Summary
Embryonic development relies on the capacity of progenitor cells to appropriately respond to inductive cues; a cellular property known as developmental competence. Here we report that epigenetic priming of enhancers signifies developmental competence during endodermal lineage diversification. Chromatin mapping during pancreatic and hepatic differentiation of human embryonic stem cells revealed the en masse acquisition of a poised chromatin state at enhancers specific to endoderm-derived cell lineages in gut tube intermediates. Experimentally, the acquisition of this poised enhancer state predicts the ability of endodermal intermediates to respond to inductive signals. Furthermore, these enhancers are first recognized by the pioneer transcription factors FOXA1 and FOXA2 when competence is acquired, while subsequent recruitment of lineage-inductive transcription factors, such as PDX1, leads to enhancer and target gene activation. Together, our results identify the acquisition of a poised chromatin state at enhancers as a mechanism by which progenitor cells acquire developmental competence.
Introduction
Embryonic development is a forward-moving process during which pluripotent cells become increasingly specialized as they develop toward a terminally differentiated state. The stepwise progression toward specific cell lineages occurs as a result of a series of inductive events. The ability of lineage intermediates to appropriately interpret inductive signals from their environment is referred to as developmental competence. A classic example of this is the induction of the neuronal lineage by mesodermal cells at the time of gastrulation. Signals from the mesoderm act on the ectoderm, causing it to form neural tissue (Linker and Stern, 2004). However, only ectoderm of a certain developmental age is capable of appropriately responding to the inductive signal (Storey et al., 1992). Thus, developmental competence is a cell-intrinsic property of the responder tissue. Furthermore, competence is not inherent to the pluripotent state but actively acquired during development. What mechanisms operate to render cells competent to respond to inductive cues with precise timing is currently unknown.
While transcription factors (TFs) are important contributors to cellular competence, they are not sufficient to explain the highly cell type-specific responses to inductive cues during development. TFs typically occupy only a small fraction of their consensus binding motifs in the genome (Carr and Biggin, 1999; Iyer et al., 2001; Yang et al., 2006), suggesting that determinants beyond DNA sequence must dictate where and when TFs bind potential targets. Emerging evidence suggests that chromatin structure represents an inherent and important determinant of accessibility of DNA to TFs (Martino et al., 2009; Shogren-Knaak et al., 2006). Of particular interest is the chromatin state at enhancers, which plays a prominent role in spatiotemporal gene regulation during development (Creyghton et al., 2010; Heintzman et al., 2009; Koch et al., 2007; Rada-Iglesias et al., 2011; Visel et al., 2009). A central feature of enhancers is their ability to function as integrated TF binding platforms, where environmental signaling cues are interpreted in a context-dependent manner (Buecker and Wysocka, 2012; Jin et al., 2011). How enhancers acquire the ability to translate signals from the extracellular environment into cell type-specific transcriptional responses during development is poorly understood.
In this study, we examined the possibility that the epigenetic state of enhancers could determine developmental competence in the context of endodermal and pancreatic development. We explored this question by generating comprehensive maps of enhancer-related chromatin modifications over a time course of human embryonic stem cell (hESC) differentiation through multiple developmental intermediates into pancreatic and hepatic cells. Through integrative analysis of these maps and further experimentation, we reveal previously insufficiently appreciated links between enhancer chromatin, TF recruitment and developmental competence. First, we show that developmental competence is encoded at the level of enhancers and is established en masse in embryonic intermediates prior to lineage induction via acquisition of poised chromatin at enhancers specific to descendant lineages. Second, we find that TF complexes assemble at lineage-specific enhancers in a stepwise fashion. TFs involved in chromatin priming are recruited early when lineage intermediates acquire competence followed by the recruitment of lineage-inductive TFs to mediate activation. Together, these findings establish a functional link between the gain of a poised enhancer chromatin state and the temporal acquisition of competence during developmental progression.
