Studying CENP-A nucleosome assembly in a cell-free system defines the role of existing CENP-A nucleosomes in centromere maintenance.
Abstract
Centromeres are defined by the presence of CENP-A nucleosomes in chromatin and are essential for accurate chromosome segregation. Centromeric chromatin epigenetically seeds new CENP-A nucleosome formation, thereby maintaining functional centromeres as cells divide. The features within centromeric chromatin that direct new CENP-A assembly remain unclear. Here, we developed a cell-free CENP-A assembly system that enabled the study of chromatin-bound CENP-A and soluble CENP-A separately. We show that two distinct domains of CENP-A within existing CENP-A nucleosomes are required for new CENP-A assembly and that CENP-A nucleosomes recruit the CENP-A assembly factors CENP-C and M18BP1 independently. Furthermore, we demonstrate that the mechanism of CENP-C recruitment to centromeres is dependent on the density of underlying CENP-A nucleosomes.
Introduction
Centromeres are epigenetically determined by the presence of the Centromere Protein-A (CENP-A) histone in chromatin. CENP-A nucleosomes are equally distributed to daughter chromosomes during DNA replication and then replenished by new CENP-A assembly during the subsequent G1 (Jansen et al., 2007; Nechemia-Arbely et al., 2012; Westhorpe and Straight, 2015). Failure to replenish CENP-A nucleosomes causes a twofold decrease in CENP-A through each cell cycle, causing centromere defects and chromosome segregation errors (Bodor et al., 2013; Fachinetti et al., 2013). Thus, how existing centromeric chromatin promotes CENP-A assembly to ensure long-term centromere integrity is a key question in chromosome biology and epigenetics.
During centromere and kinetochore formation, a group of 17 proteins, the constitutive centromere–associated network (CCAN), assembles at the site of CENP-A nucleosomes (Foltz et al., 2006; Cheeseman and Desai, 2008; Westhorpe and Straight, 2013). CCAN assembly requires direct binding of CENP-A nucleosomes by CENP-C and CENP-N and CENP-A–dependent association of CENP-I and CENP-T with centromeres via mechanisms that remain unclear (Carroll et al., 2009, 2010; Kato et al., 2013; Folco et al., 2015). Several CCAN proteins, including CENP-H,I,K, CENP-M, and CENP-C, have been implicated in maintaining centromeric chromatin, as their depletion from cells or cell extracts causes defects in CENP-A assembly (Okada et al., 2006; Erhardt et al., 2008; Hori et al., 2008; Moree et al., 2011). Forced localization of CENP-I or CENP-C to noncentromeric chromatin is sufficient to promote ectopic CENP-A assembly (Hori et al., 2013). However, it remains unclear whether the presence of CENP-C at endogenous centromeres is sufficient to seed CENP-A nucleosome assembly.
Two protein complexes, the Mis18 complex (Mis18α, Mis18β, and M18BP1 [Mis18 Binding Protein 1; KNL-2 in Caenorhabditis elegans]; Fujita et al., 2007; Maddox et al., 2007; Moree et al., 2011; Dambacher et al., 2012; Hayashi et al., 2014; Subramanian et al., 2014) and the Holliday junction recognition protein (HJURP) complex (HJURP, Npm1, CENP-A, and H4; Dunleavy et al., 2009; Foltz et al., 2009) must be recruited to centromeres for new CENP-A assembly. In metaphase, M18BP1 binds directly to CENP-C at centromeres (Moree et al., 2011; Dambacher et al., 2012), and although phosphorylation regulates the recruitment of M18BP1 to centromeres, the identity of the interacting partners that bring M18BP1 to interphase centromeres remains unclear (Silva et al., 2012; McKinley and Cheeseman, 2014). HJURP is a CENP-A–specific chaperone that binds soluble CENP-A/H4 dimers through part of the CENP-A histone fold domain, termed the CENP-A targeting domain (CATD; Black et al., 2004; Bassett et al., 2012; Yu et al., 2015). In human cells, histone H3 containing the CATD targets to centromeres (Bassett et al., 2012), and the presence of the CATD in CENP-A is required to sustain long-term centromere function (Fachinetti et al., 2013). However, it is unclear whether this simply reflects the fact that the CATD is required for the assembly of soluble CENP-A by HJURP or whether the CATD of nucleosomal CENP-A has a role in CENP-A assembly. We have previously shown that nucleosomes containing chimeric histone H3 with just the C-terminal six amino acids of CENP-A are sufficient for CENP-C recruitment and kinetochore assembly in Xenopus laevis egg extracts (Guse et al., 2011), suggesting the CATD is not required for kinetochore formation. Here, we reengineer the Xenopus egg extract/recombinant chromatin approach to establish CENP-A nucleosome assembly in a cell-free system, allowing us to test the role of existing CENP-A nucleosomes in centromere maintenance.
Results and discussion
Reconstituting CENP-A assembly in vitro
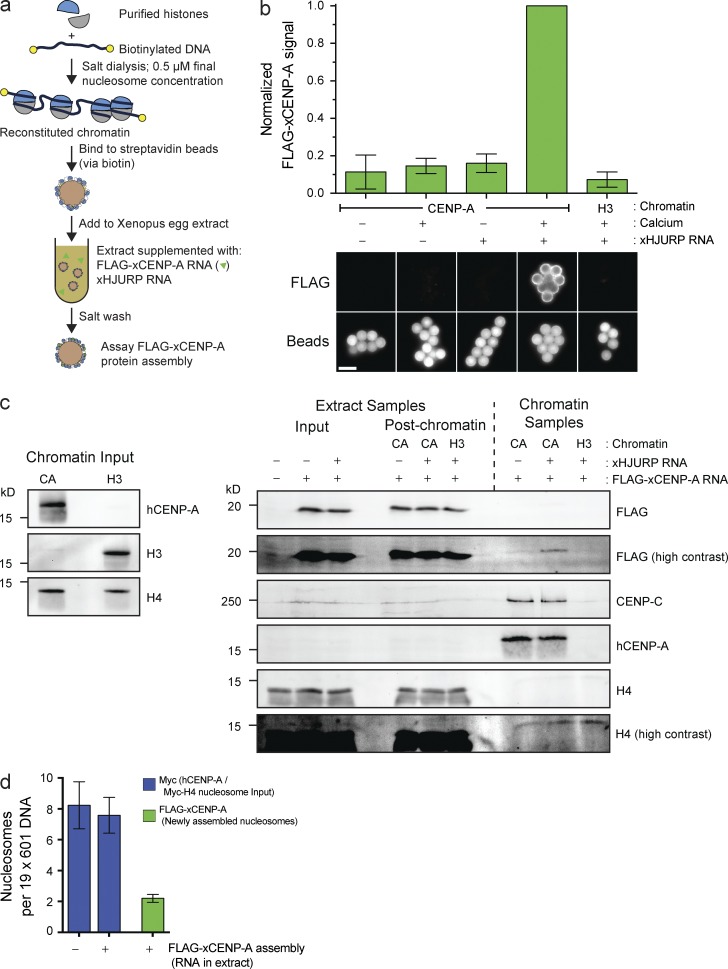
We established an in vitro CENP-A assembly system that enables us to independently manipulate CENP-A within chromatin and soluble CENP-A that is a precursor to new CENP-A nucleosomes (Fig. 1 a). Subsaturated nucleosome arrays were reconstituted in vitro on biotinylated DNA, bound to streptavidin beads, incubated in Xenopus egg extracts supplemented with FLAG–CENP-A and Xenopus HJURP, and then washed and assayed for FLAG–CENP-A assembly. Reconstituted H3 nucleosomes failed to assemble FLAG–CENP-A, whereas CENP-A nucleosomes assembled FLAG–CENP-A specifically when chromatin was incubated in interphase egg extract (Fig. 1 b). Thus, FLAG–CENP-A assembly requires release of extract from cytostatic factor (CSF) activity (CSF arrest), mimicking the cell cycle dependence of CENP-A assembly in other systems (Jansen et al., 2007; Bernad et al., 2011; Moree et al., 2011; Silva et al., 2012). Moreover, we observed robust FLAG–CENP-A assembly only when extract was supplemented with RNA encoding HJURP (Fig. 1, b and c), suggesting that this system requires the established CENP-A assembly machinery.
Figure 1.
