitment of STIL requires direct binding of SAS6
Recently, it was proposed that phosphorylation of the STIL/Ana2 STAN domain facilitates STIL binding to SAS6 (Dzhindzhev et al., 2014; Ohta et al., 2014). To test if phosphorylation of STIL S1108 and S1116 by Plk4 promotes the association of SAS6, we reconstituted SAS6 binding to STIL in vitro. GST-STIL was incubated with kinase-active or inactive Plk4 and then combined with SAS6. GST-STIL was then captured on beads and the association with SAS6 was determined by immunoblotting. Incubation with kinase-active but not kinase-dead Plk4 promoted direct binding of SAS6 to GST-STIL (Fig. 5 A). To establish if SAS6 binding was dependent on phosphorylation of STIL S1108 or S1116, we tested the ability of SAS6 to bind in vitro to WT or phosphorylation site mutants of recombinant GST-STIL C-term (aa 898–1287). Phosphorylation of GST-STIL C-term by Plk4 increased the binding of SAS-6 by more than ninefold (Fig. 5 B). Importantly, mutation of STIL S1108A and S1116A reduced SAS6 binding to 37% and 22% of that observed with WT GST-STIL C-term (aa 898–1287). To map the domain of SAS6 that interacts with STIL, cells were transfected with a series of FLAG-SUMO-SAS6 truncation constructs, and the interaction with Myc-GFP-STIL was examined in the presence of Plk4Δ24,WT-mCherry. Using this approach, STIL binding was mapped to the C-term part of SAS6 (aa 316–657; Fig. S5 A). We conclude that phosphorylation of the STIL STAN domain by Plk4 promotes direct binding of STIL to the C-term region of SAS6.
We next analyzed the requirement of STIL phosphorylation for binding to SAS-6 in cells. Deletion of the STIL STAN domain or mutation of phosphorylation sites in this region did not affect STIL binding to Plk4, or activation of Plk4 kinase activity (Fig. 5 C). Expression of kinase-active Plk4Δ24 promoted an approximately twofold increase in the binding of FLAG-SUMO-SAS6 to Myc-GFP-STIL (Fig. 5 C, lanes 2 and 3). The Myc-GFP-STIL 5A mutant associated with SAS6 at only 23% of the level observed with Myc-GFP-STIL WT (Fig. 5 C, lanes 2 and 7). Importantly, preventing phosphorylation of STIL S1116 alone also reduced SAS6 binding to a similar degree, whereas deletion of the STAN domain reduced SAS6 binding to 12% of that observed with Myc-GFP-STIL WT (Fig. 5 C, lanes 2, 5, and 8). These data indicate that Plk4-mediated phosphorylation of STIL S1116 plays a key role in promoting SAS6 binding to STIL.
Finally, we investigated how Plk4-mediated phosphorylation of STIL contributes to SAS6 recruitment to the centriole. We monitored the presence of Plk4 and SAS6 at the centriole of cells in which endogenous STIL had been functionally replaced with various Myc-GFP-STIL transgenes. Despite the fact that the centriolar abundance of the STIL transgenes varied dramatically (Fig. 4, G and H), cells contained near identical levels of endogenous centriolar Plk4 (Fig. 5 D). Because binding to Plk4 is unaffected by STIL STAN domain phosphorylation (Fig. 5 C), our data suggest that the abundance of centriolar Plk4 is mainly controlled through binding to STIL in the cytosol. In accord with previous reports, depletion of STIL dramatically reduced SAS6 recruitment to the centriole (a mean of 13% SAS6 remaining) without altering SAS6 protein levels (Figs. 5 E and S5 B). While expression of Myc-GFP-STIL WT rescued the centriole recruitment of SAS6 in cells depleted of endogenous STIL, the Myc-GFP-STIL 5A mutant failed to do so (19% centriolar SAS6 remaining; Fig. 5, E and F). Preventing phosphorylation of STIL S1108 or S1116 also reduced SAS6 recruitment (64% and 36% of centriolar SAS6 remaining, respectively). This suggests that Plk4-mediated phosphorylation of the STIL STAN domain contributes to the efficient targeting of SAS6 to the centriole.
Discussion
STIL is an in vivo regulator of Plk4 kinase activity
An important unanswered question is how Plk4 kinase activity is temporally controlled to promote centriole assembly. Our findings reveal that direct binding of Plk4 to STIL stimulates Plk4 kinase activity by promoting self-phosphorylation of the activation loop of the Plk4 kinase domain (Fig. 2, E and G). In cycling cells, STIL accumulates in late G1/early S phase and is then degraded after anaphase onset (Fig. S5 C; Tang et al., 2011; Arquint et al., 2012; Arquint and Nigg, 2014). We therefore speculate that the cell cycle–regulated accumulation of STIL could provide the trigger for activation of Plk4 at the G1/S boundary.
How STIL binding promotes Plk4 activation remains unclear. One possibility is that the binding of STIL triggers a conformational change in Plk4 that positions the Plk4 activation loop for optimal self-phosphorylation. Alternatively, STIL may promote the recruitment of an additional factor that serves to activate Plk4.
