In response to mating pheromone, the yeast MFA2 mRNA is transported to the tip of the mating projection as an RNP granule and translated; integrity of the granules is required for normal mRNA transport and for the mating process.
Abstract
Association of messenger RNAs with large complexes such as processing bodies (PBs) plays a pivotal role in regulating their translation and decay. Little is known about other possible functions of these assemblies. Exposure of haploid yeast cells, carrying mating type “a,” to “α pheromone” stimulates polarized growth resulting in a “shmoo” projection; it also induces synthesis of “a pheromone,” encoded by MFA2. In this paper, we show that, in response to α pheromone, MFA2 mRNA is assembled with two types of granules; both contain some canonical PB proteins, yet they differ in size, localization, motility, and sensitivity to cycloheximide. Remarkably, one type is involved in mRNA transport to the tip of the shmoo, whereas the other—in local translation in the shmoo. Normal assembly of these granules is critical for their movement, localization, and for mating. Thus, MFA2 mRNAs are transported to the shmoo tip, in complex with PB-like particles, where they are locally translated.
Introduction
Localization of mRNAs to specific regions of the cell can play a pivotal role in determining protein localization. Localized translation has been documented in prokaryotes (Nevo-Dinur et al., 2011) and in eukaryotes (Decker and Parker, 2012; Kong and Lasko, 2012). As an example, we and others have shown that mRNA localization plays an important role in targeting certain proteins to the daughter cell (bud) of dividing Saccharomyces cerevisiae (Darzacq et al., 2003; Aronov et al., 2007).
Exposure of haploid S. cerevisiae cells to pheromone results in cell cycle arrest, expression of mating-specific genes, and polarized growth that leads to the formation of a cell projection toward the mating partner. Through this projection, called “shmoo,” fusion will eventually occur (Chant, 1999; Bardwell, 2005; Jones and Bennett, 2011). Proteins involved in polarization, signaling, cell adhesion, and fusion are localized to the shmoo or its tip (Narayanaswamy et al., 2009). Proteins are delivered to the growing shmoo tip via secretory vesicles. Alternatively, mRNAs are transported to the shmoo tip and are locally translated therein. Recently, it was shown that some mRNAs encoding polar and mating proteins, such as Sro7, Sec3, Ste7, and Fus3, are localized to the shmoo. Delivery of these mRNAs to the shmoo is mediated by Myo4p. These mRNAs are cotransported and coregulated with cortical endoplasmic reticulum. A pheromone-activated RNA-binding protein, Scp160, is associated with the ER and is involved in localization/regulation of these specific mRNAs (Frey et al., 2001; Gelin-Licht et al., 2012). The functional role of mRNA targeting during mating, and the factors involved in this process are still not well understood.
Previous studies have demonstrated that mRNAs can be assembled with large complexes that can be detected as cytoplasmic granules. Detection of these granules is often, but not invariably, made possible during stress. In higher eukaryotes, the most characterized granules are processing bodies (PBs) and stress granules (SGs; Anderson and Kedersha, 2009; Decker and Parker, 2012). Both share common protein constituents and each carry unique components. In yeast, the first characterized granules were PBs, consisting of translationally repressed mRNAs and many proteins, including several mRNA decay factors. Consistently, the major decapping-dependent mRNA decay can be executed in these complexes (Sheth and Parker, 2003). However, PBs can represent sites where relatively stable poly(A)-containing capped mRNAs are stored (Brengues and Parker, 2007). Our view of mRNA granules is gradually changing from one in which granules are rigid and well defined (e.g., PBs and SGs) to a more dynamic view in which the protein and RNA constituents of granules can change rapidly. For example, poly(A)-containing mRNAs can move back and forth from PBs to polysomes or from polysomes to SGs (Decker and Parker, 2012). The balance of mRNA movement between the granules and polysomes is an important parameter that regulates mRNA translation and decay (Decker and Parker, 2012). The regulatory roles of PBs in mRNA translation in general, and during the yeast mating process, in particular, are still not well known.
Previously, it was shown that the yeast MFA2 mRNA, encoding the aF (a pheromone), can be detected in PBs of haploid yeast cells not exposed to any pheromone (Sheth and Parker, 2003). Because MFA2 mRNA is among the most unstable mRNAs (half-life = ∼4 min), it is quite possible that association of this mRNA with PBs leads to its degradation. Indeed, stable degradation intermediates of MFA2 mRNA can be found in PBs of optimally growing wild-type (WT) cells, whereas association of the unstable full-length MFA2 mRNA is less conspicuous (Sheth and Parker, 2003).
Here we show, that in response to αF (α pheromone), MFA2 mRNA accumulates in granules, together with standard PB markers as well as Pab1p, a poly(A) binding protein. We observed two major kinds of MFA2 mRNA–containing granules exhibiting different features: large immotile granules that localize preferentially in shmoo tips and small motile granules that are found all over the cell, yet accumulate more in the shmoo proximal portion. Data, presented here, led us to propose that small PBs are transported from the cell body to the shmoo tip and fuse with the large PBs. Remarkably, Mfa2p-RFP fusion is synthesized preferentially in the vicinity of the shmoo-localized large granule. We therefore named these structures mating bodies. We propose that MFA2 mRNA-containing granules are transported to the shmoo tip to form mating bodies, and only there is the mRNA translated.
Results
MFA2 mRNA associates with distinct granules of various intensities in response to αF
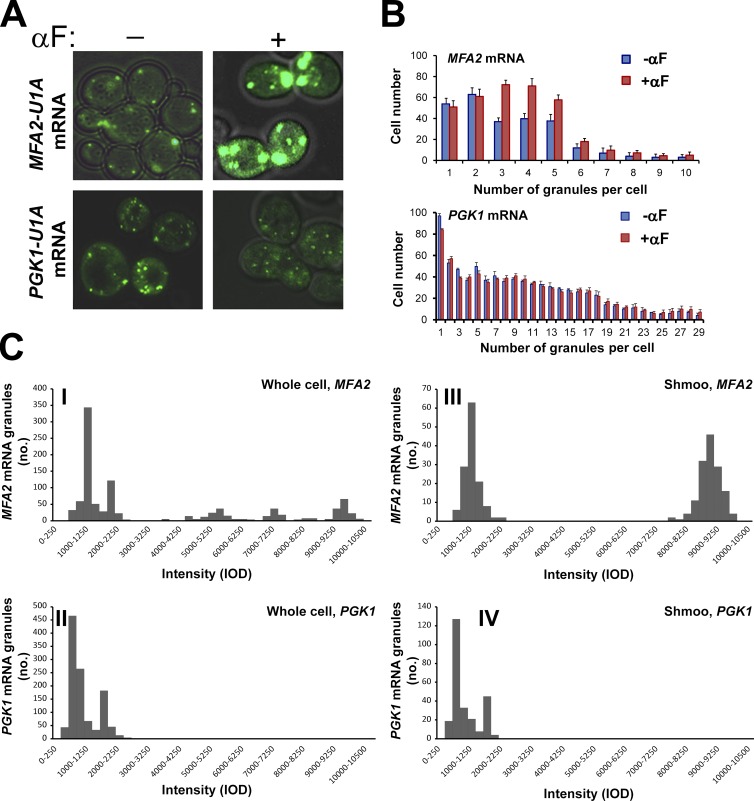
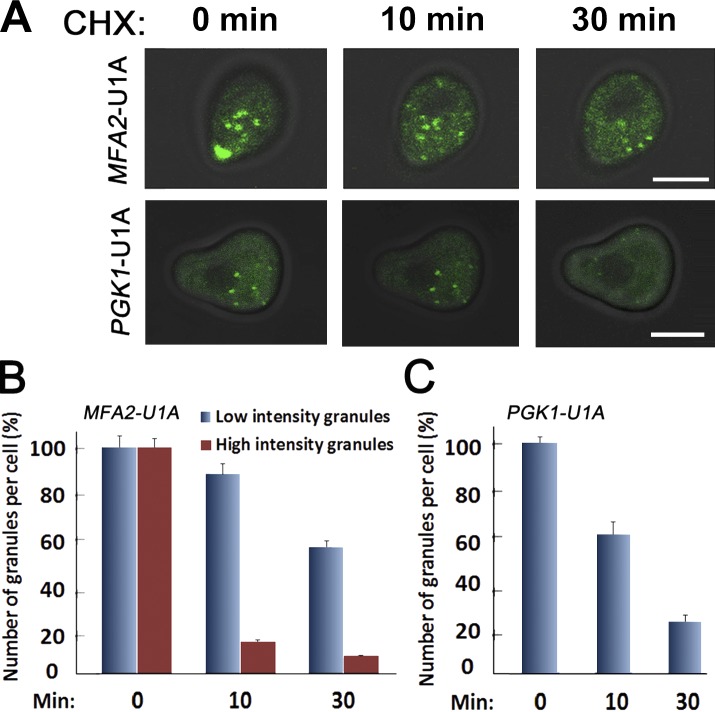
To investigate whether MFA2 mRNA localization plays any role during mating, we first analyzed the localization of MFA2 mRNA in response to αF. We used a previously developed system consisting of two plasmids encoding MFA2-U1A binding sites and U1A-GFP (Brodsky and Silver, 2000; Sheth and Parker, 2003). Plasmid expressing PGK1-U1A was used to observe mRNA localization of a housekeeping gene, PGK1, encoding 3-phosphoglycerate kinase. As expected, in untreated cells, MFA2 and PGK1 mRNAs were detected in discrete granules, defined as PBs by a previous study (Sheth and Parker, 2003). Treatment with αF increased the size (Fig. 1 A) and number (Fig. 1 B) of MFA2 mRNA-containing granules by 3–5-fold after 1 h. This effect was specific, as the granules containing the unrelated PGK1 mRNA did not respond to this treatment (Fig. 1, A and B).