Results
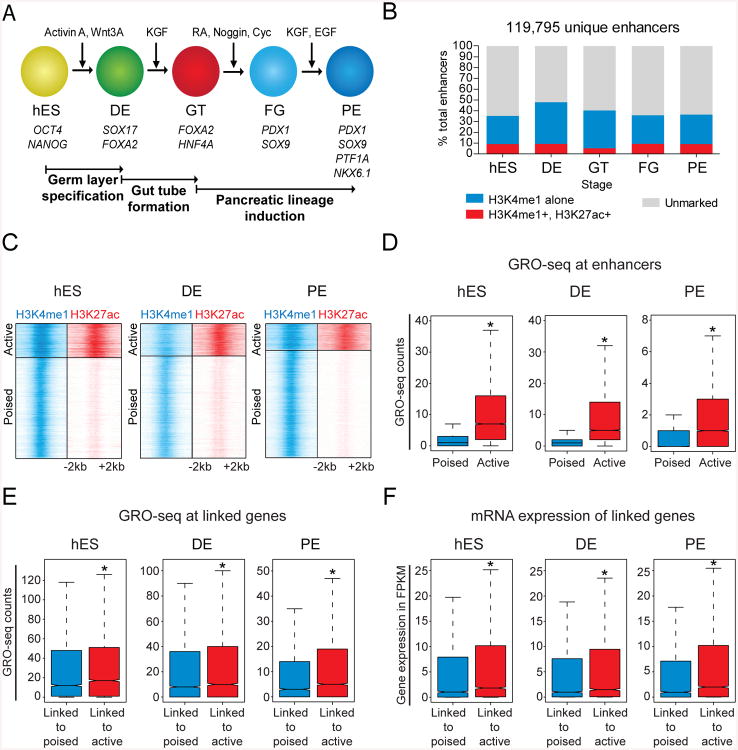
Global Identification of Enhancers during Pancreatic Differentiation of hESCs
During early embryogenesis, the pancreas, liver and lung develop from the endodermal gut tube (Wang and Sander, 2012). How and when organ-specific transcriptional programs are initiated during this developmental progression remains poorly understood. Because enhancers are important regulators of cell type-specific gene expression (Heintzman et al., 2009; Koch et al., 2007; Rada-Iglesias et al., 2011; Visel et al., 2009), we reasoned that mapping enhancers during endodermal lineage progression could provide mechanistic insight into these questions. Thus, we comprehensively mapped putative enhancers during the stepwise progression of hESCs toward the pancreatic fate using a system that accurately models early developmental processes, including gut tube formation and pancreatic lineage induction (Fig. 1A). Specifically, we analyzed enhancer-associated histone modifications genome-wide at five defined stages of differentiation: hESCs, definitive endoderm (DE), primitive gut tube (GT), posterior foregut (FG), and pancreatic endoderm (PE). These cell populations were each produced with > 90% purity (Xie et al., 2013). We identified a total of 119,795 enhancers across these five stages of pancreatic differentiation (Fig. 1B). The majority of marked enhancers at each stage are marked by H3K4me1 only, with only a small fraction also marked by H3K27ac (Fig. 1C; S1A,C; Table S1).
Figure 1. Global identification of poised and active enhancers during pancreatic differentiation of human embryonic stem cells.
(A) Human embryonic stem cell differentiation strategy. (B) Total number of candidate enhancers identified during pancreatic differentiation categorized by H3K27ac and H3K4me1 deposition. (C) Density of ChIP-seq reads for H3K4me1 and H3K27ac relative to midpoint at putative poised and active enhancers. (D) Box plots of GRO-seq counts at poised (H3K4me1 only) and active (H3K4me1 and H3K27ac) enhancers. (E) Box plots of GRO-seq counts at linked genes of poised and active enhancers. (F) Box plots of mRNA expression, measured in FPKM, at linked genes of poised and active enhancers. * P-values < 2.2e-16, wilcoxon test. hES, human embryonic stem cells; DE, definitive endoderm; GT, primitive gut tube; FG, posterior foregut; PE, pancreatic endoderm. See also Figure S1 and Table S1.
It has been suggested that H3K27ac can distinguish active from poised enhancers and that the poised state could facilitate enhancer activation (Creyghton et al., 2010; RadaIglesias et al., 2011). To obtain a direct read-out of enhancer activity, we performed global nuclear run-on sequencing (GRO-seq) of nascent transcripts. We observed a significant enrichment of transcriptionally engaged RNA polymerases indicative of enhancer RNA (eRNA) production at active (defined by H3K4me1 and H3K27ac deposition) compared to poised enhancers (defined by only H3K4me1 deposition) (Fig. 1D; S1B,D). Combined with previous studies linking eRNA production to enhancer activity (Hah et al., 2013; Kim et al., 2010; Wang et al., 2011), our results suggest that enrichment of H3K4me1 alone likely constitutes an inactive enhancer state. We also examined the transcriptional activity of putative target genes by quantifying their GRO-seq and RNA-seq counts. Consistent with higher eRNA production at active enhancers, target promoters of active enhancers exhibited higher GRO-seq and RNA-seq counts than promoters linked to poised enhancers (Fig. 1E,F; S1B,D). These results define two functionally distinct classes of putative developmental enhancers based on chromatin features.