Reconstitution of CENP-A assembly in vitro. (a) Schematic of in vitro CENP-A assembly assay. (b) In vitro FLAG–xCENP-A assembly requires HJURP and mitotic exit. Extracts were supplemented with calcium, xHJURP RNA, or both. The top graph shows the means ± SEM; n = 5. The bottom images show FLAG–xCENP-A staining (FLAG) and bead autofluorescence (Beads). Bar, 5 µm. (c) Characterization of FLAG–xCENP-A assembly. (left) The levels of CENP-A (hCENP-A), histone H3, and histone H4 on the beads before extract addition. (right) The levels of FLAG–xCENP-A, CENP-C, and histone H4 in the extract before chromatin bead addition and after bead recovery (Extract Samples), and the chromatin bead–bound proteins after their recovery from the extract (Chromatin Samples). (d) Summary of quantitative Western blots (see Fig. S1 c) estimating the number of input nucleosomes (Myc-H4 signal) and newly assembled FLAG–xCENP-A nucleosomes per chromatin array; bars show the means ± SEM; n = 5.
The test whether the FLAG–CENP-A observed on chromatin arrays represented CENP-A assembled into nucleosomes, we took advantage of the ability to control the egg extract cell cycle and the fact that CENP-A assembly is interphase specific. FLAG–CENP-A persisted on chromatin after additional incubation in CSF extract, whereas Myc-tagged HJURP was completely lost from CENP-A chromatin (Fig. S1, a and b). Moreover, an extract in which FLAG–CENP-A and HJURP were added only to the second, CSF-arrested extract, failed to assemble any FLAG–CENP-A (Fig. S1 a). As unassembled, HJURP-associated FLAG–CENP-A would be lost from chromatin after incubation in CSF; these data suggest that the new FLAG–CENP-A signal represents new CENP-A nucleosomes.
Western blotting showed that existing CENP-A nucleosomes were not lost as a consequence of FLAG–CENP-A assembly (Fig. 1 c). We measured the efficiency of CENP-A assembly by reconstituting CENP-A and H3 chromatin arrays with Myc-tagged H4 nucleosomes and used Myc-H4 to quantify the input nucleosomes. Quantitative Western blotting showed that input nucleosome arrays were 40% saturated (∼8 nucleosomes per 19 available sites; Figs. 1 d and S1 c). Consistent with this, histone H3 assembled onto the free DNA sites once the arrays were incubated in extract (Fig. S1 d). No loss of CENP-A:Myc-H4 nucleosomes was observed after FLAG–CENP-A assembly (Figs. 1 d and S1 c). A mean of about two FLAG–CENP-A nucleosomes are assembled per 19× nucleosome positioning site array. Assuming CENP-A assembly in cells is 100% efficient (every parent CENP-A nucleosome seeds one new CENP-A nucleosome), our reconstituted CENP-A assembly is ∼25–30% efficient (one new FLAG–CENP-A nucleosome every 3.5–4 input CENP-A nucleosomes). We suspect the difference in efficiency arises because our reconstituted chromatin is built on short, linear DNA, and the histones lack any modifications that occur in native centromeres. In summary, we have established the first cell-free system to study CENP-A assembly that enables the direct manipulation of CENP-A chromatin.
CENP-A nucleosomes recruit CENP-C and M18BP1 independently
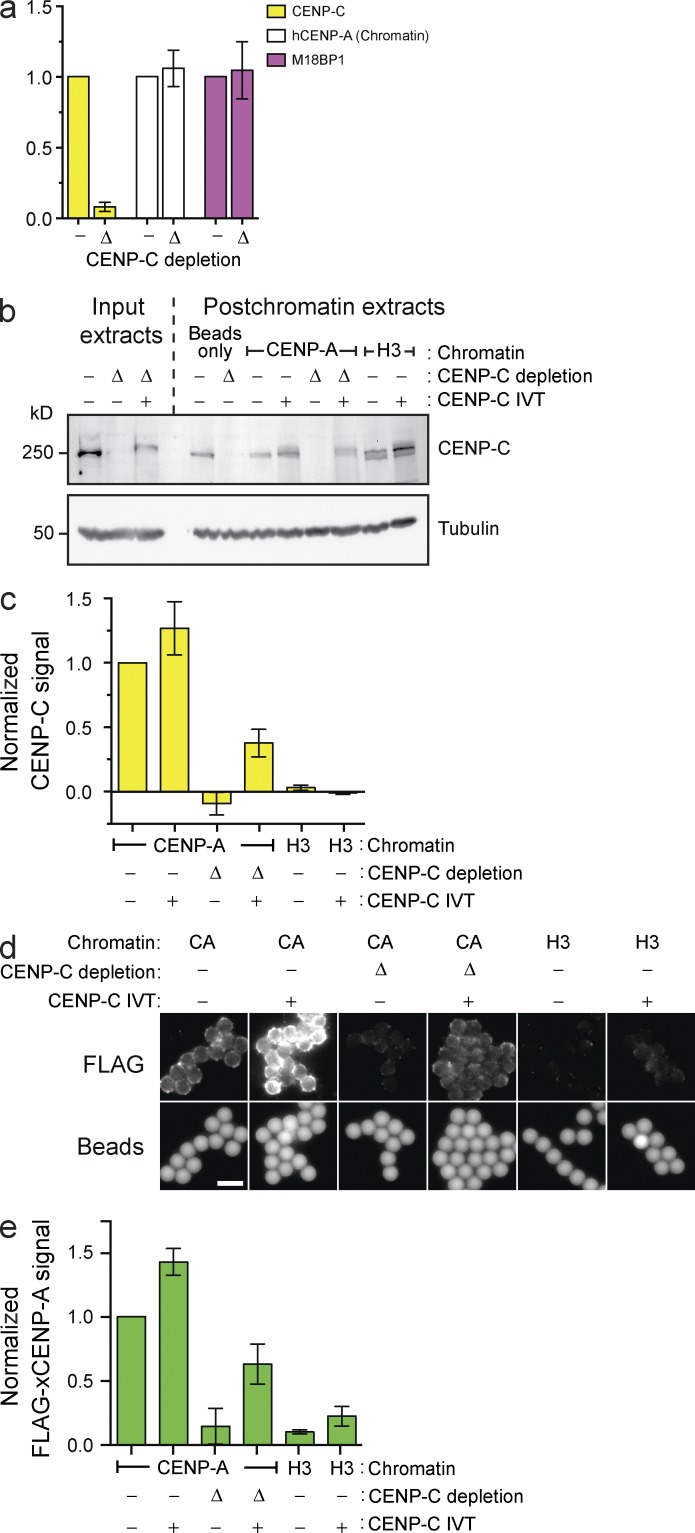
Given the proposed roles for histone modifications in CENP-A assembly (Hayashi et al., 2004; Bergmann et al., 2011; Ohzeki et al., 2012), it was unclear whether CENP-A assembly on naive chromatin would recapitulate the same dependence on CENP-A assembly factors. In addition to the requirement for HJURP in our system (Fig. 1 b), we assessed the role of CENP-C and M18BP1 by immunodepleting them from extract. Importantly, CENP-C depletion did not affect CENP-A nucleosomes on input chromatin (Fig. 2 a). Consistent with previous observations in Xenopus sperm nuclei (Moree et al., 2011), CENP-C depletion prevented M18BP1 protein recruitment to CENP-A nucleosome arrays in CSF-arrested extract (Fig. S1 e). In contrast, CENP-C depletion had no effect on M18BP1 association in interphase, during the time of CENP-A assembly (Fig. 2 a). Despite the presence of M18BP1 on chromatin in interphase, CENP-C depletion completely prevented FLAG–CENP-A assembly on reconstituted CENP-A chromatin (Fig. 2, b–e). Adding back in vitro translated (IVT) CENP-C partially complemented CENP-C depletion and partially rescued CENP-A assembly (Fig. 2, b–e). The addition of IVT CENP-C to mock-depleted extract promoted 1.5-fold more FLAG–CENP-A assembly relative to unsupplemented mock-depleted extract (Fig. 2, b–d), suggesting that CENP-C may be limiting for new CENP-A assembly. Our data show that CENP-A assembly factors, other than M18BP1, depend on the presence of CENP-C at the centromere.
Figure 2.
CENP-C depletion prevents in vitro CENP-A assembly but not M18BP1 recruitment. (a) Levels of CENP-C, hCENP-A, and M18BP1 on chromatin beads after bead incubation in mock-depleted (−) or CENP-C–depleted (Δ) extracts. All bar graphs represent means ± SEM; n = 4. (b) The levels of CENP-C protein in egg extracts before (Input extracts) or after (Postchromatin extracts) incubation of chromatin beads in the extract and CENP-A assembly. The extracts were mock depleted (−), CENP-C depleted (Δ), or complemented with CENP-C (+). Tubulin is shown as a loading control. (c) CENP-C signal on chromatin was assessed after the experiment described in b; n = 3. (d) Representative images showing FLAG–xCENP-A assembly on chromatin beads as described in b. The FLAG–xCENP-A signal (top row) and the bead autofluorescence (bottom row) are shown. Bar, 5 µm. (e) Quantification of FLAG–xCENP-A assembly assays described in d; n = 3.