Figure 1.
In response to pheromone treatment, the number, intensity, and distribution of MFA2 mRNA granules increases, whereas PGK1 mRNA granules remain unchanged. (A) The WT strain coexpressing U1A-GFP and either MFA2-U1A (ySA20) or PGK1-U1A (ySA32) was grown to the early exponential phase and treated with 3 nM αF for 60 min, as indicated. Bar, 5 µm. (B) Quantification of the number of MFA2 or PGK1 mRNA granules in treated and untreated cells 1 h after adding αF. Histograms illustrating the distribution of mRNA granules per cell are shown. Error bars represent the standard deviation of three independent experiments. n = 200–350 cells. Averaging these numbers revealed that, in response to αF, the number of MFA2 mRNA–containing granules per cell increased twofold, whereas their intensity increased threefold. No change in the number and intensity of the PGK1 mRNA–containing granules was observed in response to a similar treatment with αF. (C) Intensity histograms of MFA2 and PGK1 mRNA granules in αF-treated cells. The WT strain coexpressing U1A-GFP and either MFA2-U1A (ySA20) or PGK1-U1A (ySA32) was treated with αF for 2 h. Images were captured, threshold, and analyzed for Integrated Optical Density (IOD) using the ImageJ program. IOD distributions are depicted as histograms (see Materials and methods). 200–350 cells displaying a clear shmoo were analyzed. Distribution of intensities of the MFA2 and PGK1 mRNA granules in a whole cell (I and II) and in the shmoo region (III-IV) are shown. Note the clear distinction between the “low-intensity” granules (750–2,200 IOD) and the “high-intensity” granules (8,000–10,000 IOD). The data represent compilation of three independent experiments.
To characterize the granules, we first classified them according to their intensity. As shown in Fig. 1 C [I], MFA2 mRNA-containing granules were heterogeneous, with intensities ranging from low to high. In contrast, PGK1 mRNA-containing granules were more homogeneous and only exhibited low intensity (Fig. 1 A [II]).
High-intensity MFA2 mRNA-containing granules are preferentially localized in the shmoo
In response to αF, yeast cells grow asymmetrically and form long projections called shmoo (see Introduction). We next analyzed the intensity of the granules that were localized to the shmoo tip. Most of the shmoo-localized granules exhibited high intensity, (Fig. 1 C [III]); in contrast, most of the PGK1 mRNA-containing granules in the shmoo region exhibited low intensity (Fig. 1 C [IV]), and these granules were indistinguishable from the PKG1 mRNA-containing granules found outside the shmoo. These results demonstrate that, whereas low-intensity MFA2 mRNA granules are dispersed throughout the cell, high-intensity granules are found preferentially within the shmoo tip.
The distribution of MFA2 mRNA-containing granules was then examined over time in shmoo-growing cells after αF addition (Fig. 2 A). In 79 ± 5% of the cells, one high-intensity granule was localized asymmetrically in the shmoo tip 2–4 h after stimulation by αF. In 71 ± 5% of the cells, the granule was retained for up to 6 h. In contrast, by 2 h after αF addition, low-intensity granules were more abundant in the shmoo than in the cell body; at later time points, when the response to the pheromone decreased (Hicks and Herskowitz, 1976), these granules were evenly distributed (see also Fig. 6 C). The unrelated PGK1-containing granules were distributed evenly through the cell and were unaffected by αF.
Figure 2.
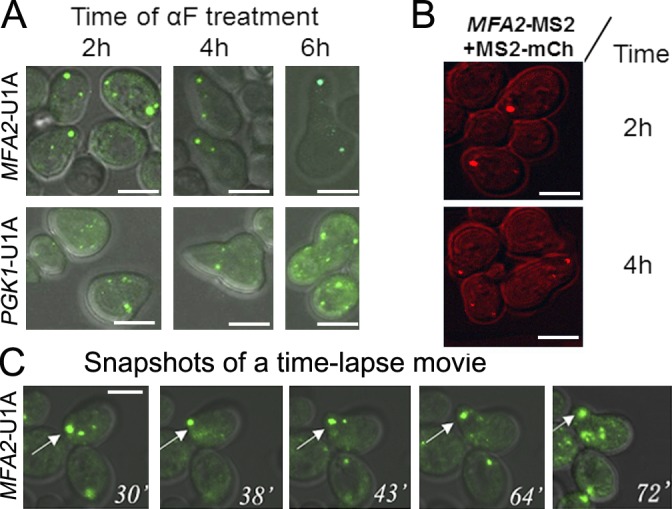
MFA2 mRNA granules, but not PGK1 mRNA granules, are preferentially localized to shmoo tips. (A) Microscopic analysis of the distribution of low- and high-intensity MFA2 and PGK1 mRNA granules during shmoo growth. The WT strain coexpressing U1A-GFP and either MFA2-U1A (ySA20) or PGK1-U1A (ySA32) was treated with αF for the indicated time. n = 200–250 from three independent experiments. (B) Shmoo localization of MFA2 mRNA granules was observed using an MS2 labeling system, instead of the U1A system. The WT strain (ySA24) expressing MFA2-MS2 (from pMC475) and MS2-mCh (from pMC522) was grown in selective medium until mid–log phase and then treated with αF. At the indicated time after αF addition, cells were inspected under confocal microscope. (C) Snapshots from time-lapse analysis of MFA2 mRNA-containing granules during shmoo formation in response to αF treatment (Video 1). The arrows point at a high-intensity MFA2 mRNA granule that was located in the shmoo tip during the growth of the shmoo (Video 1). Bars, 5 µm.
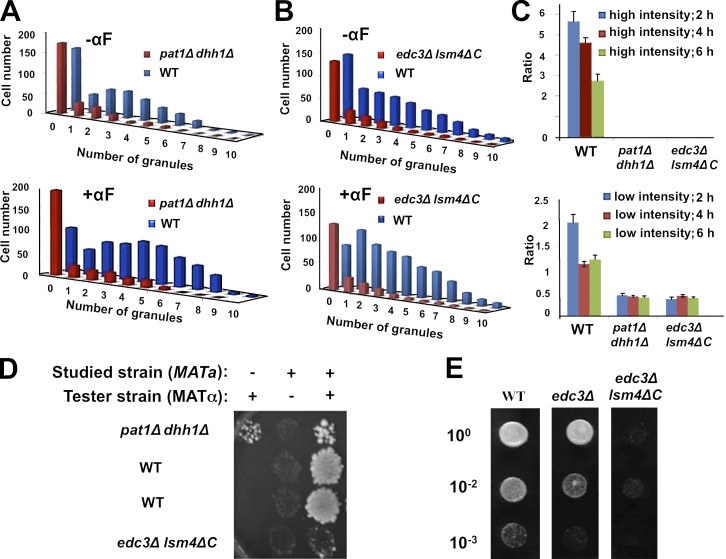
Figure 6.
PB integrity is required for the normal responses and shmoo localization of MFA2 mRNA granules to αF treatment and for efficient mating. (A and B) MFA2 mRNA was expressed in WT cells and in two strains defective in PB formation—pat1Δ dhh1Δ (ySA25; A) and edc3Δ lsm4ΔC (ySA25; B). The cells were treated with αF for 2 h, and the number of MFA2 mRNA granules were counted (n = 200–350 cells). Histograms are plotted as in Fig. 1 B. (C) Granules are preferentially localized in the shmoo-proximal portion of WT cells but not in mutant cells. WT and double deletion mutant strains, described in A and B, were treated with αF for 2, 4, or 6 h. Each type of cell was divided into two equal portions, one containing a shmoo (“shmoo-proximal”) and a “shmoo-distal” (cell body) one. MFA2 mRNA granules were counted and classified into high- and low-intensity granules. We note that the difference in the intensity between the two types of granules is ∼10-fold (see Table 1), and the distinction between them was quite apparent. The ratio between the total number of granules in shmoo-proximal region and in shmoo-distal region is indicated. The ratio of distribution for high (top)- and low (bottom)-intensity granules was calculated for the indicated strains. No high-intensity granules were observed in the double deletion mutant strains. Error bars represent the standard deviation from three independent experiments. n = 100–125 cells in each sample. (D) The studied mutant strains are sterile. The mating assay was determined with the indicated mutant strains (yMC376; yMC524) or isogenic WT (yMC375; all are MATa), as was reported previously (Michaelis and Herskowitz, 1988). The tester strain was WT (MATα; yMC134; see Table 4). After 3 h of mating on rich nonselective plate (YPD), the cells were replica plated onto appropriate selectable plate and incubated for 3 d before the photo was taken. (E) edc3Δ (yMC429) cells are only partially sterile relatively to the edc3Δ lsm4ΔC strain. Semiquantitative mating assay was performed as in D except that 10-fold dilutions were spotted, as indicated.
