Both N- and C-terminal microtubule (MT)-binding domains of CENP-F can follow depolymerizing MT ends while bearing a significant load, and the N-terminal domain prefers binding to curled oligomers of tubulin relative to MT walls by approximately fivefold, suggesting that CENP-F may play a role in the firm bonds that form between kinetochores and the flared plus ends of dynamic MTs.
Abstract
Firm attachments between kinetochores and dynamic spindle microtubules (MTs) are important for accurate chromosome segregation. Centromere protein F (CENP-F) has been shown to include two MT-binding domains, so it may participate in this key mitotic process. Here, we show that the N-terminal MT-binding domain of CENP-F prefers curled oligomers of tubulin relative to MT walls by approximately fivefold, suggesting that it may contribute to the firm bonds between kinetochores and the flared plus ends of dynamic MTs. A polypeptide from CENP-F’s C terminus also bound MTs, and either protein fragment diffused on a stable MT wall. They also followed the ends of dynamic MTs as they shortened. When either fragment was coupled to a microbead, the force it could transduce from a shortening MT averaged 3–5 pN but could exceed 10 pN, identifying CENP-F as a highly effective coupler to shortening MTs.
Introduction
Kinetochores are the principal sites of interaction between mitotic chromosomes and spindle microtubules (MTs); they enable accurate chromosome segregation. Human kinetochores are built from ∼100 different proteins. Some of these bind DNA, others bind MTs, and some bridge these domains and/or provide regulatory and signaling functions to optimize the accuracy of mitosis (DeLuca and Musacchio, 2012; Varma et al., 2013; Cheeseman, 2014). The kinetochore–MT interface has been examined by electron tomography in five organisms (VandenBeldt et al., 2006; Dong et al., 2007; McIntosh et al., 2008, 2013), and two intriguing features have emerged: (1) the ends of most kinetochore-associated MTs (KMTs) are flared as a result of outward bending by the tubulin strands that comprise the MT wall; and (2) these curling “protofilaments” appear to connect to nearby DNA through slender fibrils ∼3 nm in diameter and 45–65 nm long. Such images imply the existence of filamentous kinetochore proteins that bind preferentially to flared assemblies of tubulin, either because they interact with a domain of tubulin that is hidden in a MT or because they bind better to bent tubulin than to the straight protein in the MT wall. Such connections are likely to be important for mitosis because most chromosomes are pulled poleward in anaphase by their strong association with the pole-distal end of one or more KMTs.
Recent studies on human kinetochore proteins have identified the Ska1 complex (Hanisch et al., 2006; Gaitanos et al., 2009; Welburn et al., 2009) and shown that it binds to curling tubulin; the interaction is on a part of tubulin that faces the MT exterior, but Ska1 prefers that surface when tubulin is bent (Schmidt et al., 2012). This complex also associates with a ubiquitous kinetochore protein, the Ndc80 complex, which binds weakly to the MT wall at one of its ends and to proteins of the inner kinetochore with the other (Sundin et al., 2011). Thus, the Ska1–Ndc80 supercomplex may be one component of the fibrils seen in the electron microscope. The filamentous Centromere protein E (CENP-E) might be another because there is a kinetochore binding site near the C terminus of this long motor protein and a kinesin domain at its N terminus (Schaar et al., 1997; Wood et al., 1997). However, the critical components of the kinetochore–MT interface have not been definitively identified.
To seek additional spindle components that could bind flared MT ends we have screened mitotic proteins for those that bind better to curving tubulin oligomers than to a MT lattice. CENP-F shows this behavior. Others have found MT-binding domains at either end of this protein (Feng et al., 2006; Musinipally et al., 2013), but our data demonstrate that the N-terminal MT-binding region associates more strongly with tubulin curls than with MTs. Sites from either end of CENP-F can couple cargo to a shortening MT and transduce considerable force from the energy released as tubulin depolymerizes. These findings increase interest in CENP-F as a kinetochore protein that may play important roles in the attachment and segregation of chromosomes.
Results
Many mitotic proteins bind tubulin polymers; some prefer curls to MTs
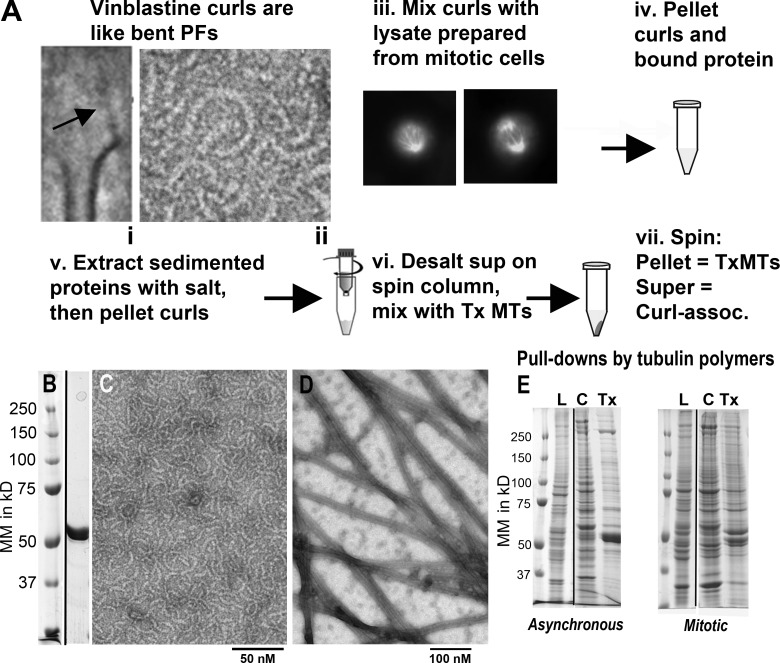
Some drugs promote the assembly of tubulin into curved oligomers: dolastatin-10 induces rings (Moores and Milligan, 2008) and vinblastine promotes curls (Jordan et al., 1986). Both of these oligomers resemble the flaring protofilaments at the ends of kinetochore MTs: their curvatures are similar and their component tubulin dimers are oriented so the surface that would normally face the MT interior is turned outward (Fig. 1 A). We used vinblastine to prepare curls from purified brain tubulin (Fig. 1, A–C), making ligands that are convenient to sediment in a preparative ultracentrifuge, pulling down from cell extracts any proteins that bind them. Given tubulin’s negative charge, such pull-downs are complex. To reduce this complexity we extracted the curl-associating proteins with 0.5 M NaCl in the presence of vinblastine, repelleted the curls, and collected curl-binding proteins from the supernatant. These were desalted in a spin column and mixed with true Taxol-stabilized MTs (Tx-MTs; Fig. 1, A and D). Another round of centrifugation brought down any proteins that associated with MT walls, whereas the proteins that preferred curls stayed in the supernatant. Gel electrophoresis revealed the complexity of these fractions (Fig. 1 E), but mass spectrometry allowed us to identify as many as 2,500 proteins in each. Using spectral counting of the tryptic peptides identifiable by amino acid sequence (Pham et al., 2010), we measured the relative abundance of every protein in each fraction prepared from either log phase cultures or cells synchronized at mitosis by Nocodazole inhibition of spindle formation, followed by mitotic shake-off.
Figure 1.
Tubulin polymers and their pull-downs from cultured cells. (A) Schematic of the protocol for finding curl-binding proteins. (i) Mean of 47 KMTs from metaphase PtK cells, showing a fibril (arrow) connecting chromatin to a bending protofilament. (ii) A tubulin curl made with vinblastine. (iii) Mitotic U2OS cells used to make cell extracts. (iv) Symbolic representation of sedimenting proteins from extracts with tubulin curls. (vi) Symbolic representation of desalting proteins in a spin column. (vii) Cellular proteins are separated into those that bind Tx-MTs (pellet) and those that prefer curls (supernatant). (B) SDS-PAGE of the tubulin used to make pull-down ligands, as eluted from a phosphocellulose column, and molecular mass markers. (C) electron micrograph of negatively stained, vinblastine-induced tubulin curls; (D) a comparable image of Tx-MTs. (E) SDS-PAGE gels of molecular mass markers, cell lysates (L) from log-phase or synchronized cells, followed by the proteins pulled down from the extracts by curls (C) or Tx-MTs (Tx), as described in A.
The tubulin-binding proteins identified and their relative abundances are given in Table S1. Many proteins bound these anionic polymers, but we have focused on cytoskeletal proteins and on known or likely kinetochore components. Our method favors the identification of big proteins, because they yield more spectral counts, but the relative amounts of the same protein in different fractions is unaffected by this bias. In some experiments 2D reverse-phase liquid chromatography was used to separate the tryptic peptides before they entered the tandem mass spectrometer. These experiments yielded far more spectral counts for each protein, so less abundant proteins became detectable. For many proteins, however, the ratios of spectral counts for curl versus Tx-MT associations were about the same with these two techniques. Table 1 presents the spectral counts for some spindle-associated proteins and Table S2 shows nonspindle, cytoskeletal proteins whose relative spectral counts suggested interesting features of their MT binding.
Table 1. Normalized spectral counts from 1D and 2D mass spectometry runs.
| Names of spindle proteins | Curls/Tx-MTs | Curls/Tx-MTs | Ratios: 1D - 2D | ||
|---|---|---|---|---|---|
| Gene name (and description) | 1D separation | 2D separation | |||
| MAP1B (MT-associated protein) | 20/3.9 | 53/20 | 5.1 - 2.6 | ||
| CKAP5 (XMAP215 MT-associated protein) | 7.0/3.9 | 39/53 | 1.8 - 0.74 | ||
| EML4 (like echinoderm MAP4) | 2.0/0 | 5.0/0 | X - X | ||
| MAP 4 (MT-associated protein) | 3.0/11.8 | 41/33 | 0.25 - 1.24 | ||
| CLIP1 (CAP-GLY domain linker protein) | 4.0/2.0 | 16.6/8.4 | 2.0 - 2.0 | ||
| CLASP1 (CLIP-associating protein 1) | 1.0/7.8 | 5.0/12.1 | 0.13 - 0.41 | ||
| KIF5B (Kinesin-1 heavy chain) | 12/7.8 | 55/29 | 1.5 - 1.9 | ||
| KIF11 (Kinesin-like protein) | 4.0/3.9 | 22/25.6 | 1.0 - 0.86 | ||
| KIF15 (Kinesin-like protein) | 2.0/0.0 | 10/1.5 | X - 6.7 | ||
| KIF20A (Kinesin-like protein) | 1.0/7.8 | 7.0/15.1 | 0.13 - 0.46 | ||
| KLC1 (Kinesin light chain 1) | 4.6/4.7 | 9.2/4.5 | 0.98 - 2.0 | ||
| KLC2 (Kinesin light chain 2) | 4.2/9.0 | 8.1/12.1 | 0.47 - 0.67 | ||
| DYNC1H1 (Dynein 1 heavy chain 1) | 0/0 | 2.0/44 | X - 0.04 | ||
| DYNC1I2 (Dynein 1 intermediate chain 2) | 1.0/3.9 | 1.0/3.0 | 0.26 - 0.33 | ||
| DCTN1 (Dynactin subunit) | 0/0 | 0/6.0 | X - 0 | ||
| ZW10 (Dynein associated at kinetochore) | 0/0 | 0/10.5 | X - 0 | ||
| STMN1 (Stathmin) | 0/0 | 14/3.0 | X - 4.7 | ||
| TPX2 (Targets a kinetochore kinesin) | 0/2.0 | 4.1/2 | 0 - 2.0 | ||
| RCC1 (RAN nucleotide exchange factor) | 1.0/0 | 11/3 | X - 3.7 | ||
| NuMA (Nuclear-mitotic apparatus protein) | 5.0/3.9 | 16/13.6 | 1.3 - 1.8 | ||
| CENP-F (Centromere protein F) | 9.0/5.9 | 42/34.7 | 1.53 - 1.2 | ||
| CENP-E (Centromere protein E, a kinesin) | 0/0 | 0/12.1 | X - 0 | ||
| NDC80 (Conserved kinetochore MT bind) | 0/0 | 1/6.0 | X - 0.17 | ||
| SPC24 (Part of the NDC80 complex) | 0/0 | 1/3.0 | X - 0.33 | ||
| SPC25 (Part of the NDC80 complex) | 0/0 | 1.0/3.0 | X - 0.33 | ||
| NUF2 (Part of the NDC80 complex) | 0/0 | 3.0/3.0 | X - 1.0 | ||
| SKA1 (Kinetochore, interacts with NDC80) | 1.0/0 | 2.0/1.5 | X - 1.3 | ||
| SKA3 (Kinetochore, interacts with NDC80) | 0/0 | 1.0/1.5 | X - 0.67 | ||
| BUB3 (Spindle assembly checkpoint protein) | 5.0/2.0 | 4.0/3.0 | 2.5 - 1.3 | ||
| BUB1 (Spindle assembly CP protein) | 0/0 | 0/1.5 | X - 0 | ||
| MAD2L1 (Spindle assembly CP protein) | 1.0/0 | 1.0/1.5 | X - 0.67 | ||
| CLTC (Clathrin heavy chain 1) | 11/2.0 | 50/19.6 | 5.5 - 2.6 |
Gene names from NCBI, with descriptions in parentheses. For more information about each protein, see the same gene name in Tables S1 or S2. Spectral counts from 1D and 2D separations were normalized by the total number of counts for all proteins in that run and rounded to two significant figures, making the 1D and 2D counts more comparable. 2D numbers are generally more accurate because the initial number of counts was larger and farther above background. X implies no value. 0 implies not above noise. Differences in ratios from 1D and 2D data indicate that these numbers are only of semi-quantitative value.