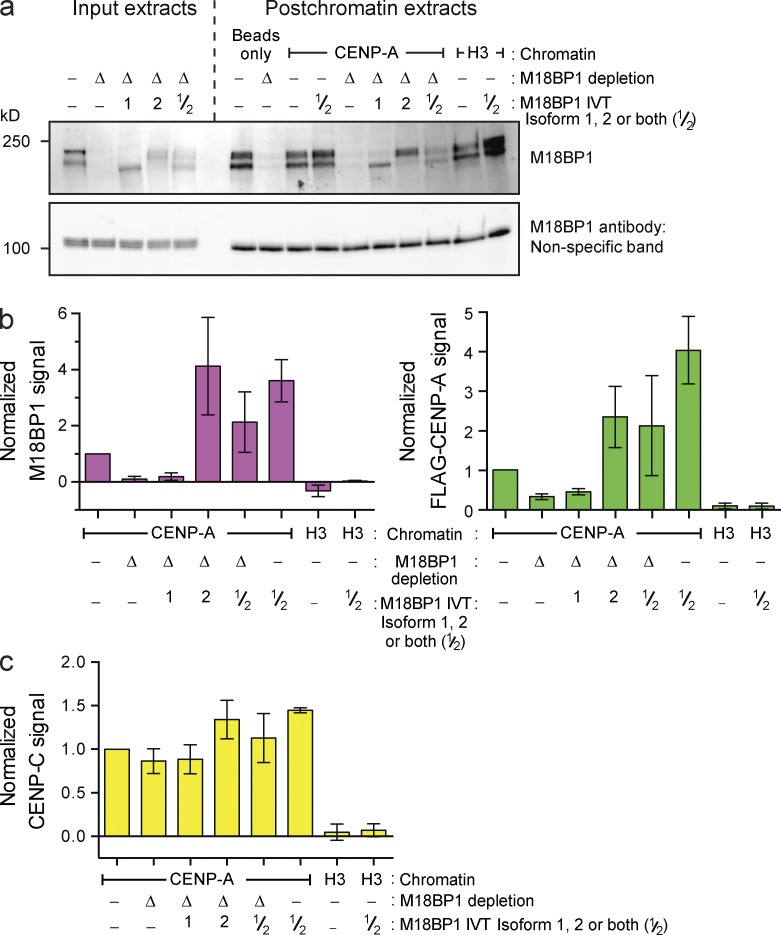
We next depleted M18BP1 from the extract (Fig. 3 a), resulting in >90% reduction of the M18BP1 protein signal bound to reconstituted CENP-A chromatin (Fig. 3 b, left). This caused a 70% decrease in FLAG–CENP-A assembly relative to mock-depleted extract (Fig. 3 b, right). Xenopus has two M18BP1 isoforms, and our M18BP1 antibody depletes both isoforms from extract (Fig. 3 a). Adding back IVT-M18BP1 isoform 1 (M18BP1-1) failed to rescue FLAG–CENP-A assembly, and M18BP1-1 did not associate with chromatin arrays (Fig. 3 b), despite the presence of this protein in extract (Fig. 3 a) and the fact this IVT protein is functional on Xenopus sperm chromatin (Moree et al., 2011). In contrast, adding back IVT-M18BP1 isoform 2 (M18BP1-2) caused a fourfold increase in total M18BP1 protein association with chromatin arrays and promoted FLAG–CENP-A assembly to about twice the levels observed in mock-depleted extract (Fig. 3 b). Moreover, addition of both M18BP1 isoforms to mock-depleted extract significantly increased FLAG–CENP-A assembly. Thus, the amount of M18BP1 protein recruited to centromeric chromatin is a major determinant of the extent of new CENP-A nucleosome formation. More CENP-C associated with the arrays under conditions that promoted greater FLAG–CENP-A assembly, likely as a result of the increase in CENP-A nucleosome number (Fig. 3 c). Finally, M18BP1 depletion had no effect on CENP-C recruitment to chromatin arrays (Fig. 3 c). In summary, our system accurately recapitulates CENP-A assembly in cells; it reveals that M18BP1 and CENP-C are recruited to chromatin independently during interphase, both are necessary for CENP-A assembly, and neither protein is sufficient for CENP-A assembly without the other.
Figure 3.
CENP-A assembly on chromatin arrays requires M18BP1 isoform 2. (a, top) M18BP1 levels in egg extract after mock depletion (−), M18BP1 depletion (Δ), or complementation with M18BP1 isoform 1 (bottom band of doublet), isoform 2 (top band of doublet) or both (1, 2, and 1/2, respectively). (bottom) A nonspecific band shown as a loading control. (b) M18BP1 (left) and FLAG–xCENP-A (right) levels on chromatin beads after M18BP1 depletion from egg extracts as described in a. All graphs show the means ± SEM, normalized to the mock-depleted CENP-A signal, n = 4. (c) CENP-C levels on chromatin beads after M18BP1 depletion as described in a.
The CATD is insufficient for soluble CENP-A assembly
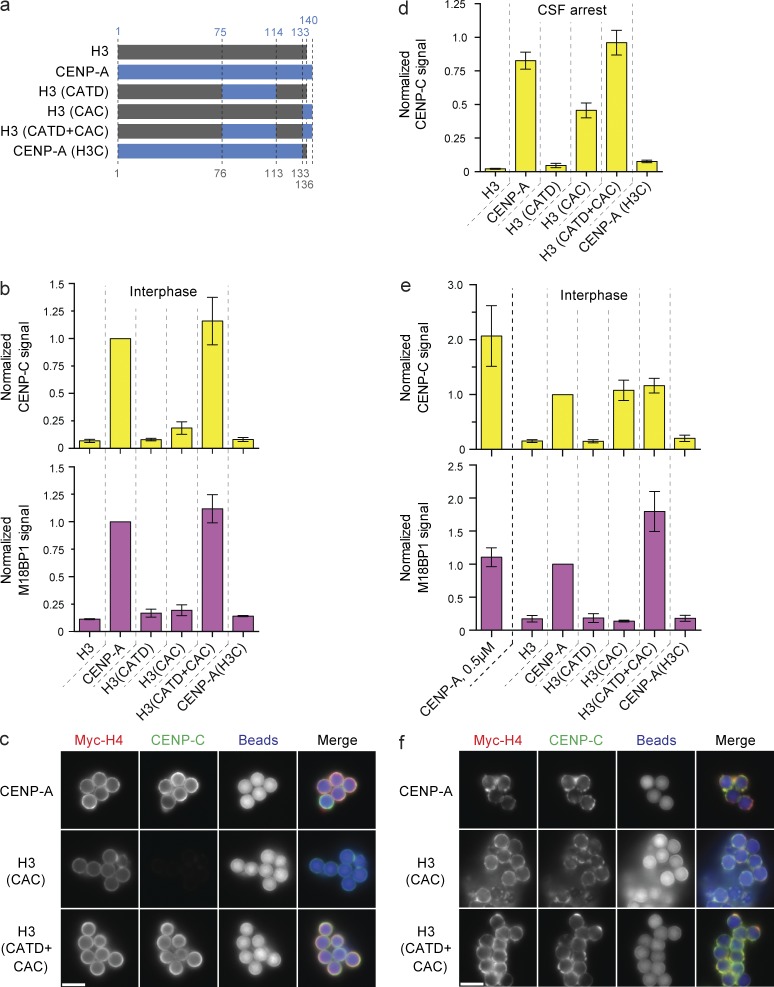
We used our ability to separate the soluble and chromatin-bound populations of CENP-A to assay the assembly of FLAG epitope–tagged CENP-A/H3 chimeras (shown in Fig. 4 a) onto reconstituted CENP-A nucleosomes. Consistent with previous observations (Black et al., 2004; Bassett et al., 2012), CENP-A assembly required the CATD but not the CENP-A C terminus (CAC; Fig. S2, a–c). Less H3(CATD) assembled into chromatin than wild-type CENP-A (Fig. S2, a–c), consistent with HJURP binding to CENP-A requiring both the CATD and Serine 68 of CENP-A (Logsdon et al., 2015; Yu et al., 2015). We suspect the conserved S68 plays similar roles in our system. These data confirm that HJURP recognition of CENP-A is not mediated solely through the CATD and show that our system recapitulates CENP-A assembly in cells.