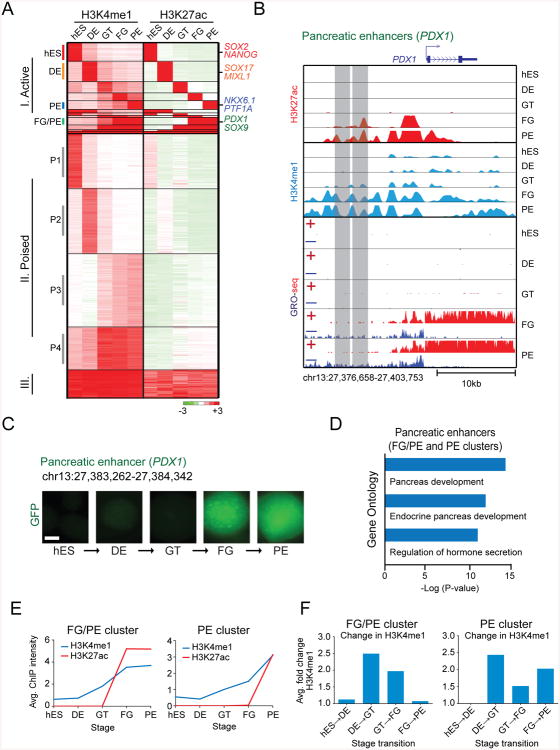
Identification of Enhancers that Drive Pancreatic Lineage Induction
Having globally predicted both poised and active enhancers across all five stages, we next sought to isolate the specific sets of enhancers that regulate developmental transitions and, in particular, pancreas induction. To do this, we clustered enhancers based on their predicted activity across all differentiation stages. This revealed three major categories: I. active in a stage-restricted manner, II. poised yet never active and III. constitutively active (Fig. 2A). As expected, when tested in enhancer-reporter assays, category I enhancers exhibited stage-specific activity according to their H3K27ac status (Fig. 2B,C; S2A-C). Illustrating their importance in developmental gene regulation, category I enhancers were predicted to regulate developmental processes specific to the embryonic stage of their activity (Fig. 2D; S2D). Of particular interest were enhancers that acquired H3K27ac during pancreatic lineage induction (FG/PE- and PE-specific clusters) (Fig. 2A). These enhancers associated with genes involved in pancreas and endocrine systems development, which include the master regulators of pancreas induction PDX1, SOX9, PTF1A, and NKX6.1 (Shih et al., 2013) (Fig. 2A,B,D). It is likely that these enhancers have roles in regulating the induction of pancreatic gene expression programs when the pancreas develops from the gut tube.
Figure 2. Identification and characterization of enhancers important for pancreatic lineage induction.
(A) K-means clustering of putative enhancers during pancreatic differentiation based on H3K4me1 and H3K27ac signal intensity. Individual genes associated with enhancer clusters are listed on the right. (B) ChIP-seq H3K4me1, H3K27ac and GRO-seq profiles of representative FG/PE-specific enhancers near PDX1. (C) In vitro GFP reporter assay for FG/PE-specific enhancer shown in B (right). GFP images of cell aggregates during pancreatic differentiation are shown. Scale bar, 100 μm. (D) Enriched Gene Ontology terms for pancreatic enhancers. (E) Average H3K4me1 and H3K27ac ChIP-seq signal intensity for pancreatic (FG/PE- and PE-specific clusters) enhancers during pancreatic differentiation. (F) Average fold change in H3K4me1 signal intensity during each stage transition for pancreatic (FG/PE- and PE-specific clusters) enhancers. hES, human embryonic stem cells; DE, definitive endoderm; GT, primitive gut tube; FG, posterior foregut; PE, pancreatic endoderm. See also Figure S2.
Pancreatic Enhancers Acquire a Poised State in Gut Tube Intermediates prior to Activation
To investigate chromatin dynamics at enhancers associated with pancreatic lineage induction, we next examined H3K4me1 and H3K27ac intensities at pancreas-specific enhancers. In both the FG/PE- and PE-specific clusters, H3K4me1 levels accumulated prior to H3K27ac (Fig. 2E), suggesting that pancreatic enhancers are poised prior to activation. Interestingly, pancreas-specific enhancers (FG/PE and PE clusters) exhibited relatively little H3K4me1 enrichment in hESCs (Fig. 2E), indicating that the poised state is actively acquired during development. By examining the average fold change in H3K4me1 levels at each transition, we observed the largest fold increase in H3K4me1 at pancreatic enhancers during the DE to GT transition (Fig. 2F), when these enhancers are still inactive as judged by H3K27ac levels and enhancer-reporter assays (Fig. 2C,E; S2B,C). Thus, enhancers associated with pancreatic lineage programs acquire a poised state during gut tube formation independent of pancreas-inductive signaling cues. These findings suggest that epigenetic bookmarking of pancreatic enhancers can pre-program gut tube intermediates to activate pancreas-specific genes when exposed to pro-pancreatic signaling cues.
