Abstract
Botulinum neurotoxins (BoNTs) are exceptionally potent inhibitors of neurotransmission, causing muscle paralysis and respiratory failure associated with the disease botulism. Currently, no drugs are available to counter intracellular BoNT poisoning. To develop effective medical treatments, cell-based assays provide a valuable system to identify novel inhibitors in a time- and cost-efficient manner. Consequently, cell-based systems including immortalized cells, primary neurons, and stem-cell derived neurons have been established. Stem cell-derived neurons are highly sensitive to BoNT intoxication and represent an ideal model to study the biological effects of BoNTs. Robust immunoassays are used to quantify BoNT activity and play a central role during inhibitor screening. In this review, we examine recent progress in physiologically relevant cell-based assays and high-throughput screening approaches for the identification of both direct and indirect BoNT inhibitors.
Keywords: Botulinum neurotoxin, Cell-based assays, Embryonic stem cells, Motor neurons, Drug discovery, High-throughput screening, High-content imaging, ELISA, MSD, Phenotypic screening
1. Introduction
Botulinum neurotoxins (BoNTs) are zinc metalloproteases that block neurotransmitter release, which results in impaired muscle function, and the potentially life-threatening respiratory arrest that is characteristic of the disease-state botulism [1]. Currently, there are no effective medical modalities to treat intoxicated patients after the toxin has been internalized by the neuron. Importantly, BoNTs, of which there are eight serotypes (designated A – H) [2,3], are among the most toxic of known biological substances, and have been weaponized [4]. As such, they are classified as category A biothreat agents by the US Centers for Disease Control and Prevention. Additionally, naturally occurring BoNTs can cause food or liquid contamination [5]. Consequently, there is a significant interest in developing novel modalities to not only counter BoNT poisoning, but also to promote neuronal recovery after toxin innervation [6,7]. Paradoxically, despite the toxicity of these enzymes, purified BoNT serotypes A and B are widely used pharmaceutically in small, localized doses to treat various neuromuscular conditions, as well as cosmetically to reduce facial wrinkles [8-10]. Therefore, in addition to the potential malicious use of these toxins, there are also elevated concerns regarding accidental overdosing in clinical settings [8].
2. Importance of cell-based assays for BoNT research and drug discovery
Current drug discovery strategies against BoNTs include both molecular and empirical approaches [11]. The molecular approach is hypothesis-driven and can be considered target-based, as the experimental objective is to obtain a small molecule inhibitor that can directly block the proteolytic activity of BoNTs. Small molecule inhibitors have traditionally been identified by high-throughput screening (HTS) in which a library of compounds are evaluated for BoNT inhibition, typically in an in vitro biochemical assay. Some of these assays and in vitro screens have been described by our group and others [12,13]. Importantly, once BoNT active site inhibitors are confirmed they are then routinely evaluated in cell-based assays to ascertain the likelihood of in vivo activity [6,14]. Specifically, cell-based testing is used to directly measure general pharmacologic properties including potency and selectivity, while also indirectly evaluating inhibitor physicochemical properties including solubility, permeability, and metabolic stability. The demonstration of cell-based activity and an absence of obvious cytotoxicity facilitate prioritization for further ADME (absorption, distribution, metabolism, and excretion)-related testing and in vivo efficacy evaluation. Whereas the molecular, target-based approach has been extensively used by academic and pharmaceutical researchers for several years, the dearth of FDA-approved products derived from this strategy has called the method into question. This may be due in part to an incomplete understanding of the molecular mechanism of action of BoNTs and other rationally selected targets.
The empirical approach, referred to as “phenotypic drug discovery” or “phenotypic screening”, relies on changes to phenotypic endpoints in response to small molecules [11,15]. Phenotypic screening requires the use of disease-relevant cell models with endpoints related to changes of the disease-related phenotype. This can help to identify known modulators of different components of biological pathway as well as new targets. A recent analysis suggested that the phenotypic approach is a more successful method for the discovery of first in class drugs [16]. Phenotypic screens for BoNT inhibitors could potentially include the evaluation of toxin amelioration, motor neuron protection, and/or the promotion of neuronal regeneration and repair. Phenotypic screening is therefore an unbiased approach for countermeasure discovery and could lead to the identification of novel pathways and targets for BoNT inhibitor research. To this end, successful phenotypic screening relies on: 1) identifying an endpoint directly related to BoNT intoxication, and 2) utilizing a cellular system that faithfully recapitulates botulism as it is manifested in the human patient.
Mechanistically, BoNT metalloendopeptidase activity induces paralysis by blocking acetylcholine neurotransmitter release at neuromuscular junctions [2]. This occurs after the holotoxin has transduced the motor neuron, undergone processing to release its catalytically active subunit (BoNT light chain), which cleavages soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins that are required for neuroexocytosis [1]. Previous studies clearly established that BoNT-mediated SNARE protein cleavage is sufficient to inhibit neurotransmitter release [17-21], indicating that SNARE proteolysis is the critical molecular event that is relevant to BoNT intoxication. Therefore, the evaluation of SNARE function is a critical endpoint for determining BoNT inhibition. This can be even further refined to develop toxin-specific or -selective assays which take advantage of the exquisite substrate specificity of the different BoNT serotypes. For example, BoNT/A and /E cleave synaptosomal-associated protein of 25 kDA (SNAP-25), i.e., one of the protein components of the SNARE complex, while BoNT/B, /D, /F, and /G cleave synaptobrevin (also called vesicle-associated membrane protein or VAMP), and serotype C proteolyses both SNAP-25 and Syntaxin1. The cleavage of SNARE proteins blocks synaptic vesicle exocytosis, thereby causing paralysis in both animal models and humans [1]. Cleavage of SNAP-25 has thus been used to evaluate BoNT/A activity in both in vitro and in vivo models [22]. While a number of bioanalytical methods are available to quantify SNAP-25 concentration, including proteomic techniques involving mass spectrometry, immunoassay platforms have become the method of choice due to their versatility in terms of throughput and amenability for both target-based and phenotypic screens [23].
Here, we review recent developments in the use of physiologically relevant cell-based systems and immunoassay technologies that are advancing BoNT research and drug discovery. These methods can be utilized for BoNT inhibitor screening as well as for research including new target identification and mechanism of action studies.
3. Mammalian cell-based assays for BoNT studies
At this critical stage in the discovery and development of novel therapeutics for BoNT poisoning, the utilization of HTS is a key strategy for identifying and characterizing novel BoNT antagonists, and for further evaluating their biological effects in a time efficient manner [6]. However, progress has been limited with respect to the development of large scale, cell-based drug screening assays for BoNT research, due in part to a lack of biologically relevant and well-characterized model systems that are applicable for high-throughput studies. Ideally, cell-based HTS assays utilize cell culture systems that are well-characterized, biologically relevant, robust, sensitive, and cost-effective. Previously, numerous cell-based assays have been established to study the biological effects of BoNTs, including mammalian neuroblastoma cells and primary spinal cord cells from rodents [6,24-28]. All of these models have strengths and limitations, which are discussed below. Recently, independent groups explored the utility of stem cell technologies for BoNT research, and established stem cell-derived neuron and motor neuron culture systems meeting the criteria indicated above [29-32].
3.1. Immortalized Cells
To identify clinically relevant lead compounds, HTS should ideally utilize cells that are naturally targeted by BoNTs, i.e, motor neurons [33]. However, until recently, mammalian motor neurons were difficult to obtain, and therefore alternative cellular systems have been utilized. Immortalized cells have been widely used in the field and have provided many advantages for BoNT studies [14]. First, these cell lines are applicable to HTS assays given that they can be generated in large quantities. Secondly, they are easy to culture and generally not costly. Third, they can be easily manipulated to overexpress BoNT light chains and/or to generate reporter lines that can be used in FRET and/or FRAP based assays [34]. Based on these advantages, previously established cell-based BoNT assays utilized various continuous lines, including Neuro-2 a cells (mouse neuroblastoma) [35,36], P19 (mouse embryonic carcinoma) [37], SH-SY5Y (human neuroblastoma) [38], SK-N-SH (human neuroblastoma) [36], NT2 (human teratocarcinoma), BE(2)-M17 (human neuroblastoma) [39], and SiMa cells (human neuroblastoma) [40]. Similarly, PC12 cells (derived from rat pheochromocytoma) have been employed in FRET based approaches to measure the biological effects of BoNTs in cells [27,34]. Human carcinoma cells (N2) have also been utilized to measure vesicle exocytosis using fluorescent activity-dependent dyes [41]. Additionally, motor neuron-like cell lines have been evaluated for their utility for BoNT research. Our group has examined the suitability of NSC-34 cells, a mouse motor neuron-like line, for BoNT studies, however these cells were not sensitive to BoNT/A at picomolar concentrations when intoxicated for 3 hours. Recently, Whitemarsh et al. reported that another motor neuron-like cell line, NG108-15, can be more sensitive than immortalized cells based on 48 hours of BoNT exposure [42]. Recent review papers provide detailed comparisons between immortalized cell lines in terms of their BoNT sensitivity [6,14].
Although immortalized cells provide simple platforms for BoNT research, these cells also have many limitations, making them less attractive for HTS. Importantly, their genetic background is different from that of native motor neurons, given that immortalized cells exhibit tumorigenic propensities. Immortalized cells are typically insensitive to BoNT intoxication and require significantly elevated doses and longer exposure times for detectable effects [6,14]. However, it is important to note that the sensitivity of these cells can be increased through various methods including the addition of ganglioside GT1b [36] or through chemical stimulation with KCl or CaCl [14]. Similarly, it was shown that co-culture of neuroblastoma cells with Schwann cells can enhance their sensitivity to BoNTs [43]. One possibility regarding the general low sensitivity of these cells is that the receptors, targets, and molecular machinery needed for BoNT action differ in immortalized cells compared to motor neurons. This is of particular importance when phenotypic screening for BoNT inhibitors is contemplated. Hence, the utilization of immortalized cells can be expedient, but cell types that better mimic in vivo conditions may provide better clinical translation.
