Abstract
Background
Members of the genera Prevotella, Veillonella and Fusobacterium are the predominant culturable obligate anaerobic bacteria isolated from periodontal abscesses. When determining the cumulative number of clinical anaerobic isolates from periodontal abscesses, ambiguous or overlapping signals were frequently encountered in 16S rRNA gene sequencing chromatograms, resulting in ambiguous identifications. With the exception of the genus Veillonella, the high intra-chromosomal heterogeneity of rrs genes has not been reported.
Methods
The 16S rRNA genes of 138 clinical, strictly anaerobic isolates and one reference strain were directly sequenced, and the chromatograms were carefully examined. Gene cloning was performed for 22 typical isolates with doublet sequencing signals for the 16S rRNA genes, and four copies of the rrs-ITS genes of 9 Prevotella intermedia isolates were separately amplified by PCR, sequenced and compared. Five conserved housekeeping genes, hsp60, recA, dnaJ, gyrB1 and rpoB from 89 clinical isolates of Prevotella were also amplified by PCR and sequenced for identification and phylogenetic analysis along with 18 Prevotella reference strains.
Results
Heterogeneity of 16S rRNA genes was apparent in clinical, strictly anaerobic oral bacteria, particularly in the genera Prevotella and Veillonella. One hundred out of 138 anaerobic strains (72%) had intragenomic nucleotide polymorphisms (SNPs) in multiple locations, and 13 strains (9.4%) had intragenomic insertions or deletions in the 16S rRNA gene. In the genera Prevotella and Veillonella, 75% (67/89) and 100% (19/19) of the strains had SNPs in the 16S rRNA gene, respectively. Gene cloning and separate amplifications of four copies of the rrs-ITS genes confirmed that 2 to 4 heterogeneous 16S rRNA copies existed.
Conclusion
Sequence alignment of five housekeeping genes revealed that intra-species nucleotide similarities were very high in the genera Prevotella, ranging from 94.3–100%. However, the inter-species similarities were relatively low, ranging from 68.7–97.9%. The housekeeping genes rpoB and gyrB1 were demonstrated to be alternative classification markers to the species level based on intra- and inter-species comparisons, whereas based on phylogenetic tree rpoB proved to be reliable phylogenetic marker for the genus Prevotella.
Introduction
Periodontal abscess is a suppurative lesion that is associated with periodontal breakdown and localised pus in the gingival wall of the periodontal pocket. Gram-negative, strictly anaerobic bacteria and partially facultative anaerobes are the predominant microorganisms that cause periodontal disease in humans [1]. Previously, we reported that the predominant culturable obligate anaerobic pathogens isolated from periodontal abscesses are members of the genera Prevotella, Veillonella and Fusobacterium [2]. These genera also comprise a portion of the indigenous microbiota of the human and animal gastrointestinal tract and oral cavity [2, 3]. In addition to dental diseases, they play a role in extraoral infections, such as cellulitis, intra-abdominal, urogenital and osteoarticular infections and bacteraemia [4–8].
Because a large number of novel species or anaerobic genera have been isolated, proposed or reclassified as Gram-negative anaerobic rods, the taxonomy has changed significantly in the recent past [9–12]. For Prevotella in particular, additional in-depth studies focused on the gut microbiome and oral diseases have led to the recent identification of large numbers of novel species [13–17]. The comparison of 16S rRNA genes (rrs) by RFLP or sequencing is widely performed to estimate the evolutionary history, provide taxonomic classification and identify clinical isolates [18–23]. Although the rrs gene is accepted as the gold standard molecular clock, use of the rrs gene has been challenged by the diversity of multiple heterogeneous copies and the low resolution of closely related species [24].
When identifying the cumulative number of anaerobic strains in our studies, we have frequently encountered ambiguous or overlapping signals in rrs gene sequencing chromatograms, even with repeated single clone isolation and sequencing. The most reasonable explanation for these results is the heterogeneity of multiple rrs genes. The high intra-chromosomal heterogeneity of rrs genes has been reported for the genus Veillonella [19, 25], but no such phenomena have been reported in other clinically relevant anaerobic bacteria. In contrast, in genome databases, all four copies of rrs in Prevotella type strains are identical, including those of P. intermedia 17, Prevotella melaninogenica ATCC 25845, Prevotella denticola F0289 and Prevotella dentalis DSM 3688.
To investigate this discrepancy in more detail, we selected 138 clinical anaerobic strains isolated from periodontal abscesses to determine whether they contained multiple heterogeneous copies of rrs and to assess the extent of intra-genomic variation. In addition, to improve the identification and phylogenetic classification of clinical Prevotella isolates, we evaluated the suitability of five conserved genes, rpoB, dnaJ, recA, hsp60 and gyrB, as alternative identification markers and molecular clocks for Prevotella in 89 clinical isolates and 18 reference species from a genomic database. Conserved housekeeping genes, such as rpoB and hsp60, are more discriminative than the rrs gene and have been suggested as possible molecular clocks for bacterial phylogenetic studies [26, 27]. Other genes such as gyrB, recA and dnaJ provide additional information that supplement 16S rRNA gene sequence analysis and have also been suggested for phylogenetic studies and multilocus sequence analysis [27–31].
Materials and Methods
Clinical anaerobic strains and reference strains
Patients who suffered from periodontal abscesses routinely undertook anaerobic bacterial culture examination and antimicrobial susceptibility tests at the Department of Stomatology of Huashan Hospital (Shanghai, China). Isolation, culturing methods and partial description of the distribution of 100 strains were previously described [2]. In detail, the abscesses were drained after decontamination of the mucosa. A sterile inoculating loop was inserted into the deep area of the fistula for 20 seconds. The loop was then immediately inoculated onto pre-reduced culture medium, specifically Anaerobe Basal Agar (Oxoid, Oxoid Ltd, UK) plates supplemented with 5% sterile defibrinated sheep blood, using quadrate section streak methods. The culture medium was immediately incubated in GENbags (bioMérieux, France) at 37°C for 2–4 days of growth. Typical anaerobic colonies with a distinct morphology were selected, cultured and preserved in our laboratory for use in oxygen tolerance tests and antimicrobial susceptibility tests. Informed written consent was obtained from each patient. The present study is approved by the Ethics Committee from Huashan Hospital, Fudan University.
A total of 138 clinical, strictly anaerobic isolates preserved in the laboratory were re-inoculated and cultured. Each strain was purified by sub-culturing a single colony.
Genome sequences of 18 reference or type strains were obtained from the GenBank database (http://www.ncbi.nlm.nih.gov/genome/). The eighteen strains included P. intermedia 17, Prevotella melaninogenica ATCC 25845, Prevotella melaninogenica D18, Prevotella nigrescens ATCC 33563, Prevotella pallens ATCC 700821, Prevotella histicola F0411, Prevotella multiformis DSM 16608, Prevotella denticola F0289, Prevotella denticola CRIS 18C-A, Prevotella veroralis F0319, Prevotella oris F0302, Prevotella salivae DSM 15606, Prevotella dentalis DSM 3688, Prevotella buccae ATCC 33574, Prevotella buccae D17, Prevotella oulorum F0390, Candidatus Prevotella conceptionensis 9403948, Alloprevotella rava F0323 and Bacteroides fragilis 638R (out-group species).
If the genome had not been annotated, housekeeping genes were predicted from genome sequences using homologous genes of the most similar species identified by BLASTN. In most Prevotella type strains, two similar but distinct homologues of gyrB were identified using BLASTN, and the annotation was not uniform in previous reports. In our study, gyrB1 was defined as the higher-similarity homologue to the only gyrB gene of B. fragilis 638R. The GenBank/EMBL/DDBJ accession numbers of the sequences studied are listed in S2 Table.