Because both MFA2 and PGK1 mRNAs were detected by the same U1A system, the different localization and behavior of these mRNAs cannot be attributed to the U1A detection system per se. Consistently, MFA2 mRNA granules that were labeled by a different mRNA detection system—the MS2 loops—with a different fluorophore, mCherry (mCh; MS2-mCh), were also localized to the shmoo tip (Fig. 2 B). This further demonstrates that the response of MFA2 mRNA granules to αF is independent of the detection system.
We used time-lapse videomicroscopy to obtain the dynamic features of these granules. Interestingly, during treatment with αF, the high-intensity and immotile granules were localized in the shmoo as early as 30 min and remained associated with the shmoo tip during its growth (Fig. 2 C and Video 1). These granules remained immotile during the entire course of the experiment, but their size first increased during the first 2 h after αF addition, and later it decreased. In contrast, the low-intensity granules were motile throughout the experiment (Table 1).
Table 1. Quantitative features of MFA2 mRNA granules in αF-treated WT cells.
| Type of mRNA | Intensity | CHX sensitivity | Intensity mean (IOD)a | Approximate | Velocityb | Type of motility |
|---|---|---|---|---|---|---|
| % | µm/s | |||||
| MFA2 mRNA | High | + | 8,000–10,000 | 12 | 0.05 ± 0.04 | Vibrational |
| Low subgroup 1 | − | 1,800–2,200 | 27 | 0.04 ± 0.03 | Vibrational | |
| Low subgroup 2 | − | 800–1,100 | 16 | 1.81 ± 0.78 | Translocational | |
| Low subgroup 3 | − | 800–2,200 | 45 | 0.45 ± 0.09 | Oscillatory with slow translocation | |
| PGK1 mRNA | Low subgroup 1 | − | 1,400–2,250 | 25 | 0.09 ± 0.08 | Vibrational |
| Low subgroup 2 | − | 500–1,200 | 75 | 0.79 ± 0.06 | Oscillatory with slow translocation |
Three independent videos were taken, using time-lapse technology with a Fast Acquisition AxioVision microscope. Videos were analyses of between 25 and 30 frames, each between 73 and 113 ms, without intervals between shots. These videos were used to determine IOD, granule velocity, and granule number per cell.
IOD refers to integrated optical density (IOD) of the fluorescent granules, obtained using the ImageJ program. For determining IOD, n = 100 random snapshots, each one contains several cells (5–9).
Tracking of individual granule was performed using Imaris program version 7.1 (Bitplane AG). Individual granules were tracked throughout the video, unless the granule appeared during a portion of the video (at least in eight consecutive frames). Average values and SD were calculated based on 10–15 granules for each group/subgroup.
Small MFA2 mRNA-containing granules are motile and are capable of directionally moving toward the shmoo tip
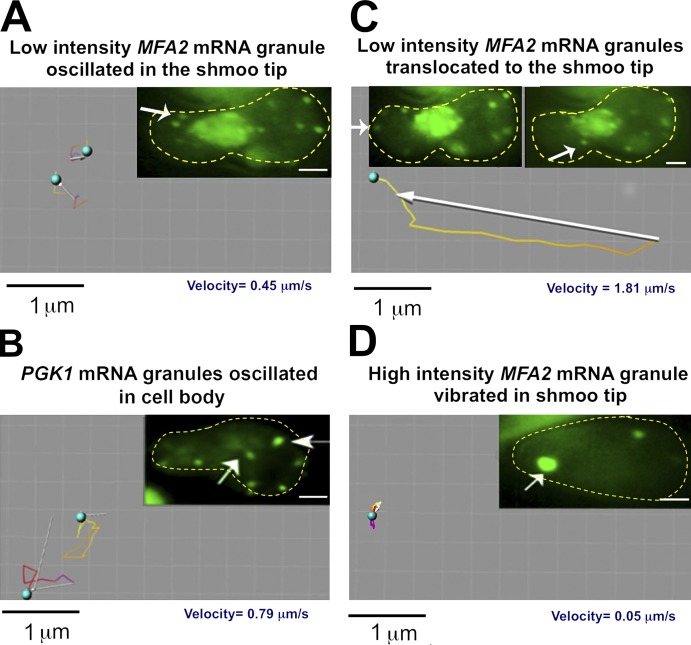
We next characterized the motility of the granules by using a short duration time-lapse videomicroscopy (see Materials and methods). We found that the high- and low-intensity MFA2 mRNA granules exhibited different types of motility that was characterized by velocity, type of movement, and the specific location in the cells. High-intensity granules (∼12% of the total number of granules) were relatively immotile during shmoo growth with an average velocity of 0.05 ± 0.04 µm/s (Fig. 3 D and Table 1). These granules were confined to the region of the shmoo tip and stayed there during shmoo growth (Fig. 2). Low-intensity granules showed three discrete types of motility: one group remained stationary, with an average velocity of 0.04 ± 0.039 µm/s (∼27% of the total number of granules; Video 2). The second group moved with an average velocity of 0.45 ± 0.095 µm/s—10 times faster than the stationary granules (∼45% of the total number of granules; Fig. 3 A and Table 1). Their movement was oscillatory with slow translocation transport. The third group of low-intensity MFA2 mRNA-containing granules moved directly and rapidly over long distances from the cell body to the shmoo tip with a velocity of 1.8 ± 0.78 µm/s (∼16% of the total number of granules; Fig. 3 C, Table 1, and Videos 2 and 3). Although this type of movement was observed early on after αF treatment (e.g., at 2 h after treatment), it was no longer observed 4 h after the treatment with αF, when the responses to the pheromone diminished (Hicks and Herskowitz, 1976). This type of movement might be responsible for accumulation of low-intensity granules in the shmoo and for depletion of these granules from the cell body observed 2 h after adding αF (see Fig. 6 C, WT).
Figure 3.
Tracking analysis of MFA2 and PGK1 mRNA granules. (A–D) Cells expressing a MFA2-U1A detection system (A, C, and D) or a PGK1-U1A detection depiction system (B) were treated with αF for 2 h and high- and low-intensity granules (marked by arrows) in the shmoo region and cell body were tracked using the Imaris program in 2D videos (see Materials and methods). Images in A and C are snapshots taken from Videos 2 and 3. The yellow dotted lines highlight the cell boundary. (A) Two low-intensity MFA2 mRNA granules located in the shmoo region of the cell were tracked. Note the oscillatory motion and slow translocation of the granules. Tracking shows movement duration of 6.6 s. (B) Two low intensity motile PGK1 mRNAs granules in the cell body were tracked; note the oscillatory motion and slow translocation of the granules. Tracking shows movement duration 6.3 s (Videos 4 and 5). (C) A low-intensity MFA2 mRNA granule that was translocated from the cell body to the shmoo tip was tracked. The right arrow points at the translocated granule, at its half-way location; the left arrow points at this granule at its final destination. The tracking shows movement duration of 4.1 s. Note the rapid translocation from the cell body to the shmoo tip. (D) A high-intensity MFA2 mRNA granule located in the growing shmoo tip was tracked. The tracking shows movement duration of 60 s. Note the vibrational motion of the granule that remains localized to the shmoo tip.
75% of PGK1 mRNA-containing granules exhibited high motility (0.79 ± 0.063 µm/s), whereas 25% exhibited slowly motion (0.09 ± 0.078 µm/s; Fig. 3 B, Table 1, and Videos 4 and 5). Their transport was characterized by oscillatory movement with slow translocation. The long distance movement, which characterizes the MFA2 mRNA granules, was not observed in the case of PGK1 mRNA granules. All these features of PGK1 granules were independent of their localization (Videos 4 and 5).