3.2. Primary neurons for BoNT research
The use of primary mammalian cells, including spinal motor neurons [28,44], dorsal root ganglion neurons [32,45], hippocampal neurons [46-48], and cortical neurons [49,50] bypasses the use of immortalized cells. Among these neuronal cell types, primary motor neurons have the greatest advantage given that molecular machinery, receptors and molecular targets for toxin uptake, processing, and activity are endogenously expressed. Importantly, motor neurons dissected from murine embryos closely resemble native motor neurons morphologically and biologically. Primary spinal neurons are also highly sensitive to BoNT intoxication [14,51], and therefore provide more physiologically relevant model systems. Another advantage is that primary neurons are terminally differentiated cells that can be maintained for long periods to study the biological effects of BoNTs without the interference of cell proliferation. The development of serum–free media simplifies drug screening by the removal of serum stimulating factors and associated serum proteins that can bind small molecules and decrease their effective concentration. The use of serum-free media also makes protocol standardization more facile and reduces cell culture costs. Finally, primary cells can be derived from transgenic and/or knock-out mice to study the roles of specific proteins in the BoNT mechanism of intoxication.
Unfortunately, these cells also have limitations that make them impractical for HTS. Primary neurons are very difficult to obtain in sufficient quantities from dissected embryos to facilitate HTS campaigns. In fact, only 5,000-15,000 spinal motor neurons per dissected murine embryo can be generated with these tedious protocols [52]. The sacrifice of timed pregnant animals is required to obtain freshly prepared neurons each time the assay is performed, while dissecting and culturing large amounts of spinal motor neurons introduces significant costs. A second technical issue involves the routine generation of homogenous cultures. Embryo-to-embryo variations are expected in each dissection in terms of motor neuron/non-neuron cell ratios of the yield, which can result in a variation from test to test. Various methods have been developed to isolate motor neurons from primary spinal cord dissections, including density based methods, and antibody-based purifications [52,53]. However, these methods yield only marginal increases in cell number, and therefore become screening liabilities.
3.3. Stem cell technologies for BoNT research
An alternative methodology to address the cellularity issue involves the directed differentiation of embryonic stem (ES) cells or induced pluripotent stem cells (iPS). Two unique characteristics of pluripotent stem cells make them attractive for large scale BoNT studies. First, these cells can be successfully differentiated into specific cell types including functional motor neurons [54-57]. Secondly, these cells possess self-renewal capacity, indicating that unlimited numbers of motor neurons can be derived from pluripotent stem cells [32]. Recent advances in stem cell technologies have therefore opened new avenues to utilize stem cell-based culture platforms.
3.3.1. Mouse embryonic stem cell-derived neuron systems
With emerging stem cell technologies promising significant potential, research efforts focused on these technologies to establish physiologically relevant neuron or motor neuron enriched culture models suitable for BoNT research. Several independent groups demonstrated that mouse ES-cell derived neurons and motor neurons can provide such a system [29,31,32]. ES cell-derived neuron and motor neuron culture systems are significantly more sensitive than established cell lines [32]. It has been shown that neuronal cell types differ in terms of their sensitivities to BoNTs. Therefore, our group has focused specifically on developing motor neuron-enriched cultures for studies of BoNT intoxication using mouse ES cell line HBG3, which includes a transgene under the control of a motor neuron (Hb9) promotor [32]. Once activated, this transgene drives eGFP expression. We and others have demonstrated that the ES-derived eGFP positive motor neurons exhibit the morphological and biological properties of in vivo motor neurons, including the expression of motor neuron markers [54,55,58]. It is also noteworthy that these neurons can form neuro-muscular junctions in vitro and integrate into chick spinal cord when transplanted [54,55,59]. As ES cell-derived neurons faithfully recapitulate all of the biological steps of the intoxication process, they could potentially be used as a surrogate system for the in vivo mouse bioassay (MBA). Currently, the MBA is the gold standard for the detection of BoNTs and assessing their potency. However, this assay can also be technically challenging, time consuming, and variable [40,60]. Bioassays performed in BoNT/A sensitive motor neurons could enable the commercial use of this platform assuming that assay standardization, optimization, and validation is conducted with the appropriate toxin standards and that cellular and animal model bridging studies are successfully completed.
3.3.2. Human stem cell-derived neuron systems
Species-related differences between pathways or drug response are frequently observed during the drug discovery process [61]. This includes translational differences between data generated using mouse versus human ES-derived neurons. For example, a recent study describing glutamate receptor agonists demonstrated a 10-fold potency difference between mouse and human [62]. However, it should be noted that any cellular model has limitations when interpolating results from a single cell to physiology relating to the whole body. Therefore, it would be ideal to understand the correlation between data sets generated with both non-human mammalian neuron systems and their human counterparts [30]. In this regard, novel techniques established in recent years made the generation of human motor neurons feasible via at least three approaches: human induced pluripotent stem (iPS) cells, human ES cells, and induced motor neurons (iMNs) [63-70]. These neurons offer an alternative model to identify and validate novel inhibitor compounds, and understand their mechanisms of action in a human neuronal system.
i. Induced pluripotent stem (iPS) cell-based systems for BoNT research
It was shown in 2006 that adult cells with mitotic propensities can be reprogrammed to establish cells displaying characteristics of pluripotent stem cells, i.e., iPS cells [71]. These cells have been successfully differentiated to various cell types, including neurons [72]. Indeed, mouse iPS cells can be utilized to generate adult mouse models, indicating that these cells can form all cell types in vivo [73]. However, the origin of the iPS cells appears to be critical [74]. Recent studies comparing human iPS and ES cells suggest that they differ in their propensities to differentiate into discrete lineages [75]. For example, a recent study demonstrated that human iPS cells are differentiated into neuroepithelial lineage with high variability and less efficiency when compared to human ES cells [76]. Similarly, the directed differentiation of human iPS cells to blood progenitor and endothelial cells also exhibited low differentiation potency and resulted in functionally compromised cell types [77,78]. Furthermore, recent studies examining the global gene expression profiles of iPS cells suggested that there might be transcriptional differences between iPS and ES cells [79,80]. Taken together, it is important to recognize that diverse iPS cell lines derived from various tissues might have significantly divergent differentiation propensities and could produce heterogeneity of the final cellular population.
Despite the potential for some modest heterogeneity, Pellett et al. have shown that neurons generated from human iPS cells are highly sensitive to BoNT intoxication and demonstrate SNARE protein cleavage [30]. This study utilized commercially available, cryopreserved neurons (Cellular Dynamics International, Madison, WI) mainly composed of glutamergic and GABAergic neurons. To the best of our knowledge, human iPS-derived motor neuron assays have not been exploited for more advanced BoNT research and may provide a good system for further evaluation.
ii. Human embryonic stem cell-derived neuron culture models
As mentioned above, although there are many similarities between human iPS and ES cells, these cells may differ in gene expression and DNA methylation [81,82], and such variation in pluripotent cells can greatly affect neuronal differentiation [81,83]. Over the past decade, there have been extensive research efforts to obtain specific neuronal cell types from human ES (hES) cells and such model systems have been successfully utilized to study neurodegenerative diseases, such as Alzheimer's disease. It is well established that human ES cells can be differentiated into functional motor neurons [63-70], and that these neurons provide excellent culture systems to understand disease mechanisms such as ALS [65,84]. In the context of BoNT research, our group evaluated the utility of human ES-derived motor neuron systems as a renewable cellular source for large scale BoNT studies (manuscript in preparation). Importantly, this system is highly sensitive to BoNTs and compatible with large-scale studies and experimentation. Similar to the iPS-derived neuron culture system, this technique also suffers from heterogeneous cultures (i.e., other cell types are present in addition to motor neurons). Therefore, the development of more robust differentiation protocols to increase the percentage of motor neurons will be required. Additionally, current human motor neuron differentiation protocols require long differentiation processes, which are expensive and laborious. However, recent efforts have focused on generating more homogenous cultures using accelerated differentiation protocols [70]. The standardization of such protocols should advance the utility of iPS and ES cell-derived motor neurons for BoNT research.
iii. Direct conversion of somatic cells to neurons
A major limitation of motor neuron generation from ES or iPS cells is the long culture time required for directed differentiation. To address this issue, it was recently shown that human fibroblasts can be directly converted to neurons (induced neurons, iNs) [85] and induced motor neurons (iMNs) [86], using defined protocols. Derivation of these neurons avoids the culture and maintenance of pluripotent stem cells, which produces savings in both time and expense. iMNs appear to have the morphological and electrophysical properties of motor neurons [85]. However, a major limitation of this approach is that the conversion efficiency of human cells is significantly lower than the differentiation of neurons from iPS or hES cells [79]. While the generation of iMNs is an exciting approach, future studies are needed to determine the functionality and mature phenotypes of these neurons. Differentiation procedures must also be optimized to increase the efficiency of conversion in order to supply sufficient cells for screening. To this end, it is important to note that a recent study demonstrated that the conversion of iNs can be achieved starting from ES or iPS cells, rather than fibroblasts. This study suggested that nearly 100% yield can be achieved within 2 weeks with the expression of a single transcription factor [87]. To best of our knowledge, the utility of iNs or iMNs for BoNT research has not been explored.