Identification of the clinical strains and analysis of the intragenomic diversity of rrs genes
Genomic DNA from all isolates was extracted as previously described [2]. The rrs gene was amplified to almost full length using the universal primers 8FLP (5’-AGTTTGATCCTGGCTCAG-3’) and 1492RPL (5’-GGTTACTTGTTACGACTT-3’) or 1527R (5’-AGAAAGGAGGTGATCCAGCC-3’) with Premix Ex-Taq enzyme (Takara, Japan). For some isolates, if the 1527R primer yielded weak or no PCR product, the primer 1492R was used instead. The PCR with Premix Ex-Taq enzyme was performed using the following program: 95°C for 5 min; followed by 35 cycles consisting of 94°C for 30 s, 55°C for 30 s and 72°C for 2 min; with a final extension period at 72°C for 5 min. The amplification products were detected using electrophoresis and sequenced using a 3730xl DNA Analyzer (Applied Biosystems, USA) by the same amplification primers. Sequence chromatograms were carefully examined, and the numbers of overlapping nucleotides and positions of base loci were recorded.
In order to confirm whether the sequence result was enzyme related or not, the rrs gene of 8 isolates, P. denticola M04, P. buccae M27-2, P. intermedia M57-2, P. buccae M70, P. melaninogenica M71, P. intermedia M83, P. oris Y04, P. denticola Y78, and one reference strain P. melaninogenica ATCC 25845 was also amplified with Q5 high-fidelity polymerase (New England Biolabs, UK) and compared with the amplification by the Premix Ex-Taq enzyme. The PCR with Q5 enzyme was performed using the following program: 98°C for 2 min; followed by 35 cycles consisting of 98°C for 15 s, 55°C for 15 s and 72°C for 1 min; with a final extension period at 72°C for 5 min.
Clean rrs gene sequences from 16S ribosomal RNA sequences (Bacteria and Archaea) and the Nucleotide collection (nr/nt) databases were analysed using the BLASTN program. Identification at the species and genus level was defined at rrs sequence similarities of >99% and >97%, respectively, with the prototype strain sequences in the databases [32]. For isolates with ambiguous signals in direct sequencing, rrs sequences were obtained from gene cloning as described below.
rrs gene cloning
To verify the rrs gene heterogeneity observed in direct sequence chromatograms, characterised by overlapping signals only in or from certain positions of the gene, rrs gene cloning was performed in 15 typical clinical Prevotella isolates that had the greatest number of overlapping chromatogram signals and 13 unidentifiable isolates (Table 1) that had mixed rrs sequencing signals. The rrs genes were re-amplified using Q5 high-fidelity polymerase (New England Biolabs, UK) by 8FLP, 1492RPL and 1527R primers under conditions described above, purified by QIAquick PCR Purification kit (Qiagen, Germany) according to the manufacturer’s instructions, inserted into a cloning vector and transformed into E. coli DH5α competent cells using the pEASY-Blunt Zero Cloning Kit (Transgen Biotech, China) according to the manufacturer’s instructions. For each strain, clones between 8 to 14 were isolated and sequenced. Alloprevotella rava was previously classified as Prevotella oral taxon 302. Although it has been separated from the genus Prevotella, this strain was analysed together with the other Prevotella spp. in this study. Sequences from each clone were compared with the direct sequencing chromatograms of the isolates. Those clones that had identical rrs sequences were counted and defined as one copy. We used copy A, B, C and D to designate different copies of rrs genes and ranked by numbers of clones.
Table 1. 16S rRNA gene cloning of 22 Prevotella spp. clinical isolates and four copies of rrs sequencing of 9 Prevotella intermedia clinical isolates.
| Specific colonies/total colonies | ||||
|---|---|---|---|---|
| Strains | Copy A | Copy B | Copy C | Copy D |
| P. intermedia HJX081-2 | 6/9 | 3/9 | - | - |
| P. intermedia HJM050 | 6/10 | 4/10 | - | - |
| P. intermedia HJM056 | 6/9 | 3/9 | - | - |
| P. intermedia HJM057-2 | 4/8 | 3/8 | 1/8 | - |
| P. intermedia HJM069 | 5/11 | 4/11 | 2/11 | - |
| P. intermedia HJM061 | 5/8 | 2/8 | 1/8 | - |
| P. intermedia HJH14 | 4/8 | 4/8 | - | - |
| P. intermedia HJH16 | 5/8 | 3/8 | - | - |
| P. buccae HJM027-2 | 5/8 | 2/8 | 1/8 | - |
| P. buccae HJM070 | 4/8 | 3/8 | 1/8 | - |
| P. denticola HJX050 | 6/8 | 1/8 | 1/8 | - |
| P. denticola HJX078 | 4/8 | 3/8 | 1/8 | - |
| P. melaninogenica HJH25 | 8/14 | 3/14 | 3/14 | - |
| P. nigrescens HJH18 | 4/8 | 2/8 | 1/8 | 1/8 |
| P. nigrescens HJX026 | 4/8 | 3/8 | 1/8 | - |
| P. nigrescens HJX052-2 | 8/12 | 4/12 | - | - |
| P. nigrescens HJM025 | 5/8 | 3/8 | - | - |
| P. nigrescens HJM032 | 7/11 | 4/11 | - | - |
| P. nigrescens HJM066 | 5/8 | 3/8 | - | - |
| P. nigrescens HJX046 | 4/8 | 4/8 | - | - |
| P. oris HJH23 | 3/8 | 2/8 | 2/8 | 1/8 |
| Alloprevotella rava HJM063 | 3/8 | 2/8 | 2/8 | 1/8 |
| Four copies of rrs | Copy A | Copy B | Copy C | |
| P. intermedia HJX081-2 | rrs-1, rrs-2 | rrs-3, rrs-4 | - | - |
| P. intermedia HJM050 | rrs-1, rrs-3 | rrs-2, rrs-4 | - | - |
| P. intermedia HJM056 | rrs-2, rrs-4 | rrs-1, rrs-3 | - | - |
| P. intermedia HJM057-2 | rrs-2, rrs-3 | rrs1 | rrn-4 | - |
| P. intermedia HJM069 | rrs-4 | rrs-2 | rrs-1 | rrs-3 |
| P. intermedia HJX070 | rrs-1, rrs-3, rrs-4 | rrs-2 | - | - |
| P. intermedia HJX057 | rrs-2, rrs-3 | rrs1 | rrn-4 | - |
| P. intermedia HJM034 | rrs-1, rrs-4 | rrs-2, rrs-3 | - | - |
| P. intermedia HJX080 | rrs-1, rrs-2 | rrs-3, rrs-4 | - | - |
Sequencing four copies of rrs-ITS genes of P. intermedia
P. intermedia has 4 copies of the rrs-ITS genes in its genome, which are highly similar. However, the flanking regions of the 4 genes are distinct. Four primers, Inter_rrn1-F, Inter_rrn2-F, Inter_rrn3-F and Inter_rrn4-F (S1 Table) specifically targeting the flanking regions of the four copies of the rrs sequences and one universal primer 23S_1-60-R (S1 Table) targeting the rrs-ITS region were designed based on the genome sequence of P. intermedia 17. Flanking region primers instead of universal primers can amplify each rrs-ITS genes separately and verify the intragenomic heterogeneity. Four copies of rrs-ITS genes were therefore amplified with Premix Ex-Taq enzyme (Takara, Japan) using Inter_rrn1-F, Inter_rrn2-F, Inter_rrn3-F, Inter_rrn4-F and 23S_1-60-R primers and the conditions are listed in S1 Table. Amplicons of approximately 2100 bp covering the entire length of the rrs and ITS regions in nine P. intermedia clinical isolates with typical overlapping chromatogram signals were sequenced. Four copies of rrs sequences were compared with the results of the direct sequencing chromatograms. For isolates which had cloning results, each rrs-ITS sequence was matched to copy A/B/C/D sequences from cloning and then a designation to copy A/B/C/D was made. For isolates which had no cloning results, copy A, B, C and D were used to designate different copies of rrs genes and ranked by copy numbers.