We used immunoblotting to reevaluate the ratios of curl versus MT binding for a few proteins that showed interesting ratios; these gave a qualitative confirmation of the binding ratios seen by mass spectrometry (Fig. S1 A).
The kinetochore protein with the greatest preference for curls was CENP-F
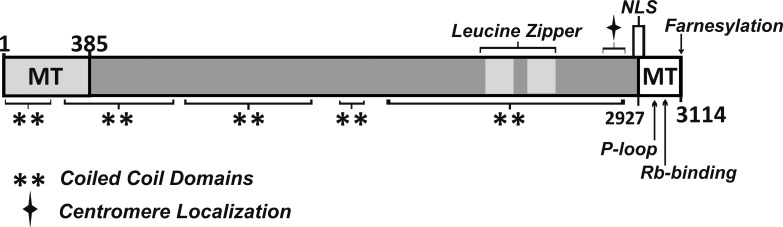
One kinetochore component with a preference for curled versus straight tubulin was CENP-F (Table 1 and Fig. 2). From work by Liao et al. (1995), CENP-F is known to be expressed during interphase and to accumulate in the nucleus. It then concentrates on the nuclear envelope, but in prophase it moves to the kinetochores (Liu et al., 2006) where it resides until anaphase, when it migrates to the midbody and is degraded (Gurden et al., 2010). cDNAs that encode human CENP-F predict a 3,114–amino acid polypeptide with a high fraction of α-helical coiled coil (Feng et al., 2006). The protein has a nuclear localization signal, a kinetochore-binding domain near its C terminus, and MT-binding domains at both its ends, a unique feature for a MT-associated protein that is not a motor. To be active in mitosis it requires farnesylation (Crespo et al., 2001; Hussein and Taylor, 2002). Depletion of CENP-F by RNA interference leads to a moderately high rate of failure in chromosome attachment to the spindle and consequent chromosome loss (Bomont et al., 2005; Holt et al., 2005; Yang et al., 2005; Feng et al., 2006). Indeed, bovine embryos that lack CENP-F do not develop beyond the 8-cell stage (Toralová et al., 2009). Interpretation of these results has, however, been difficult because CENP-F is important for the localization of other kinetochore proteins, e.g., the plus end–directed kinesin CENP-E and the minus end–directed dynein (Chan et al., 1998; Yang et al., 2005; Vergnolle and Taylor, 2007). Thus, the mitotic consequences of CENP-F depletion for chromosome behavior may be an indirect result of its absence.
Figure 2.
Diagram of the primary structure of CENP-F. Motifs identified by others are marked to show their relative positions: MT-binding sites (MT) and bipartite nuclear localization signal (NLS) are indicated, along with other motifs of interest. Numbers are amino acids from the N terminus. Redrawn from previous sources (Zhu et al., 1995; Feng et al., 2006; Evans et al., 2007).
CENP-F has been found in all vertebrates so far examined, and possible homologues have been identified in a nematode (Moore and Roth, 2001; Hajeri et al., 2008) and a deer tick (UniProt B7PYE5 by N-terminal sequence similarity); a convincing orthologue has not been found in fungi. Human CENP-F or its mouse orthologue, Lek1 (Goodwin et al., 1999), may also play roles in cellular processes beside mitosis, e.g., centrosome functions (Moynihan et al., 2009) and aspects of membrane traffic (Pooley et al., 2008). The formation of a normal mouse heart is abrogated by mutation of CENP-F (Dees et al., 2012), but this phenomenon has not yet been characterized in sufficient detail to know the cell biological problems that underlie it. Mutations in human CENP-F have been found in association with mid-gestation lethality and ciliopathic malformations; indirect evidence also implicates this protein in intraflagellar transport (Waters et al., 2015). Thus, there may be nonmitotic functions of CENP-F to consider when interpreting any whole organism phenotypes associated with this protein.
CENP-F’s two MT-binding domains are different
Given these properties and CENP-F’s preference for tubulin curls, we investigated the protein further. Several proteins that bind at or near a MT end, e.g., EB1, do so as a result of their tighter association with GTP-tubulin than with GDP-tubulin (Yajima et al., 2012). This preference allows them to bind to the end of a growing MT, but not one that is shortening. Pull-downs of CENP-F from mitotic human cell extracts by curls made with GDP-tubulin repeated CENP-F’s preference for curls over Tx-MTs (Fig. 3). Thus, CENP-F is not a conventional “TIP” (Tip-Interacting Protein).
Figure 3.
Tubulin curls made with either GTP or GDP will pull down CENP-F better than Tx-MTs. (A) RPE1 cells were synchronized, collected, and extracted as described in Materials and methods, and then mixed with either vinblastine-induced tubulin curls or Tx-MTs and pelleted. A 4× pellet volume of cell extract and equal volumes of each pellet were fractionated on 7% polyacrylamide gels. 1, cell lysate; 2, 3, and 4, pellets of GTP curls, GDP curls, and Tx-MTs, respectively; 5, molecular mass markers. (B) The gels from A blotted to nitrocellulose paper and reacted with a mouse monoclonal antibody to CENP-F (Santa Cruz Biotechnology, Inc.). Numbers above blot lanes indicate the number of microliters loaded. (C) Ratios of curls/Tx-MTs staining, as measured by densitometry. The binding ratios for GTP curls and Tx-MTs shown here are from a single representative experiment out of three repeats.
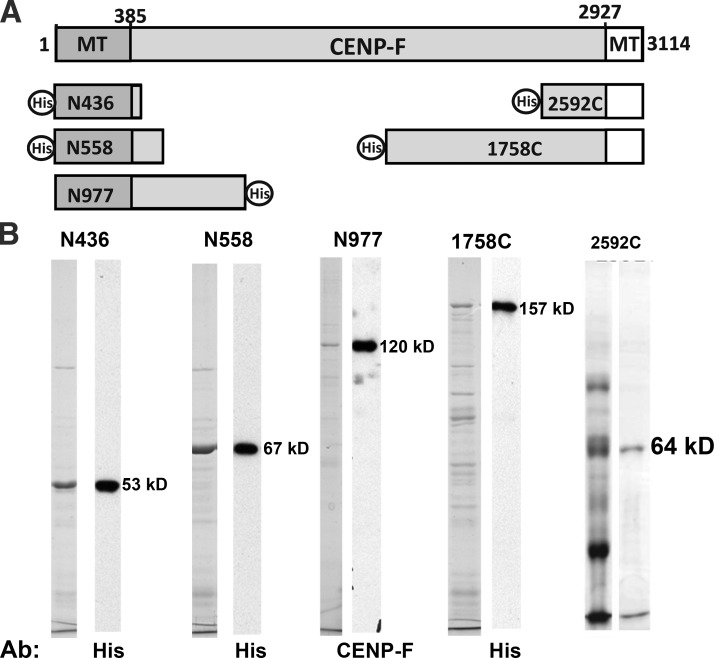
We have cloned and expressed segments of the CENP-F gene derived from cDNAs, using vectors suitable for bacteria or insect cells (Fig. 4). Each DNA segment was sequenced and a few deviations from the corresponding sequence in the National Center for Biotechnology Information (NCBI) database were identified. These have been corrected, as described in Materials and methods. Expression of these fragments with baculovirus was poor, so our work has been focused on proteins we could make in bacteria. Polypeptides from the N terminus of CENP-F bound better to curled tubulin oligomers than to MTs, whereas a fragment from the C terminus preferred Tx-MTs (Fig. 5, A and B). When curls were made with GDP- or GTP-tubulin, N558 was pulled down better by the GTP-induced oligomers (Fig. 5, A and B), suggesting that the GDP preference seen with complete CENP-F in extracts was caused either by properties of the whole protein or by unknown factors in that complex milieu.
Figure 4.
Fragments of CENP-F as expressed from cDNAs. N436, N558, 1758C, and 2592C were expressed in bacteria and N977 was expressed in SF9 cells, as described in Materials and methods. (A) The relationships between these pieces and the entire coding sequence of CENP-F. (B) For N436, N558, N977, and 1748C Coomassie-stained electrophoresis gels of the partially purified fragments and adjacent immunoblots with anti–penta-his (QIAGEN) or anti-CENP-F, confirming that the proteins isolated are the ones expressed. For 2592C, the peak fractions from Ni+2-NTA (left) and from a HiTrap SP column (right) stained with Coomassie.
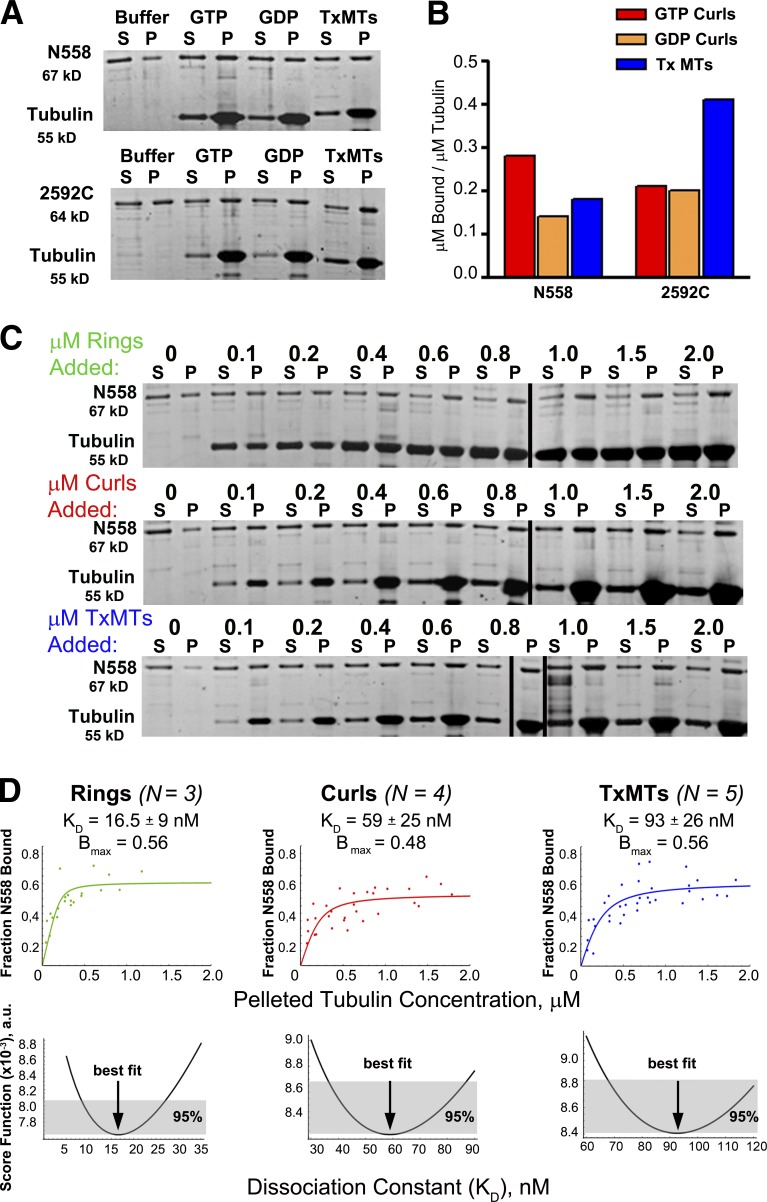
Figure 5.
An N-terminal fragment of CENP-F binds better to curls than MTs, whereas a C-terminal segment does the opposite. (A) 0.2 µM N558 or 0.4 µM 2592C were mixed with buffer alone or with buffer including 0.4 µM tubulin assembled as GTP curls, GDP curls, or Tx-MTs, and then incubated and centrifuged as described in Materials and methods. Gels of supernatant and pellet fractions were stained with SYPRO Ruby (Molecular Probes). (B) Bound protein was measured by densitometry of the pellet fractions. The fraction of protein that pelleted nonspecifically in the buffer-only lane was subtracted from the values measured for each experiment. The amounts shown are from a single representative experiment out of four repeats for N558 and two repeats for 2592C. (C) 0.2 µM N558 was mixed with various concentrations of tubulin added as rings, curls, or Tx-MTs, and then incubated, centrifuged, and analyzed by gel electrophoresis and densitometry, as described in Materials and methods. (D) Protein amounts in each lane of three to five experiments, as in C, were quantified as in Materials and methods and the results were plotted. The data were fit with a quadratic model for protein binding by the method of least squares. Normalized score functions for goodness of each fit are shown in the bottom row. A value of 0 for this function would imply a perfect fit.