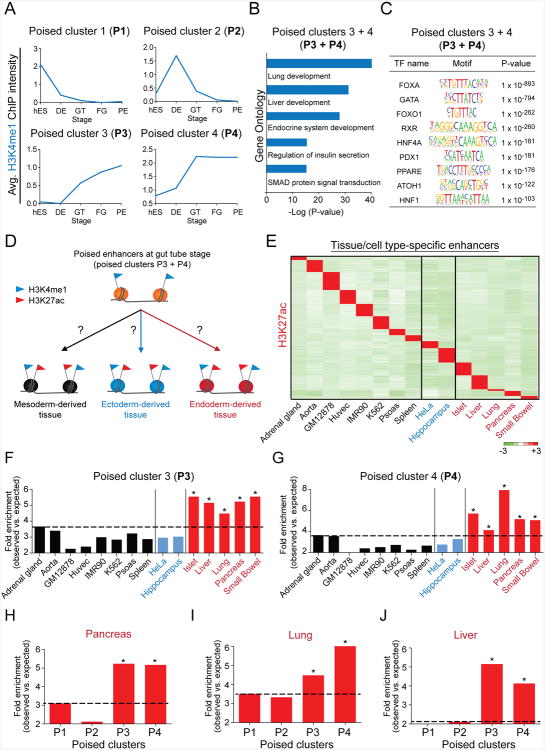
A Poised Enhancer Landscape for Multiple Endodermal Lineages Is Established in Gut Tube Intermediates
One surprising result of the enhancer cluster analysis was that the majority (66%) of predicted enhancers during pancreatic lineage progression belong to category II (Fig. 2A), meaning they acquire a poised state yet never become active. Category II enhancers are comprised of four clusters: 1. poised in hESCs (P1), 2. poised at the DE stage (P2), 3. poised with intermediate H3K4me1 levels at the GT, FG and PE stage (P3), and 4. poised with high H3K4me1 levels at the GT, FG and PE stage (P4) (Fig. 2A; 3A). Intriguingly, H3K4me1 levels in clusters P3 and P4 exhibit a dramatic gain during the DE to GT transition (Fig. 3A), as seen for pancreas-specific enhancers (Fig. 2E,F).
Figure 3. A poised enhancer landscape for multiple endodermal lineages is established in gut tube lineage intermediates.
(A) Average H3K4me1 ChIP-seq signal intensity during pancreatic differentiation for enhancers in poised clusters P1, P2, P3 and P4 (see Figure 2A). (B) Enriched Gene Ontology terms for enhancers in poised clusters P3 and P4. (C) Enriched transcription factor (TF) binding motifs with associated P-values for enhancers in poised clusters P3 and P4. (D) Experimental strategy to determine activity of poised enhancers in tissues and cell types derived from mesoderm, ectoderm and endoderm. (E) Heatmap showing H3K27ac ChIP-seq signal intensity for H3K27ac peaks specific to each listed tissue/cell type. Mesoderm-derived cells are depicted in black, ectoderm-derived cells in blue and endoderm-derived cells in red. (F, G) Enrichment of tissue- or cell type-specific H3K27ac peaks in poised enhancer clusters P3 (F, * P-values ≤ 2.7e-8, chi-square test) and P4 (G, * P-values ≤ 0.03, chi-square test). (H-J) Enrichment of pancreas-specific (H) lung-specific (I) liver-specific (J) H3K27ac peaks in poised enhancer clusters P1, P2, P3 and P4. * P-values ≤ 0.02, chi-square test. hES, human embryonic stem cells; DE, definitive endoderm; GT, primitive gut tube; FG, posterior foregut; PE, pancreatic endoderm. See Figure S3 and Table S2.
This raises the question of why P3 and P4 enhancers acquire a poised state (H3K4me1) during gut tube formation. Given that the primitive gut tube gives rise to multiple organs, including pancreas, lung and liver, we hypothesized that these poised enhancers could become active either later during terminal differentiation into mature pancreatic cells or in alternate gut tube-derived lineages. In support of this hypothesis, enhancers from clusters P3 and P4 associated with genes involved in the development and function of pancreatic islets, lung and liver (Fig. 3B). Furthermore, poised enhancers in clusters P3 and P4 were enriched for binding motifs of transcription factors known to regulate the development of endodermal organs (Fig. 3C), including FOXA, GATA, HNF4A, and HNF1 (Boj et al., 2010; Gao et al., 2008; Lango Allen et al., 2012; Lee et al., 2005; Li et al., 2000; Watt et al., 2007).
If these enhancers are indeed activated during terminal pancreatic differentiation or in alternate endodermal organs, one would expect enhancers in clusters P3 and P4 to selectively acquire H3K27ac in cells of gut tube-derived organs. To test this in an unbiased fashion, we analyzed the H3K27ac status of enhancers from clusters P3 and P4 in differentiated tissues and cells originating from all three germ layers (Fig. 3D). First, we identified distal tissue-specific H3K27ac peaks by querying data from the Roadmap Epigenomics project as well as the Encyclopedia of DNA elements consortium (ENCODE) (Fig. 3E). As expected, genes linked to tissue-specific H3K27ac peaks associated with biological processes characteristic of the respective tissue (Table S2). Next, we determined the extent to which these tissue-specific active enhancers were represented in each of the poised enhancer clusters. Enhancers in clusters P1 and P2, which are exclusively poised at the hESC or DE stage, showed no consistent overrepresentation in tissues from a specific germ layer (Fig. S3A,B). By contrast, enhancers from clusters P3 and P4, which acquire a poised state at the GT stage, were significantly enriched among active enhancers in endoderm-derived tissues, including islet, liver, lung, pancreas, and small bowel (Fig. 3F,G). Furthermore, endodermal active enhancers were only enriched in poised clusters P3 and P4 when compared to clusters P1 and P2 (Fig. 3H-J; S3C-N). Altogether, these results demonstrate that a poised chromatin landscape for enhancers linked to genes of multiple gut tube-derived lineages is gained en masse during the DE to GT transition. This suggests that instructive information for gene expression programs of multiple descendant lineages is actively acquired and becomes encoded into enhancer chromatin at this time.