Conserved gene sequences and phylogenetic analysis
Five conserved housekeeping genes and the 16S rRNA gene in members of the Prevotella genus were selected for identification and phylogenetic analysis. A partial rpoB gene (approximately 2000 bp), hsp60 (approximately 600 bp), recA (approximately 650 bp), dnaJ (approximately 850 bp) and gyrB1 (approximately 1250 bp) were amplified from 89 clinical isolates of Prevotella using the degenerate primers and conditions listed in S1 Table. PCR was also performed with Premix Ex-Taq enzyme (Takara, Japan) and the conditions are listed in S1 Table. The amplicons were detected using electrophoresis in agarose gels (1.2% w/w) and sequenced using sequencing primers (S1 Table) with a 3730xl DNA Analyzer.
Consensus sequences for all genes and clones were assembled using Lasergene SeqMan II software (DNAStar, Inc., Madison, WI) with default parameters. For 16S rRNA genes that had indels in direct sequencing, the sequence of one clone of the gene was used for alignment and phylogenetic tree construction. Five conserved housekeeping genes and an approximately 1320-bp length of rrs (nucleotides 106–1421 based on the E. coli rrs numbering system) were aligned in MEGA5 using CLUSTAL W and corrected by manual inspection. A phylogenetic tree was constructed in MEGA v5.1 by the neighbour-joining method [33]. DNA polymorphism data, mean G+C content and phylogenetic calculations, including synonymous and non-synonymous substitutions, were performed using MEGA v5.1. A total of 1000 replicate bootstrap resampling analyses were performed to test the robustness of the nodes. Sequence similarities were corrected using the Jukes–Cantor correction. Comparison of inter-species similarity values for different genes was performed with a student’s t test using SPSS v13.0.
Results
Direct sequencing of rrs genes
Among the 138 clinical strains isolated from periodontal abscesses, 125 strains were identified by direct sequencing of rrs genes; the remaining 13 strains, had continuous overlapping signals in their chromatograms, which impaired the sequencing results and were solved by gene cloning. Bacterial genera and species distributions are shown in Table 2, with the dominant isolates belonging to Prevotella (89 isolates, 66%), Veillonella (19 isolates, 14%) and Fusobacterium (12 isolates, 8.9%).
Table 2. Characteristics of intragenomic heterogeneous 16S rRNA genes in clinical anaerobic strains.
| Microorganisms | No. of strains | No. of strains with SNPs in 16S rRNA | No. of SNPs in 16S rRNA | No. strains with an indel in 16S rRNA |
|---|---|---|---|---|
| Prevotella genus | 89 | 67 | 1–18 | 0–7 |
| Prevotella intermedia | 29 | 22 | 1–12 | 0 |
| Prevotella nigrescens | 11 | 6 | 1–4 | 7 |
| Prevotella melaninogenica | 9 | 6 | 2–11 | 0 |
| Prevotella dentalis | 9 | 6 | 1–2 | 0 |
| Prevotella denticola | 9 | 8 | 4–12 | 0 |
| Prevotella buccae | 7 | 7 | 2–9 | 0 |
| Alloprevotella rava a | 3 | 3 | 11–18 | 0 |
| Prevotella oris | 2 | 2 | 6–11 | 0 |
| Prevotella pallens | 2 | 2 | 6–7 | 0 |
| Candidatus Prevotella conceptionensis | 2 | 2 | 11–12 | 0 |
| Prevotella salivae | 1 | 0 | - | 0 |
| Prevotella fusca | 1 | 0 | - | 0 |
| Prevotella histicola | 1 | 1 | 2 | 0 |
| Prevotella oulorum | 1 | 1 | 7 | 0 |
| Prevotella veroralis | 1 | 0 | - | 0 |
| Prevotella multiformis | 1 | 1 | 8 | 0 |
| Veillonella genus | 19 | 19 | 2–14 | 0–6 |
| Veillonella parvula/dispar | 15 | 15 | 3–14 | 6 |
| Veillonella sp. | 2 | 2 | 2–7 | 0 |
| Veillonella vegosae | 2 | 2 | 7–9 | 0 |
| Other genera | 30 | 14 | 1–11 | 0 |
| Fusobacterium nucleatum | 8 | 3 | 2–4 | 0 |
| Fusobacterium canifelinum | 4 | 3 | 2–3 | 0 |
| Bacteroides sp. | 3 | 2 | 2–8 | 0 |
| Actinomyces sp. | 3 | 0 | - | 0 |
| Campylobacter sp. | 2 | 0 | - | 0 |
| Dialister sp. | 2 | 1 | 1–2 | 0 |
| Peptostreptococcus sp. | 2 | 2 | 3 | 0 |
| Atopobium sp. | 1 | 0 | - | 0 |
| Bacillus sp. | 1 | 1 | 1 | 0 |
| Lachnoanaerobaculum sp. | 1 | 1 | 11 | 0 |
| Pseudoramibacter sp. | 1 | 0 | - | 0 |
| Shuttleworthia sp. | 1 | 0 | - | 0 |
| Selenomonas sp. | 1 | 1 | 2 | 0 |
| Total | 138 | 100 | 13 |
a Alloprevotella rava was previously classified as Prevotella oral taxon 302. Although it has been separated from the genus Prevotella, this strain was analysed together with the other Prevotella spp. in this study.
When the direct sequencing chromatogram data were manually inspected, surprisingly, 100 anaerobic isolates (72%) had doublet sequencing signals in multiple locations, and 13 isolates (9.4%) had continuous ambiguous signals starting from certain regions, suggesting nucleotide polymorphisms (SNPs) or insertion/deletions (Indels), respectively, in the rrs genes. Two examples of overlapping ambiguous signals in chromatograms indicating SNPs (Fig 1A) and indels (Fig 1B) are shown. In the genera Prevotella and Veillonella, 75% (67/89) and 100% (19/19) of the clinical isolates, respectively, had SNPs in the rrs gene. The numbers of the SNPs ranged from 0 to 12 (0%-0.8% divergence) and from 2 to 14 (0.13%-0.93% divergence), respectively. In addition, heterogeneity was also observed among the isolates of genera Fusobacterium, Bacteroides, Dialister, Peptostreptococcus, Bacillus, Lachnoanaerobaculum and Selenomonas (Table 2). Seven P. nigrescens and 6 Veillonella parvula/dispar isolates had Indels in the rrs genes.
Fig 1. 16S rRNA sequence trace data for the isolates Prevotella denticola HJX050 and Prevotella nigrescens HJX046, showing comparison sequence data for heterogeneous clones and direct sequencing with single ambiguous bases (A) or continuous ambiguous bases (B).
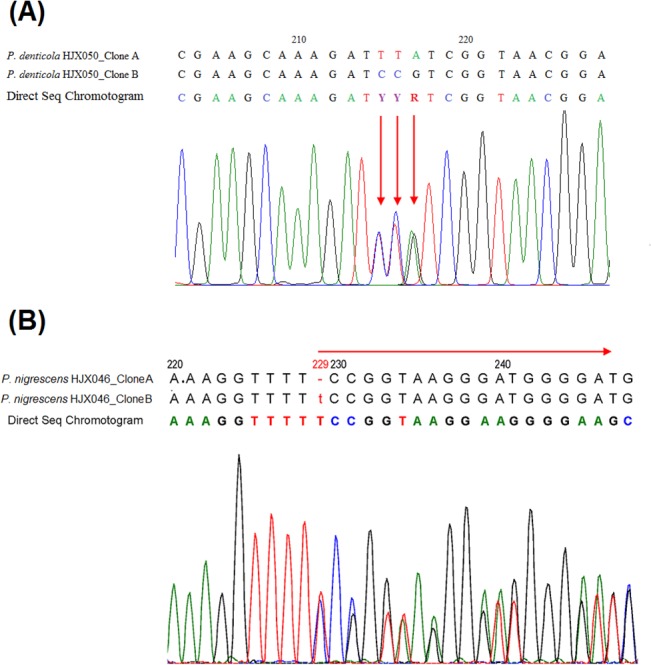
Prevotella nigrescens HJX046 clone A had a deletion at nucleotide position 229.