To test the possibility that the 6-his tag at the N terminus of these constructs was influencing the binding bias for curls, we made a version of N558 in which the tag was separated from CENP-F sequence by a cleavage site for the Tobacco Etch Virus protease. Removal of the tag reduced the protein’s solubility, but when the sedimentation tests for binding preference were performed in 0.1% Triton X-100, a similar preference for curl binding was seen (Figs. S1, C–E). From this observation and from the curl-binding preference of native CENP-F, which includes both the N- and C-terminal binding sites, we inferred that the preference of the N-terminal site for curls is strong.
An N-terminal fragment of CENP-F prefers bent tubulin oligomers but does not bind soluble tubulin
To quantify the relative strengths of N558 binding to curls versus MTs, we performed sedimentation experiments using 0.2 µM N558 mixed with varying amounts of three assembled forms of tubulin: rings induced by dolastatin-10, curls induced by vinblastine, and Tx-MTs (Fig. 5 C). The amount of N558 that pelleted with each form of assembled tubulin was quantified as described in Materials and methods, and the fraction of N558 bound as a function of tubulin polymer was plotted (Fig. 5 D, top). The data were fit with a quadratic binding equation (Musinipally et al., 2013), and estimations of binding strength were determined from the fits. The bottom row of graphs in Fig. 5 D shows scoring functions that describe the goodness of fit for each plot; the errors in the dissociation constants (KD) are expressed as 95% confidence intervals. Bmax for each curve identifies the maximal fraction of N558 bound, as estimated by the asymptote of the fitting curve.
Dolastatin-induced rings showed the tightest binding for this N-terminal piece of CENP-F; their KD is more than fivefold lower than that measured for Tx-MTs. N558 binding to vinblastine-induced curls was also stronger than to Tx-MTs, but by less than a factor of 2. We attribute this discrepancy to the greater scatter in the vinblastine experiments, which is probably a result of variation in the sizes and curvatures of curls compared with those of Dolastatin-induced rings. The heterogeneity in binding to Tx-MTs may in part be a result of variability in these polymers. Nonetheless, these data demonstrate that the N terminus of CENP-F shows a marked binding preference for bent tubulin relative to the straight tubulin in Tx-MTs. They also suggest that not all the N558 in our preparations is equally active in tubulin binding, hence the low values of the binding asymptotes for all forms of tubulin.
We then asked whether the curl preference of this moiety might be reflected in its binding to soluble tubulin bound to either GTP or GDP. N558 sedimented through a sucrose gradient with an S4,w = 4.7 S, both when alone and when in mixtures with tubulin bound either to GTP (Fig. S1 F) or GDP (not depicted). We used a calibrated gel filtration column to measure the Stokes radius of N558; the value of 6.6 nm, combined with the sedimentation velocity, yielded a solution molecular mass of 133 kD (Fig. S1 G). Because the mass predicted from N558’s primary structure is 66,734 D, this protein behaves as a soluble dimer that does not interact with tubulin dimers.
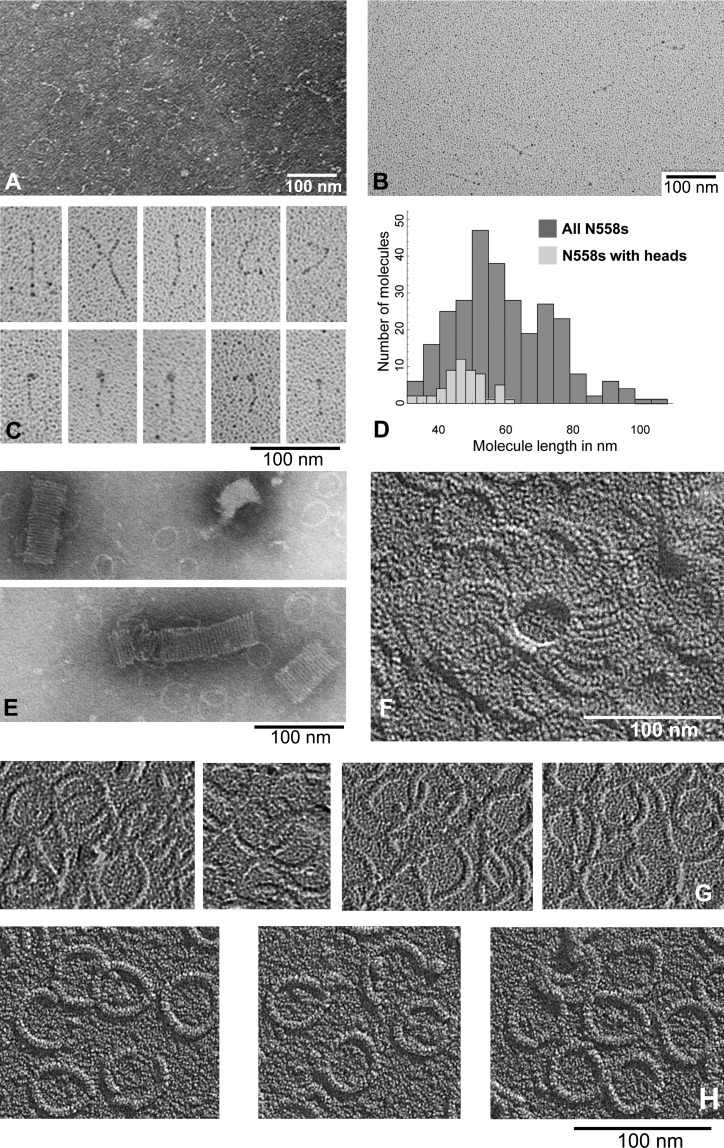
We used electron microscopy of N558, both alone and when added to complexes of curling tubulin oligomers, to visualize the protein and its interactions. By negative staining, N558 appeared elongated (Fig. 6 A), consistent with previous findings (Musinipally et al., 2013). Rotary shadowing of N558 sprayed onto mica and freeze-dried allowed us to obtain many images with which to characterize the molecule’s apparent length and shape (Fig. 6, B and C). The mean length of 286 molecules was 61 ± 18 nm (mean ± SD; Fig. 6 D), but 59 of these showed an enlargement at one end (Fig. 6 C). The mean length of this subset was 49 ± 10 nm, consistent with the idea that these fragments were folded at one end. When N558 was mixed with an equal molarity of tubulin assembled into rings by the addition of dolastatin-10, the complexes showed two deviations from ring-only controls. N558 commonly induced the rings to stack or cluster (Figs. 6, E and F), which is consistent with the idea that it has two MT-binding sites. When control images of rings alone (Fig. 6 G) were compared with ring–N558 complexes, poorly ordered material was often seen in the ring’s interior and/or there was a thickening of the ring, as see by shadowing at constant angle (Fig. 6, compare G and H). These data suggest that N558 binds to curled tubulin on a site that is on either the lateral or the outer surface of a tubulin dimer, as oriented in a MT.
Figure 6.
Electron micrographs of N558 alone and with tubulin rings formed with dolastatin-10. (A) N558 adsorbed to thin carbon films and negatively stained with uranyl acetate. (B) N558 adsorbed to mica and rotary shadowed. (C) Sample images of N558 at higher magnification. (D) Histogram of lengths of 286 images of N558, measured once. When N558 and dolastatin-10 rings are mixed and then prepared for electron microscopy, one finds ring clusters, either stacked as seen in negative staining (E) or by high resolution shadowing (F). (G) Tubulin-dolastatin rings alone are thin (note shadow lengths) and devoid of associated material. (H) When these rings are mixed with N558 before preparation for electron microscopy, the rings appear slightly thicker (longer shadows) and are associated with material that commonly clusters in the center of the ring.
The N terminus of CENP-F forms an unusual MT-binding site
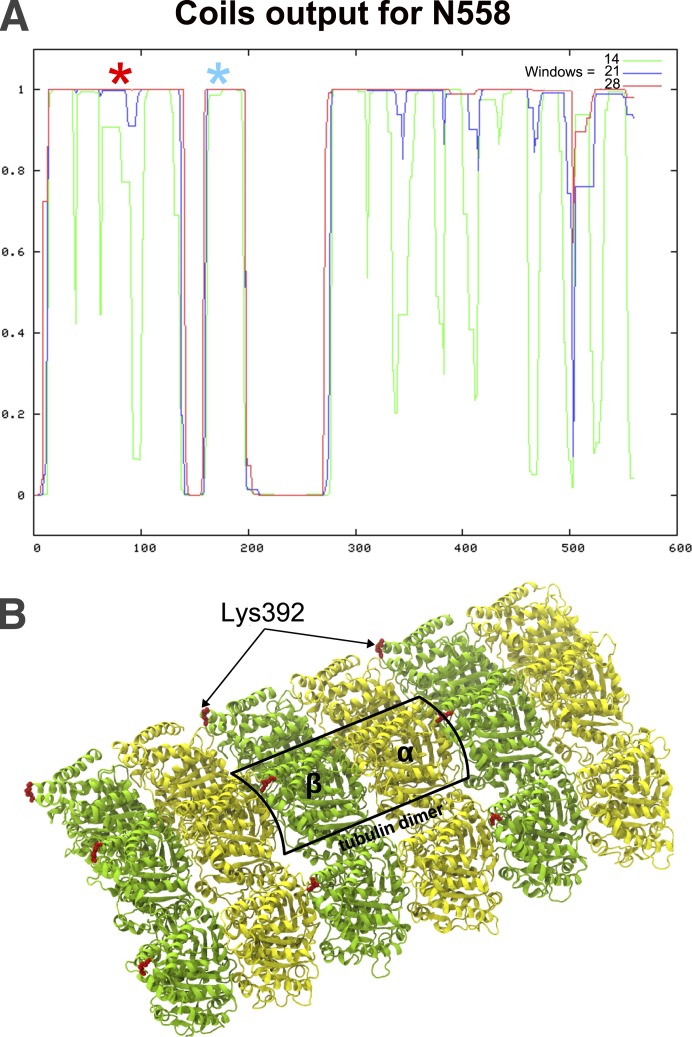
Primary structure analysis of N558 does not identify sequences like any of the well-known MT-binding sites, but it suggests that this fragment is rich in α-helical coiled coils (Fig. 7 A). The sum of the lengths of these predicted coils is 64 nm, similar to the mean length of N558 in shadowed images. However, the subset of molecules that was shorter and appear thickened at one end suggested that N558 can fold. To seek evidence about folding and to identify the part of N558 that binds to tubulin we mixed this fragment with an equal molarity of tubulin dimers assembled into rings with dolastatin-10, and then added protein cross-linkers to form covalent bonds between parts of the complex that lay close to one another. The “zero-length” cross-linker, 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide (EDC), linked E174 of N558 (Fig. 7 A, blue asterisk) to K392 of β-tubulin, which lies close to the junction between tubulin dimers (Fig. 7 B) and to the side of the domain where kinesin binds (Fig. S2 A). Disuccinimidyl suberate (DSS) linked the same residue of β-tubulin with K83 on N558 (Fig. 7 A, red asterisk), albeit with peptide coverage that was not completely unambiguous. (For a representative tandem mass spectrometry [MS/MS] spectrum of the cross-linked peptides, see Fig. S2 C.) The location of the second N558 residue linked to the same residue in tubulin supports the concept that the N-terminal half of N558 folds to make this tubulin-binding domain. In this context it is interesting that the predicted amino acid sequence for this part of CENP-F is highly conserved among members of this protein family, although it has not yet been found in any other protein.
Figure 7.
Secondary structure of N558 and cross-linking to tubulin rings. (A) Probability of coiled coils in N558 as a function of position along the protein (Lupas et al., 1991); the three colors display results for different windows sizes. The three segments with high probability of coiling should be 18, 6, and 40 nm long, respectively. Positions of amino acid residues that cross-linked to β-tubulin are marked with asterisks: blue, EDC; red, DSS.(B) Parts of three tubulin protofilaments as they would appear in a MT wall; one dimer is boxed. Image provided by V. Barsegov (University of Massachusetts, Lowell, MA; Kononova et al., 2014). The position of K392 in β-tubulin is shown in red.
When cross-linkers were applied to N558 in solution by itself, the only residues that became connected lay so close on the protein’s backbone that the results simply confirmed the dimeric structure of this fragment in solution (unpublished data). When DSS was used to cross-link N558 with tubulin in rings, however, many cross-links were seen within N558 (Fig. S2 B). The elevated concentration of links in N558 when it was complexed with tubulin rings could reflect either change in the protein’s fold, so different parts of N558 are brought into close proximity or the proximity of different N558 dimers as they bind to nearby tubulin dimers in the rings.