4.0. Cell-based immunoassays to characterize BoNT activity
Stem cell-derived motor neuron cell culture systems afford the development of new and more sensitive assays for evaluating BoNT/A intoxication in physiologically relevant cell models, and provide an ideal system for the discovery of small molecule inhibitors. As mentioned previously, immunoassay-based quantification of SNAP-25 cleavage as a measure of BoNT activity remains the most convenient and adaptable of detection techniques. Specific antibodies directed against full length, uncleaved SNAP-25 and against BoNT/A proteolyzed SNAP-25 enable the development of a variety of immunoassays. Some assays use adsorbed antibodies and require multiple steps including Western-blot analysis, standard ELISAs, and sandwich-based ELISAs with electrochemiluminescence (ECL). However, these techniques require several antibody incubation and washing steps, thus making them time and labor-intensive. Other assays are designed for higher throughput analysis and use simple cellular lysates and the addition of immunoreagents without separation steps (i.e., washing). These assays include time-resolved fluorescence (TRF-FRET) (or homogenous time resolved fluorescence, HTRF®) and the amplified luminescence proximity assay (AlphaScreen®) readouts (Table 1). Early immunoassays utilized a mouse monoclonal antibody (MAb) against the amino terminus of SNAP-25 [26] and provided a method that could reliably detect total SNAP-25. However, an immunoreagent that specifically recognized the BoNT/A cleavage site on SNAP-25 was desired, as it could be used to design screens to identify novel BoNT/A inhibitors. To this end, Nuss et al. created an antigenic peptide that spanned the BoNT/A cleavage site in SNAP-25 (Gln 197-Arg 198) to generate polyclonal antibodies that only recognized full length SNAP-25. Dual channel western blot and direct ELISA using primary chick neurons clearly denoted that a BoNT/A cleavage-sensitive (BACS) antibody was specific for full length SNAP-25 and that BoNT/A addition abrogates this interaction [23]. SNAP-25 proteolysis mediated by BoNT/E, a related serotype that cleaves 17 amino acids upstream from the BoNT/A scissile bond (between Arg 180 and Ile 181), creates a truncated peptide that also has lost the BACS antigen recognition site. BACS–mediated detection of full length SNAP-25 is also abrogated by increasing concentrations of BoNT/E using immunofluorescence assays performed in mouse motor neuron cells (data not shown). As such, the BACS antibody can be used to detect and quantify both BoNT/A and BoNT/E mediated proteolysis of SNAP-25 in multiple ELISA formats. More recently, Fernandez-Salas et al. developed a MAb (2E2A6) that recognizes BoNT/A cleaved SNAP-25 (SNAP-15197). This antibody appears to be highly specific for proteolyzed SNAP-25 and fails to detect full length SNAP-25 in western and ELISA analysis [40]. This reagent was incorporated into a robust and sensitive cell-based assay that was validated for determination of BOTOX activity in bulk drug material and product samples. This group also noted that this assay could be used for the discovery of BoNT inhibitors for human disease. It should be noted that both antibodies, cleavage-sensitive BACS and cleavage-specific 2EA6, could be used in assays that may be translatable across pre-clinical in vivo models (mouse, rat, guinea pig, rhesus monkey and humans), given that the BoNT/A cleavage site and the nine amino acids at the carboxy-terminus of SNAP-25 are identical across the highlighted species. The creation of antibody reagents that enable the quantification of BoNT/A activity is a critical factor in the development of novel immunoassays to support BoNT inhibitor research.
Table 1. Cell-based immunoassays applicable to BoNT inhibitor research.
| Immunoassay format | Detection | Throughput | Advantages | Disadvantages |
|---|---|---|---|---|
| Western blot | Luminescence | Low | Technical simplicity, standard instrumentation, gold standard for many studies | Labor intensive, highly variable, difficult to standardize |
| Cellular image-based assays | ||||
| LICOR | Fluorescence | Medium | Standard instrumentation | Low resolution imaging, typically singleplex, as labor intensive as HCI |
| High Content Imaging (HCI) | Fluorescence | Medium to high | High resolution imaging, population analysis, easy to multiplex | Labor intensive, requires specialized instrumentation, special software for analysis |
| Single-parameter well-based assays | ||||
| Enzyme-linked immunosorbent assay (ELISA) | Colorimetric, luminescence, fluorescence | Low to medium | Good dynamic range, quantitative, robust, cost-efficient | Labor intensive: multistep preparation process, low-throughput |
| Electrochemiluminescence (ECL) | Luminescence | Medium to high | Large dynamic range and sensitivity, potential for multiplexing, robust | Labor intensive, costly, requires specialized instrumentation |
| Time-resolved Fluorecence Energy Resonance Transfer (TR-FRET) | Fluorescence | High | Homogeneous, high-throughput, good dynamic range, robust, cost-efficient | Sensitive to some conditions and reagents, limitations imposed by distance for FRET |
| Amplified Luminescent Proximity Homogenous Assay Screen (AlphaScreen®) | Fluorescence | High | Homogeneous, high-throughput, large dynamic range and sensitivity, robust in variable conditions | Requires specialized instrumentation, more expensive then FRET |
4.1. Application of imaging approaches for assay development
Multiple cell-based assays have been designed using antibodies directed against different forms of SNAP-25. Immunostaining of cells with total (such as SMI-81 from Covance, Princeton, NJ) and cleavage sensitive (BACS) antibodies directed against SNAP-25 proteins allows one to visually monitor the effect of BoNT/A in neurons. The ability to discriminate the sub-cellular localization of the SNAP-25 associated signal allows the researcher to better understand the biology of BoNT intoxication of motor neurons. The same imaging approach utilizing the antibody-based detection of SNAP-25 cleavage can be applied in different formats. The readout of the fluorescently labeled secondary antibodies can be simply detected by low resolution fluorescence readers, e.g., LICOR [23]. This approach will generate a less detailed image of whole wells but can be done quickly and does not require sophisticated equipment.
Alternatively, high resolution imaging (20-40×) provides higher quality images, and therefore enables the collection of a large number of morphological endpoints (Table 2). The total number of parameters and overall utility of the imaging are governed in large part by the quality and quantity of the immunostaining signal. Additionally, the throughput of the method decreases proportionally to the increase in the amount of information extracted from the images. In practical terms, this restricts analyses to a subset of cells from within a population of cells contained within a well. Thus, when the image acquisition algorithm is designed, selection of morphological endpoints must be balanced with the speed of the overall process.
Table 2. Morphological endpoints acquired by HCI.
| Nuclei |
| Area |
| Intensity |
| Number of nuclei |
| Intensity of signal of clustered nuclei |
| Region roundness |
| Small nuclei |
| Neurites |
| Neurite length |
| Number of extremities |
| Number of nodes type 1 |
| Number of nodes type 2 |
| Number of roots |
| Number of segments |
| Region area |
| Region roundness |
| Total neurite length |
| Intensity area mean |
| Intensity area average |
| Intensity area sum |
| Select region |
| Branch level |
| Parent cell number |
| Parent segment number |
| Inner segment |
| Find Spots |
| Spot intensity |
| Spot contrast |
| Spot background intensity |
| Spot area |
| Spot region intensity |
A high-content imaging (HCI) assay can be used to quantitatively measure BoNT/A-mediated SNAP-25 cleavage in mouse ES cell-derived motor neurons by taking advantage of cleavage-sensitive BACS antibodis. As shown in Figure 1, motor neurons that transgenically express eGFP can be readily identified by their green color (Fig. 1A and E). In the absence of toxin, total SNAP-25 and full-length SNAP-25 demonstrate robust staining (Fig. 1B, C and D), whereas the addition of BoNT/A causes a concomitant decrease in full-length SNAP-25 (BACS) intensity (Fig. 1G and H) without a significant affect on the total SNAP-25 signal (Fig. 1F). Even though the overall fluorescence signal decreases in the BACS channel during BoNT/A treatment, there is some background signal associated with thick branches and neuronal bodies even after long exposure to toxin. This is possibly due to residual SNAP-25 or non-specific cross-reactivity of the polyclonal antibody with other proteins. The major advantage of the HCI platform comes from its ability to integrate relatively simple measurements, such as SNAP-25 quantification, with more complex morphological endpoints that are taken from the same biological sample [88,89]. More specifically, HCI can measure multiple cellular processes, including neurite outgrowth, in addition to the effects of BoNTs on SNARE proteolysis, within a single experiment and with a high degree of confidence. It has been previously demonstrated that BoNT intoxication of neuronal cells leads to axonal sprouting and neuromuscular junction remodeling [90]. For example, Coffield et al., monitored the effect of BoNT/A on the neuritogenesis of primary motor neurons and concluded that toxin promotes neurite outgrowth [25]. Therefore, a quantitative measurement of neurite outgrowth can potentially serve as a measurable endpoint for the assessment of BoNT activity. In general, HCI provides an excellent platform for measuring and quantifying the effects of small molecules on neurite outgrowth, branching, and other morphological changes (Fig. 2). The quantitative image analysis of neurites requires specially designed algorithms that could provide detailed and accurate detection of areas interpreted as neurons (or segmentation of neurons) and separate the points that distinguish the branches of neurons. Importantly, HCI methodologies allow for multiplexed analyses that focus on specific populations of differentiated cells and enable the precise localization of biomarkers of interest. Figure 2 illustrates how the expression of eGFP marker (Fig. 2A) can be used to specifically mask motor neurons [32] and their nuclei and branches (Fig. 2B-E). The multiplexing of the BACS signal with eGFP allows the masking of SNAP-25 only in eGFP positive motor neurons (Fig. 2F), and BoNT-mediated SNAP-25 cleavage can be quantified in these cells by measuring the loss of the BACS signal [23]. This approach is critical when evaluating cellular systems that demonstrate incomplete motor neuron differentiation or other mechanisms resulting in heterogeneous populations.