We have provided direct evidence to show that endogenous Plk4 directly phosphorylates STIL S1108 and S1116 in vivo and reveal two key roles for these phosphorylation events in promoting centriole assembly (Fig. 3, D and G). First, phosphorylation of S1108 and S1116 increased the efficiency of STIL centriole targeting (Fig. 4, G and H). This explains why Plk4 kinase activity is required for the robust targeting of STIL to the centriole (Fig. 1 F). Second, and consistent with two recent reports (Dzhindzhev et al., 2014; Ohta et al., 2014), we show that phosphorylation of STIL S1108 and S1116 is required for subsequent binding of STIL to SAS6 and for efficient recruitment of SAS6 to the centriole (Fig. 5, B and E). We speculate that STIL binding to SAS6 facilitates cartwheel assembly, leading to the stable incorporation of STIL into the centriole structure.
In human cells, SAS6 is transiently recruited to the lumen of the mother centriole in early S phase, before repositioning to the outer wall of the mother centriole to initiate cartwheel formation and procentriole assembly (Fong et al., 2014). Plk4 and STIL are both required for the release of luminal SAS6. It is therefore tempting to speculate that STIL-mediated activation of Plk4 triggers the release of luminal SAS6 and subsequent capture by STIL at a site on the wall of the mother. Further studies will be required to test this idea.
STIL localization to the centriole requires both Plk4 binding and STAN domain phosphorylation
A conserved coiled-coil domain in the central region of STIL has been shown to be necessary for Plk4 binding (Ohta et al., 2014; Kratz et al., 2015). We demonstrate that Plk4 and STIL form a complex both in vitro and in vivo in the absence of Plk4 activity (Fig. S2, A and B). This contrasts with a previous study that indicated that the binding of Plk4 to STIL required Plk4 kinase activity (Ohta et al., 2014). While the reason for this discrepancy remains unclear, we note that in C. elegans, ZYG-1/Plk4 binds directly to the coiled-coil of SAS6 independent of kinase activity (Lettman et al., 2013). It is thus tempting to speculate that while the location of the ZYG-1/Plk4 binding site differs, a conserved role of the ZYG-1/Plk4-SAS5/STIL-SAS6 module is to position ZYG-1/Plk4 for optimal phosphorylation of SAS5/STIL family proteins.
A mutant form of STIL lacking the central coiled-coil had a dramatically reduced localization to the centriole, suggesting that Plk4 may act as a centriole receptor for STIL (Fig. S3 D). In flies, Ana2/STIL localizes to the centriole in the absence of phosphorylation by Plk4 (Dzhindzhev et al., 2014). In addition, a study in human cells reported that phosphorylation of the STIL STAN domain was not required for centriole targeting of STIL (Ohta et al., 2014). How, then, do we explain the observation that Plk4 activity is also required for the localization of STIL to the centriole (Fig. 1 F)? Our evidence shows that, although not essential for STIL centriole recruitment, phosphorylation of the STAN domain by Plk4 dramatically increased the efficiency of STIL centriole targeting (Fig. 4, G and H). Importantly, we show that Plk4 binding to STIL does not require STIL STAN domain phosphorylation (Fig. 5 C), demonstrating that centriole targeting of STIL requires both Plk4 binding and phosphorylation of its STAN domain by Plk4.
How does phosphorylation of the STAN domain control the localization of STIL? We speculate that cytoplasmic STIL exists in an autoinhibited conformation that prevents recruitment to the centriole (Fig. 6). Deletion of the STIL STAN domain or phosphorylation of this region by Plk4 is proposed to release this autoinhibition to allow efficient centriole targeting. Phosphorylation of the STIL STAN domain also triggers the binding of STIL to SAS6. This interaction could promote cartwheel assembly and lead to the stable incorporation of STIL in the cartwheel structure (Fig. 6). This explains why STIL mutants that are defective in STAN domain phosphorylation have a reduced pool of protein stably bound at the centriole (Fig. 4 I).
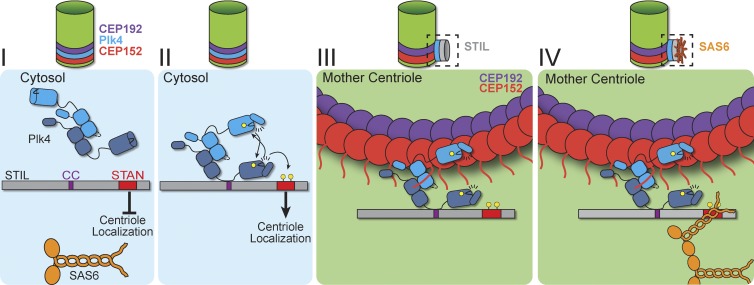
Figure 6.