Both MT-binding domains of CENP-F can interact with dynamic MTs and transduce force from depolymerizing tubulin
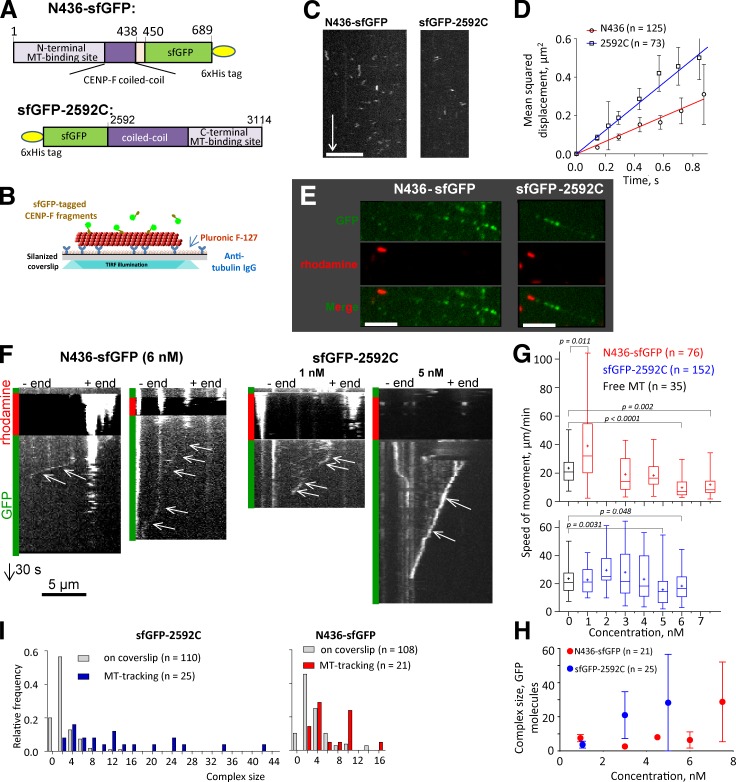
Given the unique tubulin-binding properties of CENP-F, we have studied its interactions with MTs in vitro, using fragments from either end of the protein. GFP fusions with N558 expressed in bacteria were insoluble, but a fusion of the N436 fragment with “super-folding” GFP (sfGFP; Pédelacq et al., 2006) was useful (Fig. 8 A). We introduced this protein into flow chambers with Tx-MTs and imaged the MT-associated fluorescence by total internal reflection (TIRF) microscopy (Fig. 8 B). N436-sfGFP bound to the MT lattice, as seen by MT decoration with green fluorescent spots (Fig. 8 C). At picomolar concentrations the MT-associated spots of fluorescence contained a mean of 2.6 ± 1.0 GFP molecules (mean ± SD; n = 125); they displayed a 1D diffusion coefficient of 0.16 ± 0.01 µm2/s (Fig. 8, C and D). The C-terminal fragment 2592C, fused to an N-terminal sfGFP, diffused somewhat faster (D = 0.30 ± 0.01 µm2/s) and contained a mean of 3.8 ± 1.6 GFP molecules (n = 73). By taking into account all observed binding and unbinding events, we estimated the on and off rates of these molecules with MTs; from the ratio koff/kon we have calculated KD of 1.5 µM for sfGFP-2592C and 0.97 µM for N436-sfGFP, consistent with published estimates (Musinipally et al., 2013). This affinity of N436 for Tx-MTs is, however, ∼10-fold weaker than the binding of N558 described using cosedimentation, as in Fig. 5. This difference may be a result of protein size (436 amino acids here and 385 in Musinipally et al., 2013, vs. 558 in our sedimentation work) or of the tags that were used: 6-his tag for N558 and N436 and GFP for N436.
Figure 8.
CENP-F fragments can diffuse on a MT’s surface and follow the end of a shortening MT. (A) Domain organization of sfGFP-tagged CENP-F fragments. (B) Experimental setup used to investigate 1D diffusion along stabilized MTs. (C) Kymographs showing diffusing spots of N436-sfGFP (left) and sfGFP-2592C (right). Bars: (horizontal) 5 µm; (vertical) 5 s. (D) Mean squared displacement of N436-sfGFP (circles) and sfGFP-2592C (squares) fitted with linear regressions to calculate diffusion coefficients. (E) Segmented MTs in the presence of N436-sfGFP (left) or 2592C- sfGFP (right), imaged in TIRF regimen. Bars, 5 µm. (F) Kymographs showing movement of CENP-F fragments with the ends of depolymerizing MTs. Vertical bars at the left of the kymograph color code the fluorescent channels used for imaging. Arrows point to the MT-tracking events: oblique lines that correspond to the spot movement from the MT plus end (right) toward the minus end (left). Bars: (horizontal) 5 µm; (vertical) 30 s. (G) Speed of MT disassembly in the absence of CENP-F (black) or the speed of movement of the distal fluorescent spot with increase of N436-sfGFP (red) or sfGFP-2592C (blue) concentration in the chamber. P-values are reported for an unpaired t test. The difference is not significant where the p-values are not reported. The bars show box (25–75%) and whisker (minimum to maximum) plots; horizontal line shows median; + is the mean. (H) The size of the MT-tracking complex as a function of N436- sfGFP or sfGFP-2592C concentration in the chamber. (I) Frequency distribution of the intensity of MT-tracking and coverslip-associated spots of N436-sfGFP or sfGFP-2592C expressed as the number of GFP molecules in a spot.
Binding of the sfGFP-2592C fragment to Tx-MTs was abrogated when MTs were pretreated with subtilisin (Fig. S4, A–C), suggesting a binding mechanism that involves the flexible, negatively charged C termini of either or both tubulins. MT-associated GFP fluorescence of the N436-sfGFP fragment was reduced eightfold after the treatment of MTs with subtilisin, but the residual GFP fluorescence of the digested MTs was still higher than background fluorescence without CENP-F (Fig. S3, B and C), suggesting that part of its binding does not depend on the C terminus of tubulin. Consistent with the idea of an electrostatic component in the binding of CENP-F fragments to Tx-MTs, both N436-sfGFP and sfGFP-2592C displayed gradually decreasing MT association with increases in the concentration of KCl added to BRB80 (Fig. S3, D and E).
To see whether MT-bound N436-sfGFP and sfGFP-2592C fragments could follow the end of a shortening MT we used segmented MTs made by nucleating the polymerization of dynamic tubulin from coverslip-attached, GMPCPP-stabilized MT seeds (Volkov et al., 2014). These MTs were capped with rhodamine-labeled, GMPCPP-stabilized tubulin; soluble tubulin was then washed from the flow chamber. sfGFP-tagged CENP-F fragments were added and allowed to bind the tripartite MTs (Fig. 8 E). To initiate an experiment the red fluorescent stabilizing cap was bleached, which allowed the dynamic MT to depolymerize. Thereafter, we could frequently observe a green fluorescent spot moving toward the MT seed (Fig. 8 F and Videos 1 and 2). These spots moved steadily at the speed of normal MT disassembly, translocating over distances of several micrometers (3 ± 2 µm, n = 18 for N436-sfGFP; 4 ± 3 µm, n = 21 for sfGFP-2592C). Increases in the concentration of either CENP-F fragment resulted in a decrease in the speed of fluorescent particle motion (Fig. 8 G); they also increased the apparent sizes of the moving complexes (Fig. 8 H). The fluorescence intensity of these spots did not change significantly during linear segments of movement, suggesting that the moving spots were stable and did not exchange with molecules in solution. However, coverslip-adsorbed spots of both fragments, examined in the presence of segmented MTs, consisted mainly of dimers, suggesting that bigger oligomers that were capable of MT tracking formed on the MT lattice, not in solution (Fig. 8 I). Apparently these pieces of CENP-F can form stable oligomers that will follow a shortening MT.
The ability to follow shortening MT ends has previously been reported for several kinetochore proteins that oligomerize spontaneously: Dam1 rings (Westermann et al., 2006) or Ndc80 molecules cross-linked with antibody (Powers et al., 2009) or Ska1 (Schmidt et al., 2012). It has also been seen with individual proteins that contain two distinct MT-binding sites, like CENP-E (Gudimchuk et al., 2013). These MT-tracking proteins share the ability to support cargo movement against an external load, establishing them as potential couplers of MT dynamics to cargo movements.
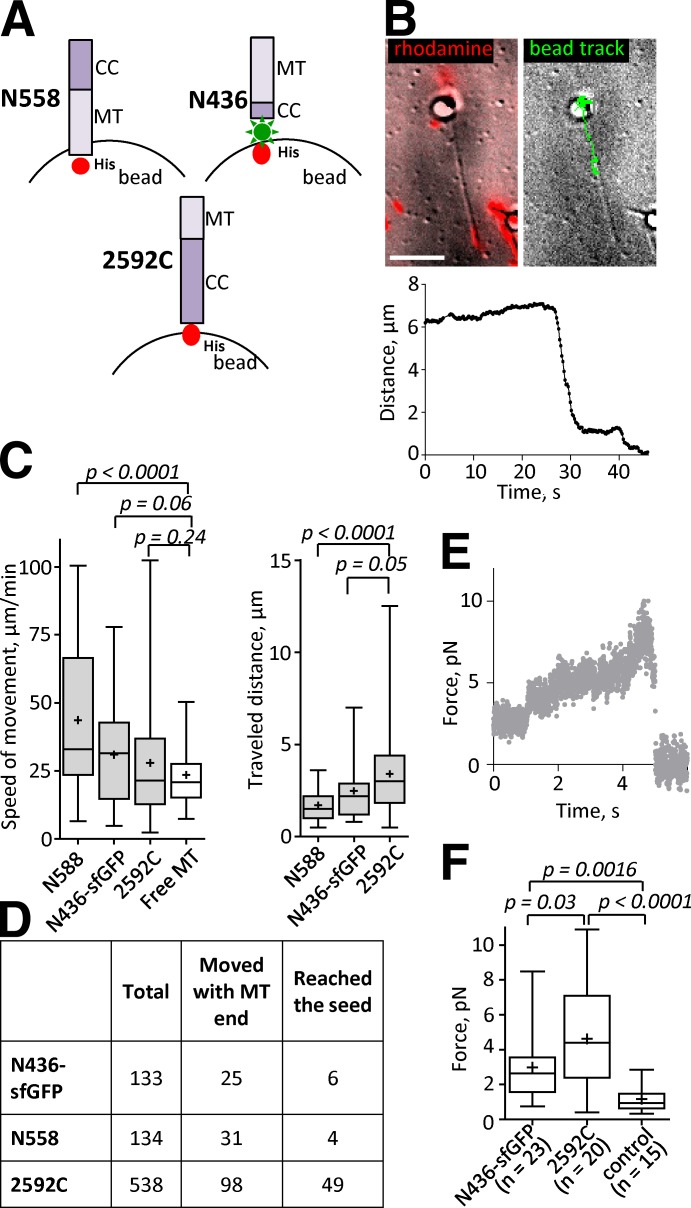
To ask whether CENP-F also showed such coupling properties, we attached its MT-binding fragments to the surface of 1-µm microbeads (Fig. 9 A). This arrangement typically resulted in CENP-F surface densities of 8,000–10,000 molecules per bead, as judged by bead fluorescence (Volkov et al., 2013). Assuming that the length of the coiled coil tether in either CENP-F fragment is ∼40 nm, this density should place up to 400 bead-bound molecules in positions to interact with a MT wall (Powers et al., 2009). Beads coated in this way were introduced into flow chambers with segmented, coverslip-anchored MTs, prepared as in Fig. 8 B. When MT depolymerization was induced by photodestruction of the stabilizing cap, the bead started to move toward the coverslip-attached seed (Fig. 9 B and Video 3). Both N-terminal fragments increased the speed of MT disassembly (Fig. 9 C), although for N436 the difference did not pass the P = 0.05 borderline for significance. These coated beads moved over only short distances, usually detaching before they reached the MT seed (Fig. 9 C, right). Beads coated with 2592C moved at the speed of free MT disassembly and frequently moved far enough to reach the MT seed (Fig. 9, C and D).
Figure 9.