Amplification of rrs genes of 8 Prevotella spp. using Premix Ex-Taq enzyme and Q5 high-fidelity polymerase showed complete sequence match which implied that the doublet sequencing signals in direct sequencing were not enzyme related. In addition, the data showed that reference strain P. melaninogenica ATCC 25845 did not have any SNPs or Indels in the rrs genes, which was consistent with its four copies homogeneous rrs genes in the genome database (Genbank No. CP002122).
rrs cloning and sequencing
Among the 13 isolates that were unidentified by direct sequencing, 6 were identified as Veillonella parvula/dispar and 7 were identified as P. nigrescens by rrs cloning. All 6 Veillonella parvula/dispar clinical isolates had the polymorphic allele “CG/AAA” at position 1137–1138 in the rrs gene (data not shown), and all 7 P. nigrescens isolates had a polymorphic T insertion at position 229 (according to the E. coli rrs numbering system) (Fig 1B). These intragenomic indels in rrs could explain the continuous doublet sequencing signals in chromatograms observed in the direct sequencing results.
To confirm the heterogeneity of the rrs genes as observed by direct PCR, we also performed rrs gene cloning for 2 Prevotella buccae, 2 Prevotella denticola, 8 Prevotella intermedia, 1 Prevotella melaninogenica, 1 Prevotella oris, 1 Alloprevotella rava and 7 P. nigrescens clinical isolates. By sequencing 8 to 14 clones of each isolates, more than one type of rrs was discovered. All intragenomic SNPs or indels revealed by cloning matched with the direct sequencing results. The data also showed that the clones of different rrs copies were unequal (Table 1). This was partially because some of the copies could be identical (see below). For example, in P. intermedia HJM057-2, rrs-2 and rrs-3 are identical (marked as copy A). Not surprisingly, the copy A was sequenced 4 out of 8 times when selection of clones, while other copies were sequenced fewer times (Table 1).
Furthermore, four copies of the rrs-ITS genes of 9 isolates of P. intermedia were separately amplified and sequenced. Unlike direct sequencing, which sequenced a mixture of all rrs copies, amplification of rrs-ITS genes directly measured each copy of rrs-ITS by flanking region primers and no doublet sequencing signals were found.
These clinical isolates contained 2 to 4 different types of rrs among 4 gene copies (Table 1), which were different from the identical rrs genes observed in the reference strains. The SNPs in the four copies of rrs were located at the same nucleotide positions observed by direct sequencing, further verifying the existence of intragenomic heterogeneity among different rrs sequences. An example of polymorphic nucleotide alignment for the 3 sequencing methods is shown in Fig 1. Totally 9 isolates of P. intermedia were sequenced by rrs-ITS separation sequencing (Table 1). Four P. intermedia isolates, HJX081-2, HJM050, HJM056 and HJM057-2, showed the same numbers of different types of rrs by both methods. For the isolate P. intermedia HJM069, rrs-ITS sequencing showed that it has 4 different copies of rrs genes, but only 3 different copies were revealed by selection of clones (Table 1). The discrepancy could be due to the limitation to only a few numbers of clones.
Conserved genes of Prevotella
Partial sequences of five highly conserved protein-coding genes, hsp60, recA, dnaJ, gyrB1 and rpoB, together with rrs were analysed and compared in 89 clinical isolates of Prevotella, 17 type strains of 14 Prevotella species and Bacteroides fragilis 638R as the out-group (S2 Table). Data from three Alloprevotella rava isolates formerly classified as Prevotella sp. Taxon 302 were preserved in the study but excluded from analysis. The sequences used for alignments and phylogenetic tree construction were 546 bp, 600 bp, 777 bp, 1107 bp, 2028 bp and 1320 bp in length for hsp60, recA, dnaJ, gyrB1, rpoB and 16S rRNA, respectively.
For all genes, the intra-species comparisons demonstrated high similarities among species where more than one strain was available, except for P. melaninogenica. Percentage sequence similarities based on pairwise comparisons for all six genes were analysed (S3 Table). The ranges of intra-species nucleotide similarities for hsp60, recA, dnaJ, gyrB1, rpoB and rrs were 94.3–100% (mean±SEM: 98.6±1.1%), 90.8–100% (98.1±1.5%), 94.9–100% (98.3±0.97%), 95.3–100% (98.5±1.1%), 93.5–100% (98.5±1.5%) and 98.6–100% (99.5±0.31%), respectively, if P. melaninogenica was divided and analysed as two subspecies as described below. The ranges of inter-species similarities for hsp60, recA, dnaJ, gyrB1, rpoB and rrs were 68.7–91.6% (mean±SEM: 79.3±4.2%), 67.7–95.3% (77.0±5.5%), 59.7–97.9% (75.5±5.6%), 69.6–90.3% (77.6±4.3%), 70.1–89.6% (79.0±4.3%), and 85.5–98.3% (91.4±2.7%) (Table 3), respectively. All inter-species similarity values for hsp60, recA, dnaJ, gyrB1 and rpoB were significantly lower than for rrs (P<0.001), and parsimony-informative sites were higher than for rrs (23.9%), resulting in greater discriminatory power in species identification compared with 16S rRNA. The average dN/dS values are shown in Table 3. All values are less than 0.2, indicating that these genes are under purifying selection pressure.
Table 3. Analysis of 106 hsp60, recA, dnaJ, gyrB1, rpoB and 16S rRNA gene sequences from Prevotella.
| Sequence information | hsp60 | recA | dnaJ | gyrB1 | rpoB | 16S rRNA |
|---|---|---|---|---|---|---|
| No. of nucleotide sites | 546 | 600 | 777 | 1107 | 2028 | 1320 |
| Mean G+C content (mol%) | 47.10% | 51.30% | 52.70% | 50.50% | 46.70% | 55.50% |
| No. of polymorphic sites | 246 | 310 | 481 | 511 | 1017 | 323 |
| No. of parsimony-informative sites | 241 (44.1%) | 299 (49.8%) | 465 (59.8%) | 506 (45.7%) | 918 (45.3%) | 315 (23.9%) |
| No. of nucleotide differences: | ||||||
| Range | 0–171 | 0–194 | 0–314 | 0–337 | 0–650 | 0–183 |
| Mean±SEM | 97.0±5.3 | 115.9±6.6 | 162.4±5.8 | 221.0±6.5 | 391.3±10.0 | 110.1±6.4 |
| Intra-species±SD | 7.6±6.0 | 11.3±9.2 | 13.1±7.57 | 16.6±11.7 | 30.7±30.8 | 6.6±4.1 |
| Inter-species±SD | 113.0±22.7 | 138.0±33.1 | 190.4±43.5 | 248.5±47.9 | 425.1±87.0 | 113.0±36.1 |
| Jukes–Cantor distance (d): | ||||||
| Overall mean±SEM | 0.209±0.014 | 0.232±0.016 | 0.389±0.023 | 0.239±0.008 | 0.229±0.007 | 0.091±0.006 |
| Transition:transversion ratio (R) | 1.8 | 1.85 | 1.26 | 1.65 | 1.82 | 1.49 |
| dS* | 1.225±0.064 | 0.990±0.116 | 0.782±0.113 | 1.103±0.117 | 1.431±0.052 | NA |
| dN* | 0.052±0.009 | 0.056±0.008 | 0.127±0.011 | 0.055±0.007 | 0.063±0.005 | NA |
| dN/dS | 0.042 | 0.057 | 0.162 | 0.05 | 0.044 | NA |
*Synonymous substitutions per site (dS) and non-synonymous substitutions per site (dN) (means±SEM) were determined by the Nei–Gojobori method using the Jukes–Cantor distance. NA, not applicable.