A model for how Plk4 and STIL cooperate to promote centriole assembly. (I) We propose that cytoplasmic STIL exists in an autoinhibited conformation that prevents recruitment to the centriole. (II) Plk4 directly binds to STIL, and this binding leads to activation of kinase activity. Plk4 activation is driven by self-phosphorylation of the activation loop (Bettencourt-Dias, personal communication). Plk4 then directly phosphorylates two sites in the STAN domain of STIL. (III) Phosphorylation of these sites releases STIL autoinhibition to promote efficient centriole targeting. (IV) In a second step, STIL STAN domain phosphorylation promotes the binding of centriolar STIL to the C-term region of SAS6. We propose that binding of STIL to SAS6 triggers cartwheel assembly and the stable binding of STIL to the centriole.
At the G1/S border, centriolar Plk4 transitions from a ring-like arrangement to a single focus on the wall of the parental centriole (Kim et al., 2013; Sonnen et al., 2013; Ohta et al., 2014). Understanding how this transition is controlled is central to understanding how a single new centriole is created on each parental centriole. Because STIL binding stimulates Plk4 kinase and subsequent destruction (Fig. 2 A), STIL recruitment may lead to the activation and destruction of Plk4 that is localized around the wall of the parental centriole. Consistently, it was recently shown that depletion of STIL prevented the formation of a single focus of Plk4 (Ohta et al., 2014). How a single focus of Plk4 is protected from self-destruction remains an important question for future studies.
Plk4 kinase activity is not required for continued growth of DLD-1 colon cancer cells
While the abundance of Plk4 is normally carefully controlled, alterations in Plk4 expression has been reported in several tumor types, prompting proposals that Plk4 inhibition may be an effective anticancer therapy (Mason et al., 2014). Surprisingly, we now show that specific inhibition of Plk4 kinase activity in a human cancer cell line with compromised p53 function results in a complete loss of centrioles and centrosomes, but only modestly reduced cell growth (Fig. 1, B and E). This demonstrates that Plk4 and centrioles are not essential for cell cycle progression, at least in transformed cells. It therefore remains to be determined whether Plk4 inhibition will be a useful strategy in cancer therapy.
Materials and methods
Antibody production
A C-term hPlk4 fragment (aa 510–970) was cloned into a pET-23b bacterial expression vector (EMD Millipore) containing a C-term 6×His tag. Recombinant protein was purified from Escherichia coli using Ni-NTA beads (QIAGEN) and used for immunization (Prosci). A STIL C-term peptide VGTFLDVKRLRQLPKLF (aa 1271–1287) was synthesized and conjugated to KLH for immunization. Rabbit immune sera were affinity-purified using standard procedures. Affinity-purified antibodies were directly conjugated to DyLight 550 and DyLight 650 fluorophores (Thermo Fisher Scientific) for use in immunofluorescence.
A synthetic phospho-peptide based on the human hSTIL sequence flanking serine 1108 [CDRSTVGL(pS)LISPN] or 1116 [CSPNNM(pS)FATKK] was synthesized, coupled to KLH, and injected into rabbits (Prosci). Polyclonal pS1108 and pS1116 antibodies were affinity-purified using the appropriate phosphopeptide coupled to a SulfoLink Coupling Resin (Thermo Fisher Scientific).
Cell culture and drug treatments
Cells were maintained at 37°C in a 5% CO2 atmosphere with 21% oxygen. Cells were grown in DMEM containing 10% fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine. 293FT cells were used in cotransfection experiments (Fig. 2, A and E–I; Fig. 5 C; Fig. S2; Fig. S3; and Fig. S5 A), while Flp-In TRex-DLD-1 cells (a gift from S. Taylor, the University of Manchester, Manchester, England, UK) were used in all other experiments. Flp-In TRex-DLD-1 cells were engineered using the Flp-In TRex Core kit (Life Technologies) to stably express the Tetracycline repressor protein and contain a single, genomic Flp recombination target site (FRT)/lacZeo site. 3MB-PP1 (Sigma-Aldrich) was dissolved in DMSO and used at a final concentration of 10 µM unless otherwise stated.
Gene targeting
Gene targeting was performed in Flp-In TRex-DLD-1 cells using CRISPR/Cas9. In brief, a gRNA targeting Plk4 (AGATAGCAATTATGTGTATC) was cloned into the PX459 expression vector that coexpresses the gRNA from a U6 promoter and SpCas9 from a CMV promoter. Cells were cotransfected with a 1:20 molar ratio of the PX459 plasmid and a 160-bp single-stranded oligonucleotide repair template. The repair template introduced the L89G mutation, a silent AflIII restriction site, and a mutation in the SpCas9 protospacer adjacent motif (PAM) to prevent recutting after homology-directed repair. Transfected cells were selected for 2 d with puromycin and single clones were isolated by limiting dilution. Genomic DNA was isolated from single clones and subjected to PCR using the following primers (forward, GCAGGAATGGTACAGAGAGTCC; reverse, GCAAAACTTTTATCCACCCAAA). PCR products were digested with AflIII for 2 h. Clones with digested PCR products were sequenced to verify insertion of the L89G mutation. Two independent homozygous L89G clones were identified and behaved identically in all assays performed.