Both kinds of MFA2 mRNA-containing granules contain Dcp2p-RFP, Lsm1p-RFP, and Pab1p-RFP
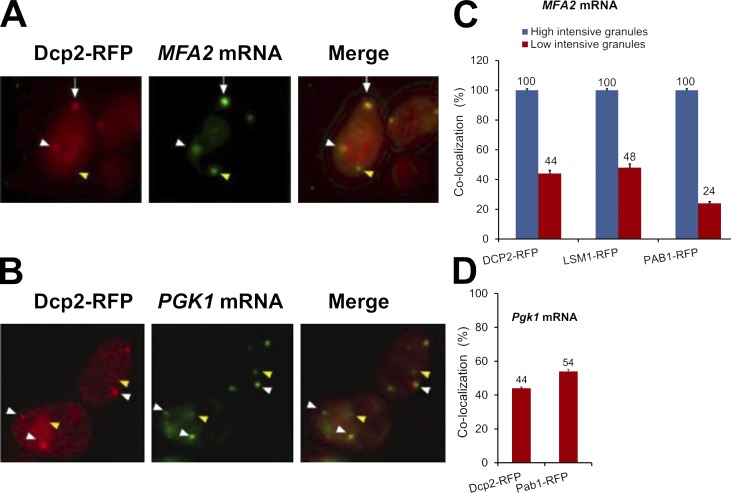
A previous study described the presence of MFA2 mRNA in PBs in untreated cells (Sheth and Parker, 2003). To determine whether the αF-responsive MFA2 mRNA-containing granules, described here, represent PBs, or PB derivatives, we investigated whether they colocalize with RFP-tagged PB markers, namely, Dcp2p and Lsm1p. Indeed, after αF treatment, in otherwise optimal conditions, about one half of the low-intensity and all of the high-intensity MFA2-containing granules colocalized with both PB markers (Fig. 4, A and C). The poly(A)-binding protein and SG marker, Pab1p-RFP, also colocalized with MFA2 mRNA in αF-treated cells (Fig. 4 C). Specifically, all high-intensity granules were colocalized with this protein, whereas 24% of low-intensity granules contained Pab1p-RFP. The capacity to bind both PB and SG markers was not specific to MFA2, as these markers also colocalized with PGK1 foci of αF-treated cells (Fig. 4, B and D). Pab1p plays an important role in translational control. It was suggested that the presence of Pab1p in mRNA granules might represent a transition state between mRNA granules and translation (Brengues and Parker, 2007). The presence of Pab1p in MFA2 and PGK1 mRNA granules suggests that, in αF-treated cells, these two mRNAs are poised to be transported to polysomes. Association of Pab1p-RFP with mRNA granules was unexpected because, in optimally proliferating cells, no Pab1p-RFP foci can be detected (Brengues and Parker, 2007).
Figure 4.
MFA2 mRNA granules colocalize with PBs in the shmoo tip. (A) Cells expressing MFA2 mRNA and the PB marker, Dcp2p-RFP (red; ySA22), were treated with αF for 2 h and analyzed by fluorescence microscopy. Both high- and low-intensity granules colocalized with the PB marker (see also Video 6). Arrows show high-intensity mRNA granules colocalized with PB marker; white arrowheads mark colocalized low-intensity granules; yellow arrowheads show low-intensity mRNA granules that did not colocalize with PB markers. (B) Colocalization of PGK1 mRNA and PB markers in αF-treated yeast. Cells expressing PGK1-U1A mRNA (green) and Dcp2p-RFP (red; ySA26) were treated with αF for 2 h, as indicated, and analyzed by fluorescence microscopy. (C) Colocalization of high- and low-intensity MFA2 mRNA granules with the indicated markers in αF-treated cells (ySA21; ySA23). (D) Colocalization of PGK1 mRNA with PB markers (ySA26; ySA33) in αF-treated cells. Error bars represent the standard deviation from three independent experiments. n = 220–250 cells. Bars, 5 µm.
To examine whether movement of MFA2 mRNA granules in αF-stimulated cells occurs in the context of the PB marker Dcp2p, we challenged cells with αF and examined movement by a short duration time-lapse analysis. We found that at early time points (30 min to 1 h) after αF addition, when movement is intense, MFA2 mRNA and Dcp2p share the same granules and move together (Fig. 5 and Video 6).
Figure 5.
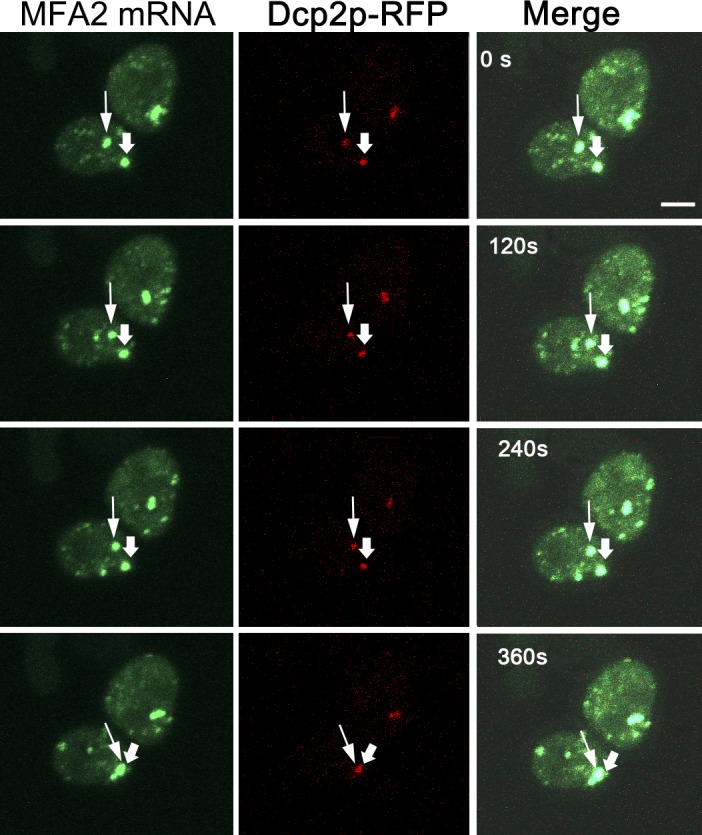
MFA2 mRNA move together with Dcp2p-RFP in αF-treated cells. Snapshots of Video 6 are shown, indicating a time-lapse analysis of early time points after αF treatment (when the stationary granule was still of low intensity) performed using z stack (six slices with 0.6-µm thickness), during 360 s. Bar, 2 µm. The thin arrowheads show one example of MFA2 mRNA granules colocalized with Dcp2p-RFP (ySA22). The thick arrowheads point at the stationary granule at the tip of the (still) small shmoo. Note that the marked low-intensity granule moved, together with Dcp2p, in the direction of the shmoo tip.
Disruptions of PB integrity, using different mutations, compromise the motility of MFA2 mRNA granules and their shmoo localization
Association of MFA2 mRNA with components of PBs prompted us to examine whether PB integrity is required for the proper localization of MFA2 mRNA to the shmoo. To address this issue, we took advantage of two kinds of mutant strains defective in PB assembly. Importantly, each mutant is defective as a result of a different mechanism. The first strain lacks DHH1 and PAT1, whose products repress translation that, in turn, stimulate assembly of the untranslated mRNA with PBs (Coller and Parker, 2005). The second strain lacks Edc3p and the C-terminal domain of Lsm4p. Edc3p serves as a PB scaffold, whereas the C-terminal domain of Lsm4p comprises a Q/N-rich “prion” domain that was proposed to bind strongly to other similar domains found in other PB constituents (Decker et al., 2007). Hence, the edc3Δ lsm4ΔC strain is defective in PB assembly. Yet, unlike the dhh1Δ pat1Δ strain, this strain is not defective in translation and exhibits a very minor defect in MFA2 mRNA decay (Decker et al., 2007). Both these mutant strains proliferate similarly to their WT counterparts (Fig. S2).
Using these mutants, we made several observations. In untreated cells, the number of granules was reduced by 70–80%, as reported previously (Coller and Parker, 2005), and no high-intensity granules were observed (Fig. 6, A and B). Second, treatment of the two mutant cells for up to 6 h with αF did not stimulate any change in the number of MFA2 mRNA granules or in their intensity (Fig. 6, A and B; and Tables 2), unlike the substantial change observed in the WT strain (Fig. 1, A and B; and Fig. 6, A and B). Motility of the low-intensity MFA2 mRNA granules was severely compromised in the mutant cells. We did not observe the translocated-type movement that characterizes some of the WT low intensity granules (e.g., Fig. 3 C). Moreover, the velocity of the oscillating granules was reduced at least 10-fold (Table 2 and Video 7). It is possible that the defect in the motility of the small granules and the defect in assembly of large granules are linked. Third, the high-intensity granules are preferentially localized in the shmoo-proximal half of the WT cells (Fig. 6 C). More specifically, ∼90% of the shmoo tips contain (mainly one) granule. Importantly, by 2 h after αF addition, the low-intensity granules also localized preferentially in the shmoo-proximal part of the WT cells (Fig. 6 C), consistent with their active transport toward the shmoo (Fig. 3 C and Videos 2 and 3). In contrast, MFA2 mRNA granules in both mutant cells (all are of low intensity) were localized in the portion of the cells distal to the shmoo, usually the one containing the nucleus (Fig. 6 C and Video 7). We observed many granules near the nucleus and surmised that the immotile granules remain in the vicinity of the nucleus, thereby contributing to the low ratio depicted in Fig. 6 C. Note that granule motility was equally defective in the two mutant cells (Table 2) and similarly distributed in these mutants (Fig. 6 C). Hence, although PB assembly is defective in both mutant strains caused by different mechanisms, motility of the MFA2 mRNA granules as well as their localization was similarly defective. Thus, it seems that PB integrity per se is required for αF-induced increased formation, motility, and shmoo localization of MFA2 mRNA-containing granules.