Although recA and dnaJ were the most informative genes (having the highest parsimony-informative sites), they had difficulty in discriminating several pairs of species. The dnaJ sequence similarities for the P. intermedia/P. nigrescens and the P. multiformis/P. denticola pairs were uncommonly high and even higher than some intra-species similarity levels for these species (S3 Table, dnaJ sheet). Similar results were observed in the P. intermedia/P. nigrescens and P. fusca/P. melaninogenica pairs for recA (S3 Table, recA sheet) and in the P. multiformis/P. denticola pair for hsp60 (S3 Table, hsp60 sheet). Nucleotide sequence similarities for gyrB1 and rpoB in particular had sufficient differences between the intra- and inter-species similarities for the accurate classification of Prevotella species, making them suitable as classification markers (S3 Table). We propose that thresholds of 93% and 91% for gyrB1 and rpoB, respectively, could be used for species classification in the Prevotella genus. The thresholds were set as the average similarity values of the maximum inter-species and the minimum intra-species.
In agreement with the alignment analysis, the phylogenetic trees constructed for each gene were very stable at the intra-species level. Isolates from the same species, except P. melaninogenica, clustered tightly together with very high bootstrap values (>95%) (Figs 2, 3, 4 and S1, S2, S3 Figs). Nevertheless, 9 clinical P. melaninogenica isolates and 2 type strains (ATCC25845 and D18) clearly clustered into 3 branches, with the first branch including P. melaninogenica HJM071-2 only, a second branch having three isolates and the third branch for the other 5 clinical isolates and the 2 type strains. The sequence similarities between these branches of hsp60, recA, dnaJ, gyrB1 and rpoB were 89–94.0%, 87.5–95.8%, 84.1–88.9%, 89.3–94.5% and 91.3–94.2%, respectively (data not shown), which are between the inter- and intra-species similarities of Prevotella. Because the phylogenetic relationships are stable (bootstrap >80%) in all trees except recA, we reclassified these isolates into 2 sub-species, specifically clades A and B, and separated strain P. melaninogenica HJM071-2 alone (Figs 2, 3, 4 and S1, S2 Figs).
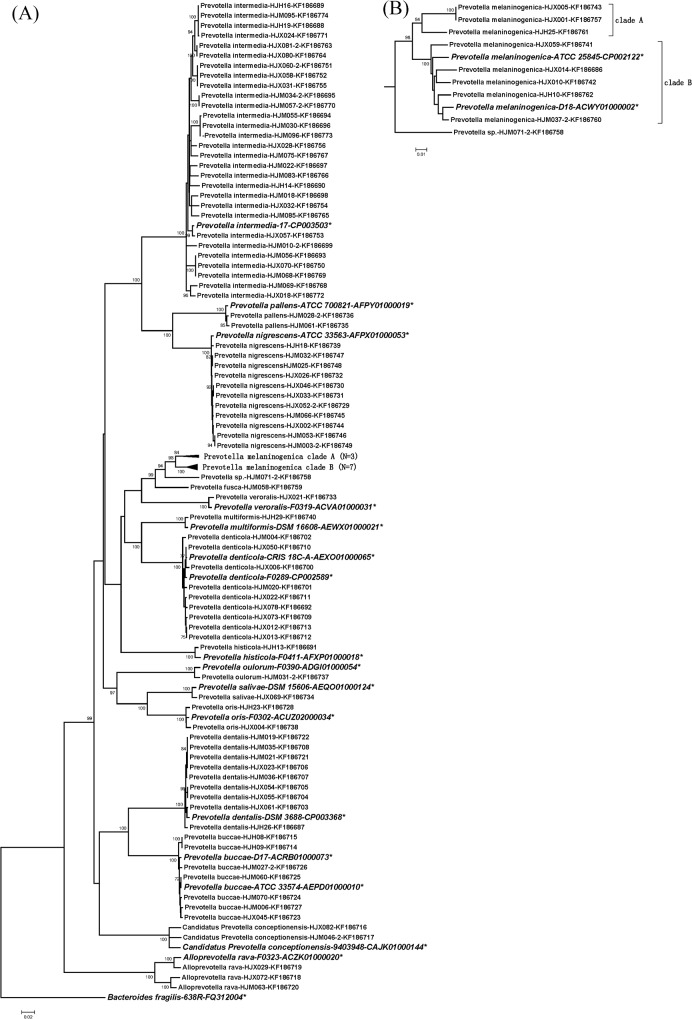
Fig 2. Phylogenetic trees based on the concatenated sequences of the gyrB1 gene.
Detailed phylogenetic trees of P. melaninogenica strains are shown in Figure (B). The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
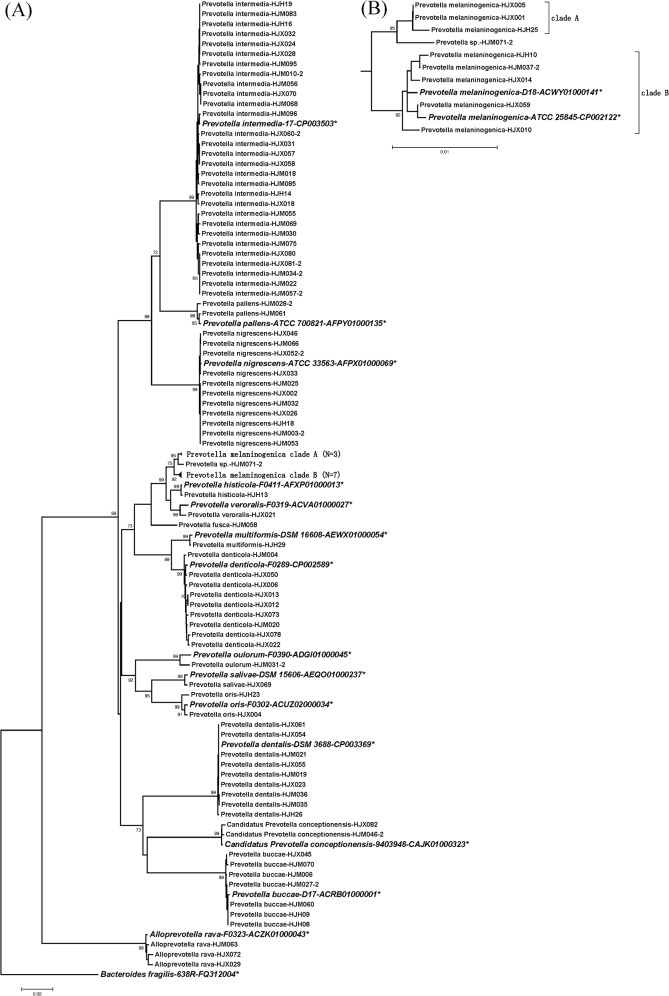
Fig 3. Phylogenetic trees based on the concatenated sequences of the 16S rRNA gene.
Detailed phylogenetic trees of P. melaninogenica strains are shown in Figure (B). The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
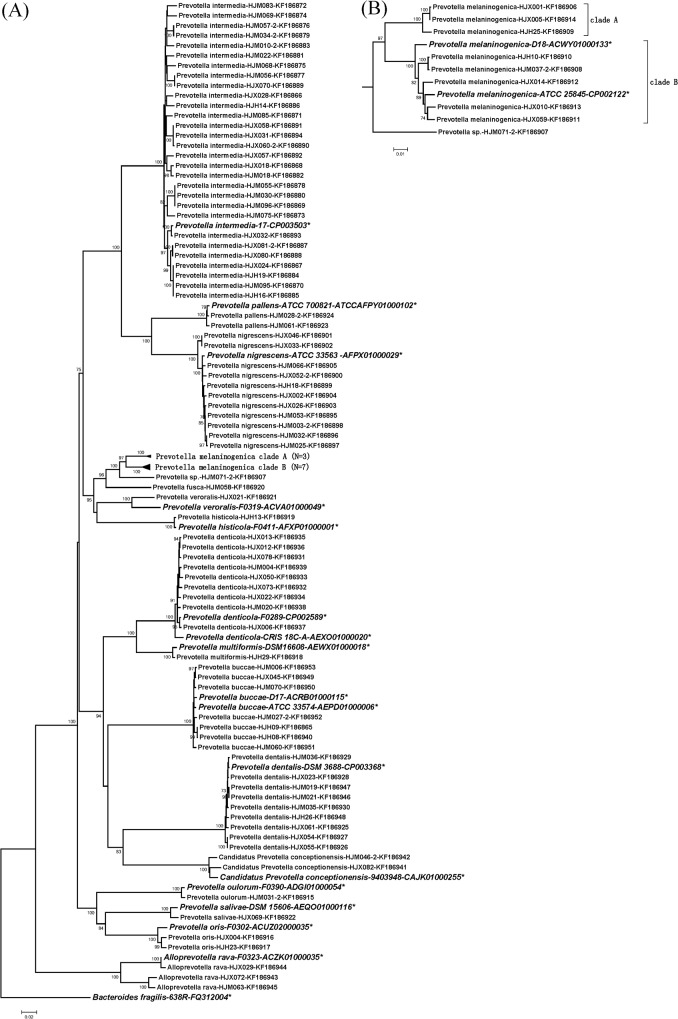
Fig 4. Phylogenetic trees based on the concatenated sequences of the rpoB gene.