Figure 1. High-throughput immunofluorescence assays using cleavage-sensitive antibodies to measure BoNT proteolytic activity in neuronal cells.
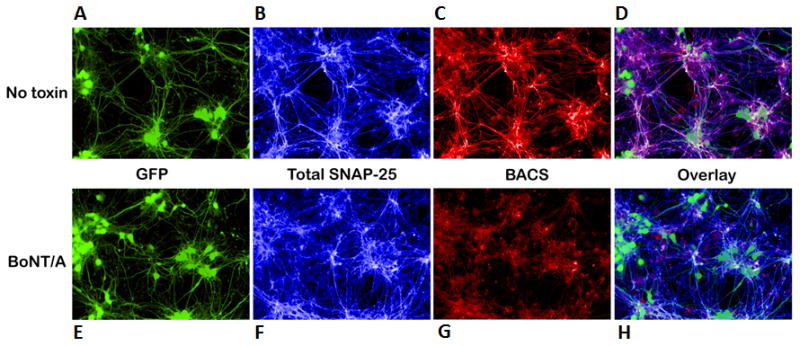
Mouse ES cell-derived motor neurons were mock treated (A-D) or intoxicated (E-H) with 1nM BoNT/A for 3 hours, fixed and stained. (A and E) eGFP positive motor neurons (green), (B and F) total SNAP-25 (blue) (SMI-81, cleavage insensitive SNAP-25 antibody), (C and G) full length SNAP-25 (red) (BACS, cleavage-sensitive SNAP-25 antibody) and (D and H) overlay of the superimposed images from all 3 channels to illustrate the loss of the SNAP-25 signal associated with its cleavage by BoNT/A.
Figure 2. Steps involved in the image analyzing algorithm for detecting endogenous SNAP-25 specifically in motor neurons.
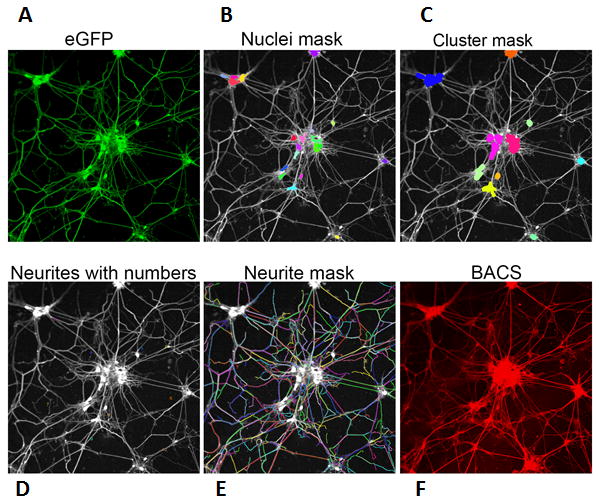
High-content imaging (HCI) can measure the effects of BoNTs and/or small molecules on neuronal morphological changes. Mouse embryonic stem cells (HBG3 line), in which eGFP expression is driven by a motor neuron specific promoter (Hb9), were differentiated into motor neurons. (A) eGFP signal was utilized to identify motor neurons. (B) The fluorescent signal from the eGFP channel was used to detect and mask nuclei and neurite outgrowth. Capella (PerkinElmer)-based nuclei and neurite detection algorithms were used to identify nuclei and neurite outgrowth, respectively. (C-E) A cluster detection module was inserted in the imaging analysis pipeline to detect nuclei clusters using nucleus masks. (F) Without the toxin exposure, BACS antibody signal exhibits total SNAP-25 in eGFP+ cells.
Laboratory automation is a critical component of every successful HCI screening campaign, as the overall process can be labor-intensive and time consuming. A robust HCI assay requires cell plating and differentiation, BoNT/A intoxication, immunostaining, image acquisition, and data analysis (Fig. 3). Automating this operation into an HTS-compatible process became possible only with the development of sophisticated robotic equipment capable of handling the different operations in the protocol. The best example of a fully automated system for handling such an HCI assay is the Cell-explorer platform (designed by PerkinElmer). The different combinations of manual and automated handling of plates provided by this system can improve the throughput of an otherwise very slow operation. In our laboratory, automated compound management and compound dispensing systems have been added to further expedite the process. Briefly, the multiple steps of immunostaining are performed using a Hudson Robotics Plate Crane for plate loading on a Biotech Elx 405 washer and Thermo Multidrop dispenser in semi-automated mode. Plates ready for imaging are loaded on the Opera in automated mode by a PE Plate-handler II robotic arm and read. Image acquisition and data analysis have also been facilitated by the use of commercially available automated confocal and wide field microscopy systems [91]. As briefly indicated above, our method has integrated an Opera (PerkinElmer) confocal plate reader into the BoNT/A HCI screen (Fig. 3). The most advanced configuration of this instrument features 4 lasers (405nm, 488nm, 562nm and 640nm), an arc lamp for UV spectroscopy and 4 CCD cameras for parallel or sequential detection of images. This configuration allows for the detection of emitted fluorescent light at several wavelengths: excited DNA-binding Hoechst staining at 405 nm, reporter–associated GFP signal at 488, and two additional channels at 561 and 640 nm for detection of antibodies for full-length SNAP-25 and β-III tubulin. One limitation of the HCI assay is the number of cells detected per field image. Unfortunately, the resolution acquired with the 20× objective requires the capture of several images from each well (at least 6 or more) to accumulate a sufficient number of images for statistical analysis. Image analysis can be accomplished using commercially available Acapella neurite algorithms (Fig. 3).
Figure 3. Integration of high-content imaging (HCI) assays with embryonic stem cell-derived motor neurons as an analytical platform to measure biological effects of BoNTs.
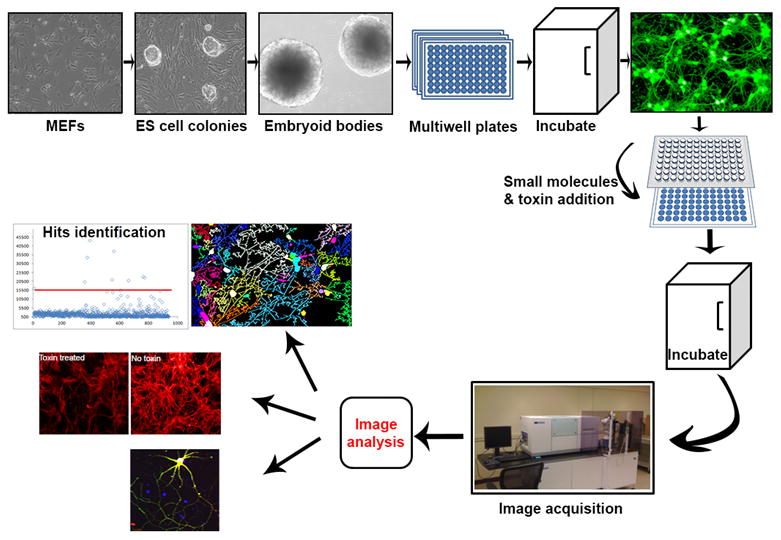
Workflow showing HCI assay steps: derivation of motor neurons from ES cells, image acquisition, image analysis, and data analysis. Mouse ES cells are cultured on mouse embryonic fibroblasts and differentiated to motor neurons. These neurons can be plated in 96-well plates (or greater) and utilized in HCI assays.
Clearly, significant assay development and optimization are needed to create a high quality HCI screening assay. Once a standardized protocol is obtained, it must be further evaluated for statistical robustness, with certain criteria being met: (i) the production of statistically robust data with an acceptable assay window (3-fold or greater) and acceptable Z' (>0.5), and (ii) demonstration of plate to plate and day to day reproducibility using well characterized controls. During the optimization process for our HCI assay, several key variables were found to profoundly influence the quality of the assay and included cell culturing and BoNT intoxication conditions, antibody titering and fixation methods, and data analysis techniques. A representative example of the optimized HCI protocol quantifying SNAP-25 cleavage in mouse ES cell-derived motor neurons is shown in Figure 4. BoNT/A-dependent proteolysis of SNAP-25 has improved substantially, and the dose-dependent increase in SNAP-25 cleavage in response to increasing concentrations of BoNT/A at both 4 and 24 hours after intoxication is evident (Fig. 4).
Figure 4. HCI of SNAP-25 cleavage in mouse ES-derived neuron cultures.
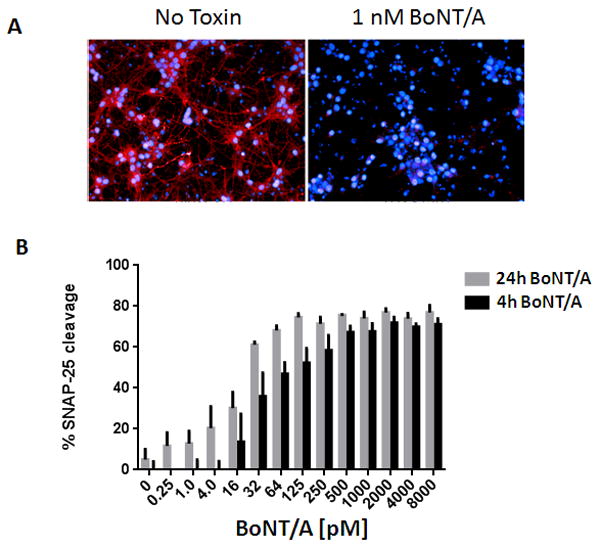
Motor neuron cultures were treated with increasing concentrations of BoNT/A for 4 and 24 hours to evaluateSNAP-25 cleavage efficacy. SNAP-25 was detected with BACS antibodies. In another channel (not shown), neurons were detected with βIII-tubulin antibody to create masks of neurons for SNAP-25 image analysis. (A) Representative image of one field (from 7 taken) for wells either untreated (no toxin, left) or treated with 1nM BoNT/A for 24h (right). Blue indicates nuclei stained with Hoechst 3339 and red is the BACS signal. (B) The data was analyzed using the Columbus (PerkinElmer) algorithm and the values for the signal associated with the cleavage sensitive BACS was normalized to the total βIII-tubulin signal. Error bars represent standard deviation. The percent cleavage of SNAP-25 was plotted using GraphPad Prism (n=3 for each point).