CENP-F fragments transport cargos and transduce significant forces from tubulin depolymerization. (A) Schematics of attaching CENP-F fragments to the surface of beads (not to scale). Note that N558 is attached with the MT-binding domain (MT) facing the bead, whereas N436-sfGFP is attached with coiled coil (CC) facing the bead. (B) A bead coated with N436-sfGFP is attached to a segmented MT. Left image shows an overlay of an average of 18 differential interference contrast images before MT depolymerization was triggered (grayscale) and the rhodamine fluorescence (red). Right image shows an overlay of the first frame of the image sequence and the track of the bead’s motion after the depolymerization was triggered. See also Video 3. (bottom) Graph shows a single representative dependence of the distance from the N436-sfGFP–coated bead to the MT seed as a function of time (total n = 98). Bar, 5 µm. (C) Mean speeds (left) and distances (right) traveled by the CENP-F–coated beads. (D) Statistics of the MT-associated beads. Total: number of the beads analyzed; Moved with MT end: number of beads that started processive motion; Reached the seed: number of the beads that processively followed MT ends until they reached the coverslip-bound MT seeds. (E) A representative example of the MT-generated force transduced to the bead by 2592C (total n = 20). (F) Mean forces measured with N436-sfGFP, 2592C, and control beads coated with anti-DIG IgG and attached to DIG-labeled disassembling MTs. The bars show box (25–75%) and whisker (minimum to maximum) plots; horizontal line shows median; + is the mean.
To ask whether CENP-F can form load-bearing attachments with a shortening MT end, we captured beads coated with N436-sfGFP or 2592C in a laser trap. When a segmented MT bound to a trapped bead was induced to disassemble, the shortening MT end displaced the bead from the center of the trap, allowing us to measure the MT-generated force (Fig. 9 E). Forces measured with N436-sfGFP and 2592C were 3.0 ± 2.0 pN (n = 23) and 4.6 ± 2.9 pN (n = 20), respectively (Fig. 9 F). Both of these fragments developed forces that were statistically higher than control beads coated with anti-digoxigenin (DIG) IgG, attached to DIG-labeled disassembling MTs (1.2 ± 0.7 pN, n = 15; Fig. 9 F). Maximal forces of 8.5 and 10.9 pN were recorded from N436-sfGFP and CENP-F-2592C, respectively.
The number of monomers in MT-tracking oligomers of N436-sfGFP and sfGFP-2592C (Fig. 8 H) was much smaller than the maximum number of bead-bound molecules that could interact with the MT wall in laser trap experiments. To estimate the minimum surface density of CENP-F that could bear a load we decreased the CENP-F concentration on the bead surface by attaching 1:10 and 1:100 mixtures of anti-6His IgG and BSA to the beads and then saturating the beads with CENP-F (Fig. S4 A). Decrease in the density of N436-sfGFP down to ∼20 molecules/bead did not decrease the measured force (Fig. S4 B). However, decrease in the density of sfGFP-2592C down to 500–1,000 or fewer bead-bound molecules resulted in a decrease of force to control levels (Fig. S4 B).
Discussion
The pole-distal tips of KMTs are of great physiological interest because of their ability to bind tenaciously to kinetochores, even as the polymers gain and lose tubulin subunits. There are kinetochore-associated TIP proteins that bind preferentially to the ends of growing MTs (Jiang and Akhmanova, 2011), but the ability to bind MT ends as they shorten is rare. The ring-forming Dam1 complex of yeast kinetochores can do this by surrounding the MT wall and being pushed by bending protofilaments as the MT shortens, in spite of its rather tight association with polymerized tubulin (Volkov et al., 2013). CENP-E can use its motor activity to move toward a MT plus end, but if that end begins to shorten, a second MT-binding site can maintain MT attachment, tracking the MT tip by diffusing on the MT wall (Gudimchuk et al., 2013). The Ndc80 complex too can follow the end of a depolymerizing MT (McIntosh et al., 2008), but it is a comparatively weak coupler (Powers et al., 2009). Exactly how strong a coupler is needed when a chromosome binds multiple KMTs has not been established. However, Ndc80’s contribution to the kinetochore–spindle attachment probably depends in part on a binding partner, like the Dam1 complex in yeasts (Tien et al., 2010) or the Ska1 complex in multicellular organisms (Schmidt et al., 2012).
Here, we have sought additional proteins that might serve as kinetochore–MT couplers by looking for those that prefer binding to tubulin in the curled configuration that is characteristic of a shortening MT end. Although many proteins display this property (Table S1), we have confined our attention to proteins of the mitotic spindle (Table 1). Among these, CENP-F stood out as an interesting candidate for the fibrils that appear to make chromosome–MT connections (McIntosh et al., 2008). Our studies have distinguished the two MT-binding sites previously discovered on CENP-F (Feng et al., 2006), identifying the N-terminal one as the source of the protein’s preference for tubulin curls. N558 shows an approximately fivefold preference for rings relative to Tx-MTs. Its ∼16-nM KD from bent tubulin in our best measurement was significantly lower than has been reported for the human Ska complex (∼2 µM in Schmidt et al. (2012)) or Ndc80 (230 nM, even at comparatively low salt, in Powers et al. (2009)). This high affinity may allow CENP-F to contribute to the ability of a kinetochore to capture the ends of dynamic MTs in prometaphase. Because both MT-binding domains of CENP-F will tether a microbead to a dynamic MT end in a way that supports significant force, this protein may also contribute to a kinetochore’s ability to remain bound to shortening MTs during anaphase.
Microbeads coated with N-terminal fragments of CENP-F sped up MT depolymerization. An acceleration of MT shortening by protein-coated beads was previously seen with Dam1-coated beads in the absence of soluble Dam1 (Grishchuk et al., 2008). This was explained by evidence that such beads roll along the MT lattice, probably gathering tubulin protofilaments as they go. Several other bead-bound proteins have exhibited analogous behavior (McIntosh et al., 2010), all when conjugated directly to a bead’s surface. A different behavior was seen when Dam1 was tethered by long coiled coils (Volkov et al., 2013) or when Dam1 was both on the beads and in solution (Grishchuk et al., 2008); the latter constructs actually slowed the rate of MT shortening. The similarity of our data on beads coated with the CENP-F N terminus to the non-tethered beads coated with Dam1 suggests that the length of the endogenous coiled-coil tether in the N558 and N436 fragments is insufficient to allow the end-on configuration achieved with Dam1 bound to the rod segment from myosin, which is ∼100 nm (Volkov et al., 2013).
Although we have sometimes measured 10 or more piconewtons of force for sfGFP-2592C, the effective coupling of this CENP-F fragment to dynamic MTs requires a high a protein density. N436-sfGFP has not provided such high forces, perhaps because its shorter length prevented reorientation relative to the axis of the shortening MT (Volkov et al., 2013). Nonetheless, this protein continued to transduce significant force from shortening MTs, even at high dilution, probably because of its high affinity for curled tubulin oligomers. The force values reported here make CENP-F one of the most effective couplers to dynamic MTs currently known. Although the specific mitotic roles of CENP-F in vivo are still poorly understood, given the multiplicity of proteins with which it binds, its MT binding alone draws our attention to this protein as an interesting component of the protein assembly that attaches kinetochores to the pole-distal ends of dynamic spindle MTs. Its considerable length may contribute to the efficiency of kinetochore–MT attachment during prometaphase.
Our cross-linking and structural studies of N558 suggest that this dimeric fragment can fold, and that its tubulin binding site is formed by amino acid residues in the folded domain. A folded conformation for the tubulin-binding domain is consistent with the observations that pieces of the N-terminal domain truncated before L385, or pieces that extend to residue L385 but are deprived of the N-terminal 100 amino acids, cannot bind MTs (Feng et al., 2006). A folded conformation for MT binding might account for the two morphologies seen when N558 was sprayed on mica and visualized by shadowing. It may also account for the low values of Bmax seen in our sedimentations studies of N558 binding. It is intriguing that the amino acid sequence of this region shows considerable sequence conservation; informaticists at NCBI call it the CENP-F domain, and it is not found outside of CENP-F. On current evidence we propose that this sequence forms an unusual tubulin-binding site that interacts better with bent tubulin than with straight. The DSS cross-links formed within N558 when it was in complex with dolastatin-tubulin rings may reflect the folding that accompanies formation of this binding site or they may have arisen while different copies of this dimer lay next to one another on the tubulin polymer.
Our cross-linking studies identified a region of β-tubulin in the neighborhood of K392 that interacts with N558 (Figs. 7 B and S2). This region of tubulin lies near the interface between adjacent dimers in a MT protofilament and proximal to an adjacent protofilament. This position may explain the curl-binding preference of N558; a site near the boundary between tubulin dimers is likely to be affected strongly by protofilament bending. The effect on N436-sfGFP binding of increasing salt or of tubulin digestion with subtilisin suggests, however, that the amino acids in tubulin that interact with CENP-F’s N terminus are distributed over a significant part of tubulin’s surface.
The approximately fivefold stronger binding of N558 to curled versus straight tubulin would give this part of CENP-F a clear preference for binding near the end of a KMT where the flared configuration is found. This binding preference may be part of the molecular mechanism that leads kinetochores to bind firmly to KMT ends, rather than to the MT walls that are often their initial sites of interaction with the spindle (Roos, 1976; Kitamura et al., 2007). Certainly, some kinetochore proteins that prefer MT walls, like the Ndc80 complex, are important for achieving kinetochore–MT interaction. Moreover, the number of tubulins in spindle MT walls, versus the number at MT tips, makes initial binding to the walls very likely. The preference for walls shown by Ndc80 may explain why this complex is so important for attaching chromosomes to the spindle (DeLuca et al., 2006). Once kinetochores are MT bound, however, tubulin dynamics and kinetochore enzymes like CENP-E can convey them to the plus ends. Kinetochore proteins that prefer structures at the MT tip may then be important for establishing the dynamic, force-bearing attachment that is characteristic of both metaphase and anaphase. The concept of proteins binding to sites on tubulin that are found only at the polymer’s end is a simple and plausible explanation for this important aspect of mitotic physiology.
We do not yet know how CENP-F’s 3,114 amino acids are folded in vivo. On a 1D map, an epitope near CENP-F’s C terminus lies at about the position of the N-terminal part of the Ndc80 complex, whereas an epitope near its middle is ∼45 nm farther from the centromere (Wan et al., 2009). Without more information one cannot propose realistic models for the position of the N-terminal MT-binding site. Nonetheless, it is intriguing to think about the roles that such a site might play during chromosome attachment and congression while the spindle fiber attachment is forming. Further studies on both the biochemistry and cell biology of this kinetochore component should yield valuable insight into the molecular mechanisms of chromosome–spindle interactions in cells with big kinetochores.
Materials and methods
Preparation of assembled tubulin
MT protein was prepared from cow brains by two polymerization cycles (Weingarten et al., 1974), frozen in assembly buffer (100 mM MES, pH 6.4, 1 mM EGTA, and 1 mM MgSO4, plus 1.5 mM GTP), and stored at −80°C until needed. Tubulin was purified from this protein mixture by phosphocellulose chromatography (Williams and Detrich, 1979). In brief, resin was prepared according to the manufacturer’s instructions, brought up in PME buffer (100 mM Pipes, pH 6.9, 2 mM EGTA, and 1 mM Mg SO4) plus 2 mM dithiothreitol and 0.2 mM GTP, and then packed into a column. MT protein was added to the column, eluted with the same buffer, aliquoted, snap frozen, and again stored at −80°C. Tx-MTs were made by supplementing PME that contained at least 50 µM of purified tubulin with 25% glycerol and 1 mM MgGTP, and then warming to 37°C for 25 min, adding 20 µM Taxol, and incubating for 5 min more at 37°C to ensure MT stability. Tubulin curls were made by adding 15 µM vinblastine sulfate to 10 µM phosphocellulose-purified tubulin in 100 mM MES buffer, pH 6.4, or 80 mM Pipes, pH 6.9, containing 1 mM MgCl2, EGTA, and DTT, 0.1 mM EDTA, plus 1 mM MgGTP or MgGDP, and then warming to 37°C for 25 min. The purity of the tubulin and electron micrographs of two forms into which it assembled are shown in Fig. 1 (B–D).
Preparation of cell extracts
U2OS cells were grown in McCoy’s 5A medium (Life Technologies) supplemented with 10% fetal calf serum and 100 U/ml penicillin and 0.1 mg/ml streptomycin. RPE1 cells were grown in DMEM with High Glucose (Invitrogen) with 10% fetal calf serum and penicillin/streptomycin, as for the U2OS cells. Interphase cells were scraped from the bottom of 18 T175 flasks (Corning), collected by centrifugation at 1,750 g for 8 min, washed twice in Dulbecco’s phosphate-buffered saline (Sigma-Aldrich), and then once in PME buffer with 1 mM DTT and 0.1 mM EDTA plus the following protease inhibitors: 10 µg/ml pepstatin, 10 µg/ml leupeptin, 50 µg/ml antipain, and 250 µg/ml pefabloc SC. Cell lysis was accomplished by the addition of 0.5% Triton X-100 and rapid passage of the liquid through a 26-gauge needle. This homogenate was clarified by spins first at 1,000 g for 10 min in an Eppendorf microfuge, and then at ∼190,000 g for 60 min in a Type 65 rotor in a preparative ultracentrifuge (L65; Beckman Coulter).