L89G donor oligonucleotide: CTGAATTTTTGTATATTTTAATTTATTATGCCCTTTCACATTTCAGCTTTATAACTATTTTGAAGATAGCAATTACGTGTATCTAGTAGGAGAAATGTGCCATAATGGAGAAATGAACAGGTATCTAAAGAATAGAGTGAAACCCTTCTCAGAAAATGAAG
Cloning
All DNA constructs were cloned into a pcDNA5/FRT/TO vector backbone (Life Technologies) and expressed from a CMV promoter under the control of two tetracycline operator sites. All constructs were full-length proteins unless otherwise noted.
Generation of stable cell lines and siRNA treatment
Stable, isogenic cell lines expressing Myc-GFP-STIL from a CMV promoter under the control of two Tetracycline operator sites were generated according to the manufacturer’s recommendation using the FRT/Flp-mediated recombination in Flp-In TRex-DLD-1 cells (Flp-In TRex Core kit; Life Technologies). Expression of Myc-GFP-STIL was induced with 1 µg/ml Tetracycline (Sigma-Aldrich). For RNA interference, 2 × 105 cells were seeded in a 6-well plate and duplexed siRNAs were introduced using RNAiMax (Life Technologies). siRNA directed against STIL (5′-GCUCCAAACAGUUUCUGCUGGAAU-3′) was purchased from GE Healthcare and control siRNA (Universal Negative Control #1) was purchased from Sigma-Aldrich. 24 h after transfection, tetracycline was added to induce expression of Myc-GFP-STIL. Cells were harvested and processed for immunoblotting or fixed for immunofluorescence 24 h later.
Cell biology
To prepare cells for flow cytometry, cell pellets were fixed in cold 70% EtOH for 24 h, washed once in PBS, and suspended in PBS supplemented with 0.5 mg/ml RNase A and 50 mg/ml propidium iodide (PI). Samples were incubated at room temperature for 30 min and analyzed on a flow cytometer (FACSCalibur; BD). For metaphase spreads, cells were treated for up to 4 h with 3.3 µM nocodazole, then incubated in 0.45% hypotonic buffer (32 mM KCl, 16 mM Hepes, and 0.5 mM EDTA, pH 7.4) at 37°C for 20 min. Cells were fixed in methanol/acetic acid (3:1) and stored at –20°C overnight. Fixed cells were dropped onto acetic acid–coated slides and air-dried. Chromosomes were stained with Hoechst, mounted, and imaged. For clonogenic assays, 500 cells were seeded in a 10-cm2 culture dish and left to grow for ∼2 wk until colonies were visible by eye. Cells were fixed in methanol for 10 min at room temperature and colonies were stained with crystal violet (Sigma-Aldrich).
Coimmunoprecipitation
2 × 106 293-FT cells were seeded into 10-cm2 dishes and 24 h later transfected with 2 µg of plasmid DNA. 48 h later, transfected cells were lysed in lysis buffer (10 mM Tris, pH 7.5, 0.2% Triton X-100, 300 mM NaCl, 1 mM EDTA, 50 mM NaF, 50 mM β-glycerophosphate, 1 mM DTT, 500 nM microcystin, 1 mM PMSF, and EDTA-free protease inhibitor tablet [Roche]) and sonicated, then soluble extracts were prepared. The supernatant was incubated with beads coupled to GFP-binding protein (Rothbauer et al., 2008). Alternatively, 2 µg of anti-mCherry antibody (rabbit, a gift from J. Soek-Han, Ludwig Institute for Cancer Research, La Jolla, CA) was added per sample and collected using Affi-Prep Protein A (Bio-Rad Laboratories; Fig. 2 G). Beads were washed three times in lysis buffer and immunopurified protein was analyzed by immunoblotting. For lambda phosphatase treatment, cells were lysed in lambda phosphatase buffer (New England Biolabs, Inc.) and soluble lysates were incubated with 2 µl of Lambda Protein Phosphatase (New England Biolabs, Inc.) for 60 min at 30°C.
Immunoblotting and immunofluorescence
For immunoblot analysis, protein samples were separated by SDS-PAGE, transferred onto nitrocellulose membranes with a Trans-Blot Turbo Transfer System (Bio-Rad Laboratories), and then probed with the following antibodies: DM1A (mouse anti–α-tubulin, T6199, 1:5,000; Sigma-Aldrich), STIL (rabbit, A302-441A, 1:2,500; Bethyl Laboratories), FLAG M2 (mouse, F1804, 1:1,000; Sigma-Aldrich), Myc 4A6 (mouse, 1:1,000; EMD Millipore), SAS6 (mouse, sc-81431, 1:1,000; Santa Cruz Biotechnology, Inc.), Plk4 pT170 (rabbit, 1:1,000; a gift from M. Takekawa, The University of Tokyo, Tokyo, Japan; Nakamura et al., 2013), Plk4 (rabbit, 1:3,200; this study), mCherry (rabbit, 1:1,000; a gift from J. Soek-Han), and STIL pS1116 (rabbit, this study, 1:250).