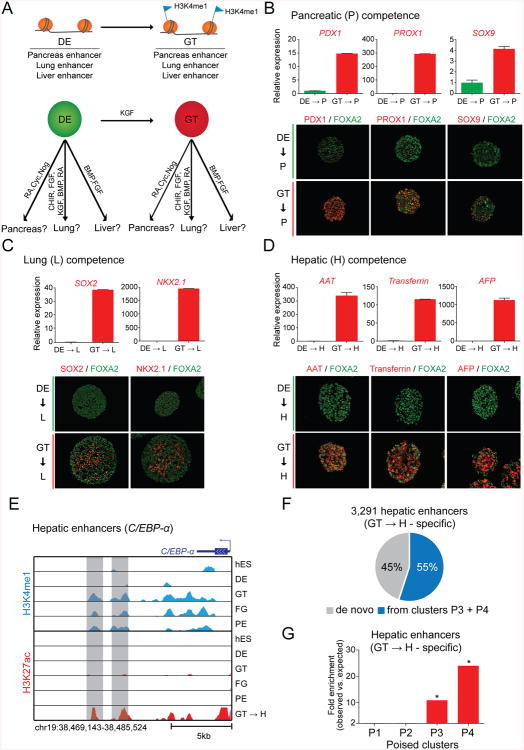
Acquisition of a Poised State at Lineage-specific Enhancers Is Indicative of Developmental Competence
Developmental competence refers to a cell's ability to respond to inductive signals during development (Waddington, 1940). We hypothesized that “poising” lineage-specific enhancers in GT intermediates could be a mechanism by which cells become competent to adopt endodermal organ fates in response to organ-inductive signals. To test this idea, we exposed DE and GT stage cells briefly to pancreas-, lung- or liver-inductive cues and determined whether early lineage markers are induced (Fig. 4A). Because pancreas, lung and liver enhancers become poised in GT intermediates, we predicted that GT stage cells, but not DE stage cells, will readily respond to appropriate organ-inductive cues. To first test pancreatic competence, we exposed DE and GT cells to pancreas-inductive factors (RA, Cyc, Nog) for two days. We found that expression of early pancreatic lineage markers, including PDX1, PROX1 and SOX9, was readily induced from GT cells (Fig. 4B; S4A). In contrast, this was not the case for DE cells (Fig. 4B; S4A). Second, to test lung competence, we cultured DE and GT cells for three days in the presence of lung-inductive factors (CHIR, FGF, KGF, BMP, and RA) (Huang et al., 2014). Analogous to our findings for pancreas, only GT stage cells responded by activating expression of the lung markers SOX2 and NKX2.1 (Fig. 4C). Finally, a similar experiment testing hepatic competence showed that GT stage but not DE stage cells quickly responded by expressing hepatic markers, such as AAT, Transferrin and AFP, when exposed to the liver-inductive factors BMP and FGF (Wang and Sander, 2012) for three days (Fig. 4D; S4B). The result that lung and liver could be induced from GT stage cells was unexpected, because the differentiation protocol has been specifically developed for production of pancreatic cells. Together, these findings establish a clear temporal connection between the acquisition of a poised enhancer state at organ-specific enhancers and the gain of developmental competence for the respective lineage.
Figure 4. The acquisition of a poised enhancer state coincides with gain of developmental competence.
(A) Experimental strategy to test the competence of endodermal intermediates to activate pancreatic (P), lung (L) or hepatic (H) genes in response to their respective organinductive signals. (B-D) Quantitative reverse transcription PCR analysis and immunofluorescence staining for the early pancreas markers PDX1, PROX1 and SOX9 in DE and GT cells treated with RA, Cyc, Nog for 2 days (B), the early lung markers SOX2 and NKX2.1 in DE and GT cells treated with CHIR, FGF, KGF, BMP, and RA for 3 days (C), and the early liver markers alpha 1-antitrypsin (AAT), transferrin and alpha-fetoprotein (AFP) in DE and GT cells treated with BMP and FGF for 3 days (D). Data are shown as average ± S.E.M. (E) H3K27ac and H3K4me1 ChIP-seq profiles of enhancers in poised clusters P3 and P4 near the hepatic gene C/EBP-α. (F) Percentage of hepatic enhancers (specific H3K27ac peaks in GT cells treated with BMP and FGF for 3 days; GT->H cells) that are poised in GT intermediates (clusters P3 + P4 in Figure 2A). (G) Enrichment of hepatic enhancers (GT->H cell-specific H3K27ac peaks) in poised enhancer clusters P1, P2, P3, and P4 (see Figure 2A). * P-values < 2.2e-16, chi-square test. RA, retinoic acid; Cyc, cyclopamine; Nog, noggin; CHIR, CHIR99021; FGF, fibroblast growth factor; KGF, keratinocyte growth factor; BMP, bone morphogenetic protein; hES, human embryonic stem cells; DE, definitive endoderm; GT, primitive gut tube; FG, posterior foregut; PE, pancreatic endoderm. See also Figure S4 and Table S3.