Table 2. Quantitative features of MFA2 mRNA granules in αF-treated mutant strains that carry defective PBs.
| Strain | Intensity | Intensity mean (IOD) | Number of granules per cell | Velocity | Type of motility |
|---|---|---|---|---|---|
| µm/s | |||||
| pat1dhh1Δ | Low | 400–600 | 1.7 ± 0.85 | 0.021 ± 0.06 | Oscillatory |
| edc3Δ lsm4ΔC | Low | 450–650 | 2.3 ± 0.49 | 0.025 ± 0.04 | Oscillatory |
This table is based on analyses that are described in Table 1.
PB mutants are sterile
If indeed PB assembly is essential for MFA2 mRNA transport to the shmoo tip, which is, in turn, important for the mating pathway, PB mutants should be sterile. We found that both mutant cells sensed and responded to the pheromone, as demonstrated by the capacity of αF to induce transcription of the pheromone responsive FIG1 gene (Fig. S1 A; Erdman et al., 1998). However, we found that they were sterile (Fig. 6, D and E) and that only a portion of these cells (∼25%) were able to form a shmoo, which was relatively small. We note that pat1Δ dhh1Δ strain and edc3Δ lsm4ΔC strains are severely defective in PB assembly as a result of different and unrelated mechanisms. edc3Δ (carrying WT LSM4) is partially defective in PB formation (Decker et al., 2007). Interestingly, we found a direct correlation between PB assembly and the capacity to mate (Fig. 6 E). In general, edc3Δ cells do not have a translation defect nor do they have a defect in MFA2 mRNA decay (Decker et al., 2007). Collectively, the results shown in Fig. 6 (D and E) suggest that the sterile phenotype is caused by defects in PB assembly per se and not to defect in mRNA translation or decay. However, we cannot rule out the possibility that these three different mutants are sterile as a result of unrelated causes.
The high-intensity MFA2 mRNA granules dissociate rapidly in response to cycloheximide (CHX)
mRNAs can move back and forth between polysomes and mRNA-containing granules (Brengues et al., 2005; Teixeira et al., 2005; Brengues and Parker, 2007). Addition of CHX is proposed to trap the mRNA in the polysomes, without affecting its movement away from the granules. Consequently, PBs and SGs are rapidly depleted of mRNPs after CHX addition and dissociate (Teixeira et al., 2005). Addition of CHX to αF-treated cells led to a rapid dissociation of the shmoo-localized high-intensity granules (Fig. 7, A–C; and Table 3). This result argues against the possibility that translation of MFA2 mRNAs occurs in the granules themselves and suggests that MFA2 mRNAs shuttle between the polysomes and the granules. CHX had a slower effect on the integrity of low-intensity and mobile granules, suggesting that the mRNPs in these granules do not move to polysomes as rapidly as the mRNPs of high-intensity granules. By 10 min after CHX addition, 91% of the small granules remained visible. Only after 30 min in the presence of CHX did the number of the small granules decline to 57%. This late decline can be an indirect effect of blocking protein synthesis in general. The relative resistance of these low-intensity granules to CHX was unexpected because, in optimally proliferating cells (not challenged with αF), all MFA2 mRNA–containing granules disappeared in response to CHX (Teixeira et al., 2005). We propose that, in response to αF, MFA2 mRNA–containing granules change their cross talk with the ribosomes.
Figure 7.
The large granules of MFA2 mRNA dissociate rapidly in response to CHX. (A) Cells expressing MFA2-U1A (ySA20) and PGK1-UA1 mRNA (ySA32) detection system were treated with αF for 2 h. 100 µg/ml cycloheximide (CHX) was then added, and cells were inspected microscopically 10 or 30 min later (top). Bars, 5 µm. (B and C) The percentage of high- and low-intensity MFA2 or PGK1 mRNAs granules was determined at the indicated time points after CHX addition. The percentage is plotted relative to t = 0 (before CHX addition), which was arbitrarily defined as 100%. Error bars represent the standard deviation of three independent experiments.
Table 3. Effect of CHX treatment on the fluorescence associated with MFA2 and PGK1 mRNAs in αF-treated cells.
| Type of mRNA | Intensity | CHX sensitivity | |
|---|---|---|---|
| 30 min | 10 min | ||
| MFA2 mRNA | High | +++ | +++ |
| Low | + | − | |
| PGK1 mRNA | Low | +++ | + |
These results were obtained from three independent experiments. n = 200–230 cells (see Fig. 7).
These observations raise the possibility that MFA2 mRNA in the small granules is translationally repressed. Regardless, these results further illustrate the difference between the two kinds of MFA2 mRNA-containing granules (Fig. 7, A–C). PGK1 mRNA granules in αF-treated cells responded more slowly to CHX than the high-intensity MFA2 mRNA granules but faster than the low-intensity MFA2 mRNA granules. It leads to a dissociation of 38% of all low-intensity PGK1 mRNA granules after 10 min and to a dissociation of 78% after 30 min. These results demonstrated that PGK1 mRNA granules were more sensitive to CHX than the low-intensity MFA2 mRNA granules. Perhaps the reason for such sensitivity was that more than half of the PGK1 mRNA granules are translationally competent. Thus, in αF-treated cells, both types of MFA2 mRNA granules seem to be different than PGK1 mRNA granules.
MFA2 mRNA is translated in the vicinity of the large granules in the shmoo tip
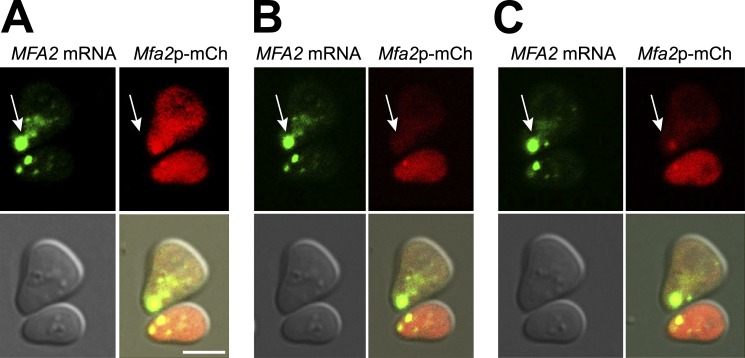
The rapid disappearance of the shmoo tip–localized MFA2 mRNA granules in response to CHX suggested that the mRNA in these granules is translated. To further examine this possibility, we created an MFA2-mCh-U1A chimeric gene. The transcript of this gene contains U1A binding sites and encodes Mfa2p fused to the fluorescent mCh tag, allowing for the detection of both mRNA and its fluorescent protein product. Addition of the mCh tag did not affect the number and distribution of the RNA-containing granules in response to αF treatment (unpublished data). In untreated cells, mCh-tagged Mfa2p protein was distributed evenly throughout the cell (not depicted), whereas it accumulated in the shmoo tip in response to αF treatment (Fig. 8 A). To detect newly synthesized Mfa2p protein, red fluorescence was bleached from one of the two cells shown in Fig. 8 B. Reappearance of the red fluorescence was detected shortly thereafter (∼10 min) in the vicinity of the shmoo tip, clearly demonstrating that MFA2 mRNA is translated there (Fig. 8 C). Because the red fluorescence of the entire cell was bleached, all the observed fluorescence that was reappeared after 10 min could only be caused by de novo translation. Note that the green signal was unaffected by bleaching of the red fluorescence, demonstrating that the red signal shown in Fig. 8 (A and C) is not a spillover of the green signal. We note that the red chromophore might have acquired its fluorescent conformation after bleaching; accordingly, the red signal, shown in Fig. 8 C, is caused by a protein that had been synthesized earlier. To further demonstrate that the reappearance of the red signal in the shmoo tip is caused by de novo translation, we added CHX and immediately thereafter bleached the red fluorescence. As expected (Fig. 7), the high intensity MFA2 mRNA-containing granules disappeared gradually after CHX treatment, demonstrating that the drug was effective. The red signal could not be detected during the recovery from bleaching, suggesting that the red signal is caused by a protein that was synthesized after bleaching. We conclude that MFA2 mRNA-Pab1p-Dcp2p-Lsm1p–containing granules are transported to the shmoo tip where the mRNA is subsequently released and translated.
Figure 8.
Local translation of MFA2 mRNA in the vicinity of the high-intensity granule in the shmoo. (A–C) Mfa2p-mCh protein colocalizes with its mRNA in the shmoo tip after αF treatment. WT yeast cells cotransformed with MFA2p-mCh-UA1 and UA1p-GFP (ySA147) were grown to early logarithmic phase and treated with αF for 2 h. The upper cell was photobleached, and fluorescence recovery was monitored by time-lapse microscopy. (A) Representative images from the time-lapse video before photobleaching; bar, 5 µm. The arrows point at high-intensity MFA2 mRNA granules (green)/protein (red) accumulated in the shmoo tip. (B) Immediately after photobleaching. (C) 10 min after recovery.
Discussion
Our view of mRNA granules has evolved from a simple one involving clearly defined PBs and SGs to a more complex one in which mRNP granules can change RNA and protein partners—forming various different combinations, possibly a continuum, of complexes. Three main classes of proteins are known to accommodate these granules: mRNA decay factors and translation factors as well as structural and/or regulatory factors. Indeed, it is by now an established view that these granules can regulate mRNA decay, storage, or translation (Anderson and Kedersha, 2009; Decker and Parker, 2012).