Even though many neuronal outgrowth assays have become commonplace, they still represent an operational challenge for some laboratories. All critical (rate-limiting) reagents should be available in sufficient quantities to supply the entire screening campaign. The comprehensive protocol should be cost-efficient as well. In general, image-based HTS assays could be extremely valuable for BoNT screening, but also very challenging to establish and implement.
4.2 ELISA/ECL-based assays
The development of specific and high binding affinity monoclonal antibodies against SNARE components led to the development of enzyme linked immunosorbent (ELISA) assays originally used to evaluate SNARE protein regulation in severe mental disorders, including schizophrenia [92]. To design a standard ELISA for SNAP-25, simple cell lysates containing SNAP-25 have been adhered to ELISA plates and then probed with anti-SNAP-25 antibodies. The next generation sandwich ELISA improved upon the standard technique by utilizing two individual antibodies (Fig. 5A). SNAP-25 is first bound by a capture antibody that recognizes any form of the protein. The second antibody recognizes a distinct epitope of SNAP-25 and is conjugated with a reporter enzyme (horse radish peroxidase or alkaline phosphatase) or directly labeled with a fluorescent dye which allows for detection of the complex. Our laboratory was the first describe a SNAP-25 sandwich ELISA using the BACS antibody for the characterization of BoNT/A-mediated proteolysis of SNAP-25 in primary motor neurons [23]. The sandwich ELISA was found to have better a signal/background ratio than the standard ELISA, and could be used to measure the percentage of full-length SNAP-25 in experimental samples while using recombinant SNAP-25 as a standard [23].
Figure 5. Principles of ELISA (A), HTRF® (B) and AlphaScreen® (C).
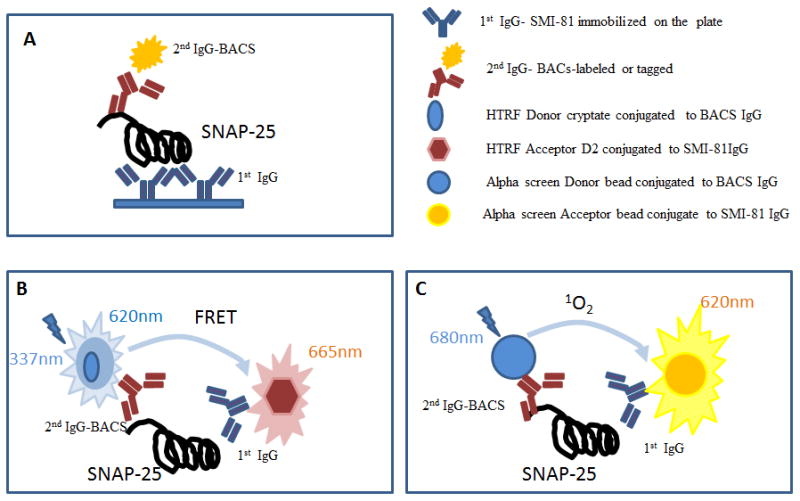
Illustration of the principles for three assays using pairs of specific antibodies recognizing distinct epitopes of SNAP-25 (BACS recognizes only full-length SNAP-25; SMI-81 recognizes total SNAP-25). In the ELISA assay (A), the antibodies are absorbed on the plate. For the HTRF® (B) and AlphaScreen® (C) assays, antibodies are either labeled directly by conjugation or through the species-specific antibodies containing the respective donor and acceptor moieties.
A significant improvement over the sandwich ELISA can be made by switching to an electro-chemiluminescence (ECL) system. The ECL platform, as developed by Meso Scale Discovery (MSD, Rockville, MD), has additional advantages over classical ELISA, such as greater sensitivity and dynamic range, the potential for multiplexing, and smaller quantities of sample used during testing. ECL utilizes a detection system that emits light when stimulated electrochemically. Signal is induced by subjecting a ruthenium complex (Sulfo-Tag) attached to a detection antibody to an electric field generated from a specialized carbon ink electrode plate by the interaction between antibody and analyte. In the presence of tripropylamine coreactant, a redox reaction occurs and leads to the emission of light.
Fernandez-Salas et al., using SiMa cells, were the first to report a cell-based BoNT/A potency assay utilizing an ECL format to measure BoNT/A-dependent intracellular increases of cleaved SNAP-25 [40]. The novelty of the assay included the use of a specific monoclonal antibody (2E2A6), which recognizes only cleaved SNAP-25. As such, this assay produces an increasingly larger signal when increasing concentrations of BoNT/A are added to cells. However, due to the nature of the 2E2A6 antibody, screens using this reagent analyze as decrease in ECL signal in response to increasing BoNT/A inhibition. In order to take advantage of the high sensitivity and dynamic range of the ECL platform, our group converted the previously established BACS sandwich ELISA [23] into an ECL assay - to create an assay that detects only the full length form of SNAP-25. Optimization procedures with respect to plate type, blocking buffers, antibody combinations and concentrations were performed to obtain a robust assay featuring excellent plate statistics. As seen in Figure 6A, mouse motor neurons intoxicated with increasing concentrations of BoNT/A demonstrate a near complete degradation of endogenous SNAP-25, with and EC50 of 5.84 pM/L (95% confidence intervals: 5.063- 6.743). This assay as currently configured is statistically robust and provides sufficient throughput and cost-effectiveness to support secondary testing of compounds arising from our targeted molecular approach for identification of proteolytic, active site BoNT/A inhibitors (Fig. 6B). However, this is still a labor intensive and expensive assay to use for the phenotypic screening of BoNT/A inhibitors. For this type of screen, an alternative immunoassay format with greater throughput will be required.
Figure 6. BoNT/A intoxication evaluated by electrochemiluminescence.
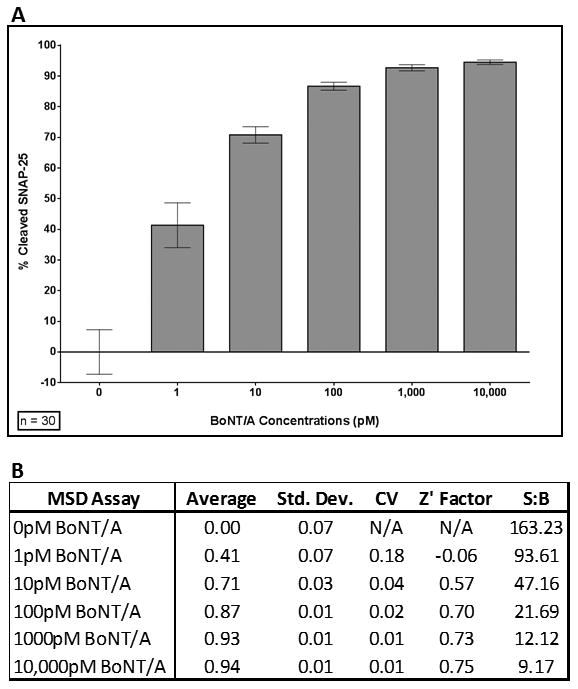
Mouse ES cell-derived motor neurons were treated with different concentrations of BoNT/A for 24 hours. Cells were lysed in a lysis buffer including protease inhibitor cocktail and the lysates were analyzed using an MSD assay. The results were normalized to percent cleaved SNAP-25 and plotted (A). The summary table (B) contains average cleaved SNAP-25, standard deviation (Std. Dev.), coefficient of variance (CV), Z' Factor, and signal to background ratio (S:B) for measurement at different concentrations of BoNT/A. Each data point represents samples collected in 3 independent experiments from 5 wells (on 2 plates in each experiment), total n=30 for each data point.
4.3. Homogeneous immunoassays
Alternative formats for immunoassays have been developed by pharmaceutical HTS groups to increase the throughput of protocols, improve the reproducibility of the results, and reduce total operational steps. The key difference provided by these assays is their ability to detect the analyte in a cell lysate without the separation and washing steps associated with ELISAs (Fig. 5A) and HCI immunoassays. There are several alternative homogeneous immune-detection approaches; our laboratory utilizes two systems: TR-FRET or HTRF® (homogenous time resolved fluorescence) and AlphaScreen® [93]. Both assays feature a robust detection system that is dependent upon the distance (proximity) between two labels (donor and acceptor) and is due to the ability of the signal to travel the short distance from donor to acceptor (Fig. 5B and C). In the case of HTRF®, the donor is a Europium-cryptate (Eu 3+ cryptate) or a Terbium-cryptate (Tb-cryptate) molecule that can transfer ions with a specific fluorescent pattern (Fig. 5B). The energy can be absorbed by red pigment derivatives (XL665 or d2) that emit in the far red spectrum (665nm). The distance between donor and acceptor is critical for the ability of HTRF® to occur; if the distance is too great, the transfer will not occur. The sensitivity to the proximity of two detector molecules allows the use of two different antibodies that recognize two distinct epitopes of SNAP-25, with SMI-81 recognizing total SNAP-25 and the BACS antibody being sensitive to only full-length SNAP-25. In the absence of BoNT/A, or successful BoNT/A inhibition, both antibodies will bind SNAP-25 and the energy transfer from donor to acceptor will transpire after illumination of the sample. Conversely, in the presence of active BoNT/A (no inhibition), SNAP-25 is cleaved and the cleavage-sensitive BACS antibody does not recognize the proteolyzed SNAP-25. Complex formation will not occur and no signal will be generated. The FRET principle has been utilized previously to create an in vitro assay for evaluating SNAP-25 proteolysis by BoNT/A via labeling both ends of a synthetic substrate [34]. However this type of FRET uses an artificial substrate analog that does not accurately reflect the physiological substrate within motor neurons.