Detailed phylogenetic trees of P. melaninogenica strains are shown in Figure (B). The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
At the genus level, the phylogenetic trees constructed for different genes gave similar results. P. intermedia, P. nigrescens and P. pallens were consistently clustered in one group for all genes (all bootstrap values >94%). P. oris, P. salivae and P. oulorum had smaller genetic distances and thus tended to cluster together according to the gyrB1 (Fig 2), 16S rRNA (Fig 3), rpoB (Fig 4) and hsp60 (S1 Fig) genes. P. melaninogenica, P. fusca, P. veroralis and P. histicola were closely related in the hsp60 (S1 Fig), 16S rRNA (Fig 3) and rpoB (Fig 4) trees. There was a small difference between genes regarding the phylogenetic position of P. fusca, as this strain was close to P. melaninogenica in all gene trees except the 16S rRNA tree (Fig 3). Another group consisted of P. buccae, P. dentalis and Candidatus Prevotella conceptionensis, but this group was rather unstable. P. multiformis and P. denticola were closely clustered in all trees except for the recA tree (S3 Fig). The grouping results could explain the uncommonly high sequence similarity values for some of the aforementioned genes for the P. intermedia/P. nigrescens, P. fusca/P. melaninogenica and P. multiformis/P. denticola pairs (S3 Table, recA and dnaJ sheets), suggesting that gene recombination was more likely to have occurred in closely grouped taxa. The phylogenetic analysis also indicated a higher evolutionary rate for the five housekeeping gene sequences compared to the rrs gene sequence.
Discussion
The Prevotella genus was systematically reclassified from Bacteroides in 1990 by Shah and Collins [34]. With increasing discoveries of novel Prevotella species and in-depth studies of intestinal and oral flora, accurate classification and identification of these causative strictly anaerobic pathogens has gradually received greater attention [35, 36]. Currently, clinical identification of anaerobic bacteria mainly relies on conventional biochemical phenotyping [37], rrs sequencing [21, 23] or bacterial fingerprint identification by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS) [38–41]. Sequence analysis of rrs remains inevitable in rrs-based methods, gold or reference methods and comparison studies as well as for difficult-to-identify isolates.
In our study, many clinical opportunistic anaerobic pathogens were shown to have intra-species rrs heterogeneity, contrasting with the 4 homogeneous rrs genes in the genomes of the reference strains. With the exception of Veillonella [19, 25], this is the first report of this variability. We believe the discrepancy between the reference strains may comprise two aspects. The data showed that 22 of 29 P. intermedia had SNPs in their 16S rRNA genes (Table 2). In other words, there are still a certain number of isolates whose rrs genes are homogeneous. Second, differences in geography, site of infection and antibiotic usage could contribute to the discrepancy. The percentage and the type (Indel or SNP) of heterozygosity of rrs genes will directly affect the accuracy and feasibility of the classification and identification of strains. In this study, the intra-genomic 16S rRNA sequence similarity of most heterozygous Prevotella strains, such as P. denticola, P. conceptionensis and P. intermedia, was only 99.1%. For Alloprevotella rava stains, the similarity was even lower. In diagnostic practice, continuous doublet signals caused by indels, noted in our study in 7 P. nigrescens and 6 Veillonella parvula/dispar isolates, are a challenge not only in terms of the feasibility of direct PCR-sequencing methods and automated clinical instruments but also to the researcher who might repeatedly suspect contamination of the strains. If indels appear to be stable at certain positions, a compromise could be to use other shorter viable regions of rrs and avoid heterogeneous regions. The identification accuracy of other commonly utilised microbiological methods, such as PCR-TTGE and PCR-RFLP, would also be affected by heterozygosity, with results consisting of multiple erroneous bands. For microbial community methods, such as PGGE and 16S rDNA high-throughput sequencing, the heterogeneity of rrs would result in overestimation of the diversity of the population, particularly for those strains with higher intra-species diversity than inter-species diversity [42]. Heterogeneity of rrs genes appears to be correlated with the complexity of the microbiota environment and may contribute to survival under stress [43]. Periodontal abscesses in clinical patients usually have multiple bacterial infections and are under stress from antibiotics or host immune responses. It appears rational to propose that clinical strains isolated from microflora infections such as periodontitis are more likely evolve or mutate their rrs genes to adapt to the environment [25, 44–46]. This study performed rrs cloning to confirm the heterogeneity of the rrs gene. The method could not determine accurate copy numbers, whether there were 4 copies of rrs, such as in Prevotella intermedia, or greater or fewer copies. Selection of clones satisfies the binomial distribution; therefore, limitation to only a few clones cannot predict the distribution of different copies in the isolates. Genomic sequencing and analysis is needed to study the exact numbers and the distribution of rrs genes.
In this study, in Prevotella, 75% of 89 isolates exhibited intra-genomic heterogeneity, which included multiple species. Isolates of the genera Streptomyces and Aeromonas were previously shown to possess only 21% and 6.9% intra-genomic heterogeneity for rrs, respectively, based on sequencing patterns and RFLP analysis [43, 47]. Although different methods were used, Prevotella and the clinically relevant anaerobes examined here demonstrated the highest occurrences of intra-genomic heterogeneity compared with these previously described genera.
Previously Sakamoto et al. analysed hsp60 sequences in 48 Prevotella strains from 38 species [48] and concluded that hsp60 could be an alternative phylogenetic marker in Prevotella. Our study analysed the recA, dnaJ, gyrB1, hsp60 and rpoB genes in a greater number of clinical isolates and type strain sequences, which permitted better evaluation and comparison of classification markers. Although hsp60 sequences could accurately classify most species of Prevotella, the high similarity of P. multiformis and P. denticola isolates impeded its accuracy in classification. Instead, gyrB1 and rpoB showed to be better markers for classification in Prevotella based on intra- and inter-species comparisons. However, based on phylogenetic tree rpoB proved to be reliable phylogenetic marker for the genus Prevotella.
We recognise several limitations to this study. All clinical strains were isolated from one clinical centre, and some species, such as P. fusca, comprised only a limited number of clinical isolates. Although we added certain type strains that were available from databases to the study, some findings and conclusions must be verified using additional clinical isolates and strains from other locations. Although we verified the homogeneous rrs genes of the only available reference strain, P. melaninogenica ATCC 25845, we could not test the others reference strains due to the limitation of resources. The homogeneous rrs genes of the others reference strains are needed to be further verified. Furthermore, because the nucleotide database for housekeeping genes such as gyrB1, rpoB and hsp60 is quite small, particularly for newly classified Prevotella species, identification and classification using these genes should be supported by a larger database. Hopefully, when the whole genome of a bacteria is sequenced, all housekeeping genes can be acquired simultaneously. With the expansion of the Human Microbiome Project and the advent of bacterial whole genomic sequencing, inadequate database problems will gradually be diminished. For instance, in this study, we did not amplify or sequence the housekeeping genes of any reference strains ourselves; instead, these genes were analysed and extracted from the genome database.