Our work uncovers an additional function for yeast RNA granules in establishing mRNA localization within the cell. We show that, in response to αF, MFA2 mRNA accumulates in granules, together with standard PB markers as well as with Pab1p, a poly(A) binding protein. Unlike PGK1 mRNA granules whose intensities follow a normal distribution, MFA2 mRNA granules can be functionally classified into two major types. The first type constitutes low-intensity granules (possibly small granules that contain one or two mRNAs) that are highly motile, dispersed throughout the cell, and do not respond to CHX treatment (Fig. 7). Significantly, components that maintain PB integrity are required for their composition, motility, and for transport of their constituents (most notably their mRNAs; Fig. 6, A–C; Table 2; and Video 7). The second type constitutes high-intensity granules that are immotile and preferentially localize to the shmoo tip; these granules disappear rapidly in response to CHX and translation of Mfa2p-mCh occurs in their vicinity. We observed that MFA2 mRNA were transported, localized, and accumulated in shmoo tips of αF-treated cells and were therein locally translated. Unrelated PGK1 mRNA distributed similarly all over cells. Its translation probably occurs equally well all over the cells without specific localization preferences.
We propose that the low-intensity granules are used to transport MFA2 mRNP toward the shmoo tip, based on the following observations: (a) Most fluorescence emanating from MFA2 mRNA is found in the form of granules. (b) The low-intensity granules are the only motile granules; in addition to the local movement, we have observed also a direct type of movement of the granules from cell body to the shmoo tip (e.g., Fig. 3, A–C; and Videos 2 and 3). Consistently, these low intensity granules are localized preferentially in the shmoo-containing portion of the cells; when cells start to acclimate to the unproductive αF treatment (>2 h after αF addition), this preferential accumulation diminishes (Fig. 6 C, WT). (c) The low-intensity granules respond slowly to CHX treatment, suggesting that they do not release their mRNAs to polysomes. (d) The high intensity granules in the shmoo tip disappear rapidly in response to CHX treatment, suggesting that their mRNP constituent rapidly moves to polysomes. Indeed, translation of MFA2-mCh-U1A mRNA occurs in the vicinity of these granules. Therefore, to maintain a steady-state level, a constant flow of mRNPs needs to replenish the otherwise diminishing high-intensity granules. The most likely candidates to contribute these mRNPs are the low-intensity granules. Note that the fastest granules (1.8 µm/s) exhibit the lowest fluorescent intensity (Table 1), implying that they contain one MFA2 mRNA.
Based on our results, we propose the following scenario, illustrated schematically in Fig. 9: In response to αF, MFA2 mRNA, together with Pab1p and PB components, associates with low-intensity and highly motile granules. MFA2 mRNA in these granules is translationally silent, as evident by the low sensitivity of the granules to CHX and the absence of nascent Mfa2p-mCh in the vicinity of these granules. The motile granules are transported to the shmoo tip where they fuse to form a high and relatively immotile granule. This mobility requires some key PBs components. Indeed, deleting the key components Dhh1p and Pat1p, or Edc3p- and the C-terminal domain of Lsm4p diminished movement of the low-intensity granules, resulting in their accumulation in the shmoo-distal portion of the cell, near the nucleus (Fig. 6 C). This led to the disappearance of the high-intensity granules.
Figure 9.
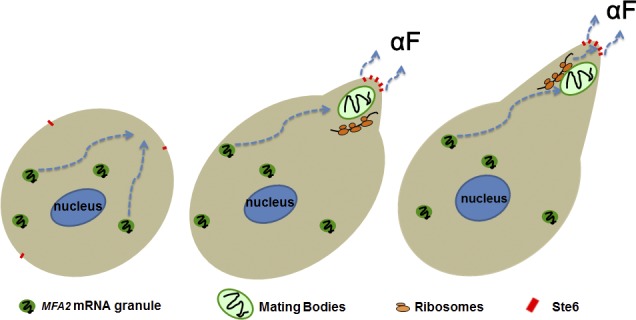
A model of local translation of MFA2 mRNA in the shmoo tip of an αF-treated cell. Shortly after αF treatment, low-intensity MFA2-containing granules begin to be transported to a region in the cell that is destined to form a shmoo (left cell). MFA2 mRNA in these granules might be partially translationally repressed (and its granules are relatively resistant to CHX—see the “low-intensity granules” in Fig. 7 B). Later, the shmoo begins to form, and several low-intensity granules are merged to form a high-intensity granule “mating body.” At the same time, an aF transporter, Ste6p, is transported to the shmoo tip (middle cell; see Discussion). MFA2 mRNA is released for translation in the vicinity of the mating body, and the peptide is processed and secreted by Ste6p in the shmoo tip (middle and right cells). The shmoo continues to grow until it is fused with the shmoo of the mating partner (not depicted). In our experiments, all cells were MATa, and no mating occurred. Therefore, at later time points (>2 h), the transport of low-intensity granules subsided, and the mating body gradually disassembled, in preparation for later cell division (not depicted).
It is quite possible that the low-intensity granules use some motor machinery, probably using actin cables, consistent with the observation of directed movement along most of the cell body toward the shmoo (Fig. 3 C and Videos 2 and 3). In contrast, the high-intensity granules that are located in the shmoo tip are immotile. It is currently unclear why they are stationary. The simplest model proposes that their large size impedes their motility (e.g., if the cytoskeleton average pore size is smaller than the granule size). It is also plausible that the high-intensity granules lose grip of the motor machinery as a result of specific modifications, as is the case for ASH1 mRNA (Beach et al., 1999) or that they are specifically anchored by a tethering molecule. Unlike the low-intensity motile granules, the high-intensity granules are capable of releasing MFA2 mRNAs to the translation apparatus, resulting in their local translation in the shmoo tip. Time-lapse analysis revealed that a high-intensity MFA2 mRNA-containing PBs can be found in the vicinity of the site where asymmetric growth begins; it later remains confined to the tip of the resulting shmoo as it grows (Fig. 2 C and Video 1). It is tempting to speculate that these unique granules and their location play a role in shmoo growth, in addition to their role in synthesizing the pheromone there. We speculate that these granules contain mRNAs whose products are required for growth of the cell membrane and the cell wall. Local synthesis of these proteins may result in local, and thus asymmetric, cell growth. Consistently, cells defective in PB assembly (i.e., dhh1Δ pat1Δ or edc3Δ lsm4Δc) are defective also in asymmetric growth, as only ∼25% of them exhibit a (rather small) shmoo. Their sterile phenotype is also consistent with this model.
Mutant cells that are incapable of producing aF are sterile. Interestingly, addition of aF to the medium is not sufficient to rescue the sterility of these mutants (Michaelis and Herskowitz, 1988), suggesting that the context and location of the secreted pheromone is critical for mating (or that the mature aF is not the only relevant signal). A plausible scenario involves a dialog between the shmoos of the two mating partners that involves local pheromone secretion. Proper dialog directs their growth toward each other in a manner that eventually permits fusion. Jackson and Hartwell (1990a,b) have elegantly shown that, when the mating process is initiated, the two mating types communicate with each other by secreting the pheromones. The ability of MATa cells to respond to the αF by producing their own aF is important for their ability to mate. Remarkably, MATa cells can choose among several potential nearby partners the one that secretes the highest level of αF (Jackson and Hartwell, 1990b). During the mating process, the shmoo tips grow toward each other. Therefore, once a mating partner is selected, the most effective way to secrete the pheromone and “convince” the selected cell to accept the mating offer would be in the tip of the shmoo. In this way, the local concentration that the partner senses would be the highest. Why have cells evolved a mechanism to prefer a partner capable of secreting the highest level of pheromone? We propose that the capacity to produce effective levels of aF in the shmoo tip in response to αF is correlated with the cell quality. How can a simple peptide signal the genotypic and phenotypic quality of the mating partner? SGs and PBs are regulated by a surprisingly large variety of genes (Ohn et al., 2008; Mitchell et al., 2013), integrating many aspects of cellular metabolism with mRNA translation, decay (Ohn et al., 2008), and transport (this work). Therefore, to produce high levels of effective aF in the vicinity of the partner, the cell has to efficiently assemble MFA2 mRNA in PBs, using this large variety of gene products, and transport them to the shmoo tip, where the mRNA is released and translated. After translation, at least five enzymes, including proteases a carboxymethylase and a farnesylase, process the peptide (Michaelis and Barrowman, 2012). Finally, to initiate the process, the cells have to sense the pheromone by a complex process, which depends on the proper execution of signaling pathways (Jones and Bennett, 2011). Evidently, then, the cell needs to efficiently execute several processes involving a large number of genes, while executing and integrating many aspects of cellular metabolism. Thus, the ability to produce high levels of aF, specifically in the shmoo tip, is correlated with both genotypic and phenotypic quality. Interestingly, transport of the αF to the shmoo tip is mediated by Golgi and lipid vesicles, a process that uses many genes not involved in the localized secretion of aF (Jones and Bennett, 2011). Thus, each of the two mating partners tests the quality of a distinct set of large number of genes, so that the diploid cell is guaranteed to have at least one WT set of homologues. We propose that, during evolution, this intricate process of pheromone production has been selected as it delivers information about the quality of the potential mate. Remarkably, then, it seems that a tiny peptide of 13 amino acids can reliably provide information about the genotype and phenotype of a mating partner.