Figure 4.
CAC-mediated CENP-C recruitment is dependent on high nucleosome density. (a) Schematic of the histone chimeras used and the amino acid residues of human CENP-A (blue) and histone H3 (gray), respectively. (b) CENP-C (top) and M18BP1 (bottom) levels on low saturation chimeric nucleosome arrays in interphase. Signals are compared with the amount of recruitment to wild-type CENP-A beads and internally normalized in each sample to the levels of Myc-H4 on the beads. Bars in all panels represent the means ± SEM; n = 3. (c) Fluorescent images of CENP-C recruitment to low-saturation CENP-A, H3(CAC), and H3(CATD+CAC) chromatin arrays. The Myc-H4, CENP-C, and bead autofluorescence signals are shown. (d) CENP-C levels on CENP-A/H3 chimeric nucleosome arrays reconstituted at 0.5 µM nucleosome concentration in CSF-arrested extract. Signals are normalized as in b. (e) CENP-C (top) and M18BP1 (bottom) levels on high-saturation chimeric nucleosome arrays. Signals are normalized as in b. The signal on low-saturation CENP-A arrays (CENP-A, 0.5 µM) are shown for comparison. (f) Fluorescence images of CENP-C levels on high-saturation CENP-A, H3(CAC), and H3(CATD+CAC) chromatin arrays. Images labeled as in c. Bars, 5 µm.
CENP-A nucleosome density specifies distinct mechanisms of CENP-C recruitment
An advantage of this system is the ability to manipulate CENP-A nucleosomes while bypassing any effect on the soluble CENP-A pool. Thus, we used chromatin arrays assembled with recombinant chimeric CENP-A/H3 histones (Fig. 4 a) to test the ability of chimeric arrays to recruit the CENP-A assembly machinery.
Interestingly, we found that CENP-C required both the CATD and CAC to associate with reconstituted chromatin in interphase (Fig. 4, b and c; and Fig. S2 d) and that the CAC did not fully support CENP-C binding in metaphase (Fig. 4 d). These data were inconsistent with our previous observations of kinetochore assembly on reconstituted chromatin in metaphase-arrested Xenopus extract, where CENP-C recruitment required only the CAC (Guse et al., 2011). In this study, we used subsaturated chromatin arrays assembled at 0.5 µM nucleosomes, whereas our previous experiments assaying kinetochore formation used chromatin arrays assembled at higher concentration (2 µM nucleosomes; Guse et al., 2011). Assembly at 0.5 µM nucleosomes resulted in no more than 40% CENP-A nucleosome saturation (Fig. 1 d), whereas 2 µM histone and nucleosome positioning site resulted in higher nucleosome saturation (Fig. S3 a). Saturated nucleosome arrays bound streptavidin beads more heterogeneously than subsaturated arrays, resulting in areas of high nucleosome density (Fig. S3 b).
We compared CENP-C protein recruitment to high versus low saturation nucleosome arrays. Strikingly, H3(CAC) nucleosomes, when assembled at high saturation, recruited CENP-C to the same level as highly saturated wild-type CENP-A nucleosomes (Fig. 4, e and f; and Fig. S2 d), consistent with our previous observations (Guse et al., 2011). This indicates that a higher density of CENP-A C termini is sufficient to recruit CENP-C. In contrast, at low nucleosome saturation, both the CATD and the CAC are required to recruit CENP-C.
We found that the CENP-C to CENP-A nucleosome ratio was twice as high on low saturation nucleosome arrays (Fig. 4 e and Fig. S2 d). Thus, more CENP-C is recruited by both the CAC and CATD than the CAC alone. The CATD maypromote CENP-C recruitment only at lower CENP-A nucleosome density if the interaction between adjacent CENP-A nucleosomes masks the CATD at high nucleosome density. Together, our data suggest that the density of CENP-A nucleosomes influences how proteins recognize centromeric chromatin.
The CATD and CAC recruit M18BP1 in a CENP-C–independent manner
We tested the ability of the CENP-A/H3 chimeric arrays to recruit M18BP1 in interphase, during the time of CENP-A assembly. M18BP1 recruitment was not supported by the CATD or CAC alone, whereas the combination of both domains (H3(CATD+CAC)) fully supported M18BP1 recruitment (Fig. 4 b). In contrast to CENP-C, M18BP1 recruitment required both the CATD and CAC irrespective of underlying nucleosome saturation, and M18BP1 levels did not change between low- and high-density nucleosome arrays on a per nucleosome basis (Fig. 4, b and c). Depletion of CENP-C had no effect on M18BP1 recruitment to any CENP-A/H3 chimera in interphase extracts, demonstrating that M18BP1 binding to CENP-A chromatin does not require CENP-C, except in mitosis (Fig. S1 e). The cell cycle dependence may be caused by phosphorylation of M18BP1 as previously observed in human cells (Silva et al., 2012; McKinley and Cheeseman, 2014). In all conditions, CENP-A lacking the CAC (CENP-A(H3C)) failed to recruit M18BP1 (Fig. 4, b and d). As recruitment of M18BP1 to centromeres requires the CAC but does not require CENP-C, these data uncover a new role for the C-terminal domain of CENP-A in recruiting M18BP1 to interphase centromeres, a role independent of direct CENP-C recognition of that same domain. Establishing whether M18BP1 and CENP-C compete for access to the CAC will require further mechanistic insight into how M18BP1 is recruited to centromeres.
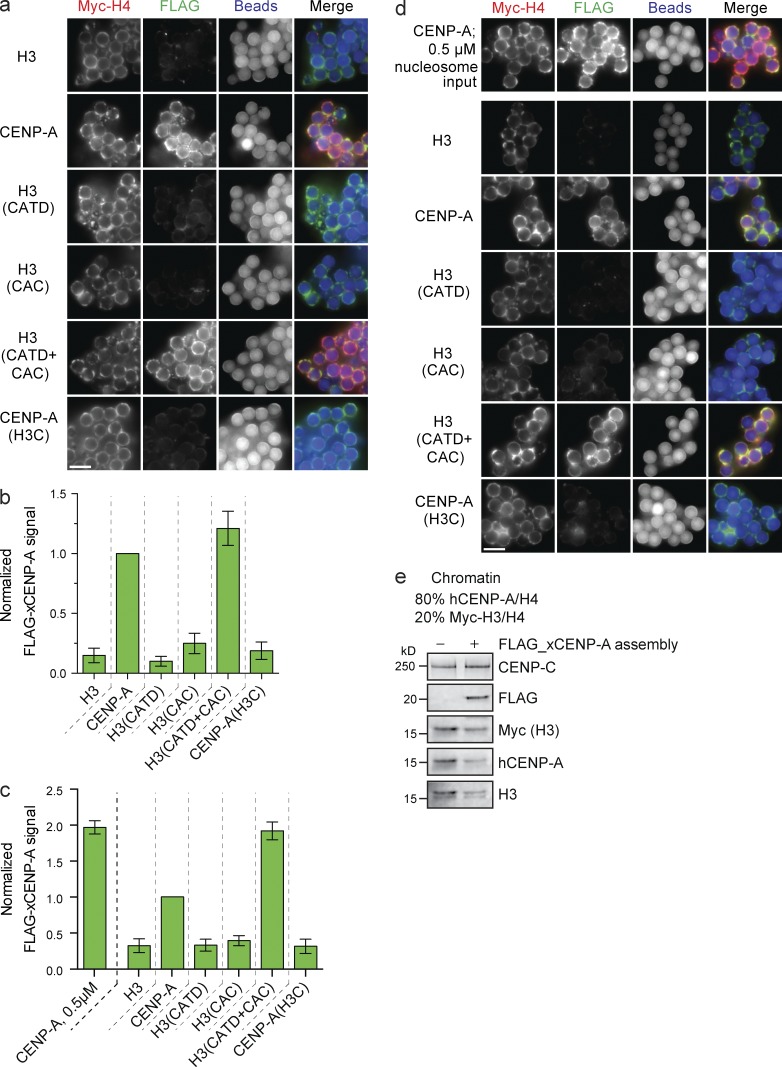
FLAG–CENP-A assembly requires both the CATD and the CAC
Using low saturation arrays, we assayed FLAG–CENP-A assembly on chimeric CENP-A/H4 nucleosome templates. New FLAG–CENP-A nucleosomes were only assembled when either wild-type CENP-A or H3 containing both the CATD and CAC (H3(CATD+CAC)) was used as the reconstituted chromatin substrate (Fig. 5, a and b). Importantly, FLAG–CENP-A assembly required the CATD and the CAC on both high- and low-density nucleosome arrays but was less efficient on high-density nucleosome arrays (Fig. 5, c and d). Thus, although CENP-C is recruited to high-density H3(CAC) nucleosome arrays, this CENP-C cannot support FLAG–CENP-A assembly. This is likely because H3(CAC) does not recruit M18BP1. Thus, our data show that CENP-C recruitment alone does not lead to new CENP-A nucleosome deposition. Furthermore, the assembly of new CENP-A into nucleosomes requires the CATD and CAC irrespective of substrate nucleosome saturation.