Given the rapid induction of hepatic genes from GT stage cells after exposure to BMP and FGF, we postulated that liver-inductive cues convert poised enhancers into an active state. To test this contention, we mapped active enhancers specific to GT-derived hepatic cells (Table S3). Consistent with the cells' hepatic identity, enhancers activated upon hepatic induction were proximal to key hepatic regulators (i.e. C/EBP-α), associated with genes for liver-specific biological processes and were enriched for recognition motifs of known hepatic TFs (Fig. 4E; S4C,D). Strikingly, 55% of active enhancers specific to GT-derived hepatic cells belonged to poised clusters P3 and P4, which gain H3K4me1 in GT intermediates (Fig. 4F). A similar enrichment was not seen for poised clusters P1 and P2, which gain H3K4me1 at earlier stages (Fig. 4G). These results demonstrate that the majority of enhancers that are activated in response to liver-inductive cues acquire a poised state at the endoderm to gut tube transition.
Identification of Transcription Factors that Regulate Enhancers during Competence Establishment and Lineage Induction
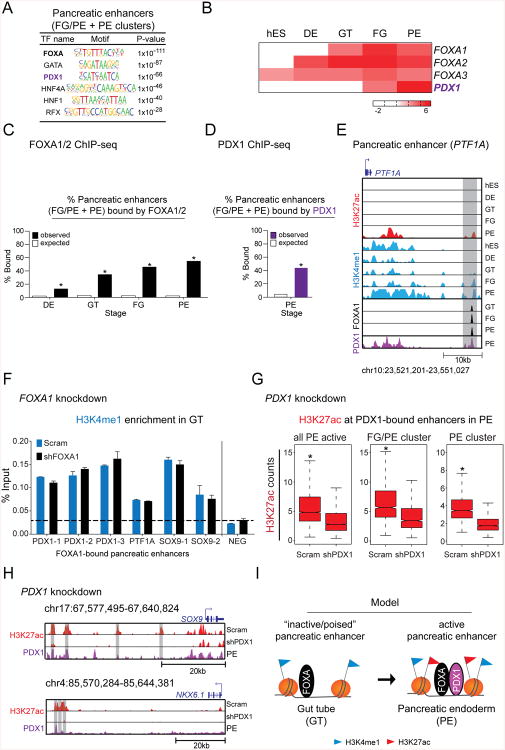
Because enhancers contain clusters of TF binding sites (Buecker and Wysocka, 2012), we reasoned that important transcriptional regulators of developmental competence and lineage induction could be identified by analyzing lineage-specific enhancers. Therefore, to identify potential regulators of pancreatic competence and lineage induction, we performed MOTIF enrichment analysis for pancreatic enhancers (FG/PE and PE clusters). This analysis revealed enrichment for FOXA, GATA, PDX1, HNF4A, HNF1, and RFX motifs (Fig. 5A), which bind TFs with documented functions in pancreas development (Boj et al., 2010; Gao et al., 2008; Jonsson et al., 1994; Lango Allen et al., 2012; Smith et al., 2010).
Figure 5. FOXA transcription factors and PDX1 are sequentially recruited to pancreatic enhancers.
(A) Enriched transcription factor (TF) binding motifs with associated P-values for pancreatic enhancers (FG/PE- and PE-specific clusters). (B) Heatmap showing mRNA expression levels, measured in FPKM, of FOXA family TFs and PDX1 during pancreatic differentiation. (C) Percentage of pancreatic enhancers versus random genomic regions bound by FOXA1 or FOXA2 at the DE, GT, FG, and PE stages. * P-values < 2.2e-16, chi-square test. (D) Percentage of pancreatic enhancers versus random genomic regions bound by PDX1 in pancreatic endoderm. * P-value < 2.2e-16, chi-square test. (E) H3K27ac, H3K4me1, FOXA1, and PDX1 ChIP-seq profiles at a candidate enhancer near PTF1A during pancreatic differentiation. (F) H3K4me1 enrichment by ChIP-qPCR at FOXA1-bound pancreatic enhancers in scrambled control (Scram) and FOXA1 knockdown (shFOXA1) cells differentiated to GT. (G) Box plots of H3K27ac ChIP-seq counts at all enhancers active in PE, FG/PE-specific, and PE-specific enhancers in scrambled control (Scram) and PDX1 knockdown (shPDX1) cells differentiated to PE. * P-values < 2.2e-16, wilcoxon test. (H) H3K27ac ChIP-seq profiles at enhancers near SOX9 and NKX6.1 in Scram and shPDX1 cells differentiated to PE as well as the PDX1 ChIP-seq profile at the same enhancers in PE. (I) Model for the stepwise activation of pancreatic enhancers. Pioneer TFs, such as FOXAs, associate with poised enhancers in gut tube intermediates. Lineage-specific TFs, such as PDX1, subsequently regulate the transition from a poised to an active enhancer state. hES, human embryonic stem cells; DE, definitive endoderm; GT, primitive gut tube; FG, posterior foregut; PE, pancreatic endoderm. See also Figure S5 and Tables S4-S6.