Materials and methods
Yeast strains and growth conditions
The S. cerevisiae strains and plasmids used in this study are listed in Tables 4 and 5. Cells were grown at 25–30°C in synthetic medium under appropriate selection to the early exponential phase. To express mCh fluorescent protein (MS2-mCh) from the inducible MET25 promoter, the cells were switched to medium lacking methionine for 1.5 h at 26°C before αF treatment and further analyses.
Table 4. S. cerevisiae strains used in the study.
| Strains | Genotype | Source |
|---|---|---|
| yMC375 | MATa, ura3, his3, leu2 | R. Parkera; Coller and Parker, 2005 |
| yMC376 | MATa, ura3, his3, leu2, dhh1::NEO, pat1::NEO | R. Parkera; Coller and Parker, 2005 |
| yMC524 | MATa leu2-3.112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG lsm4ΔC::NEO edc3::NEO | R. Parkera; Decker et al., 2007 |
| yMC134 | MATα, can2 (arg-), ura4, gal/2 | S. Michaelisb; Michaelis and Herskowitz, 1988 |
| yMC429 | MATa; his3Δ1 leu2Δ0 met15Δ0 ura3; Δedc3 | Euroscarf |
| yMC430 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| ySA196 | MATa leu2-3.112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG lsm4ΔC::NEO edc3::NEO [pMC B470] | This work |
| ySA20 | MATa, ura3, his3, leu2; [pMC314] [pMC313] | This work |
| ySA3 | MATa, ura3, his3, leu2; [pMC314] [pMC409] | This work |
| ySA21 | MATa, ura3, his3, leu2; [pMC314] [pMC313] [pMC413] | This work |
| ySA22 | MATa, ura3, his3, leu2; [pMC314] [pMC313] [pMC414] | This work |
| ySA23 | MATa, ura3, his3, leu2; [pMC314] [pMC313] [pMC389] | This work |
| ySA24 | MATa, ura3, his3, leu2; [pMC522] [pMC475] | This work |
| ySA25 | MATa, ura3, his3, leu2, dhh1::NEO, pat1::NEO ; [pMC314] [pMC313] | This work |
| ySA28 | MATa leu2-3.112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG lsm4 C:: NEO edc3::NEO [pMC314] [pMC313] | This work |
| ySA26 | MATa, ura3, his3, leu2; [pMC314] [pMC409] [pMC414] | This work |
| ySA33 | MATa, ura3, his3, leu2; [pMC314] [pMC409] [pMC389] | This work |
| ySA147 | MATa, ura3, his3, leu2, [pMC413] [pSA01] | This work |
Howard Hughes Medical Institute, University of Colorado, Boulder, CO.
Johns Hopkins University School of Medicine, Baltimore, MD.
Table 5. Plasmids used in the study.
| Plasmidsa | Genotype | Yeast marker | Source |
|---|---|---|---|
| pMC314b | GPDp-U1A-GFP | LEU2 | P. Silverc; Brodsky and Silver, 2002 |
| pMC313 | GPDp-MFA2-U1Ax16-3′UTR of MFA2 | URA3 | This work |
| pSA01 | GPDp-MFA2-mCHERRY- U1Ax16-3′UTR of MFA2 | URA3 | This work |
| pMC409 | PGK1p-PGK1-U1Ax16-3′UTR of PGK1 | URA3 | R. Parker; Sheth and Parker, 2003 |
| pMC414 | DCP2p-DCP2-RFP | HIS3 | This work |
| pMC413 | LSM1p-LSM1-RFP | HIS3 | This work |
| pMC389 | PAB1p-PAB1-RFP | HIS3 | This work |
| pMCB470 | EDC3p-EDC3-mCh | TRP1 | R. Parker; Buchan et al., 2008 |
| pMC522 | MET25p-MS2-mCh | HIS3 | D. Ravehd; Lavut and Raveh, 2012 |
| pMC475b | GPDp-MFA2-MS2 × 2 | URA3 | R. Parker; Sheth and Parker, 2003 |
All plasmids, unless otherwise indicated, contain CEN and therefore are present in one or two copies per cell.
Multicopy 2μ plasmid.
Harvard Medical School, Boston, MA.
Ben-Gurion University of Negev, Beer-Sheva, Israel.
Plasmids
Centromeric plasmids carrying either an MFA2, whose 3′-UTR contains 16 repeats of UA1-binding site (pRP1193), or PGK1, whose 3′-UTR also contains 16 repeats of UA1-binding site (pPS2037), pU1A-GFP (pRP1187), or DCP2-RFP (pRP1152) or LSM1-RFP (pRP1084) were gifts from R. Parker (Table 5). pMFA2-mCh-U1A was constructed by inserting a PCR fragment carrying mCh flanked by MFA2 3′ UTR into Bam H1 digest of pMC313 using homologous recombination in vivo: forward primer, 5′-CTATATAATCAAGGGCCTCTTCTGGGATCCCGCCTGTGTTATCGCTatggtgagcaagggcgag-3′, where capital letters are MFA2 ORF excluding stop and small letters are mCh (ATG is the first codon); reverse primer, 5′-GGGGGAATTCCtGATCAAATCAATATttacttgtacagctcgtcc-3′, where capital letters are MFA2 3′ UTR and small letters are 3′ end of mCh ORF (stop is underlined). The sequence was verified by Sanger sequencing analysis.
αF treatment
Stock solution of αF (Sigma-Aldrich) was 1 mg/ml in methanol. Cells were grown in synthetic complete medium to early exponential phase and treated with 3 nM αF for 1–6 h. In some cases, the culture was collected by centrifugation and resuspended in sixfold smaller volume just before treatment.
Preparation of cells for confocal microscopy
The cells were grown overnight until they reached an early exponential phase and treated with αF. Only fresh cell samples of low concentration (a few cells per field) were inspected under the microscope (<15 min) to prevent adverse effects (e.g., starvation, hypoxia) of incubating them in between the slides. For the time-lapse analysis, the cells were loaded on special slides with a thin layer of 1% agar containing synthetic medium and 3 nM αF.
Image processing and analysis
The images and long time-lapse videos were taken at room temperature (∼23°C) by confocal microscope (LSM 510 Meta; Carl Zeiss). The following wavelengths were used: for GFP, excitation at 480 nm and emission at 530 nm; for mCh or monomeric RFP, excitation at 545 nm and emission at 560–580 nm. For the coexpression experiments, sequential screening was used to avoid overlapping with the LSM image. Examiner software was used for observation. Images were presented as a z series compilation of 10 photographs in a stack by using the LSM image browser program, except for the colocalization analysis, where images result from a single z section. The 3D Reconstruction feature of the MetaMorph software package (Universal Imaging Corporation) was used to generate a composite image of several time points (Fig. 3 B). The function controls were set to create a single image with 0° of rotation of a projection of the brightest points from a sequence of images. This produced a time-lapse composite image representing the distribution of the MFA2 mRNA/DCP2-RFP over a period of time.
The short time-lapse videos and the analyses of the mRNA granules intensity, tracking, and velocity were performed at room temperature (∼23°C), using the inverted motorized fluorescent microscope (using a 63×/1.40 NA objective; Axiovert 200 M; Carl Zeiss) and the Colibri led illumination light source and high speed camera (Zeiss HS; Carl Zeiss). The videos were made using AxioVision Rel. 4.8 imaging program (Carl Zeiss). The properties of the mRNA granules, tracking, and volume were processed using the Imaris software (Bitplane AG). The intensity of granules was also analyzed using the ImageJ program (National Institutes of Health) and compared with the analysis done by the Imaris program. Data analysis was performed in Excel (Microsoft). The final images were prepared with Photoshop CS (Adobe).
Measurement of mRNA and protein localization in vivo
To obtain quantitative data on the localization of mRNA, cells with visible shmoos were observed and counted manually for the localization of U1A-GFP–containing mRNA granules. We defined shmoo-localized mRNA as one that forms green particles that associate with the tips of small and emerging shmoos in a cell sample in which >80% of cells produced shmoo projections after 2 h of αF treatment. Usually, 250–300 cells were observed and counted in each experiment. Images are representative of three independent experiments.
Tracking analysis of MFA2 mRNA or PGK1 mRNA granules
The velocity of mRNA granules was analyzed using the Imaris program version 7.1 on 2D videos that were acquired using a microscope (Fast Acquisition AxioVision; Carl Zeiss) at room temperature (∼23°C). Velocity was calculated according to the intensity and position of the granules in the cells (low-intensity granules in mother and shmoo regions, and the high-intensity granules localized in shmoo). The velocity analysis for the low-intensity granules was restricted to granules with track length longer than 0.4 µm and for high-intensity granules with track length longer than 0.1 µm. The duration, length, and types of the tracking were analyzed on the cells according to intensity and position of the mRNA granules in the cell using the same program.