Figure 5.
FLAG–xCENP-A assembly requires the CATD and CAC. (a) Fluorescence images of FLAG–xCENP-A assembly on low saturation chimeric chromatin. Myc-H4, FLAG–xCENP-A, and bead autofluorescence are shown. (b) Quantification of FLAG–xCENP-A assembly as shown in a; all bars represent means ± SEM normalized to the signal on CENP-A arrays; n = 4. (c) FLAG–xCENP-A assembly on high saturation CENP-A/H3 chimeric chromatin. Normalized as in b. n = 4. (d) Fluorescence images of FLAG–xCENP-A assembly on high saturation CENP-A/H3 chimeric chromatin. Labeled as in a. (e) Protein levels of CENP-C, FLAG–xCENP-A, Myc-H3, hCENP-A, and H3 with son saturated chromatin arrays containing 80% hCENP-A nucleosomes and 20% Myc-H3 nucleosomes after incubation in extract supplemented with buffer (−) or with FLAG_xCENP-A and xHJURP RNA (+). Bars, 5 µm.
The CENP-A assembly machinery can remove existing nucleosomes from chromatin
It is not known whether the CENP-A assembly machinery can replace existing nucleosomes or simply add new nucleosomes to nucleosome-free regions. To assess whether CENP-A assembly can displace existing nucleosomes, we used saturated CENP-A arrays doped with a subpopulation of Myc-H3 nucleosomes to measure changes in the H3 nucleosome content during assembly (Fig. S3 d). In the absence of doping in H3 nucleosomes, subsaturated CENP-A chromatin arrays assembled more CENP-A than saturated CENP-A arrays (Fig. 5 c). When we used mixed arrays, FLAG–CENP-A assembly was readily detectable on saturated chromatin containing CENP-A and H3. This assembly caused a loss of both CENP-A and H3 from substrate chromatin when compared with chromatin incubated in extract not supplemented with FLAG–CENP-A or xHJURP RNA (Fig. 5 e). Thus, the CENP-A assembly machinery can remove both CENP-A and H3 nucleosomes from chromatin to deposit new CENP-A nucleosomes.
Displacement of H3 nucleosomes is consistent with the observation that H3.3 nucleosomes are incorporated into centromeric chromatin during DNA replication (Dunleavy et al., 2011), yet CENP-A nucleosomes are highly stable on centromeric chromatin in cells and thus unlikely to exchange during new CENP-A assembly (Jansen et al., 2007; Bodor et al., 2013; Fachinetti et al., 2013). As we do not detect any loss of existing CENP-A nucleosomes after CENP-A assembly on chromatin arrays containing 40% CENP-A nucleosomes (Fig. 1 d), and CENP-A nucleosomes represent a minority of nucleosomes in centromeric chromatin (Bodor et al., 2014), we suspect that cells rarely encounter the situation where CENP-A assembly, initiated by one CENP-A nucleosome, displaces an adjacent CENP-A nucleosome. In addition, MgcRacGAP stabilizes CENP-A nucleosomes in chromatin after replication (Lagana et al., 2010). These two considerations may account for the measured stability of CENP-A at endogenous centromeres.
Conclusions
CENP-C binds to CENP-A nucleosomes by interacting with both the C-terminal tail of CENP-A and the acidic patch on histones H2A and H2B (Carroll et al., 2010; Kato et al., 2013). CENP-C does not bind to H3/CENP-A chimeric nucleosomes containing only the CATD domain (Carroll et al., 2010). We found that both the CAC and the CATD are required to recruit CENP-C when CENP-A nucleosomes are sparsely distributed. Therefore, although the CAC binds CENP-C directly, the CATD may indirectly recruit CENP-C. In budding yeast, CENP-CMif2 interacts with the CENP-NChl4 protein that binds to the CATD domain of the CENP-ACse4 nucleosome (Carroll et al., 2009; Hinshaw and Harrison, 2013), thus CENP-N may bridge the CATD and CENP-C. However, in Drosophila melanogaster, which lack CENP-N, the CATD is required for CENP-C targeting to ectopic chromatin and, in human cells, CENP-C requires the CATD to associate with artificially tethered CENP-A chimeras independently of CENP-N (Logsdon et al., 2015).
An alternative model is that the CATD may influence higher-order chromatin structure by changing the properties of the CENP-A nucleosome (Panchenko et al., 2011; Sekulic and Black, 2012; Miell et al., 2013; Geiss et al., 2014). When CENP-A nucleosomes are in close proximity, either through higher-order chromatin compaction or in blocks of adjacent CENP-A nucleosomes suggested from fiber spreading experiments (Blower et al., 2002; Sullivan and Karpen, 2004), the high local concentration of CENP-A nucleosomes may enable CENP-C to bind via the CAC alone. One interesting idea is that chromosome condensation during mitosis may change the mode of CENP-C binding to CENP-A nucleosomes to only require the CAC. Consistent with this, we observed more CENP-C bound to H3(CAC) nucleosomes in CSF-arrested versus interphase extract (Fig. 4, b vs. d). Moreover, the CATD is dispensable for kinetochore formation on saturated CENP-A chromatin in metaphase Xenopus extracts; CENP-C recruitment is supported by the CAC alone (Guse et al., 2011). CENP-N’s association with human centromeres decreases during mitosis (Hellwig et al., 2011), and in chicken cells, CENP-C depends on the presence of the CENP-H complex in interphase but not in mitosis (Kwon et al., 2007).
Our observation that both the CATD and CAC are required for CENP-C recruitment when CENP-A nucleosomes are sparsely distributed provides a good explanation for recent observations made in human cells expressing lacI-CENP-A/H3 chimeras, where robust CENP-C recruitment to noncentromeric LacO arrays required both the CATD and CAC (Logsdon et al., 2015; Tachiwana et al., 2015). We speculate the local concentration of CENP-A nucleosomes was insufficient to recruit CENP-C via the CAC alone. It would be interesting to determine whether this dependency changed in mitotic cells.
We show that, in chromatin incubated in interphase extracts, CENP-A nucleosomes must contain both the CATD and CAC to promote soluble CENP-A assembly. In human cells, chimeric nucleosomes containing the CATD and either the CENP-A N-terminal tail or the CAC rescue the viability of a CENP-A knockout (Fachinetti et al., 2013). Thus, human centromere function can be sustained without the CAC that directly binds CENP-C. In contrast, we observe that CENP-A lacking only the CAC (CENP-A(H3C)) is not functional for CENP-C or M18BP1 recruitment, CENP-A assembly, or kinetochore function. The CAC may be required for de novo centromere formation but be dispensable at an already existing centromere that had wild-type CENP-A nucleosomes before CENP-A knockout. Consistent with this, gene replacement of endogenous human CENP-A with a mutant lacking the CAC causes a 75% reduction in CENP-C, but the remaining CENP-C is sufficient to maintain functional centromeres (Fachinetti et al., 2013). An alternative possibility stems from the observation that CENP-B can stabilize CENP-C at centromeres independently of the CAC (Fachinetti et al., 2015). Xenopus lack a CENP-B protein, and thus, centromere maintenance and kinetochore assembly may depend more on the recognition of the CATD and C-terminal domains of the CENP-A histone in frogs.
In conclusion, fully reconstituting the CENP-A assembly process is an important goal for understanding the epigenetic mechanisms that maintain eukaryotic centromeres. The cell-free system we have developed provides a key step toward this goal by enabling CENP-A assembly on well-defined chromatin templates. We have shown that both the CATD and CAC play essential roles in bringing the CENP-A assembly machinery to centromeres. Because assembly of new CENP-A nucleosomes requires cooperation between CENP-C–dependent and CENP-C–independent processes, identifying the additional factors that depend upon these unique features of CENP-A nucleosomes is central to understanding centromere maintenance.