Mitotic cell extracts were prepared from cultures greatly enriched for cells in division. 18 T175 flasks of U2OS or RPE1 cells at ∼40% confluence were supplemented with 20 µM Nocodazole and allowed to continue growth under normal conditions for 12 h. Mitotic cells were selected by shake-off, and then the medium was returned to the flasks, allowing the accumulation of additional mitotic cells for another 6 h. Each of these harvests, which contained ∼90% dividing cells, was immediately washed by the same procedure used for interphase cells and monitored for viability by Trypan blue exclusion. Different collections ranged from 70% to 85% viable by this assay. These cells were lysed and clarified, as described for interphase cells, to make mitotic cell extracts.
Pull-downs of tubulin-interacting proteins
Cell extract containing 1 mM MgGTP and 20 µM of either Taxol or vinblastine was mixed with either Tx-MTs or vinblastine-induced tubulin curls, both at a final concentration of 0.125 mg/ml in BRB80 buffer (80 mM Pipes, pH 6.9, 1 mM MgCl2, EGTA, and DTT). After 15-min incubation at room temperature, these samples were spun in an Airfuge (A-100/18; Beckman Coulter) at 20 psi for 30 min (∼84,000 rpm or 95,000 g). The supernatants were collected and the pellets were resuspended in Laemmli sample buffer for electrophoretic analysis.
For several of the preparations analyzed by mass spectrometry, we used a serial sedimentation strategy to compare the proteins that bound to the vinblastine curls versus the Tx-MTs. In these experiments, cell extract was first mixed with vinblastine curls and sedimented though a cushion of 20% glycerol in PME buffer plus 1 mM MgGTP and 20 µM vinblastine in a Type 75 Ti rotor at 250,000 g for 90 min at 23°C. The resulting pellet was resuspended in PME buffer containing 1 mM MgGTP and 20 µM vinblastine, supplemented with 0.5 M NaCl to reduce the strength of binding between tubulin and its associated proteins. The vinblastine curls were then pelleted away from the associated proteins in an AirFuge, as described in the previous paragraph. The supernatant liquid was collected, desalted on a Zeba Desalting Spin Column (Thermo Fisher Scientific) that had been equilibrated with PME buffer, mixed with Tx-MTs in the presence of 1 mM MgGTP and 20 µM Taxol, and pelleted again in the AirFuge. Samples were taken from the supernatant and pellet to be assessed by either electrophoresis (with or without immunoblotting) or mass spectrometry.
Mass spectrometry
Samples were prepared for mass spectrometry by reduction with 3 mM dithiothreitol, alkylation with 14 mM iodoacetamide, neutralization by a second treatment with 3 mM dithiothreitol, followed by a change of buffer into 200 mM ammonium bicarbonate, pH 8.0, with 1 mM CaCl2, using a Zeba spin column. They were then digested by overnight treatment with 2% (wt/wt) Trypsin (Promega) in the same buffer at 37°C. The resulting peptides were separated by liquid chromatography using a nanoAcquity UPLC system (Waters) coupled to a Velos or LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific). For 1D analysis, a BEH C18 reversed phase column (25 cm × 75 µm i.d., 1.7 µm, 100 Å; Waters) was used for the analytical separation. Peptide mixtures (10 µl and 22 µg) were loaded onto the column maintained at 40°C and separated by a linear gradient from 90% buffer A2 (0.1% formic acid) to 40% buffer B2 (0.1% formic acid and 80% acetonitrile) over 120 min at a flow rate of 300 nl/min. For 2D online separations before mass spectrometry analysis, samples were injected at pH 10.0 onto a 300-µm × 50-mm XBridge C18, 130-Å, 5-µm column, eluting peptides in six fractions corresponding to 10%, 15%, 20%, 30%, 40%, and 60% buffer B (buffer A1: 20 mM ammonium formate, pH 10.0; and buffer B1: 100% acetonitrile). Steps were eluted from the high pH column at 20 µl/min onto a 180-µm × 20-mm C18, 100-Å, 5-µm trap column, which was then switched in-line with the analytical column and eluted as in the 1D method.
MS/MS data were collected by an enabling monoisotopic precursor and charge selection settings. Ions with unassigned charge state or charge state equal to 1 were excluded. For each mass spectrometry scan, the 10 most intense ions were targeted with dynamic exclusion 30 s, 1 D exclusion width, and repeat count equal to 1. The maximum injection time for Orbitrap parent scans was 500 ms, allowing 1 microscan and automatic gain control of 106. The maximal injection time for the LTQ MS/MS was 250 ms, with 1 microscan and automatic gain control of 104. The normalized collision energy was 35%, with activation Q of 0.25 for 30 ms.
Database search
MS/MS data were searched against a human protein database (IPI v.3.65) using MASCOT v.2.2 (Matrix Sciences), with ion score thresholds set to false discovery rate of 0.01 at the peptide level using inverted database searching. Parent ion tolerances were set to 50 ppm on the monoisotopic peak (A0) and the first isotopic peak (A1) and the fragment ion tolerance was set to 0.6 D, allowing two missed cleavages.
Analyses of binding ratios
Protein assembly and label-free quantification by spectral counting were performed using Isoform Resolver (Meyer-Arendt et al., 2011). In brief, to assemble protein assignments from peptide identifications, an in silico digest of the human protein database was performed using the same trypsin digestion rules specified in the MS/MS database search. From this digest one can generate a mapping from a protein to all its tryptic peptide sequences. Inferences of protein identity are then done using a maximum parsimony approach on the set of peptides identified in the database search and the in silico digest. Spectral counts are tabulated as the total number of peptide-spectrum matches assigned to peptides that map uniquely to each given protein. For each set of peptides that map to multiple proteins, the spectral count for the set was distributed to the proteins of that set with unique peptide evidence, according to the model described in Meyer-Arendt et al. (2011).
Binding ratios determined by immunoblotting
As a check on the validity of binding ratios determined by mass spectrometry and the relative binding of various CENP-F constructs, we used immunoblots with antibodies against selected proteins to probe extracts from mitotic human cells and the proteins pulled down from those extracts with either Tx-MTs or vinblastine-induced curls. Samples from the pellets and supernatant liquids were fractionated by electrophoresis on 7% polyacrylamide gels, blotted to nitrocellulose paper, and reacted with primary antibodies. CENP-F was identified with a mouse monoclonal antibody to a peptide that included amino acids 209–381 of this protein (Santa Cruz Biotechnology, Inc.); IQGAP was identified with a rabbit polyclonal antibody raised against amino acids 314–422 from the human protein (Santa Cruz Biotechnology, Inc.). Talin was probed with a rabbit polyclonal antibody to talin purified from human blood platelets in the laboratory of K. Burridge (University of North Carolina, Chapel Hill, NC). Vinculin was identified with a mouse monoclonal antibody (Sigma-Aldrich; Fig. S1, A and B). The relative amounts of protein were estimated by detecting the primary antibody with an appropriate secondary antibody labeled for detection by chemiluminescence with SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific). X-Ray film was exposed for several intervals, the appropriate image was scanned, and the area under the peak was assessed by ImageJ.
Cloning and expression of cDNA for CENP-F
Three cDNAs that covered the structural gene for CENP-F were obtained from T. Yen (Fox Chase Cancer Center, Philadelphia, PA). The CENP-F constructs used in this study were N436 (amino acids 1–436), N558 (amino acids 1–558), N977 (amino acids 1–977), 1758C (amino acids 1758–3114), and 2592C (amino acids 2592–3114). N436 was cloned into the BamHI and PstI sites of pQE-80L (QIAGEN) under the control of the T5 promoter; N558 was cloned into the BamHI and HindIII sites of pQE-80L; N977 was cloned into the SmaI and KpnI site of pQE-TriSystem (QIAGEN); and 1758C was cloned into the KpnI site of pQE-81L (QIAGEN). 2592C was amplified from 1758C/pQE81L and cloned into the BamHI and XhoI sites of pET28a (EMD Millipore) under a T7 promoter, and then sfGFP was inserted into the BamHI site. N436-sfGFP was produced by subcloning the sfGFP sequence into NcoI and XhoI sites in pET28a, also under T7 promoter, and then inserting the N436 sequence amplified from N436/pQE80L into the BamHI and NcoI sites. Clones were sequence verified and mistakes were corrected using the QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent Technologies). Clones were expressed in Rosetta (DE3) or Rosetta Blue (DE3) pLacI (EMD Millipore) and induced with 0.4 mM IPTG for 16 h at 23°C (or 2 h at 37°C for sfGFP fusions and 2592C); the cells were harvested by centrifugation and the pellets were frozen at −80°C.
Purification of CENP-F constructs
Cell pellets were resuspended in lysis buffer (50 mM NaH2PO4, pH 6.9, 300 mM NaCl, 10 mM imidazole, and 250 µg/ml Pefabloc SC). When samples were not used for mass spectrometry we also added a protease inhibitor tablet (Roche), lysed with 1 mg/ml lysozyme, and performed sonication. 0.25% Tween-20 was added and the lysate was centrifuged at 10,000 g for 20 min at 4°C. Clarified lysates were mixed with Ni-NTA resin and after 1.5 h the resin was transferred to a column and washed with buffer (50 mM NaH2PO4, pH 6.9, 500 mM NaCl, and 20 mM imidazole), and the proteins were eluted with elution buffer (50 mM NaH2PO4, pH 6.9, 300 mM NaCl, and 250 mM imidazole). Eluted fractions were desalted on a Zeba Desalting Spin Column that had been equilibrated with BRB80 buffer plus 150 mM NaCl. Appropriate fractions were pooled, aliquoted, and frozen. Cells containing expressed 2592C fragments were lysed using B-PER (Thermo Fisher Scientific) supplemented with 0.5 M NaCl, 10 mM imidazole, 1 mg/ml lysozyme, 2 U/ml DNase I, and a protease inhibitor tablet (Roche). For in vitro fluorescence and bead motility experiments, CENP-F fragments were additionally purified using ion-exchange chromatography. Ni-NTA fractions were desalted into 50 mM NaH2PO4, pH 6.6 (N436-sfGFP) or 7.0 (2592C and sfGFP-2592C), with 0.1 M NaCl; applied to a 1-ml HiTrap SP HP column (GE Healthcare); washed with 10 ml of the same buffer; and eluted with a linear gradient from 0.1 to 1 M NaCl in the same buffer. Peak fractions were snap frozen in liquid nitrogen.
Binding curves and KD determination
Binding of N558 at 0.2 µM in the presence of varying amounts of Tx-MTs, vinblastine-induced curls, or dolastatin-induced rings, all prepared in BRB80 buffer, was initiated as in the previous paragraph and sedimented in a S100-AT3 rotor at 162,000 g for 30 min at 23°C. Samples from the pellets and supernatants were fractionated by electrophoresis on 4–12% Nupage Bis-Tris polyacrylamide gels, stained with SYPRO Ruby protein stain (Molecular Probes), and imaged with a Fotodyne gel system. The amounts of protein in pellet and supernatant fractions were quantified by ImageJ. For each set of experiments, standards of known protein concentration and of N558 centrifuged without added tubulin were run on the same gel. The fraction of N558 that pelleted in buffer only was used to correct each sedimentation experiment by multiplying it times the total amount of N558 in that experiment (supernatant + pellet) and subtracting this amount from the observed pellet. The fraction of N558 bound to tubulin was then calculated from the corrected intensity of N558 in the pellet divided by the sum of intensities measured for the supernatant and pellet. This fraction was plotted as a function of the amount of pelleted tubulin (note that soluble tubulin does not bind N558 so its concentration was not included).
Binding data were fit with the model that assumes A + B <−> C with dissociation constant KD and A + C = A0 and B + C = B0, where A0 and B0 are total N558 and tubulin concentrations, respectively. The fraction of bound ligand (C/A0) can now be found as function of A0, B0, KD, and saturation level Bmax:
To obtain the optimal values of the fitting parameters KD and Bmax a score function was calculated as the sum of squared residuals between model prediction and experimental data. Values of KD and Bmax that resulted in the minimal score function value were taken as optimal. To estimate the accuracy of the fitting, the score function was calculated as a function of the parameter KD. Areas where the score function differed from the optimal value by 5% were highlighted on the plots and used to obtain the fitting error of the parameter KD.
Hydrodynamic characterization of the CENP-F N558 fragment
Sucrose gradient sedimentation and gel filtration chromatography in BRB80 buffer were used to measure the molecular mass as previously described (Grissom et al., 2009). Sedimentation of N558 along with a set of sedimentation standards was performed on 5–20% sucrose gradients at 36 krpm (SW41 rotor; Beckman Coulter) for 20 h. Gel filtration chromatography of N558 was performed on a Superdex 200 10/300 GL column using an ÄKTA FPLC system (GE Healthcare). Gel filtration molecular mass makers (Bio-Rad Laboratories) were run on the same column and the –logKav0.5 versus Stokes radius for the standards was plotted and used to measure the Stokes radius for N558. The partial specific volume of N558 was calculated from the mean partial specific volumes of the individual amino acids (Perkins, 1986) and the native molecular mass was calculated from the Svedberg equation (Fig. S1 G), as described previously (Siegel and Monty, 1966).