Conclusions
In conclusion, 16S rRNA gene heterogeneity was apparent in clinical, strictly anaerobic oral bacteria, particularly in the genera Prevotella and Veillonella, which could interfere with classification and identification methods based on 16S rRNA genes. The housekeeping gene sequences rpoB and gyrB1 were demonstrated to be alternative classification markers to the species level based on intra- and inter-species comparisons, whereas based on phylogenetic tree rpoB proved to be reliable phylogenetic marker for the genus Prevotella. We propose nucleotide similarity thresholds for species classification for rpoB and gyrB1 as 91%, and 93%, respectively.
Supporting Information
The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
(TIF)
The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
(TIF)
The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
(TIF)
(DOCX)
(DOCX)
Green, pink, blue, yellow and open-shaded rectangles indicate >50%, >60%, >70%, >80% and >90% sequence similarity, respectively. *** indicates that data are not available because there is only one strain in the genus.
(XLSX)
Acknowledgments
The work was supported all by the National Natural Science Foundation of China (81101226 and 81471987) and the Foundation of Fudan University.
Data Availability
The GenBank/EMBL/DDBJ accession numbers of the sequences studied are listed in S2 Table.
Funding Statement
The work was supported all by the National Natural Science Foundation of China 443 (81101226 and 81471987) and the Foundation of Fudan University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Robertson D, Smith AJ. The microbiology of the acute dental abscess. J Med Microbiol. 2009;58(Pt 2):155–62. Epub 2009/01/15. 10.1099/jmm.0.003517-058/2/155 [pii]. . [DOI] [PubMed] [Google Scholar]
- 2. Xie Y, Chen J, He J, Miao X, Xu M, Wu X, et al. Antimicrobial resistance and prevalence of resistance genes of obligate anaerobes isolated from periodontal abscesses. J Periodontol. 2014;85(2):327–34. 10.1902/jop.2013.130081 . [DOI] [PubMed] [Google Scholar]
- 3. Jaramillo A, Arce RM, Herrera D, Betancourth M, Botero JE, Contreras A. Clinical and microbiological characterization of periodontal abscesses. J Clin Periodontol. 2005;32(12):1213–8. Epub 2005/11/05. doi: CPE839 [pii] 10.1111/j.1600-051X.2005.00839.x . [DOI] [PubMed] [Google Scholar]
- 4. Alauzet C, Mory F, Teyssier C, Hallage H, Carlier JP, Grollier G, et al. Metronidazole resistance in Prevotella spp. and description of a new nim gene in Prevotella baroniae. Antimicrob Agents Chemother. 2010;54(1):60–4. Epub 2009/10/07. 10.1128/AAC.01003-09AAC.01003-09 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang JJ, Kwon TY, Seo MJ, Nam YS, Han CS, Lee HJ. 16S ribosomal RNA identification of Prevotella nigrescens from a case of cellulitis. Ann Lab Med. 2013;33(5):379–82. Epub 2013/09/05. 10.3343/alm.2013.33.5.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boo TW, Cryan B, O'Donnell A, Fahy G. Prosthetic valve endocarditis caused by Veillonella parvula. J Infect. 2005;50(1):81–3. Epub 2004/12/18. doi: S0163445303002238 [pii] 10.1016/j.jinf.2003.11.008 . [DOI] [PubMed] [Google Scholar]
- 7. Marriott D, Stark D, Harkness J. Veillonella parvula discitis and secondary bacteremia: a rare infection complicating endoscopy and colonoscopy? J Clin Microbiol. 2007;45(2):672–4. Epub 2006/11/17. doi: JCM.01633-06 [pii] 10.1128/JCM.01633-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhatti MA, Frank MO. Veillonella parvula meningitis: case report and review of Veillonella infections. Clin Infect Dis. 2000;31(3):839–40. Epub 2000/10/06. doi: CID994276 [pii] 10.1086/314046 . [DOI] [PubMed] [Google Scholar]
- 9. Sakamoto M, Suzuki M, Umeda M, Ishikawa I, Benno Y. Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology. 2002;52(3):841–9. 10.1099/ijs.0.01945-0 [DOI] [PubMed] [Google Scholar]
- 10. Downes J, Dewhirst FE, Tanner AC, Wade WG. Description of Alloprevotella rava gen. nov., sp. nov., isolated from the human oral cavity, and reclassification of Prevotella tannerae Moore et al. 1994 as Alloprevotella tannerae gen. nov., comb. nov. Int J Syst Evol Microbiol. 2013;63(Pt 4):1214–8. Epub 2012/07/04. 10.1099/ijs.0.041376-0ijs.0.041376-0 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kitahara M, Sakamoto M, Tsuchida S, Kawasumi K, Amao H, Benno Y, et al. Bacteroides stercorirosoris sp. nov. and Bacteroides faecichinchillae sp. nov., isolated from chinchilla (Chinchilla lanigera) faeces. Int J Syst Evol Microbiol. 2012;62(Pt 5):1145–50. Epub 2011/07/05. 10.1099/ijs.0.032706-0ijs.0.032706-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 12. Mashima I, Kamaguchi A, Miyakawa H, Nakazawa F. Veillonella tobetsuensis sp. nov., an anaerobic, gram-negative coccus isolated from human tongue biofilms. Int J Syst Evol Microbiol. 2013;63(Pt 4):1443–9. Epub 2012/07/31. 10.1099/ijs.0.042515-0ijs.0.042515-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 13. Takada K, Hayashi K, Sato Y, Hirasawa M. Prevotella dentasini sp. nov., a black-pigmented species isolated from the oral cavity of donkeys. Int J Syst Evol Microbiol. 2010;60(Pt 7):1637–9. Epub 2009/09/01. 10.1099/ijs.0.017020-0ijs.0.017020-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 14. Sakamoto M, Suzuki N, Okamoto M. Prevotella aurantiaca sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2010;60(Pt 3):500–3. Epub 2009/08/06. 10.1099/ijs.0.012831-0ijs.0.012831-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 15. Downes J, Wade WG. Prevotella fusca sp. nov. and Prevotella scopos sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2011;61(Pt 4):854–8. Epub 2010/05/25. 10.1099/ijs.0.023861-0ijs.0.023861-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 16. Hedberg ME, Israelsson A, Moore ER, Svensson-Stadler L, Wai SN, Pietz G, et al. Prevotella jejuni sp. nov., isolated from the small intestine of a child with coeliac disease. Int J Syst Evol Microbiol. 2013;63(Pt 11):4218–23. Epub 2013/06/25. 10.1099/ijs.0.052647-0ijs.0.052647-0 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alauzet C, Mory F, Carlier JP, Marchandin H, Jumas-Bilak E, Lozniewski A. Prevotella nanceiensis sp. nov., isolated from human clinical samples. Int J Syst Evol Microbiol. 2007;57(Pt 10):2216–20. Epub 2007/10/04. doi: 57/10/2216 [pii] 10.1099/ijs.0.65173-0 . [DOI] [PubMed] [Google Scholar]
- 18. Ciantar M, Newman HN, Wilson M, Spratt DA. Molecular identification of Capnocytophaga spp. via 16S rRNA PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 2005;43(4):1894–901. Epub 2005/04/09. doi: 43/4/1894 [pii] 10.1128/JCM.43.4.1894-1901.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchandin H, Teyssier C, Simeon De Buochberg M, Jean-Pierre H, Carriere C, Jumas-Bilak E. Intra-chromosomal heterogeneity between the four 16S rRNA gene copies in the genus Veillonella: implications for phylogeny and taxonomy. Microbiology. 2003;149(Pt 6):1493–501. Epub 2003/06/05. . [DOI] [PubMed] [Google Scholar]
- 20. Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, et al. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol. 2003;41(5):1996–2001. Epub 2003/05/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wellinghausen N, Kochem AJ, Disque C, Muhl H, Gebert S, Winter J, et al. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J Clin Microbiol. 2009;47(9):2759–65. Epub 2009/07/03. 10.1128/JCM.00567-09JCM.00567-09 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haag H, Locher F, Nolte O. Molecular diagnosis of microbial aetiologies using SepsiTest in the daily routine of a diagnostic laboratory. Diagn Microbiol Infect Dis. 2013;76(4):413–8. Epub 2013/06/12. 10.1016/j.diagmicrobio.2013.04.027S0732-8893(13)00266-6 [pii]. . [DOI] [PubMed] [Google Scholar]