Materials and methods
Protein and DNA expression and purification
Histones constituting wild-type H3 octamers and CENP-A/H3 chimeras lacking the CATD were purified using a denaturing preparation, refolded with H4, and gel filtered as previously described (Guse et al., 2012). In brief, a single BL21(DE3) Codon Plus RIPL Escherichia coli (230280; Agilent Technologies) bacterial colony harboring a plasmid for histone expression (pET3a vector) was grown in 2 liters of 2× YT medium (20 g/liter tryptone, 10 g/liter yeast extract, and 5 g/liter NaCl) and induced with 0.25 mM IPTG when the OD600 reached 0.6. After 3 h further growth, washed bacteria were lysed in 20 mM lysis buffer (K-phosphate buffer, pH 6.8, 1 M NaCl, 5 mM 2-mercaptoethanol, 1 mM PMSF, 1 mM benzamidine, 0.05% NP-40, and 0.2 mg/ml lysozyme) and homogenized using an EmulsiFlex-C5 (Avestin, Inc.), and the soluble lysate was removed from the insoluble pellet after centrifugation (18,000 g, 20 min, 4°C). The washed insoluble pellet containing insoluble histone was resuspended in unfolding buffer (7 M guanidine-HCl, 20 mM Tris-HCl, pH 7.5, and 10 mM DTT), and the supernatant after additional centrifugation was dialyzed into urea buffer (6 M deionized urea, 200 mM NaCl, 10 mM Tris-HCl, pH 8, 1 mM EDTA, 5 mM 2-mercaptoethanol, and 0.1 mM PMSF). Histones were purified by running the dialyzed solution through a HiTrap Q column followed by a HiTrap S column. Histones were eluted from the S column with urea buffer containing 1 M NaCl, dialyzed into water, and lyophilized for future use. Histones were refolded together by dialysis into 2 M NaCl, 10 mM Tris-HCl, pH 7.6, 1 mM EDTA, and 5 mM 2-mercaptoethanol, and complexes were purified by size-exclusion chromatography.
CENP-A and CENP-A/H3 chimeras containing the CATD were coexpressed with H4 and purified as soluble tetramers, as previously described (Guse et al., 2012). All histone tetramers were expressed using the pST39 multicistronic vector (Tan, 2001). Bacteria were lysed in lysis buffer and homogenized as described above for wild-type H3 octamers. Soluble protein containing histone tetramer was isolated from the pellet by centrifugation (rotor [Type 45Ti; Beckman Coulter], 20,000 g, 20 min at 4°C) and run through a preequilibrated hydroxyapatite (HA) column (type II 20 µM HA; Bio-Rad Laboratories). Bound protein was washed with HA buffer (20 mM potassium phosphate, pH 6.8, 1 M NaCl, and 5 mM 2-mercaptoethanol) and then eluted over a 2–column volume gradient with HA buffer containing 3.5 M NaCl. Eluted fractions were pooled and dialyzed into 10 mM Tris-HCl, pH 7.4, 0.75 M NaCl, 10 mM 2-mercaptoethanol, and 0.5 mM EDTA. Dialyzed protein was bound to a 1 ml HiTrap SP FastFlow column, washed in dialysis buffer, and purified histone tetramer eluted over a 20–column volume gradient into dialysis buffer containing 2 M NaCl. Positive fractions were pooled, aliquoted, snap frozen in liquid nitrogen, and stored at −80°C until required. Human CENP-A was used for nucleosome reconstitution throughout this study, as Xenopus CENP-A tetramers cannot be purified effectively.
Biotinylated DNA was purified as previously described (Guse et al., 2012). 2 liters of pUC18 containing 19 repeats of the 601 nucleosome positioning sequence was grown in SURE2 bacteria in Luria Broth media and purified with a Gigaprep kit (QIAGEN). The plasmid was digested with EcoRI, XbaI, DraI, and HaeII and purified by polyethylene glycol (PEG) precipitation, during which 0.5% incremental step increases in PEG concentration (separated by 10 min, 5,000 g centrifugations) facilitated pelleting of the larger 19 × 601 sequence from smaller plasmid backbone fragments. PEG precipitates containing 19 × 601 were dialyzed into 10 mM Tris, pH 8, and 0.5 mM EDTA, the DNA was concentrated to ∼2.5 mg/ml by ethanol precipitation, and the overhangs from digestion were filled in using Klenow fragment (3′→5′ exo; New England Biolabs, Inc.), biotin-14-dATP (Invitrogen) dCTP α-thiο-dGTP, and α-thio-dTTP (ChemCyte).
Nucleosome array reconstitution
Nucleosome arrays were assembled by salt dialysis as previously described (Guse et al., 2012). In brief, biotinylated DNA and recombinant histones were combined in a high-salt buffer (2 M NaCl, 10 mM Tris-HCl, pH 7.5, and 0.25 mM EDTA) and dialyzed in a dialysis button into a low-salt buffer (2.5 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 0.25 mM EDTA) over a 36-h period. To determine optimal conditions for assembly, the concentration of tetramer (or octamer for H3) was titrated versus DNA at a fixed ratio of 2.2 H2A/H2B dimers per tetramer. Nucleosome array assemblies were performed at a final DNA concentration of 0.5 or 2 µM 601 sequence to promote undersaturation or saturation of arrays, respectively. For all CENP-A/H3 chimera experiments, in which different nucleosome arrays were compared, tetramer with Myc-H4 was used, and the Myc signal on beads was used to normalize protein signal. Before using chromatin arrays in extract, 60 ng DNA from all nucleosome arrays was assessed for saturation by AvaI digestion and native PAGE on a 5% acrylamide native gel, stained with 5 µl SyBr Gold (Life Technologies).
Xenopus extract preparation
CSF-arrested Xenopus egg extracts were prepared as previously described (Desai et al., 1999; Guse et al., 2012). In brief, Xenopus eggs were washed in MMR buffer (5 mM Na-Hepes, pH 7.8, 0.1 mM EDTA, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 2 mM CaCl2) and then dejellied in MMR + 2% l-cysteine. Dejellied eggs were washed in CSF-XB buffer (100 mM KCl, 50 mM sucrose, 2 mM MgCl2, 0.1 mM CaCl2, 10 mM K-Hepes, pH 7.7, and 5 mM K-EGTA, pH 7.7) and washed in CSF-XB + protease inhibitor buffer (CSF-XB + 10 µg/ml LPC [leupeptin/pepstatin A/chymostatin]). Eggs were placed in a 13 × 51–mm polyallomer tube (Beckman Coulter) and packed by low-speed spin in a table top clinical centrifuge for 45 s. After removal of excess buffer, eggs were centrifuged in a rotor (SW55Ti; Beckman Coulter) for 15 min at 10,000 rpm. The soluble cytoplasmic material was removed from the centrifuge tube and supplemented with energy mix (7.5 mM creatine phosphate, 1 mM ATP, and 1 mM MgCl2), 50 mM sucrose, 10 µg/ml LPC, and 10 µg/ml cytochalasin D.
Immunodepletions of CENP-C and M18BP1 from extracts were prepared as previously described (Moree et al., 2011). In brief, depletions were achieved using affinity-purified antibodies bound to protein A beads (Dynabeads; Invitrogen). For 100 µl extract, 2.5 µg α-xM18BP1 antibody or 0.6 µg α-CENP-C antibody was bound to 33 µl of beads in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Triton X-100 for ≥30 min at 4°C. Whole-rabbit IgG was used for control depletions. The beads were then washed and resuspended in Xenopus extract for 1 h at 4°C. Beads were removed from the extract by two 5-min exposures to a magnet.
IVT protein was prepared from as previously described (Moree et al., 2011). In brief, IVT proteins were produced by incubating pCS2+ plasmids (encoding an SP6 promoter upstream of coding sequence for the protein to be produced) in the SP6 TNT Quick-Coupled Transcription/Translation (Promega) following the manufacturer’s instructions. 2 µl 6×Myc–CENP-C IVT protein was added per 20 µl extract. 2 µl M18BP1-1, M18BP1-2 IVT protein was added per 20 µl extract, or 1 µl of each isoform when both isoforms were added together. 1 µl of 3×FLAG–CENP-A or 3×FLAG xCENP-A/H3 chimeras was used per 20-µl extract in the experiment described in Fig. S2 (a–c). In all experiments, IVT protein was added at the same time as chromatin arrays.
RNA preparation
FLAG–xCENP-A and HJURP RNA, used for translation in extract, were prepared with the SP6 mMessage mMachine kit (Life Technologies). In brief, pCS2 plasmids containing either 3×FLAG–xCENP-A or xHJURP (both full length and wild type) under the control of the SP6 promoter were linearized with NotI, which cuts after the transgene sequence. The transcription reaction was performed according to manufacturer’s instructions (except that double the amount of DNA was used) and purified using RNeasy mini columns (QIAGEN).