Statistical analysis
Statistical analysis was performed using Excel (Microsoft) and R (http://www.R-project.org). To access any significant enrichment of granules in shmoo tip in WT versus mutant strains, the one-sided Fisher’s exact test was performed on 2-by-2 contingency tables, and p-values were calculated for each of the 18 comparisons (WT vs. two mutants and mutant vs. mutant; for each one of three time points and for each type of granules). To adjust for multiple testing, the Bonferroni correction was used, and adjusted p-values <0.05 were considered significant.
Cell growth and RNA extraction
The yeast WT (yMC375) and pat1Δ dhh1Δ (yMC376) strains were grown in the rich medium (YPD), at 25°C, into 15-ml tubes, ON. The next day, the cells in the mid–log stage, the comparable amounts of cells (5–10 OD 600 equivalents) were collected by centrifugation at 3,000 g for 5 min at 4°C. Cell aliquots were cooled to ∼5°C rapidly by placing the tubes in liquid nitrogen with vigorous shaking for 10–15 s. Cells were then rapidly harvested, washed with water, resuspended in 0.4–1 ml of ice-cold extraction buffer (0.2 M Tris-HC1, pH 7.5, 0.5 M NaC1, 10 mM EDTA, and 0.1% diethylpyrocarbonate; Finley et al., 1989), in a screw-capped Eppendorf tube, and immediately frozen in liquid nitrogen. Later on, 400 µl of glass beads and 400 µl of phenol-chloroform (1:1 vol/vol) were added, whereas the cell suspensions were still frozen and kept on ice. All tubes were vortexed together, by using a multiple sample head (VWR) at the highest speed for 2.5 rains at room temperature (it takes from 10 to 20 s of vortexing for the frozen suspension to thaw). Samples were centrifuged at 4°C for 8 min, and the aqueous phases were extracted with phenol-chloroform (1:1 vol/vol) and ethanol precipitated. RNA pellets were dissolved in 40 µl of H2O, and RNA concentrations were measured by OD 260.
Northern blot analysis
Polyacrylamide Northern analysis was performed as described previously (Lotan et al., 2005). In brief, RNA (5 µg/lane) was run in a 20 × 20 × 0.15–cm gel of 6% polyacrylamide and 7 M urea in TBE (Tris borate and EDTA) buffer. The RNA was then electrotransferred onto GeneScreen plus membrane (PerkinElmer) in 0.5× TBE at 30 V for 12–15 h at 4°C and hybridized as described previously (Lotan et al., 2005).
Online supplemental material
Fig. S1 shows that both WT (yMC375) and mutant cells pat1Δ dhh1Δ (yMC376) sensed and responded to the pheromone, as demonstrated by the capacity of αF to induce transcription of the pheromone responsive FIG1 gene. Fig. S2 shows the proliferation rate of the WT and the mutant strains (pat1Δ dhh1Δ, edc3Δ lsm4ΔC, edc3Δ, and lsm4ΔC). Fig. S3 demonstrated that CHX treatment prevents local translation of MFA2 mRNA in the vicinity of the high-intensity granule in the shmoo using WT strain cotransformed with MFA2p-mCh-UA1 and UA1p-GFP. Video 1 shows that the low-intensity MFA2 mRNA-containing granules are distributed all over the cell, whereas a high-intensity granule localizes to the shmoo before, and during its growth. Video 2 shows the distribution and the motility features of MFA2 mRNA–containing granules. Video 3 shows the directed movement of the low intensity granule from the cell body to the shmoo region by tracking analysis. Video 4 demonstrated the distribution and the motility features of PGK1 mRNA-containing granules in αF-treated cells. Video 5 shows the movement of PGK1 mRNA-containing granules by tracking analysis. Video 6 shows the colocalization of MFA2 mRNA and Dcp2p-RFP during movement in αF-treated cells. Video 7 demonstrated the reduction in the intensity, number, and motility features of MFA2 mRNA–containing granules in α-treated pat1Δ dhh1Δ mutant cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408045/DC1.
Supplementary Material
Acknowledgments
We thank Sussan Michaelis and Roy Parker for the generous gift of plasmids and strains, Polina Geva and Enav Halperin for help with experiments, and Maya Schuldiner for her invaluable comments on the manuscript.
This work was supported partially by the Israel Science Foundation 716/11 (granted to M. Choder) and Ariel University Internal grant (S. Aronov).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CHX
- cycloheximide
- IOD
- integrated optical density
- mCh
- mCherry
- PBs
- processing bodies
- SG
- stress granule
- WT
- wild type
References
- Anderson P., and Kedersha N.. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Aronov S., Gelin-Licht R., Zipor G., Haim L., Safran E., and Gerst J.E.. 2007. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:3441–3455. 10.1128/MCB.01643-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L. 2005. A walk-through of the yeast mating pheromone response pathway. Peptides. 26:339–350. 10.1016/j.peptides.2004.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D.L., Salmon E.D., and Bloom K.. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9:569–578. 10.1016/S0960-9822(99)80260-7 [DOI] [PubMed] [Google Scholar]
- Brengues M., and Parker R.. 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:2592–2602. 10.1091/mbc.E06-12-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., and Parker R.. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 310:486–489. 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky A.S., and Silver P.A.. 2000. Pre-mRNA processing factors are required for nuclear export. RNA. 6:1737–1749. 10.1017/S1355838200001059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky A.S., and Silver P.A.. 2002. A microbead-based system for identifying and characterizing RNA-protein interactions by flow cytometry. Mol. Cell. Proteomics. 1:922–929. 10.1074/mcp.T200010-MCP200 [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Muhlrad D., and Parker R.. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183:441–455. 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J. 1999. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15:365–391. 10.1146/annurev.cellbio.15.1.365 [DOI] [PubMed] [Google Scholar]
- Coller J., and Parker R.. 2005. General translational repression by activators of mRNA decapping. Cell. 122:875–886. 10.1016/j.cell.2005.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Powrie E., Gu W., Singer R.H., and Zenklusen D.. 2003. RNA asymmetric distribution and daughter/mother differentiation in yeast. Curr. Opin. Microbiol. 6:614–620. 10.1016/j.mib.2003.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., and Parker R.. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4:a012286 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., and Parker R.. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437–449. 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S., Lin L., Malczynski M., and Snyder M.. 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140:461–483. 10.1083/jcb.140.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Bartel B., and Varshavsky A.. 1989. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 338:394–401. 10.1038/338394a0 [DOI] [PubMed] [Google Scholar]
- Frey S., Pool M., and Seedorf M.. 2001. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 276:15905–15912. 10.1074/jbc.M009430200 [DOI] [PubMed] [Google Scholar]
- Gelin-Licht R., Paliwal S., Conlon P., Levchenko A., and Gerst J.E.. 2012. Scp160-dependent mRNA trafficking mediates pheromone gradient sensing and chemotropism in yeast. Cell Reports. 1:483–494. 10.1016/j.celrep.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J.B., and Herskowitz I.. 1976. Evidence for a new diffusible element of mating pheromones in yeast. Nature. 260:246–248. 10.1038/260246a0 [DOI] [PubMed] [Google Scholar]
- Jackson C.L., and Hartwell L.H.. 1990a Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 63:1039–1051. 10.1016/0092-8674(90)90507-B [DOI] [PubMed] [Google Scholar]
- Jackson C.L., and Hartwell L.H.. 1990b Courtship in Saccharomyces cerevisiae: an early cell-cell interaction during mating. Mol. Cell. Biol. 10:2202–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.K. Jr, and Bennett R.J.. 2011. Fungal mating pheromones: choreographing the dating game. Fungal Genet. Biol. 48:668–676. 10.1016/j.fgb.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., and Lasko P.. 2012. Translational control in cellular and developmental processes. Nat. Rev. Genet. 13:383–394. 10.1038/nrg3184 [DOI] [PubMed] [Google Scholar]
- Lavut A., and Raveh D.. 2012. Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet. 8:e1002527 10.1371/journal.pgen.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R., Bar-On V.G., Harel-Sharvit L., Duek L., Melamed D., and Choder M.. 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 19:3004–3016. 10.1101/gad.353205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S., and Barrowman J.. 2012. Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol. Mol. Biol. Rev. 76:626–651. 10.1128/MMBR.00010-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S., and Herskowitz I.. 1988. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell. Biol. 8:1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.F., Jain S., She M., and Parker R.. 2013. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20:127–133. 10.1038/nsmb.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R., Moradi E.K., Niu W., Hart G.T., Davis M., McGary K.L., Ellington A.D., and Marcotte E.M.. 2009. Systematic definition of protein constituents along the major polarization axis reveals an adaptive reuse of the polarization machinery in pheromone-treated budding yeast. J. Proteome Res. 8:6–19. 10.1021/pr800524g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Dinur K., Nussbaum-Shochat A., Ben-Yehuda S., and Amster-Choder O.. 2011. Translation-independent localization of mRNA in E. coli. Science. 331:1081–1084. 10.1126/science.1195691 [DOI] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P.. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10:1224–1231. 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., and Parker R.. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808. 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., and Parker R.. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382. 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.