In the case of AlphaScreen® assay, the underlying principal is the same as TR-FRET and relates to the proximity of the donor and acceptor labels (Fig. 5C). In this method, the two antibodies recognizing total and full length SNAP-25 are bound to chemically coated donor and acceptor beads. Donor beads coated with a photosensitive reagent, which, after activation by a laser, converts ambient oxygen into singlet oxygen molecules. The singlet oxygen can diffuse up to 200nm and react with the acceptor bead. In the presence of full length SNAP-25, a complex is formed. The thioxen derivatives coated on the acceptor bead interact with the singlet oxygen and induce a chemiluminescent reaction. The luminescent emission at 370nm excites the fluorophore on the same acceptor bead to emit a signal for detection at 620nm.
A common drawback for both technologies is their sensitivity to sample contaminants that could disrupt the transfer of the signal from donor to acceptor. This is why both technologies are very robust for pure in vitro enzymatic assays, but may be less robust for cellular or tissue samples. However, both methods are designed and optimized for the ability to detect signal in cellular lysate. The AlphaScreen® approach has a better dynamic range than HTR® (CisBio) or TRF-FRET (PE), especially when an analysis requires cellular lysates. The bead-based technology can also be costly if one considers the large reagent quantity required for screening campaigns, as well as the need for a special reader that has a laser capable of exciting the acceptor beads. Both technologies allow for the conversion of multiple-step ELISAs into a format with limited reagent additions and without washing steps, which shortens the process time by at least 4-fold. In both cases, assay performance time is determined primarily by the length of incubation with the detection reagents, which makes these technologies desirable for in vitro HTS. The same assay can be used for cell-based phenotypic screens of BoNT/A inhibitors and to potentially evaluate different pathways that are involved in the cellular regulation of BoNT activity. For example, successful BoNT/A intoxication of motor neurons requires neurotoxin internalization involving ganglioside binding and internal processing with the endosome to release the catalytically active light chain [6]. Compounds that interfere with these processes can function as BoNT/A antagonists. Motor neuron treatment with Triticum vulgaris lectin competes with BoNT/A for ganglioside binding, thereby preventing BoNT/A endocytosis [94]. Likewise, Toosendanin will arrest BoNT/A light chain translocation with nanomolar potency and can block BoNT/A activity [95]. Both compounds will protect SNAP-25 against BoNT/A mediated proteolysis in mouse motor neurons (data not shown). As such, phenotypic screens for small molecules that prevent BoNT/A uptake and processing can be built using SNAP-25 expression as quantified by homogeneous immunoassays as a functional endpoint.
B. Expert Commentary
Recent studies have established the utility of stem cells to generate clinically and biologically relevant motor neurons, and have demonstrated that these cells provide highly sensitive models for BoNT studies. These neurons can be generated with well-established protocols and can be used in high-throughput BoNT assays [32]. In general, mouse ES-derived motor neurons are easier and cheaper to differentiate and can provide high yields. On the other hand, although requiring more effort, motor neurons derived from human ES or iPS cells afford researchers the unique ability to conduct mechanism, screening, and validation experiments with a species-relevant system.
These systems can support target-based screening approaches that first identify active site proteolytic inhibitors of BoNTs and confirm their activity in cell-based functional assays. Alternatively, these cell systems can be used for empirical or “phenotypic screening” in which neuronal cells are treated with molecules which may block BoNT/A-mediated intoxication or promote regenerative pathways that can rescue or repair neuronal damage. Assays employing human motor neurons that directly measure neuron function may provide novel mechanisms to counter BoNTs, and the resulting countermeasures can be quickly developed for human use. Once small molecule BoNT inhibitors are identified, the corresponding targets can be isolated using human motor neurons, and the absolute confirmation of binding/inhibition of the targets can be determined via advanced studies. Importantly, human motor neurons provide unique tools for the dissection of cellular pathway that are important for BoNT intoxication and or neuronal recovery. The identification of such pathways and key enzymes/proteins may provide alternative targets for BoNT drug development efforts. Overall, the convergence of advanced cellular models and high throughput and multiplexed immunoassay technologies provides a promising conduit for BoNT research and drug discovery that can potentially promote the identification and characterization of novel therapeutics.
C. Five Year View
Modeling human botulism in robust and physiologically relevant cell-based assays is crucial for the advancement of BoNT research and drug discovery. In the near term, it is anticipated that neuronal differentiation protocols, particularly those involving human-derived cells, will continue to be optimized to improve neuronal homogeneity and accelerate the time course of differentiation. Additionally, novel techniques may also be introduced that enable the rapid and efficient production of human motor neurons from a non-controversial cellular source.
The completion of both molecularly directed and phenotypic HTS campaigns should unambiguously demonstrate if active site proteolytic inhibitors of BoNT are possible to design, as well as identify and validate alternative targets that may be useful in countering BoNT/A intoxication after neuronal uptake. The identification and validation of novel targets against BoNT/A is particularly exciting, as this approach may bring forward molecular targets with a more straightforward development path, particularly if they are associated with a druggable class such as kinases or G-protein coupled receptors. However, it should be noted that the deconvolution of phenotypic screening hits can be a complex process. Nevertheless, the availability of advanced neuronal models will be beneficial to this effort.
In terms of high-throughput screens, we anticipate that most assay formats will be routinely multiplexed so as to enable the capture of additional endpoints during target interrogation. Additionally, as our understanding of BoNT intoxication and the pathways involved continue to grow, it is likely that new biomarkers and/or diagnostic antigens will become available to better gauge intoxication or monitor neuronal regeneration and repair. Biomarker development will play an important role in determining the dosing regimens and in determining efficacy during preclinical BoNT inhibitor studies. As such, the next 5 years should witness major advancements in the therapeutic discovery and development of BoNT inhibitors, as well as breakthroughs in understanding of the BoNT mechanism of intoxication and the development of treatments that promote neuronal recovery.
D. Key Issues.
Pluripotent human ES and iPS cells possess self-renewal capacity and the ability to differentiate into motor neurons, the natural target of BoNTs, and thereby offer a unique renewable cell source for BoNT studies.
Novel technologies combining physiologically relevant cell-based assays and high-throughput screening offer improved opportunities for identifying drug candidates to treat Botulism (for which there is currently no therapeutic available for treating post-neuronal intoxication).
Human motor neuron-based assays can be utilized to determine the mechanism of action of identified lead compounds.
Physiologically relevant human motor neuron systems can be also utilized to increase our understanding of host cellular pathways involved in either BoNT intoxication and/or recovery, which is critical for developing novel methods to treat botulism.
It is well established that BoNT serotypes A, B and E are responsible for the majority of human botulism cases, therefore drug screening efforts should be designed to identify compounds that can inhibit multiple serotypes.
Immunoassays that quantify SNAP-25 cleavage are sensitive and specific means of evaluating BoNT activity in cellular environments and can be used to drive both molecular (direct) and empirical (indirect) BoNT/A screening approaches.
The implementation of high-throughput homogenous assays may accelerate the discovery and development of effective BoNT/A inhibitors.
Acknowledgments
We thank Laura M. Wanner, Glenn Y. Gomba and Hao T. Du for their assistance with figures. This work was supported by grants from the Defense Threat Reduction Agency and National Institutes of Health (1 R21 AI101387-01 and 5 U01AI082051-05).
Footnotes
Disclaimer: The views, findings, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Department of Health and Human Services or the U.S. Army.