Structural characterization of CENP-F fragments and tubulin binding by electron microscopy
N558 was viewed on its own or in complex with tubulin oligomers by electron microscopy. For negative staining, N558 was allowed to adsorb to copper grids coated with a thin film of carbon, rinsed with buffer, then with 1% uranyl acetate, blotted, dried, and imaged with a T12 Spirit (FEI). For shadow imaging, N558 was diluted into 100 mM ammonium acetate buffer, pH 7.0, with 50% glycerol, applied to freshly cleaved mica, rapidly frozen, freeze-dried in a Bal Tec BAP 060 freeze-etch instrument and rotary shadowed with a thin layer of platinum-carbon, and imaged on the same microscope. For studies of N558 with curving tubulin, tubulin was induced to form rings, rather than curls, by mixing phosphocellulose-purified protein at 20 µM with 40 µM dolastatin-10, following the procedure of Moores and Milligan (2008). Rings were mixed with recombinant proteins at a molarity equal to that of tubulin, incubated at room temperature for 10 min, and applied to a freshly glow-discharged carbon-coated Formvar grid. Negatively stained samples were prepared as described at the beginning of this paragraph and examined in an electron microscope (CM10; Philips) operating at 80 KeV. Shadowed samples were prepared and imaged at the electron microscopy laboratory in ETH Zurich. Images were recorded on charge coupled device cameras and studied without further image processing.
Cross-linking of protein mixtures
Equimolar N558 and tubulin in the form of dolastatin rings were incubated in MES buffer with 40 µM dolastatin-10 at 23°C for 15 min. 1 mM EDC was added and the mixture was cross-linked at 23°C for 1 h. The reaction was stopped with the addition of 20 mM 2-Mercaptoethanol. The sample was precipitated with trichloroacetic acid, centrifuged at 16,000 g for 25 min at 4°C, washed with −20°C acetone, and spun again in the same way. The dried pellet was resuspended in SDS-PAGE buffer and loaded onto a 6% polyacrylamide gel. The band that reacted with both CENP-F and tubulin antibodies was excised for in-gel digestion. Cross-linking with Disuccinimidyl Suberate (DSS-H12/D12; Creative Molecules Inc.) was performed as described for EDC except the buffer used was BRB20 (same as BRB80 except with 20 mM Pipes), DSS was added to 500 µM at 23°C for 30 min, and the reaction was quenched with 50 mM ammonium bicarbonate at 23°C for 15 min. The DSS cross-linked samples were then trypsin digested in solution. DSS cross-linked protein samples were washed three times with 8 M urea on a Vivacon 500 spin column (Sartorius Stedim), reduced with 5 mM Tris(2-carboxyethyl)phosphine hydrochloride for 30 min at 23°C, and then alkylated with 10 mM iodoacetamide in the dark for 30 min at 23°C. Excess iodoacetamide was removed by buffer exchange with 8 M urea and 50 mM Tris, pH 8.0, on the aforementioned spin column. The samples were digested first with LysC in a ratio of 1:20 wt/wt LysC/protein (in 2 M urea and 50 mM Tris, pH 8.0) for 2 h at 37°C and then with trypsin in a ratio of 1:50 wt/wt trypsin/protein (in 1 M urea and 50 mM Tris, pH 8.0) overnight at 37°C. 1% formic acid and 5% acetonitrile were added to the digested samples before purification on C18 spin columns (Thermo Fisher Scientific) following the manufacturer’s protocol using formic acid in place of trifluoroacetic acid. Proteins were eluted from the C18 columns with 70% acetonitrile, dried, and resuspended in 1% formic acid for mass spectrometry analysis.
In-gel digestion of proteins
Proteins were digested and extracted from excised gel bands according to the method of Shevchenko et al. (2007), with the following modifications: sulfhydryl reduction was performed in-gel with 10 mM DTT and alkylated with 55 mM iodoacetamide; gel pieces were incubated with 0.6 µg each of chymotrypsin and endopeptidase Arg-C in 200 mM NH4CO3, pH 8.0, for 30 min at 4°C, and then overnight at 23°C. The protease buffer was removed and peptides were extracted from the gel pieces with the addition of 100 µl 50% acetonitrile/2.5% formic acid. Extracted peptides were lyophilized in a SpeedVac, resuspended in 50 µl 0.1% trifluoroacetic acid, and desalted using a C18 spin column (Thermo Fisher Scientific) according to the manufacturer’s instructions. Desalted peptides were lyophilized and resuspended in 20 µl 0.1% TFA. A 4-µl sample was loaded for direct injection onto the analytical column for liquid chromatography MS/MS analysis with an LTQ-Orbitrap.
Identification of cross-linked proteins with StavroX
Thermo.Raw files were converted to Mascot Generic File format with MSConvert, with MS level set from 1–2. FASTA files corresponding to bovine tubulin β3 (tubb3) and the first 558 residues of human CENP-F (N558) were obtained from UniProtKB and used as reference protein sequences. Data were interrogated for cross-linked peptides corresponding to linkages between reference proteins using StavroX v3.2.2 (Götze et al., 2012). For EDC-linked proteins, variable modifications were limited to oxidized methionine with a maximum of 2; carbamidomethyl-cysteine was set as a fixed modification. Protease sites were set as phenylalanine, tryptophan, tyrosine, and arginine, blocked by C-terminal proline, with a maximum of two missed cleavages. The cross-linker mass was set to −18.01056 (loss of H2O from carboxy-amine linkage). For both cross-linked protein datasets, precursor precision was set to 3.0 ppm, with 0.8-D fragment ion precision, 200-D lower mass limit, and 5,000-D upper mass limit, and only y and b ions were considered.
Biophysics of interactions between CENP-F fragments and MTs in vitro
Tubulin was purified from cow brains by two cycles of polymerization in 0.33 M Pipes followed by depolymerization in cold 50 mM MES with the addition of 1 mM CaCl2 as described previously (Castoldi and Popov, 2003). Tubulin was then additionally cycled or labeled with rhodamine (Molecular Probes), HyLite Fluor-647 (AnaSpec), or DIG (Molecular Probes) as described previously (Hyman et al., 1991). Single-molecule diffusion with TIRF illumination was investigated as described previously (Volkov et al., 2014). In brief, Taxol-stabilized, rhodamine-labeled MTs were attached with anti-tubulin antibodies to the silanized coverslips passivated with Pluronic F-127. A solution of CENP-F fragment in BRB80 supplemented with 2 mM dithiothreitol, 0.1 mg/ml glucose oxidase, 68 µg/ml catalase, 20 mM glucose, 0.5% 2-mercaptoethanol, and 10 µM taxol was continuously pumped through the flow chamber at 10–20 µl/min. Flow chambers were equilibrated at 23°C during the acquisition. Images were acquired every 0.13 s using an AxioImager Z1 microscope (Carl Zeiss) equipped with α-Plan Apochromat NA 1.46 objective and a Cascade II electron multiplying charge coupled device (EMCCD) camera (Photometrics). Images were analyzed using custom-written software in Wolfram Mathematica and ImageJ as described previously (Zaytsev et al., 2015). In brief, kymographs of diffusing spots were fitted with a Gaussian in each frame to determine the position of the spot. Kon and koff were obtained by integrating the number and duration of binding events, respectively. KD was calculated as the ratio koff/kon.
To visualize binding of CENP-F to subtilisin-treated MTs (S-MTs), Tx-MTs were labeled with DIG and either rhodamine (dye/tubulin molar ratio 1:30) or HyLite (dye/tubulin molar ratio 1:20) and then incubated with 200 µg/ml subtilisin (Sigma-Aldrich) for 1 h at 30°C (Skiniotis et al., 2004). The reaction was stopped by the addition of 5 mM PMSF. MTs were centrifuged for 15 min at 25,000 g and resuspended in BRB80. A mixture of HyLite-labeled S-MTs and rhodamine-labeled untreated MTs (or rhodamine-labeled S-MTs and HyLite-labeled untreated MTs) were bound to coverslip-adsorbed anti-DIG IgG in a flow chamber with silanized coverslips, as described in the previous paragraph, followed by perfusion of constant concentration of sfGFP-tagged CENP-F fragments.
Experiments with dynamic MTs were performed in flow chambers at 32°C as described previously (Volkov et al., 2014). In brief, DIG-labeled, GMPCPP-stabilized MT seeds were attached to the silanized, passivated coverslips with anti-DIG antibodies (Roche), elongated using cycled unlabeled tubulin in the presence of 1 mM GTP, and then capped in the presence of rhodamine-labeled tubulin and GMPCPP. sfGFP-tagged CENP-F fragments were introduced into the flow chamber and allowed to bind to the MTs for 5 min. MT disassembly was triggered using a 532-nm laser, and then GFP fluorescence was imaged using a 473-nm laser in epifluorescence or TIRF regimen using an AxioImager Z1 microscope equipped with α-Plan Apochromat NA 1.46 objective and a Cascade II EMCCD camera.
1-µm glass COOH-modified beads (Bangs Laboratories) were coated with streptavidin (Thermo Fisher Scientific) and then biotinylated with anti-Penta His antibodies (QIAGEN) or biotinylated anti-GFP antibodies (Rockland) as described previously (Grishchuk et al., 2008). 6xHis-tagged CENP-F fragments were then attached to these beads followed by four washes. The beads were introduced into the flow chambers and allowed to bind to the segmented MTs. MT disassembly was triggered by illumination with HBO100 lamp filtered for rhodamine excitation. Bead movements were followed by differential interference contrast optics using an AxioImager Z1 microscope equipped with EC Plan-Neofluar NA 1.3 objective and a Cascade II EMCCD camera. MT-generated forces were measured with a custom-built laser trap as described previously (Volkov et al., 2013). In brief, displacement of the bead from the center of the laser beam induced by MT disassembly was measured using quadrant photo-detector. Force was then obtained as a product of displacement and trap stiffness, with correction for the nonlinear response of the trapping force as a function of bead displacement from the trap center.
Online supplemental material
Tables S1 and S2 describe the binding of many proteins from cultured human cell extracts to tubulin, assembled into either curls or Tx-MTs. GO terms and other descriptions of each protein are included. Fig. S1 (A and B) shows immunoblots of mitotic cell extracts pulled down by curls or Tx-MTs probed with antibodies to four proteins of interest for comparison with the mass spectrometry data. C, D, and E confirm that N558 without a 6-His tag binds better to curls than to Tx-MTs. F and G show biochemical data for the dimeric condition of N558 in solution. Fig. S2 presents additional evidence on the cross-linking of N558 to tubulin. Fig. S3 compares that binding of fragments from CENP-F with tubulin before and after its cleavage by the protease subtilisin. Fig. S4 evaluates the effect of protein dilution on the ability of fragments from CENP-F to transduce force from shortening MTs. Videos 1–3 display the action of dynamic MTs on small clusters of N436-sfGFP or sfGFP-2592C and on microbeads coated with the same CENP-F fragments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408083/DC1.
Supplementary Material
Acknowledgments
We thank Ryther Anderson, Mary Morphew, and Andreas Hoenger of the University of Colorado for help with electron microscopy; Evgenia Glazunova and Ekaterina Panasenko (Center for Theoretical Problems of Physicochemical Pharmacology, Russian Academy of Sciences [CTP PCP RAS]) for help with cloning and protein purification; and Fazly Ataullakhanov, Nikita Gudimchuk, and other members of the Ataullakhanov (CTP PCP RAS) laboratory for stimulating discussions.