To begin to define the specific roles of candidate TFs at pancreas-specific enhancers, we examined mRNA levels of TFs corresponding to the enriched binding motifs. Our analysis revealed two general patterns: 1. TF expression coincides with pancreatic gene induction at the FG stage and 2. TF expression is initiated in GT intermediates or prior (Fig. S5A). As expected, the regulator of early pancreatic development PDX1 (Ahlgren et al., 1996; Jonsson et al., 1994) belonged to the group of TFs that was first expressed at the point of pancreas induction (Fig. 5B). Interestingly, expression of the FOXA family of TFs was initiated prior to pancreas induction (Fig. 5B). FOXAs are known to function as pioneer TFs, which can actively open chromatin and facilitate binding of other TFs (Zaret and Carroll, 2011). Based on their temporal expression pattern, we speculated that FOXAs might have a role in establishing competence at poised enhancers, whereas PDX1 might be involved in activating poised enhancers. To test these predictions, we first examined occupancy of pancreatic enhancers by FOXAs and PDX1 using ChIP-seq (Tables S4-6). We found that 34.8% (1943/5581; compared to an expected 2.8% by random chance) of pancreatic enhancers were indeed occupied by FOXA1 or FOXA2 at the GT stage prior to enhancer activation (Fig. 5C). After pancreatic induction, 44.7% (2497/5581; compared to an expected 3.6% by random chance) of pancreatic enhancers showed binding of PDX1 (Fig. 5D). Among the enhancers that were sequentially occupied by FOXAs and PDX1 was an enhancer for the early pancreatic regulator PTF1A (Fig. 5E). Mutations in this enhancer have been linked to familial pancreatic agenesis (Weedon et al., 2014).
To next investigate whether FOXAs have a regulatory role at poised enhancers, we delivered shRNAs targeting FOXA1 to hESCs and differentiated these cells towards the pancreatic lineage. shRNA-mediated FOXA1 knockdown caused a ∼2-fold reduction in FOXA1 transcript levels at the GT stage (Fig. S5B). When FOXA1-depleted GT intermediates were further differentiated toward pancreas, we observed a reduction in mRNA levels for early pancreatic markers, including PDX1, SOX9, NKX6.1, and PTF1A (Fig. S5B), showing that FOXA1 is necessary for the proper expression of early pancreatic genes. Given that the temporal pattern of FOXA recruitment to pancreatic enhancers mirrors H3K4me1 levels (Fig. 5C; 2E), we tested whether FOXA1 is required for H3K4me1 deposition at pancreatic enhancers. Examination of H3K4me1 levels by ChIP-qPCR analysis at multiple FOXA1-bound pancreatic enhancers did not reveal a noticeable difference in H3K4me1 enrichment between FOXA1 knockdown and control cells at the GT stage (Fig. 5F). These results suggest that FOXA1 is not directly involved in methylating histone 3 at lysine 4, but instead might recognize a poised enhancer state and help facilitate subsequent activation. This result is consistent with prior findings in different cell lines (Lupien et al., 2008).
We next investigated whether PDX1 is necessary for the activation of pancreatic enhancers by examining the effect of PDX1 inhibition on expression of pancreatic genes and histone acetylation at enhancers. Consistent with the role of PDX1 in mice and humans (Jonsson et al., 1994; Stoffers et al., 1997), PDX1 knockdown cells failed to initiate expression of important early pancreatic genes upon directed pancreatic differentiation (Fig. S5C,D). ChIP-seq analysis of H3K27ac in PDX1 knockdown cells after pancreas induction revealed a significant decrease in H3K27ac intensity at PDX1-bound pancreatic enhancers (Fig. 5G). For example, PDX1-bound enhancers near the genes encoding the pancreatic TFs SOX9 and NKX6.1 exhibited a drastic reduction in H3K27ac signal (Fig. 5H). Thus, PDX1 occupies pancreatic enhancers and is required for their activation. Together, our findings suggest a model whereby pancreatic enhancers assemble sequentially. In the primitive gut tube, pancreatic enhancers acquire a poised state and become occupied by FOXA TFs. Exposure to pancreas-inductive signaling cues subsequently leads to PDX1 induction, its recruitment to pre-marked pancreatic enhancers and histone acetylation (Fig. 5I).