G. Reference Anotations
* of interest
**of outstanding interest
- 1.Tighe AP, Schiavo G. Botulinum neurotoxins: mechanism of action. Toxicon. 2013;67:87–93. doi: 10.1016/j.toxicon.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Montal M. Botulinum neurotoxin: a marvel of protein design. Annu Rev Biochem. 2010;79:591–617. doi: 10.1146/annurev.biochem.051908.125345. [DOI] [PubMed] [Google Scholar]
- 3.Dover N, Barash JR, Hill KK, Xie G, Arnon SS. Molecular Characterization of a Novel Botulinum Neurotoxin Type H Gene. J Infect Dis. 2013 doi: 10.1093/infdis/jit450. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Arnon SS, Schechter R, Inglesby TV, et al. Botulinum toxin as a biological weapon: medical and public health management. Jama. 2001;285(8):1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 5.Wein LM, Liu Y. Analyzing a bioterror attack on the food supply: the case of botulinum toxin in milk. Proc Natl Acad Sci U S A. 2005;102(28):9984–9989. doi: 10.1073/pnas.0408526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hakami RM, Ruthel G, Stahl AM, Bavari S. Gaining ground: assays for therapeutics against botulinum neurotoxin. Trends Microbiol. 2010;18(4):164–172. doi: 10.1016/j.tim.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Burnett JC, Henchal EA, Schmaljohn AL, Bavari S. The evolving field of biodefence: therapeutic developments and diagnostics. Nat Rev Drug Discov. 2005;4(4):281–297. doi: 10.1038/nrd1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S. Clinical uses of botulinum neurotoxins: current indications, limitations and future developments. Toxins (Basel) 2012;4(10):913–939. doi: 10.3390/toxins4100913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellett S. Learning from the past: historical aspects of bacterial toxins as pharmaceuticals. Curr Opin Microbiol. 2012;15(3):292–299. doi: 10.1016/j.mib.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Carruthers A, Carruthers J. Botulinum toxin products overview. Skin Therapy Lett. 2008;13(6):1–4. [PubMed] [Google Scholar]
- 11.Schenone M, Dancik V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9(4):232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt JJ, Stafford RG, Millard CB. High-throughput assays for botulinum neurotoxin proteolytic activity: serotypes A, B, D, and F. Anal Biochem. 2001;296(1):130–137. doi: 10.1006/abio.2001.5236. [DOI] [PubMed] [Google Scholar]
- 13.Burnett JC, Schmidt JJ, Stafford RG, et al. Novel small molecule inhibitors of botulinum neurotoxin A metalloprotease activity. Biochem Biophys Res Commun. 2003;310(1):84–93. doi: 10.1016/j.bbrc.2003.08.112. [DOI] [PubMed] [Google Scholar]
- 14.Pellett S. Progress in cell based assays for botulinum neurotoxin detection. Curr Top Microbiol Immunol. 2013;364:257–285. doi: 10.1007/978-3-642-33570-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JA, Chu S, Willard FS, et al. Open innovation for phenotypic drug discovery: The PD2 assay panel. J Biomol Screen. 2011;16(6):588–602. doi: 10.1177/1087057111405379. [DOI] [PubMed] [Google Scholar]
- 16.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10(7):507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 17.Kalandakanond S, Coffield JA. Cleavage of SNAP-25 by botulinum toxin type A requires receptor-mediated endocytosis, pH-dependent translocation, and zinc. J Pharmacol Exp Ther. 2001;296(3):980–986. [PubMed] [Google Scholar]
- 18.Apland JP, Adler M, Oyler GA. Inhibition of neurotransmitter release by peptides that mimic the N-terminal domain of SNAP-25. J Protein Chem. 2003;22(2):147–153. doi: 10.1023/a:1023423013741. [DOI] [PubMed] [Google Scholar]
- 19.Blasi J, Chapman ER, Link E, et al. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature. 1993;365(6442):160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- 20.Apland JP, Biser JA, Adler M, et al. Peptides that mimic the carboxy-terminal domain of SNAP-25 block acetylcholine release at an Aplysia synapse. J Appl Toxicol. 1999;19(Suppl 1):S23–26. doi: 10.1002/(sici)1099-1263(199912)19:1+<s23::aid-jat609>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 21.Bajohrs M, Rickman C, Binz T, Davletov B. A molecular basis underlying differences in the toxicity of botulinum serotypes A and E. EMBO Rep. 2004;5(11):1090–1095. doi: 10.1038/sj.embor.7400278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellett S, Tepp WH, Toth SI, Johnson EA. Comparison of the primary rat spinal cord cell (RSC) assay and the mouse bioassay for botulinum neurotoxin type A potency determination. J Pharmacol Toxicol Methods. 2010;61(3):304–310. doi: 10.1016/j.vascn.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Nuss JE, Ruthel G, Tressler LE, et al. Development of cell-based assays to measure botulinum neurotoxin serotype A activity using cleavage-sensitive antibodies. J Biomol Screen. 2010;15(1):42–51. doi: 10.1177/1087057109354779. [DOI] [PubMed] [Google Scholar]
- 24.Sheridan RE, Smith TJ, Adler M. Primary cell culture for evaluation of botulinum neurotoxin antagonists. Toxicon. 2005;45(3):377–382. doi: 10.1016/j.toxicon.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 25*.Coffield JA, Yan X. Neuritogenic actions of botulinum neurotoxin A on cultured motor neurons. J Pharmacol Exp Ther. 2009;330(1):352–358. doi: 10.1124/jpet.108.147744. This study demonstrates the possibility of determining the neuritogenic effects of BoNTs on motor neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl AM, Ruthel G, Torres-Melendez E, et al. Primary cultures of embryonic chicken neurons for sensitive cell-based assay of botulinum neurotoxin: implications for therapeutic discovery. J Biomol Screen. 2007;12(3):370–377. doi: 10.1177/1087057106299163. [DOI] [PubMed] [Google Scholar]
- 27.Dong M, Tepp WH, Johnson EA, Chapman ER. Using fluorescent sensors to detect botulinum neurotoxin activity in vitro and in living cells. Proc Natl Acad Sci U S A. 2004;101(41):14701–14706. doi: 10.1073/pnas.0404107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellett S, Tepp WH, Clancy CM, Borodic GE, Johnson EA. A neuronal cell-based botulinum neurotoxin assay for highly sensitive and specific detection of neutralizing serum antibodies. FEBS Lett. 2007;581(25):4803–4808. doi: 10.1016/j.febslet.2007.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellett S, Du ZW, Pier CL, et al. Sensitive and quantitative detection of botulinum neurotoxin in neurons derived from mouse embryonic stem cells. Biochem Biophys Res Commun. 2010;404(1):388–392. doi: 10.1016/j.bbrc.2010.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Whitemarsh RC, Strathman MJ, Chase LG, et al. Novel application of human neurons derived from induced pluripotent stem cells for highly sensitive botulinum neurotoxin detection. Toxicol Sci. 2012;126(2):426–435. doi: 10.1093/toxsci/kfr354. The study demonstrates the applicability of iPS-derived neuronal cultures for BoNT studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNutt P, Celver J, Hamilton T, Mesngon M. Embryonic stem cell-derived neurons are a novel, highly sensitive tissue culture platform for botulinum research. Biochem Biophys Res Commun. 2011;405(1):85–90. doi: 10.1016/j.bbrc.2010.12.132. [DOI] [PubMed] [Google Scholar]
- 32.Kiris E, Nuss JE, Burnett JC, et al. Embryonic stem cell-derived motoneurons provide a highly sensitive cell culture model for botulinum neurotoxin studies, with implications for high-throughput drug discovery. Stem Cell Res. 2011;6(3):195–205. doi: 10.1016/j.scr.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grumelli C, Verderio C, Pozzi D, et al. Internalization and mechanism of action of clostridial toxins in neurons. Neurotoxicology. 2005;26(5):761–767. doi: 10.1016/j.neuro.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Basavanna U, Muruvanda T, Brown EW, Sharma SK. Development of a Cell-Based Functional Assay for the Detection of Clostridium botulinum Neurotoxin Types A and E. Int J Microbiol. 2013;2013:593219. doi: 10.1155/2013/593219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eubanks LM, Hixon MS, Jin W, et al. An in vitro and in vivo disconnect uncovered through high-throughput identification of botulinum neurotoxin A antagonists. Proc Natl Acad Sci U S A. 2007;104(8):2602–2607. doi: 10.1073/pnas.0611213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yowler BC, Kensinger RD, Schengrund CL. Botulinum neurotoxin A activity is dependent upon the presence of specific gangliosides in neuroblastoma cells expressing synaptotagmin I. J Biol Chem. 2002;277(36):32815–32819. doi: 10.1074/jbc.M205258200. [DOI] [PubMed] [Google Scholar]
- 37.Tsukamoto K, Arimitsu H, Ochi S, et al. P19 embryonal carcinoma cells exhibit high sensitivity to botulinum type C and D/C mosaic neurotoxins. Microbiol Immunol. 2012;56(10):664–672. doi: 10.1111/j.1348-0421.2012.00490.x. [DOI] [PubMed] [Google Scholar]
- 38.Purkiss JR, Friis LM, Doward S, Quinn CP. Clostridium botulinum neurotoxins act with a wide range of potencies on SH-SY5Y human neuroblastoma cells. Neurotoxicology. 2001;22(4):447–453. doi: 10.1016/s0161-813x(01)00042-0. [DOI] [PubMed] [Google Scholar]
- 39.Hale M, Oyler G, Swaminathan S, Ahmed SA. Basic tetrapeptides as potent intracellular inhibitors of type A botulinum neurotoxin protease activity. J Biol Chem. 2011;286(3):1802–1811. doi: 10.1074/jbc.M110.146464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40**.Fernandez-Salas E, Wang J, Molina Y, et al. Botulinum neurotoxin serotype a specific cell-based potency assay to replace the mouse bioassay. PLoS One. 2012;7(11):e49516. doi: 10.1371/journal.pone.0049516. Authors utilized a monoclonal antibody that specifically detects BoNT/A cleaved SNAP-25 fragment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tegenge MA, Bohnel H, Gessler F, Bicker G. Neurotransmitter vesicle release from human model neurons (NT2) is sensitive to botulinum toxin A. Cell Mol Neurobiol. 2012;32(6):1021–1029. doi: 10.1007/s10571-012-9818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Whitemarsh RC, Pier CL, Tepp WH, Pellett S, Johnson EA. Model for studying Clostridium botulinum neurotoxin using differentiated motor neuron-like NG108-15 cells. Biochem Biophys Res Commun. 2012;427(2):426–430. doi: 10.1016/j.bbrc.2012.09.082. The study demonstrates the applicability of iPS-derived neuronal cultures for BoNT studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong WS, Young EW, Tepp WH, Johnson EA, Beebe DJ. A microscale neuron and schwann cell coculture model for increasing detection sensitivity of botulinum neurotoxin type a. Toxicol Sci. 2013;134(1):64–72. doi: 10.1093/toxsci/kft082. [DOI] [PubMed] [Google Scholar]