For immunofluorescence, cells were grown on 18-mm glass coverslips and fixed in 100% ice cold methanol for 10 min. Cells were blocked in 2.5% FBS, 200 mM glycine, and 0.1% Triton X-100 in PBS for 1 h. Antibody incubations were conducted in the blocking solution for 1 h. DNA was detected using DAPI and cells were mounted in Prolong Antifade (Invitrogen). Staining was performed with the following primary antibodies: GTU-88 (mouse anti–γ-tubulin, 1:250; Abcam), Centrin (mouse, 04-1624, 1:1,000; EMD Millipore), CNAP (guinea pig, raised against the CNAP peptide sequence SPTQQDGRGQKNSDAKC, 1:1000; a gift from O. Stemmann, University of Bayreuth, Bayreuth, Germany,), CEP152 (rabbit, A302-479A, 1:5,000; Bethyl Laboratories, Inc.), Plk4-650 (directly labeled rabbit, 1:1,000, this study), STIL-550 (directly labeled rabbit, 1:1,000, this study), STIL pS1108 (rabbit, 1:250, this study), CEP135 (rabbit, raised against CEP135 aa 695–838, 1:1,000; a gift from A. Hyman, Max Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany), CEP192-Cy3 (directly labeled rabbit, raised against CEP192 aa 1–211, 1:1,000; a gift from K. Oegema, Ludwig Institute for Cancer Research, La Jolla, CA), SAS6-Cy3 (directly labeled rabbit, raised against SAS6 aa 501–657, 1:1,000; a gift from K. Oegema), CPAP-Cy3 (1:1,000; directly labeled rabbit, a gift from K. Oegema), and CENP-F (sheep, raised against CENP-F aa 1363–1640, 1:1000; a gift from S. Taylor). Secondary donkey antibodies were conjugated to Alexa Fluor 488, 555, or 650 (Life Technologies).
For the cell cycle analysis of STIL levels shown in Fig. S3 F, cells were pulsed with EdU for 1 h before fixation in 100% ice cold methanol at −20°C for 10 min. Cells were washed three times with 0.1% Triton X-100 in PBS and stained using a Click-It EdU Alexa Fluor 555 imaging kit (Life Technologies) according to the manufacturer’s recommendations. Cells were blocked in 2.5% FBS, 200 mM glycine, and 0.1% Triton X-100 in PBS for 1 h, and immunofluorescence microscopy was performed using the following antibodies: CENP-F, GTU-88, and STIL-550. G1 phase cells were classified as CENP-F and EdU negative, S phase cells were classified as EdU positive, and G2 phase cells were classified as CENP-F positive and EdU negative. The γ-tubulin staining was used to define the position of the centrosome.
Immunofluorescence images were collected using a DeltaVision Elite system (GE Healthcare) controlling a Scientific CMOS camera (pco.edge 5.5). Acquisition parameters were controlled with the SoftWoRx suite (GE Healthcare). Images were collected at room temperature using an Olympus 60× 1.42 NA or Olympus 100× 1.4 NA oil objective lens with 0.2 µM z sections and subsequently deconvolved in the SoftWoRx suite. Images were acquired using Applied Precision immersion oil (n = 1.516). For quantitation of signal intensity at the centrosome, deconvolved 2D maximum intensity projections were saved as 16-bit TIF images. Signal intensity was determined using ImageJ, by drawing a circular region of interest (ROI) around the centriole (ROI S). A larger concentric circle (ROI L) was drawn around ROI S. ROI S and L were transferred to the channel of interest and the signal in ROI S was calculated using the following formula: IS – [(IL-IS/AL-AS) x AS]. A, area; I, integrated pixel intensity.
Fluorescence recovery after photobleaching
Cells were seeded into four-chamber, 35-mm glass bottom culture dishes (Greiner) and maintained in cell culture medium at 37°C and 5% CO2 in an environmental control station. Images were collected using a 40× 1.4 NA Plan-Apochromat oil-immersion objective lens (Carl Zeiss) on a confocal microscope (LSM 780; Carl Zeiss) equipped with a solid-state 488-nm laser and a spectral GaAsP detector. Images were acquired using an immersion oil lens (n = 1.518; Carl Zeiss). Acquisition parameters, shutters, and focus were controlled by Zen black software (Carl Zeiss). 10 × 0.5 µM z sections were acquired for EGFP at each time point. Two consecutive prebleach scans were collected at 5% of the maximum ATOF value. Centrosome-localized EGFP-STIL was bleached within a circular region encompassing the centrosome (∼3 µM in diameter) at 100% laser power with a 100-µs dwell time. Post-bleach scans were performed at 20-s time intervals for a total period of 400 s. Maximum intensity projections were created using Zen black (Carl Zeiss). The integrated intensity value within a circular ROI in the cytosol of the cell was subtracted from an identically sized region of interest drawn around the bleached centrosome. Recovery values were plotted relative to the difference between the fluorescence before and after bleaching.