This work was supported in part by National Institutes of Health grant GM033787 to J.R. McIntosh and grants 13-04-40188-H, 13-04- 40190-H, and 15-04-04467 from the Russian Fund for Basic Research and from the Presidium of Russian Academy of Sciences Molecular and Cell Biology program to F.I. Ataullakhanov, plus grants 14-04-31061 and 14-04-00057 from the Russian Fund for Basic Research and a Postdoctoral Fellowship from Dmitry Zimin Dynasty Foundation to V.A. Volkov.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CENP
- centromere protein
- DIG
- digoxigenin
- DSS
- disuccinimidyl suberate
- EDC
- 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide
- KMT
- kinetochore-associated MT
- MS/MS
- tandem mass spectrometry
- MT
- microtubule
- TIRF
- total internal reflection fluorescence
- Tx-MT
- Taxol-stabilized MT
References
- Bomont P., Maddox P., Shah J.V., Desai A.B., and Cleveland D.W.. 2005. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. EMBO J. 24:3927–3939. 10.1038/sj.emboj.7600848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M., and Popov A.V.. 2003. Purification of brain tubulin through two cycles of polymerization–depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88. 10.1016/S1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Chan G.K., Schaar B.T., and Yen T.J.. 1998. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143:49–63. 10.1083/jcb.143.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M. 2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6:a015826 10.1101/cshperspect.a015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo N.C., Ohkanda J., Yen T.J., Hamilton A.D., and Sebti S.M.. 2001. The farnesyltransferase inhibitor, FTI-2153, blocks bipolar spindle formation and chromosome alignment and causes prometaphase accumulation during mitosis of human lung cancer cells. J. Biol. Chem. 276:16161–16167. 10.1074/jbc.M006213200 [DOI] [PubMed] [Google Scholar]
- Dees E., Miller P.M., Moynihan K.L., Pooley R.D., Hunt R.P., Galindo C.L., Rottman J.N., and Bader D.M.. 2012. Cardiac-specific deletion of the microtubule-binding protein CENP-F causes dilated cardiomyopathy. Dis. Model. Mech. 5:468–480. 10.1242/dmm.008680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., and Musacchio A.. 2012. Structural organization of the kinetochore–microtubule interface. Curr. Opin. Cell Biol. 24:48–56. 10.1016/j.ceb.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K.J., Meng X., Khodjakov A., and McEwen B.F.. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522. 10.1038/ncb1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H.J., Edwards L., and Goodwin R.L.. 2007. Conserved C-terminal domains of mCenp-F (LEK1) regulate subcellular localization and mitotic checkpoint delay. Exp. Cell Res. 313:2427–2437. 10.1016/j.yexcr.2007.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Huang H., and Yen T.J.. 2006. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 115:320–329. 10.1007/s00412-006-0049-5 [DOI] [PubMed] [Google Scholar]
- Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., and Nigg E.A.. 2009. Stable kinetochore–microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 28:1442–1452. 10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin R.L., Pabón-Peña L.M., Foster G.C., and Bader D.. 1999. The cloning and analysis of LEK1 identifies variations in the LEK/centromere protein F/mitosin gene family. J. Biol. Chem. 274:18597–18604. 10.1074/jbc.274.26.18597 [DOI] [PubMed] [Google Scholar]
- Götze M., Pettelkau J., Schaks S., Bosse K., Ihling C.H., Krauth F., Fritzsche R., Kühn U., and Sinz A.. 2012. StavroX—a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 23:76–87. 10.1007/s13361-011-0261-2 [DOI] [PubMed] [Google Scholar]
- Grishchuk E.L., Efremov A.K., Volkov V.A., Spiridonov I.S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J.R., and Ataullakhanov F.I.. 2008. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA. 105:15423–15428. 10.1073/pnas.0807859105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom P.M., Fiedler T., Grishchuk E.L., Nicastro D., West R.R., and McIntosh J.R.. 2009. Kinesin-8 from fission yeast: a heterodimeric, plus-end–directed motor that can couple microtubule depolymerization to cargo movement. Mol. Biol. Cell. 20:963–972. 10.1091/mbc.E08-09-0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudimchuk N., Vitre B., Kim Y., Kiyatkin A., Cleveland D.W., Ataullakhanov F.I., and Grishchuk E.L.. 2013. Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 15:1079–1088. 10.1038/ncb2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurden M.D., Holland A.J., van Zon W., Tighe A., Vergnolle M.A., Andres D.A., Spielmann H.P., Malumbres M., Wolthuis R.M., Cleveland D.W., and Taylor S.S.. 2010. Cdc20 is required for the post-anaphase, KEN-dependent degradation of centromere protein F. J. Cell Sci. 123:321–330. 10.1242/jcs.062075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajeri V.A., Stewart A.M., Moore L.L., and Padilla P.A.. 2008. Genetic analysis of the spindle checkpoint genes san-1, mdf-2, bub-3 and the CENP-F homologues hcp-1 and hcp-2 in Caenorhabditis elegans. Cell Div. 3:6 10.1186/1747-1028-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A., Silljé H.H., and Nigg E.A.. 2006. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 25:5504–5515. 10.1038/sj.emboj.7601426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S.V., Vergnolle M.A., Hussein D., Wozniak M.J., Allan V.J., and Taylor S.S.. 2005. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J. Cell Sci. 118:4889–4900. 10.1242/jcs.02614 [DOI] [PubMed] [Google Scholar]
- Hussein D., and Taylor S.S.. 2002. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 115:3403–3414. [DOI] [PubMed] [Google Scholar]
- Hyman A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., and Mitchison T.. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485. [DOI] [PubMed] [Google Scholar]
- Jiang K., and Akhmanova A.. 2011. Microtubule tip-interacting proteins: a view from both ends. Curr. Opin. Cell Biol. 23:94–101. 10.1016/j.ceb.2010.08.008 [DOI] [PubMed] [Google Scholar]
- Jordan M.A., Margolis R.L., Himes R.H., and Wilson L.. 1986. Identification of a distinct class of vinblastine binding sites on microtubules. J. Mol. Biol. 187:61–73. 10.1016/0022-2836(86)90406-7 [DOI] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Kitamura Y., and Tanaka T.U.. 2007. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 21:3319–3330. 10.1101/gad.449407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononova O., Kholodov Y., Theisen K.E., Marx K.A., Dima R.I., Ataullakhanov F.I., Grishchuk E.L., and Barsegov V.. 2014. Tubulin bond energies and microtubule biomechanics determined from nanoindentation in silico. J. Am. Chem. Soc. 136:17036–17045. 10.1021/ja506385p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Winkfein R.J., Mack G., Rattner J.B., and Yen T.J.. 1995. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 130:507–518. 10.1083/jcb.130.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-T., Rattner J.B., Jablonski S.A., and Yen T.J.. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53. 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., and Stock J.. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- McIntosh J.R., Grishchuk E.L., Morphew M.K., Efremov A.K., Zhudenkov K., Volkov V.A., Cheeseman I.M., Desai A., Mastronarde D.N., and Ataullakhanov F.I.. 2008. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 135:322–333. 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., Volkov V., Ataullakhanov F.I., and Grishchuk E.L.. 2010. Tubulin depolymerization may be an ancient biological motor. J. Cell Sci. 123:3425–3434. 10.1242/jcs.067611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., O’Toole E., Zhudenkov K., Morphew M., Schwartz C., Ataullakhanov F.I., and Grishchuk E.L.. 2013. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J. Cell Biol. 200:459–474. 10.1083/jcb.201209154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Arendt K., Old W.M., Houel S., Renganathan K., Eichelberger B., Resing K.A., and Ahn N.G.. 2011. IsoformResolver: A peptide-centric algorithm for protein inference. J. Proteome Res. 10:3060–3075. 10.1021/pr200039p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L.L., and Roth M.B.. 2001. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153:1199–1208. 10.1083/jcb.153.6.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores C.A., and Milligan R.A.. 2008. Visualisation of a kinesin-13 motor on microtubule end mimics. J. Mol. Biol. 377:647–654. 10.1016/j.jmb.2008.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan K.L., Pooley R., Miller P.M., Kaverina I., and Bader D.M.. 2009. Murine CENP-F regulates centrosomal microtubule nucleation and interacts with Hook2 at the centrosome. Mol. Biol. Cell. 20:4790–4803. 10.1091/mbc.E09-07-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musinipally V., Howes S., Alushin G.M., and Nogales E.. 2013. The microtubule binding properties of CENP-E’s C-terminus and CENP-F. J. Mol. Biol. 425:4427–4441. 10.1016/j.jmb.2013.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq J.D., Cabantous S., Tran T., Terwilliger T.C., and Waldo G.S.. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88. 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- Perkins S.J. 1986. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur. J. Biochem. 157:169–180. 10.1111/j.1432-1033.1986.tb09653.x [DOI] [PubMed] [Google Scholar]
- Pham T.V., Piersma S.R., Warmoes M., and Jimenez C.R.. 2010. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics. 26:363–369. 10.1093/bioinformatics/btp677 [DOI] [PubMed] [Google Scholar]
- Pooley R.D., Moynihan K.L., Soukoulis V., Reddy S., Francis R., Lo C., Ma L.J., and Bader D.M.. 2008. Murine CENPF interacts with syntaxin 4 in the regulation of vesicular transport. J. Cell Sci. 121:3413–3421. 10.1242/jcs.032847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., and Asbury C.L.. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875. 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos U.P. 1976. Light and electron microscopy of rat kangaroo cells in mitosis. Chromosoma. 54:363–385. 10.1007/BF00292816 [DOI] [PubMed] [Google Scholar]
- Schaar B.T., Chan G.K., Maddox P., Salmon E.D., and Yen T.J.. 1997. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139:1373–1382. 10.1083/jcb.139.6.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980. 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., and Mann M.. 2007. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Siegel L.M., and Monty K.J.. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 112:346–362. 10.1016/0926-6585(66)90333-5 [DOI] [PubMed] [Google Scholar]
- Skiniotis G., Cochran J.C., Müller J., Mandelkow E., Gilbert S.P., and Hoenger A.. 2004. Modulation of kinesin binding by the C-termini of tubulin. EMBO J. 23:989–999. 10.1038/sj.emboj.7600118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin L.J., Guimaraes G.J., and Deluca J.G.. 2011. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore–microtubule attachment in mitosis. Mol. Biol. Cell. 22:759–768. 10.1091/mbc.E10-08-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., and Davis T.N.. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723. 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toralová T., Susor A., Nemcová L., Kepková K., and Kanka J.. 2009. Silencing CENPF in bovine preimplantation embryo induces arrest at 8-cell stage. Reproduction. 138:783–791. 10.1530/REP-09-0234 [DOI] [PubMed] [Google Scholar]
- VandenBeldt K.J., Barnard R.M., Hergert P.J., Meng X., Maiato H., and McEwen B.F.. 2006. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr. Biol. 16:1217–1223. 10.1016/j.cub.2006.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Wan X., Cheerambathur D., Gassmann R., Suzuki A., Lawrimore J., Desai A., and Salmon E.D.. 2013. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 202:735–746. 10.1083/jcb.201304197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle M.A., and Taylor S.S.. 2007. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 17:1173–1179. 10.1016/j.cub.2007.05.077 [DOI] [PubMed] [Google Scholar]
- Volkov V.A., Zaytsev A.V., Gudimchuk N., Grissom P.M., Gintsburg A.L., Ataullakhanov F.I., McIntosh J.R., and Grishchuk E.L.. 2013. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc. Natl. Acad. Sci. USA. 110:7708–7713. 10.1073/pnas.1305821110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V.A., Zaytsev A.V., and Grishchuk E.L.. 2014. Preparation of segmented microtubules to study motions driven by the disassembling microtubule ends. J. Vis. Exp. 2014:51150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684. 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.M., Asfahani R., Carroll P., Bicknell L., Lescai F., Bright A., Chanudet E., Brooks A., Christou-Savina S., Osman G., et al. 2015. The kinetochore protein, CENPF, is mutated in human ciliopathy and microcephaly phenotypes. J. Med. Genet. 52:147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten M.D., Suter M.M., Littman D.R., and Kirschner M.W.. 1974. Properties of the depolymerization products of microtubules from mammalian brain. Biochemistry. 13:5529–5537. 10.1021/bi00724a012 [DOI] [PubMed] [Google Scholar]
- Welburn J.P., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., Yates J.R. III, and Cheeseman I.M.. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Wang H.W., Avila-Sakar A., Drubin D.G., Nogales E., and Barnes G.. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 440:565–569. 10.1038/nature04409 [DOI] [PubMed] [Google Scholar]
- Williams R.C. Jr., and Detrich H.W. III. 1979. Separation of tubulin from microtubule-associated proteins on phosphocellulose. Accompanying alterations in concentrations of buffer components. Biochemistry. 18:2499–2503. 10.1021/bi00579a010 [DOI] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz R., Goldstein L.S., and Cleveland D.W.. 1997. CENP-E is a plus end–directed kinetochore motor required for metaphase chromosome alignment. Cell. 91:357–366. 10.1016/S0092-8674(00)80419-5 [DOI] [PubMed] [Google Scholar]
- Yajima H., Ogura T., Nitta R., Okada Y., Sato C., and Hirokawa N.. 2012. Conformational changes in tubulin in GMPCPP and GDP-taxol microtubules observed by cryoelectron microscopy. J. Cell Biol. 198:315–322. 10.1083/jcb.201201161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Guo J., Chen Q., Ding C., Du J., and Zhu X.. 2005. Silencing mitosin induces misaligned chromosomes, premature chromosome decondensation before anaphase onset, and mitotic cell death. Mol. Cell. Biol. 25:4062–4074. 10.1128/MCB.25.10.4062-4074.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev A.V., Mick J.E., Maslennikov E., Nikashin B., DeLuca J.G., and Grishchuk E.L.. 2015. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. Cell. 26:1829–1844. 10.1091/mbc.E14-11-1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Chang K.H., He D., Mancini M.A., Brinkley W.R., and Lee W.H.. 1995. The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J. Biol. Chem. 270:19545–19550. 10.1074/jbc.270.33.19545 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.