Preparation of chromatin-coated magnetic beads
Chromatin-coated magnetic beads were prepared starting with M-280 streptavidin-coated Dynabeads (Invitrogen). Beads were washed in Bead buffer (50 mM Tris, pH 7.4, 75 mM NaCl, 0.25 mM EDTA, 0.05% Triton X-100, and 2.5% polyvinyl alcohol). Reconstituted nucleosome arrays were added to the beads in bead buffer at a ratio of 2.6 fmol chromatin array per microgram of beads and allowed to bind for 30 min at room temperature with constant agitation. Beads and arrays were washed in bead buffer before addition of Xenopus egg extract.
CENP-A assembly assays
For assaying CENP-A assembly on reconstituted chromatin, FLAG–xCENP-A and HJURP RNA were added to extract to a final concentration of 20 and 40 ng/µl, respectively, and allowed to translate for 45 min in an 18°C water bath. After the translation period, cycloheximide was added to the extract to a final concentration of 0.1 mg/ml to stop translation. Extract was added to the washed chromatin-coated beads to a final bead concentration of 25 µg beads/100 µl extract. Extract was released from CSF arrest by addition of CaCl2 to 0.75 mM and then incubated for 75 min in an 18°C water bath, with gentle mixing every 15 min. In histone chimera experiments, in which equivalent release from CSF arrest was required across multiple reactions, a master extract was released with calcium, incubated at 18°C for 10 min, and then aliquoted to beads.
At the end of the incubation, the extract was diluted with CSF-XBT (100 mM KCl, 50 mM sucrose, 2 mM MgCl2, 0.1 mM CaCl2, 10 mM K-Hepes, pH 7.7, 5 mM K-EGTA, pH 7.7, and 0.05% Triton X-100). For the experiment in Fig. S1 (a and b), the bead population was split into two, with one population incubated in CSF-arrested extract for 1 h before processing. For immunoblotting analysis, beads were boiled in SDS sample buffer (50 mM Tris, pH 6.8, 15 mM EDTA, 1 M β-mercaptoethanol, 3.3% SDS, 10% glycerol, and 1 µg/ml Bromophenol blue). For immunofluorescence analysis, beads were washed three times in CSF-XBT and fixed for 5 min in CSF-XBT containing 2% formaldehyde. For experiments in which protein localization to chromatin-coated beads was assessed in the absence of CENP-A loading, the same procedure was used except for addition of RNA, translation, and cycloheximide addition, which did not apply. For experiments in which low and highly saturated nucleosome arrays were compared, the same total amount of DNA was added to beads. FLAG–CENP-A assembly and protein recruitment were normalized to the signal from Myc-H4 nucleosomes, which constitute the chromatin input. No significant difference in Myc-H4 signal was observed across the panel of chimeras.
Immunofluorescence
After fixation, beads were washed in AbDil (20 mM Tris-HCl, pH 7.4, 150 mM NaCl with 0.1% Triton X-100, and 2% bovine serum albumin), pipetted onto poly(l-lysine)-coated coverslips and allowed to adhere for ≥30 min. Coverslips were stained in primary antibody diluted in AbDil for ≥20 min and then washed in AbDil. Primary antibodies used were 2 µg/ml FLAG (F7425 [rabbit] and F1804 [mouse], both obtained from Sigma-Aldrich), 0.25 µg/ml Myc (4A6; EMD Millipore) 1 µg/ml xCENP-C (rabbit, raised and purified against xCENP-C207–296; Milks et al., 2009), and 1.5 µg/ml xM18BP1 (rabbit, raised against GST-xM18BP1-2 amino acids 161–415 and purified against xM18BP1-1161–375; Moree et al., 2011). Coverslips were then stained in secondary antibodies diluted in AbDil for ≥20 min, and then washed in AbDil. Secondary antibodies used were Alexa Fluor 488–conjugated donkey anti–rabbit (Jackson ImmunoResearch Laboratories, Inc.), Alexa Fluor 647–conjugated donkey anti–rabbit (Jackson ImmunoResearch Laboratories, Inc.), and Alexa Fluor 647–conjugated goat anti–mouse (Life Technologies) all at 2 µg/ml. Coverslips were washed in PBST and PBS, gently blotted with filter paper, and mounted in 90% glycerol, 10 mM Tris, pH 8.8, 0.5% p-phenylenediamine. Coverslips were sealed to a slide with clear nail polish.
Microscopy
Imaging was performed at room temperature on a microscope (IX70; Olympus) with a DeltaVision core system (Applied Precision) with a 100× 1.4 NA oil immersion objective (Olympus), a Sedat quad-pass filter set (Semrock), and monochromatic solid-state illuminators, controlled via softWoRx 4.1.0 software (Applied Precision). Coverslips were mounted onto glass slides with 0.5% p-phenylenediamine, 20 mM Tris, pH 8.8, and 90% glycerol. Images were acquired with a charge-coupled device camera (CoolSNAP HQ; Photometrics) and digitized to 12 bits. Z sections were taken at 0.4-µm intervals over a 6-µm total axial distance (the beads have a 3-µm diameter).
Automated image analysis
Images were analyzed using custom ruby software based on earlier methods (Guse et al., 2012). In brief, a single plane of a multi–z-section image was used to segment the beads using bead autofluorescence in the 605-nm emission channel. The segmentation used Otsu’s method (Otsu, 1979) to threshold the images followed by recursively thresholding within any regions larger than the known size of a bead (Xiong et al., 2006) until all regions were bead size or smaller. Regions smaller than one third the size of a bead were discarded. After calculating the centroid of each region, a bead-sized circle was drawn around the centroid, and overlapping circles were separated by excluding pixels equidistant from the centroids of two regions. Images were maximum-intensity projected, and then the signal for each bead in each channel was calculated as the mean pixel intensity for the region corresponding to that bead. Source code for the bead segmentation and quantification is provided as supplemental material.
Panels of representative images were compiled first using freely available custom software (panelize). Panelize automates the process of mounting several muticolor microscope images into a grid for display. It automatically scales images linearly within each color channel to maximize visibility while maintaining uniform scaling across different images. Image panels were sometimes further processed with Photoshop (Adobe) but always linearly and uniformly across conditions. The γ values were not altered in any image.
Immunoblotting
Immunoblots were performed as described previously (Moree et al., 2011), except that a fluorescence imager (VersaDoc; Bio-Rad Laboratories, Inc.) was used for fluorescence detection in some experiments and that a tertiary antibody was not required for detection of M18BP1. Primary antibodies used in this study were 1.5 µg/ml xCENP-C, 5 µg/ml xM18BP1 (xCENP-C and xM18BP1 antibodies further described in Immunofluorescence Materials and methods section), 2 µg/ml xHJURP (rabbit, raised against GST-xHJURP, purified against 6His-xHJURP42–194; Moree et al., 2011), 1 µg/ml hCENP-A (rabbit, raised and purified against GST-hCENP-A1–42; Carroll et al., 2009), Ndc80 (1:10,000, rabbit, gift from T. Stukenberg, University of Virginia, Charlottesville, VA), 1 µg/ml FLAG (F1804; Sigma-Aldrich), 0.25 µg/ml Myc (4A6; EMD Millipore), 0.5 µg/ml Tubulin (DM1A; Sigma-Aldrich), 2 µg/ml H4 (7311; Abcam), and 1 µg/ml H3 (1791; Abcam). Alexa Fluor 647–conjugated goat anti–rabbit or goat anti–mouse secondary antibodies were used at 2 µg/ml (Life Technologies). Quantitative Western blots were performed using a 3×His, 2×Thrombin, 3×FLAG, 2×Prescission, 3×Myc-xCENP-A protein (HTFPM_xCENP-A) as a standard for FLAG–xCENP-A assembly, as FLAG-CENP-A assembled was translated from RNA encoding 3×FLAG–xCENP-A. HTFPM_xCENP-A was purified from inclusion bodies as a histone, described in the Protein and DNA expression and purification Materials and methods section.
Online supplemental material
Fig. S1 shows characterization of in vitro CENP-A assembly on reconstituted chromatin arrays. Fig. S2 shows that assembly of soluble FLAG–CENP-A onto centromeric chromatin depends on the CATD and does not require the CAC. Fig. S3 shows characterization of highly saturated, 2 µM nucleosome reconstitutions and FLAG–CENP-A assembly on untagged CENP-A/H4 nucleosome arrays. Supplemental material also includes a ZIP file containing source code for automated image analysis (find_beads) and image processing (panelize). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503132/DC1.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Annika Guse for preliminary work and other Straight laboratory members for helpful discussion. We thank T. Stukenberg for reagents and B. Black for sharing unpublished results.