Recombinant protein expression and purification
GFP-binding protein (GBP) and recombinant His-hPlk4 (aa 1–416) were expressed and purified from E. coli (strain Rosetta [DE3]) using standard procedures. Recombinant GST-hPlk4, GST-hSAS6, GST-hSTIL, and GST-hSTIL C-term (aa 898–1287) were expressed and purified from High Five insect cells (Invitrogen) using the Bac-to-Bac expression system (Invitrogen). Infected cell pellets were suspended in lysis buffer (10 mM PO43−, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10% glycerol, 2 mM MgCl2, 5 mM DTT, 100 nM Microcystin, 1 mM Na3VO4, 250 U of Benzonase nuclease [Sigma-Aldrich], 1 mM PMSF, and EDTA-free protease inhibitor tablet [Roche]) and lysed by sonication. After centrifugation at 15,000 rpm for 30 min, the supernatant was supplemented with 110 mM KCL and 0.1% Triton X-100, and incubated with Glutathione Sepharose beads (GE Healthcare) for 4 h at 4°C. Beads were washed extensively in wash buffer (10 mM PO43−, pH 7.4, 237 mM NaCl, 2.7 mM KCl, 10% glycerol, 5 mM DTT, 0.1% Triton X-100, 1 mM PMSF, and EDTA-free protease inhibitor tablet [Roche]), and protein was eluted in elution buffer (10 mM PO43−, pH 7.4, 137 mM NaCl, 2.7 mM KCl, and 10% glycerol, with 40 mM reduced glutathione and 5 mM DTT). Protein was dialyzed into a final buffer of 10 mM PO43−, pH 7.4, 137 mM NaCl, 2.7 mM KCl, and 10% glycerol. When necessary, the GST tag was removed by overnight incubation with GST-PreScission protease (GE Healthcare).
In vitro kinase assay
In vitro kinase assays were performed for 30 min at 30°C in 20 mM Tris, pH 7.5, 25 mM KCl, 1 mM MgCl2, and 1 mM DTT, in the presence of 10 µM MgCl2 and 100 µM ATP. For radioactive kinase assays, reactions took place in the presence of 100 µM ATP and 0.03 µCi γ-[32P]ATP. 2 µg of substrate was incubated with 1 µg of of His-hPlk4 (aa 1–416). Kinase reactions were stopped with sample buffer and analyzed by SDS-PAGE.
In vitro binding assay
Recombinant GST-hSTIL or GST-hSTIL C-term (aa 898–1287) were incubated with kinase-active or kinase-dead His-hPlk4 (aa 1–416) in kinase buffer with or without cold ATP as described in the preceding paragraph. Reactions were then supplemented with 500 µl of binding buffer (50 mM Na-Hepes, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1% Triton X-100, 100 nM Microcystin [EMD Millipore], and 0.5 mg/ml BSA) and incubated at 4°C for 1 h. Glutathione Sepharose beads (GE Healthcare) were incubated with the protein for a further 1 h at 4°C. Beads were washed three times in binding buffer without BSA and proteins were eluted in SDS sample buffer.
Mass spectrometry
In-solution protein digestion was performed using the filter-assisted sample preparation (FASP) method (Wiśniewski et al., 2009). Data-dependent tandem mass spectrometry (MS/MS) analysis of peptides was performed on the LTQ-Orbitrap Velos (Thermo Fisher Scientific) interfaced with Eksigent 2D nanoflow liquid chromatography system (SCIEX). Peptides were enriched on a 2-cm trap column (YMC gel ODS-A S-10 µm), fractionated on a 75 µm × 15 cm column packed with 5 µm, 100-Å Magic AQ C18 material (Michrom Bioresources), and electrosprayed through a 15-µm emitter (PF3360-75-15-N-5; New Objective). Reversed-phase solvent gradient consisted of 0.1% formic acid with increasing levels of 0.1% formic acid, 90% acetonitrile over a period of 90 min. LTQ Orbitrap Velos was set at 2.0 kV spray voltage, full MS survey scan range was set at 350–1800 m/z, and data-dependent HCD MS/MS analysis was set for top 8 precursors with minimum signal of 2,000. Other parameters include peptide isolation width of m/z 1.9; dynamic exclusion limit 30 s and normalized collision energy 35; precursor and the fragment ions resolutions were 30,000 and 15,000, respectively. Internal mass calibration was applied using lock mass ion m/z = 371.101230.
Mass spectrometry raw files were automatically processed through Proteome Discoverer 1.4 software (Thermo Fisher Scientific). Raw MS and MS/MS data were isotopically resolved with deconvolution and de-isotoping using Xtract (Thermo Fisher Scientific) and MS2 processor software in addition to a default spectrum selector node. The data were selected from Refseq human entries using the Mascot (v2.2.6; Matrix Sciences) search engine interfaced with different processing nodes of Proteome Discoverer 1.4. Mass tolerances on precursor and fragment masses were set to 15 ppm and 0.03 D, respectively. The peptide validator node was used for identification confidence, and a 1% false discovery rate cutoff was used to filter the peptides. Phosphorylation site probability was analyzed using the phosphoRS 3.0 node in Proteome discoverer software (Taus et al., 2011).