- 23. Grif K, Heller I, Prodinger WM, Lechleitner K, Lass-Florl C, Orth D. Improvement of detection of bacterial pathogens in normally sterile body sites with a focus on orthopedic samples by use of a commercial 16S rRNA broad-range PCR and sequence analysis. J Clin Microbiol. 2012;50(7):2250–4. Epub 2012/05/04. 10.1128/JCM.00362-12JCM.00362-12 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, Gerz EA, et al. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 2010;76(12):3886–97. Epub 2010/04/27. 10.1128/AEM.02953-09AEM.02953-09 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Michon AL, Aujoulat F, Roudiere L, Soulier O, Zorgniotti I, Jumas-Bilak E, et al. Intragenomic and intraspecific heterogeneity in rrs may surpass interspecific variability in a natural population of Veillonella. Microbiology. 2010;156(Pt 7):2080–91. 10.1099/mic.0.038224-0 . [DOI] [PubMed] [Google Scholar]
- 26. Ko KS, Kuwahara T, Haehwa L, Yoon YJ, Kim BJ, Lee KH, et al. RNA polymerase beta-subunit gene (rpoB) sequence analysis for the identification of Bacteroides spp. Clin Microbiol Infect. 2007;13(1):48–54. Epub 2006/12/23. doi: CLM1553 [pii] 10.1111/j.1469-0691.2006.01553.x . [DOI] [PubMed] [Google Scholar]
- 27. Weng FY, Chiou CS, Lin PH, Yang SS. Application of recA and rpoB sequence analysis on phylogeny and molecular identification of Geobacillus species. J Appl Microbiol. 2009;107(2):452–64. Epub 2009/05/12. 10.1111/j.1365-2672.2009.04235.xJAM4235 [pii]. . [DOI] [PubMed] [Google Scholar]
- 28. Sakamoto M, Ohkuma M. Identification and classification of the genus Bacteroides by multilocus sequence analysis. Microbiology. 2011;157(Pt 12):3388–97. 10.1099/mic.0.052332-0 . [DOI] [PubMed] [Google Scholar]
- 29. Carrasco G, Valdezate S, Garrido N, Villalon P, Medina-Pascual MJ, Saez-Nieto JA. Identification, typing, and phylogenetic relationships of the main clinical Nocardia species in spain according to their gyrB and rpoB genes. J Clin Microbiol. 2013;51(11):3602–8. 10.1128/JCM.00515-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takeda K, Kang Y, Yazawa K, Gonoi T, Mikami Y. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J Med Microbiol. 2010;59(Pt 2):165–71. 10.1099/jmm.0.011346-0 . [DOI] [PubMed] [Google Scholar]
- 31. Alexandre A, Laranjo M, Young JP, Oliveira S. dnaJ is a useful phylogenetic marker for alphaproteobacteria. Int J Syst Evol Microbiol. 2008;58(Pt 12):2839–49. 10.1099/ijs.0.2008/001636-0 . [DOI] [PubMed] [Google Scholar]
- 32. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38(10):3623–30. Epub 2000/10/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. Epub 1987/07/01. . [DOI] [PubMed] [Google Scholar]
- 34. Shah HN, Collins DM. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol. 1990;40(2):205–8. Epub 1990/04/01. . [DOI] [PubMed] [Google Scholar]
- 35. Wade WG. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol. 2011;38 Suppl 11:7–16. 10.1111/j.1600-051X.2010.01679.x . [DOI] [PubMed] [Google Scholar]
- 36. Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M, et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. Journal of microbiological methods. 2009;79(3):266–71. 10.1016/j.mimet.2009.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez-Cavallini E, Vargas P, Rodriguez C, Quesada-Gomez C, Gamboa-Coronado MM. Phenotypic identification of over 1000 isolates of anaerobic bacteria recovered between 1999 and 2008 in a major Costa Rican hospital. Clin Microbiol Infect. 2011;17(7):1043–7. Epub 2011/07/05. 10.1111/j.1469-0691.2010.03419.x . [DOI] [PubMed] [Google Scholar]
- 38. Garner O, Mochon A, Branda J, Burnham CA, Bythrow M, Ferraro M, et al. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK((R)) MS system. Clin Microbiol Infect. 2014;20(4):335–9. Epub 2013/08/10. 10.1111/1469-0691.12317 . [DOI] [PubMed] [Google Scholar]
- 39. La Scola B, Fournier PE, Raoult D. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe. 2011;17(3):106–12. Epub 2011/06/16. doi: 0.1016/j.anaerobe.2011.05.010S1075-9964(11)00099-0 [pii] 10.1016/j.anaerobe.2011.05.010 . [DOI] [PubMed] [Google Scholar]
- 40. Wybo I, Soetens O, De Bel A, Echahidi F, Vancutsem E, Vandoorslaer K, et al. Species identification of clinical Prevotella isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50(4):1415–8. Epub 2012/02/04. 10.1128/JCM.06326-11JCM.06326-11 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagy E, Becker S, Kostrzewa M, Barta N, Urban E. The value of MALDI-TOF MS for the identification of clinically relevant anaerobic bacteria in routine laboratories. J Med Microbiol. 2012;61(Pt 10):1393–400. Epub 2012/06/16. 10.1099/jmm.0.043927-0jmm.0.043927-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 42. Sun DL, Jiang X, Wu QL, Zhou NY. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol. 2013;79(19):5962–9. Epub 2013/07/23. 10.1128/AEM.01282-13AEM.01282-13 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol. 1999;181(1):78–82. Epub 1998/12/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jensen S, Frost P, Torsvik VL. The nonrandom microheterogeneity of 16S rRNA genes in Vibrio splendidus may reflect adaptation to versatile lifestyles. FEMS microbiology letters. 2009;294(2):207–15. 10.1111/j.1574-6968.2009.01567.x . [DOI] [PubMed] [Google Scholar]
- 45. Lopez-Lopez A, Benlloch S, Bonfa M, Rodriguez-Valera F, Mira A. Intragenomic 16S rDNA divergence in Haloarcula marismortui is an adaptation to different temperatures. Journal of molecular evolution. 2007;65(6):687–96. 10.1007/s00239-007-9047-3 . [DOI] [PubMed] [Google Scholar]
- 46. Satokari RM, Vaughan EE, Akkermans AD, Saarela M, de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67(2):504–13. 10.1128/AEM.67.2.504-513.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graf J. Diverse restriction fragment length polymorphism patterns of the PCR-amplified 16S rRNA genes in Aeromonas veronii strains and possible misidentification of Aeromonas species. J Clin Microbiol. 1999;37(10):3194–7. Epub 1999/09/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sakamoto M, Ohkuma M. Usefulness of the hsp60 gene for the identification and classification of Gram-negative anaerobic rods. J Med Microbiol. 2010;59(Pt 11):1293–302. Epub 2010/07/31. 10.1099/jmm.0.020420-0jmm.0.020420-0 [pii]. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
(TIF)
The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
(TIF)
The trees were constructed by the neighbour-joining (NJ) method. The numbers at nodes indicate the percentage bootstrap values of 1000 replicates (>70%). Bars indicate the expected nucleotide substitutions per site. * represents reference strains.
(TIF)
(DOCX)
(DOCX)
Green, pink, blue, yellow and open-shaded rectangles indicate >50%, >60%, >70%, >80% and >90% sequence similarity, respectively. *** indicates that data are not available because there is only one strain in the genus.
(XLSX)
Data Availability Statement
The GenBank/EMBL/DDBJ accession numbers of the sequences studied are listed in S2 Table.