This study was funded by the National Institutes of Health (R01 GM074728 to A.F. Straight) and a Stanford Dean’s Fellowship to F.G. Westhorpe.
The authors declare no competing financial interests.
Author contributions: F.G. Westhorpe and C.J. Fuller performed experiments. C.J. Fuller designed and wrote data analysis software. F.G. Westhorpe, C.J. Fuller, and A.F. Straight designed experiments and wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- CAC
- CENP-A C terminus
- CATD
- CENP-A targeting domain
- CCAN
- constitutive centromere–associated network
- CSF
- cytostatic factor
- HA
- hydroxyapatite
- HJURP
- Holliday junction recognition protein
- IVT
- in vitro translated
- PEG
- polyethylene glycol
References
- Bassett E.A., DeNizio J., Barnhart-Dailey M.C., Panchenko T., Sekulic N., Rogers D.J., Foltz D.R., and Black B.E.. 2012. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell. 22:749–762. 10.1016/j.devcel.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann J.H., Rodríguez M.G., Martins N.M., Kimura H., Kelly D.A., Masumoto H., Larionov V., Jansen L.E., and Earnshaw W.C.. 2011. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30:328–340. 10.1038/emboj.2010.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R., Sánchez P., Rivera T., Rodríguez-Corsino M., Boyarchuk E., Vassias I., Ray-Gallet D., Arnaoutov A., Dasso M., Almouzni G., and Losada A.. 2011. Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol. 192:569–582. 10.1083/jcb.201005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L. Jr., and Cleveland D.W.. 2004. Structural determinants for generating centromeric chromatin. Nature. 430:578–582. 10.1038/nature02766 [DOI] [PubMed] [Google Scholar]
- Blower M.D., Sullivan B.A., and Karpen G.H.. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330. 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Valente L.P., Mata J.F., Black B.E., and Jansen L.E.. 2013. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell. 24:923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., and Jansen L.E.. 2014. The quantitative architecture of centromeric chromatin. eLife. 3:e02137 10.7554/eLife.02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., and Straight A.F.. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., and Straight A.F.. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Dambacher S., Deng W., Hahn M., Sadic D., Fröhlich J., Nuber A., Hoischen C., Diekmann S., Leonhardt H., and Schotta G.. 2012. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 3:101–110. 10.4161/nucl.18955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., and Walczak C.E.. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G.. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497. 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Almouzni G., and Karpen G.H.. 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G₁ phase. Nucleus. 2:146–157. 10.4161/nucl.2.2.15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., and Straight A.F.. 2008. Genome-wide analysis reveals a cell cycle–dependent mechanism controlling centromere propagation. J. Cell Biol. 183:805–818. 10.1083/jcb.200806038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D., Folco H.D., Nechemia-Arbely Y., Valente L.P., Nguyen K., Wong A.J., Zhu Q., Holland A.J., Desai A., Jansen L.E., and Cleveland D.W.. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15:1056–1066. 10.1038/ncb2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D., Han J.S., McMahon M.A., Ly P., Abdullah A., Wong A.J., and Cleveland D.W.. 2015. DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell. 33:314–327. 10.1016/j.devcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco H.D., Campbell C.S., May K.M., Espinoza C.A., Oegema K., Hardwick K.G., Grewal S.I., and Desai A.. 2015. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 25:348–356. 10.1016/j.cub.2014.11.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., and Cleveland D.W.. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484. 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., and Yanagida M.. 2007. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev. Cell. 12:17–30. 10.1016/j.devcel.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Geiss C.P., Keramisanou D., Sekulic N., Scheffer M.P., Black B.E., and Frangakis A.S.. 2014. CENP-A arrays are more condensed than canonical arrays at low ionic strength. Biophys. J. 106:875–882. 10.1016/j.bpj.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Carroll C.W., Moree B., Fuller C.J., and Straight A.F.. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 477:354–358. 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Fuller C.J., and Straight A.F.. 2012. A cell-free system for functional centromere and kinetochore assembly. Nat. Protoc. 7:1847–1869. 10.1038/nprot.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., and Yanagida M.. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 118:715–729. 10.1016/j.cell.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Ebe M., Nagao K., Kokubu A., Sajiki K., and Yanagida M.. 2014. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes Cells. 19:541–554. 10.1111/gtc.12152 [DOI] [PubMed] [Google Scholar]
- Hellwig D., Emmerth S., Ulbricht T., Döring V., Hoischen C., Martin R., Samora C.P., McAinsh A.D., Carroll C.W., Straight A.F., et al. . 2011. Dynamics of CENP-N kinetochore binding during the cell cycle. J. Cell Sci. 124:3871–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S.M., and Harrison S.C.. 2013. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Reports. 5:29–36. 10.1016/j.celrep.2013.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. . 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052. 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60. 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., and Cleveland D.W.. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805. 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., and Bai Y.. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 340:1110–1113. 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., and Fukagawa T.. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168. 10.1091/mbc.E07-01-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A., Dorn J.F., De Rop V., Ladouceur A.M., Maddox A.S., and Maddox P.S.. 2010. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 12:1186–1193. 10.1038/ncb2129 [DOI] [PubMed] [Google Scholar]
- Logsdon G.A., Barrey E.J., Bassett E.A., DeNizio J.E., Guo L.Y., Panchenko T., Dawicki-McKenna J.M., Heun P., and Black B.E.. 2015. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 208:521–531. 10.1083/jcb.201412011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Hyndman F., Monen J., Oegema K., and Desai A.. 2007. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176:757–763. 10.1083/jcb.200701065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., and Cheeseman I.M.. 2014. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 158:397–411. 10.1016/j.cell.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miell M.D., Fuller C.J., Guse A., Barysz H.M., Downes A., Owen-Hughes T., Rappsilber J., Straight A.F., and Allshire R.C.. 2013. CENP-A confers a reduction in height on octameric nucleosomes. Nat. Struct. Mol. Biol. 20:763–765. 10.1038/nsmb.2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks K.J., Moree B., and Straight A.F.. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B., Meyer C.B., Fuller C.J., and Straight A.F.. 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194:855–871. 10.1083/jcb.201106079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechemia-Arbely Y., Fachinetti D., and Cleveland D.W.. 2012. Replicating centromeric chromatin: spatial and temporal control of CENP-A assembly. Exp. Cell Res. 318:1353–1360. 10.1016/j.yexcr.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J., Bergmann J.H., Kouprina N., Noskov V.N., Nakano M., Kimura H., Earnshaw W.C., Larionov V., and Masumoto H.. 2012. Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 31:2391–2402. 10.1038/emboj.2012.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. III, Desai A., and Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Otsu N. 1979. Threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9:62–66. 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Panchenko T., Sorensen T.C., Woodcock C.L., Kan Z.Y., Wood S., Resch M.G., Luger K., Englander S.W., Hansen J.C., and Black B.E.. 2011. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc. Natl. Acad. Sci. USA. 108:16588–16593. 10.1073/pnas.1113621108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N., and Black B.E.. 2012. Molecular underpinnings of centromere identity and maintenance. Trends Biochem. Sci. 37:220–229. 10.1016/j.tibs.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.C., Bodor D.L., Stellfox M.E., Martins N.M., Hochegger H., Foltz D.R., and Jansen L.E.. 2012. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell. 22:52–63. 10.1016/j.devcel.2011.10.014 [DOI] [PubMed] [Google Scholar]
- Subramanian L., Toda N.R., Rappsilber J., and Allshire R.C.. 2014. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 4:140043 10.1098/rsob.140043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., and Karpen G.H.. 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H., Müller S., Blümer J., Klare K., Musacchio A., and Almouzni G.. 2015. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Reports. 11:22–32. 10.1016/j.celrep.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Tan S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli . Protein Expr. Purif. 21:224–234. 10.1006/prep.2000.1363 [DOI] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 25:334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2015. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 7:a015818 10.1101/cshperspect.a015818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Zhou X., Ji L., Bradley P., Perrimon N., and Wong S.. 2006. Segmentation of Drosophila RNAI fluorescence images using level sets. IEEE Image Processing, 2006 IEEE International Conference. 2006:73–76. 10.1109/ICIP.2006.312365 [DOI] [Google Scholar]
- Yu Z., Zhou X., Wang W., Deng W., Fang J., Hu H., Wang Z., Li S., Cui L., Shen J., et al. . 2015. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev. Cell. 32:68–81. 10.1016/j.devcel.2014.11.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.