Online supplemental material
Fig. S1 shows that inhibition of Plk4 kinase activity leads to a progressive loss of centrioles, that de novo centrioles are created when Plk4 kinase activity is restored in centriolar cells, and that chronic inhibition of Plk4 activity does not alter clonogenic survival, but leads to increased levels of aneuploidy. Fig. S2 shows that Plk4 binds to STIL regardless of kinase activity in vivo and in vitro, and that aa 715–850 of STIL are sufficient for Plk4 binding. Fig. S3 demonstrates that the conserved coiled-coil domain of STIL (aa 721–747) is required for Plk4 binding, the centriole recruitment of STIL, and centriole duplication. Fig. S4 shows that Plk4 phosphorylates STIL in vitro, that STIL pS1108 and pS1116 antibodies are phosphospecific, and that phosphorylation of STIL S1108 and S1116 is required for centriole duplication. Fig. S5 shows that STIL binds to the C terminus of SAS6, that SAS6 levels remain unaffected after STIL knockdown, and that centriolar STIL levels fluctuate in a cell cycle–dependent manner. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502088/DC1.
Supplementary Material
Acknowledgements
The authors would like to thank Stephen Taylor, Mutsuhiro Takekawa, Joo Soek Han, Anthony Hyman, Olaf Stemmann, and Karen Oegema for providing reagents for this study. We thank the Johns Hopkins Center for Sensory Biology/Hearing and Balance imaging center for use of their confocal microscope. We thank Michelle Levine and Randall Reed for comments on the manuscript.
This work was supported by a W.W. Smith Charitable Trust Research grant (#C1304), a March of Dimes Basil O’Conner Scholar Award (5-FY14-83), a Pew Scholar Award (27348), a Kimmel Scholar Award (90059259), and an R01 research grant (GM 114119).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AS
- analogue-sensitive
- C-term
- C terminal
- FRT
- Flp recombination target site
- MPD
- multi-phosphodegron
- ROI
- region of interest
- WT
- wild type
References
- Arquint C., and Nigg E.A.. 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 24:351–360. 10.1016/j.cub.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Arquint C., Sonnen K.F., Stierhof Y.D., and Nigg E.A.. 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125:1342–1352. 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., and Raff J.W.. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042. 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H., and Anderson K.V.. 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl. Acad. Sci. USA. 111:E1491–E1500. 10.1073/pnas.1400568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E., Dominguez P., and Gonzalez C.. 2008. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr. Biol. 18:1209–1214. 10.1016/j.cub.2008.07.029 [DOI] [PubMed] [Google Scholar]
- Chan J.Y. 2011. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 7:1122–1144. 10.7150/ijbs.7.1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19:43–49. 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Bento I., Pimenta-Marques A., Jana S.C., Lince-Faria M., Duarte P., Borrego-Pinto J., Gilberto S., Amado T., Brito D., et al. . 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23:2245–2254. 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Müller-Reichert T., Pelletier L., Habermann B., Desai A., and Oegema K.. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829. 10.1016/j.devcel.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Debec A., Sullivan W., and Bettencourt-Dias M.. 2010. Centrioles: active players or passengers during mitosis? Cell. Mol. Life Sci. 67:2173–2194. 10.1007/s00018-010-0323-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Leidel S., Wani K., Baumer K., Bamat J., Schnabel H., Feichtinger R., Schnabel R., and Gönczy P.. 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6:656–664. 10.1038/ncb1146 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., and Glover D.M.. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24:2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong C.S., Kim M., Yang T.T., Liao J.C., and Tsou M.F.. 2014. SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication. Dev. Cell. 30:238–245. 10.1016/j.devcel.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho S.A., Picone R., Burute M., Dagher R., Su Y., Leung C.T., Polyak K., Brugge J.S., Théry M., and Pellman D.. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 510:167–171. 10.1038/nature13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Guderian G., Westendorf J., Uldschmid A., and Nigg E.A.. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 123:2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., and Stearns T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffert J.D., Pisitkun T., Wang G., Shen R.F., and Knepper M.A.. 2006. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc. Natl. Acad. Sci. USA. 103:7159–7164. 10.1073/pnas.0600895103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., and Cleveland D.W.. 2014. Polo-like kinase 4 inhibition: a strategy for cancer therapy? Cancer Cell. 26:151–153. 10.1016/j.ccr.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., and Cleveland D.W.. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Fachinetti D., Zhu Q., Bauer M., Verma I.M., Nigg E.A., and Cleveland D.W.. 2012. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 26:2684–2689. 10.1101/gad.207027.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein D., and Taylor S.S.. 2002. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 115:3403–3414. [DOI] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., and Gygi S.P.. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 143:1174–1189. 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo D., Wang W.J., Uryu K., and Tsou M.F.. 2014. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Reports. 8:957–965. 10.1016/j.celrep.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.F., Stewart K.D., Woods K.W., Giranda V.L., and Luo Y.. 2007. Pharmacological and functional comparison of the polo-like kinase family: insight into inhibitor and substrate specificity. Biochemistry. 46:9551–9563. 10.1021/bi7008745 [DOI] [PubMed] [Google Scholar]