- 44*.Restani L, Giribaldi F, Manich M, et al. Botulinum neurotoxins A and E undergo retrograde axonal transport in primary motor neurons. PLoS Pathog. 2012;8(12):e1003087. doi: 10.1371/journal.ppat.1003087. This study utilizes motor neurons to provide insights into how BoNTs travel inside the cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch MJ, Purkiss JR, Foster KA. Sensitivity of embryonic rat dorsal root ganglia neurons to Clostridium botulinum neurotoxins. Toxicon. 2000;38(2):245–258. doi: 10.1016/s0041-0101(99)00153-1. [DOI] [PubMed] [Google Scholar]
- 46.Sun S, Suresh S, Liu H, et al. Receptor binding enables botulinum neurotoxin B to sense low pH for translocation channel assembly. Cell Host Microbe. 2011;10(3):237–247. doi: 10.1016/j.chom.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong M, Yeh F, Tepp WH, et al. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312(5773):592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 48.Verderio C, Grumelli C, Raiteri L, et al. Traffic of botulinum toxins A and E in excitatory and inhibitory neurons. Traffic. 2007;8(2):142–153. doi: 10.1111/j.1600-0854.2006.00520.x. [DOI] [PubMed] [Google Scholar]
- 49.Holtje M, Schulze S, Strotmeier J, et al. Exchanging the minimal cell binding fragments of tetanus neurotoxin in botulinum neurotoxin A and B impacts their toxicity at the neuromuscular junction and central neurons. Toxicon. 2013;75:108–121. doi: 10.1016/j.toxicon.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Kroken AR, Karalewitz AP, Fu Z, Kim JJ, Barbieri JT. Novel ganglioside-mediated entry of botulinum neurotoxin serotype D into neurons. J Biol Chem. 2011;286(30):26828–26837. doi: 10.1074/jbc.M111.254086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keller JE, Cai F, Neale EA. Uptake of botulinum neurotoxin into cultured neurons. Biochemistry. 2004;43(2):526–532. doi: 10.1021/bi0356698. [DOI] [PubMed] [Google Scholar]
- 52.Wiese S, Herrmann T, Drepper C, et al. Isolation and enrichment of embryonic mouse motoneurons from the lumbar spinal cord of individual mouse embryos. Nat Protoc. 2009;5(1):31–38. doi: 10.1038/nprot.2009.193. [DOI] [PubMed] [Google Scholar]
- 53.Camu W, Henderson CE. Purification of embryonic rat motoneurons by panning on a monoclonal antibody to the low-affinity NGF receptor. J Neurosci Methods. 1992;44(1):59–70. doi: 10.1016/0165-0270(92)90114-s. [DOI] [PubMed] [Google Scholar]
- 54.Miles GB, Yohn DC, Wichterle H, et al. Functional properties of motoneurons derived from mouse embryonic stem cells. J Neurosci. 2004;24(36):7848–7858. doi: 10.1523/JNEUROSCI.1972-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 56.Wichterle H, Peljto M, Nedelec S. Xenotransplantation of embryonic stem cell-derived motor neurons into the developing chick spinal cord. Methods Mol Biol. 2009;482:171–183. doi: 10.1007/978-1-59745-060-7_11. [DOI] [PubMed] [Google Scholar]
- 57.Wichterle H, Peljto M. Differentiation of mouse embryonic stem cells to spinal motor neurons. Curr Protoc Stem Cell Biol. 2008;Chapter 1:Unit 1H 1 1–1H 1 9. doi: 10.1002/9780470151808.sc01h01s5. [DOI] [PubMed] [Google Scholar]
- 58.Soundararajan P, Miles GB, Rubin LL, Brownstone RM, Rafuse VF. Motoneurons derived from embryonic stem cells express transcription factors and develop phenotypes characteristic of medial motor column neurons. J Neurosci. 2006;26(12):3256–3268. doi: 10.1523/JNEUROSCI.5537-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harper JM, Krishnan C, Darman JS, et al. Axonal growth of embryonic stem cell-derived motoneurons in vitro and in motoneuron-injured adult rats. Proc Natl Acad Sci U S A. 2004;101(18):7123–7128. doi: 10.1073/pnas.0401103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adler S, Bicker G, Bigalke H, et al. The current scientific and legal status of alternative methods to the LD50 test for botulinum neurotoxin potency testing. The report and recommendations of a ZEBET Expert Meeting. Altern Lab Anim. 2010;38(4):315–330. doi: 10.1177/026119291003800401. [DOI] [PubMed] [Google Scholar]
- 61**.Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110. This is an extremely interesting study demonstrating that species-specific differences between mouse and human systems are critical for basic research and drug discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McNeish J, Roach M, Hambor J, et al. High-throughput screening in embryonic stem cell-derived neurons identifies potentiators of alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate-type glutamate receptors. J Biol Chem. 2010;285(22):17209–17217. doi: 10.1074/jbc.M109.098814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li XJ, Hu BY, Jones SA, et al. Directed differentiation of ventral spinal progenitors and motor neurons from human embryonic stem cells by small molecules. Stem Cells. 2008;26(4):886–893. doi: 10.1634/stemcells.2007-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu BY, Zhang SC. Directed differentiation of neural-stem cells and subtype-specific neurons from hESCs. Methods Mol Biol. 2009;636:123–137. doi: 10.1007/978-1-60761-691-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3(6):637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 66.Dimos JT, Rodolfa KT, Niakan KK, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 67.Lee H, Shamy GA, Elkabetz Y, et al. Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons. Stem Cells. 2007;25(8):1931–1939. doi: 10.1634/stemcells.2007-0097. [DOI] [PubMed] [Google Scholar]
- 68.Singh Roy N, Nakano T, Xuing L, et al. Enhancer-specified GFP-based FACS purification of human spinal motor neurons from embryonic stem cells. Exp Neurol. 2005;196(2):224–234. doi: 10.1016/j.expneurol.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 69.Nizzardo M, Simone C, Falcone M, et al. Human motor neuron generation from embryonic stem cells and induced pluripotent stem cells. Cell Mol Life Sci. 2010;67(22):3837–3847. doi: 10.1007/s00018-010-0463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Amoroso MW, Croft GF, Williams DJ, et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J Neurosci. 2013;33(2):574–586. doi: 10.1523/JNEUROSCI.0906-12.2013. This is an excellent study demonstrating the generation of high-yield motor neurons from pluripotent cells in as short as 3 weeks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 72.Marchetto MC, Brennand KJ, Boyer LF, Gage FH. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum Mol Genet. 2011;20(R2):R109–115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boland MJ, Hazen JL, Nazor KL, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461(7260):91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 74.Zhao XY, Li W, Lv Z, et al. Viable fertile mice generated from fully pluripotent iPS cells derived from adult somatic cells. Stem Cell Rev. 2010;6(3):390–397. doi: 10.1007/s12015-010-9160-3. [DOI] [PubMed] [Google Scholar]
- 75.Narsinh KH, Plews J, Wu JC. Comparison of human induced pluripotent and embryonic stem cells: fraternal or identical twins? Mol Ther. 2011;19(4):635–638. doi: 10.1038/mt.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu BY, Weick JP, Yu J, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107(9):4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng Q, Lu SJ, Klimanskaya I, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28(4):704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 78.Narsinh KH, Sun N, Sanchez-Freire V, et al. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest. 2011;121(3):1217–1221. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sandoe J, Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat Neurosci. 2013;16(7):780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 80.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9(1):17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 81.Bock C, Kiskinis E, Verstappen G, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boulting GL, Kiskinis E, Croft GF, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(3):279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marchetto MC, Muotri AR, Mu Y, et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3(6):649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 85.Yang N, Ng YH, Pang ZP, Sudhof TC, Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell. 2011;9(6):517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Son EY, Ichida JK, Wainger BJ, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9(3):205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y, Pak C, Han Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78(5):785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buchser W, Collins M, Garyantes T, et al. Assay Development Guidelines for Image-Based High Content Screening, High Content Analysis and High Content Imaging. In: Sittampalam GS, Gal-Edd N, Arkin M, et al., editors. Assay Guidance Manual. Bethesda (MD): 2004. [Google Scholar]
- 89.Eglen RM, Reisine T. New insights into GPCR function: implications for HTS. Methods Mol Biol. 2009;552:1–13. doi: 10.1007/978-1-60327-317-6_1. [DOI] [PubMed] [Google Scholar]
- 90.de Paiva A, Meunier FA, Molgo J, Aoki KR, Dolly JO. Functional repair of motor endplates after botulinum neurotoxin type A poisoning: biphasic switch of synaptic activity between nerve sprouts and their parent terminals. Proc Natl Acad Sci U S A. 1999;96(6):3200–3205. doi: 10.1073/pnas.96.6.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bickle M. The beautiful cell: high-content screening in drug discovery. Anal Bioanal Chem. 2010;398(1):219–226. doi: 10.1007/s00216-010-3788-3. [DOI] [PubMed] [Google Scholar]
- 92.Honer WG, Falkai P, Bayer TA, et al. Abnormalities of SNARE mechanism proteins in anterior frontal cortex in severe mental illness. Cereb Cortex. 2002;12(4):349–356. doi: 10.1093/cercor/12.4.349. [DOI] [PubMed] [Google Scholar]
- 93.Degorce F, Card A, Soh S, et al. HTRF: A technology tailored for drug discovery - a review of theoretical aspects and recent applications. Curr Chem Genomics. 2009;3:22–32. doi: 10.2174/1875397300903010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bakry N, Kamata Y, Simpson LL. Lectins from Triticum vulgaris and Limax flavus are universal antagonists of botulinum neurotoxin and tetanus toxin. J Pharmacol Exp Ther. 1991;258(3):830–836. [PubMed] [Google Scholar]
- 95.Fischer A, Nakai Y, Eubanks LM, et al. Bimodal modulation of the botulinum neurotoxin protein-conducting channel. Proc Natl Acad Sci U S A. 2009;106(5):1330–1335. doi: 10.1073/pnas.0812839106. [DOI] [PMC free article] [PubMed] [Google Scholar]