- Kemp C.A., Kopish K.R., Zipperlen P., Ahringer J., and O’Connell K.F.. 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell. 6:511–523. 10.1016/S1534-5807(04)00066-8 [DOI] [PubMed] [Google Scholar]
- Kettenbach A.N., Wang T., Faherty B.K., Madden D.R., Knapp S., Bailey-Kellogg C., and Gerber S.A.. 2012. Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem. Biol. 19:608–618. 10.1016/j.chembiol.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., and Rieder C.L.. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153:237–242. 10.1083/jcb.153.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. . 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert T., Oegema K., Grill S., and Hyman A.A.. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587. 10.1016/S0092-8674(03)00117-X [DOI] [PubMed] [Google Scholar]
- Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M.C., Flückiger I., Gönczy P., and Steinmetz M.O.. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell. 144:364–375. 10.1016/j.cell.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Buster D.W., Nguyen A.L., Swatkoski S., Gucek M., Rusan N.M., and Rogers G.C.. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 23:2255–2261. 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz A.S., Bärenz F., Richter K.T., and Hoffmann I.. 2015. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open. 4:370–377. 10.1242/bio.201411023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer R., Forrest B., Li S.W., Liu Y., Sampson P., Edwards L., Lang Y., Awrey D.E., Mao G., Plotnikova O., et al. . 2013. The discovery of PLK4 inhibitors: (E)-3-((1H-Indazol-6-yl)methylene)indolin-2-ones as novel antiproliferative agents. J. Med. Chem. 56:6069–6087. 10.1021/jm400380m [DOI] [PubMed] [Google Scholar]
- Leidel S., and Gönczy P.. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 4:431–439. 10.1016/S1534-5807(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., and Gönczy P.. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125. 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Lettman M.M., Wong Y.L., Viscardi V., Niessen S., Chen S.H., Shiau A.K., Zhou H., Desai A., and Oegema K.. 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev. Cell. 25:284–298. 10.1016/j.devcel.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J.M., Lin D.C., Wei X., Che Y., Yao Y., Kiarash R., Cescon D.W., Fletcher G.C., Awrey D.E., Bray M.R., et al. . 2014. Functional characterization of CFI-400945, a Polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell. 26:163–176. 10.1016/j.ccr.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Saito H., and Takekawa M.. 2013. SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat. Commun. 4:1775 10.1038/ncomms2752 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- O’Connell K.F., Caron C., Kopish K.R., Hurd D.D., Kemphues K.J., Li Y., and White J.G.. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558. 10.1016/S0092-8674(01)00338-5 [DOI] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., Ozlü N., Hannak E., Cowan C., Habermann B., Ruer M., Müller-Reichert T., and Hyman A.A.. 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14:863–873. 10.1016/j.cub.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Roberts D.M., Peifer M., and Rogers S.L.. 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239. 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M., and Cimini D.. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J.H., Pütz M., Daly O., Morrison C.G., Dunning M., Kilmartin J.V., and Gergely F.. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 203:747–756. 10.1083/jcb.201309038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Gabryjonczyk A.M., Anselm E., Stierhof Y.D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Stevens N.R., Dobbelaere J., Brunk K., Franz A., and Raff J.W.. 2010a Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188:313–323. 10.1083/jcb.200910016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Roque H., and Raff J.W.. 2010b DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell. 19:913–919. 10.1016/j.devcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur S., Pagliarini R., Bunz F., Rago C., Diaz L.A. Jr., Kinzler K.W., Vogelstein B., and Papadopoulos N.. 2009. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc. Natl. Acad. Sci. USA. 106:3964–3969. 10.1073/pnas.0813333106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow C.J., Ko M.A., Siddiqui N.U., Hudson J.W., and Dennis J.W.. 2005. Sak/Plk4 and mitotic fidelity. Oncogene. 24:306–312. 10.1038/sj.onc.1208275 [DOI] [PubMed] [Google Scholar]
- Tang C.J.C., Lin S.Y., Hsu W.B., Lin Y.N., Wu C.T., Lin Y.C., Chang C.W., Wu K.S., and Tang T.K.. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30:4790–4804. 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K.. 2011. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10:5354–5362. 10.1021/pr200611n [DOI] [PubMed] [Google Scholar]
- Tsou M.F., and Stearns T.. 2006. Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 18:74–78. 10.1016/j.ceb.2005.12.008 [DOI] [PubMed] [Google Scholar]
- van Breugel M., Hirono M., Andreeva A., Yanagisawa H.A., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I.O., Robinson C.V., et al. . 2011. Structures of SAS-6 suggest its organization in centrioles. Science. 331:1196–1199. 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- van Breugel M., Wilcken R., McLaughlin S.H., Rutherford T.J., and Johnson C.M.. 2014. Structure of the SAS-6 cartwheel hub from Leishmania major. eLife. 3:e01812 10.7554/eLife.01812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., and Krämer A.. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362. 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., and Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.