To examine whether this model of stepwise enhancer assembly applies more generally to endodermal lineages, we analyzed motifs of hepatic enhancers to make predictions about which TFs could poise and which could activate hepatic enhancers during liver differentiation. We reasoned that by comparing motifs at hepatic enhancers that emerge from poised enhancers at the GT stage, to motifs at de novo active hepatic enhancers, we could identify TFs with potential roles in “poising” or “activating” enhancers. This analysis revealed a specific enrichment for FOXA motifs at those hepatic enhancers which are poised in GT intermediates (clusters P3 and P4) (Fig. S5E). 54.2% of active hepatic enhancers (981/1810; compared to an expected 3.0% by random chance) that are poised in GT were indeed occupied by either FOXA1 or FOXA2 prior to hepatic induction (Fig. S5F,G). Thus, FOXAs associate with a poised enhancer landscape for multiple endodermal organ lineages prior to lineage induction. The requirement of FOXAs for pancreas (Fig. S5B) and liver (Lee et al., 2005) development suggests that this early association of FOXAs with lineage-specific enhancers helps prepare poised enhancers for future activation. Similar to PDX1 in pancreas, enhancer activation in liver likely requires additional TFs. Comparison of TF recognition motifs at enhancers in poised clusters P3 and P4 that become active in GT-derived hepatic cells to motifs at enhancers in clusters P3 and P4 not active in hepatic cells revealed overrepresentation of binding motifs for HNF4A, HNF1 and TEAD (Fig. S5H), suggesting that these TFs could be involved in the activation of hepatic enhancers.
Enhancers for Islet Cell Functional Genes Are Poised prior to Terminal Differentiation
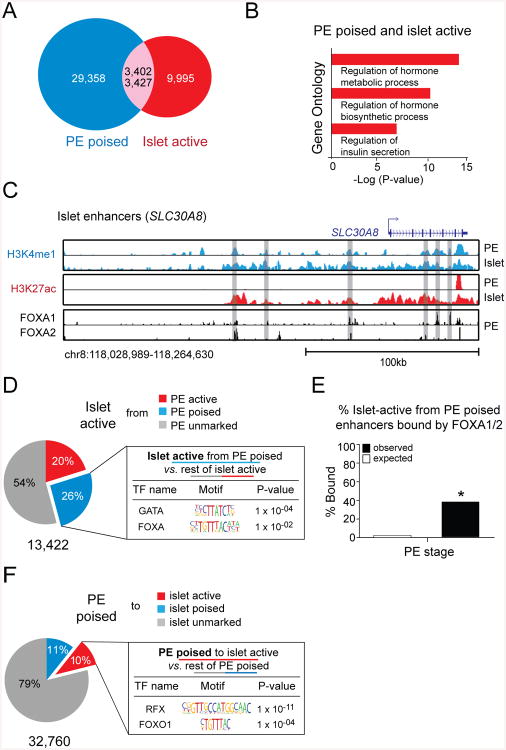
An important characteristic of hESC-derived PE is its competency to differentiate into functional endocrine cells after subcutaneous engraftment into mice (Kroon et al., 2008; Xie et al., 2013). Based on our observation that a poised enhancer state indicates developmental competence during endodermal organ lineage induction, we postulated that a similar priming of enhancer chromatin could render pancreatic progenitor cells competent to activate endocrine functional genes in response to extrinsic signals. To test this, we first identified active enhancers in human cadaveric islets (Fig. S6A,B; Table S7) and examined the extent to which these active islet enhancers are poised in PE prior to terminal differentiation. Strikingly, 25.5% (3,427/13,422) of all active islet enhancers are already poised in PE (Fig. 6A). This is remarkable considering that the differentiation of pancreatic progenitor cells into functional islet cells requires multiple developmental steps and considerable time. Furthermore, the select group of poised enhancers is relevant for the regulation of genes associated with vital islet-specific cellular properties, such as hormone biosynthesis and insulin secretion (Fig. 6B). One such example is the type II diabetes-associated gene SLC30A8 (van Hoek et al., 2008), for which we identified six associated enhancers that are poised in PE and active in islets (Fig. 6C). This analysis shows that enhancers for important genes involved in the regulation of endocrine cell function are pre-marked in pancreatic progenitor cells. Thus, instructive information for gene expression programs of islet function appears to be programmed into enhancer chromatin prior to terminal differentiation.
Figure 6. Enhancers linked to important islet cell genes are poised in pancreatic progenitor cells.
(A) Overlap of poised enhancers in pancreatic endoderm (PE) and active enhancers in cadaveric human islets. 3,402 out of 32,760 poised enhancers in PE become active in islets. 3,427 out of 13,422 active enhancers in islets are poised in PE. (B) Enriched Gene Ontology terms for enhancers that are poised in PE and become active in islets. (C) H3K4me1, H3K27ac, FOXA1, and FOXA2 ChIP-seq profiles in PE and islets at enhancers near SLC30A8. (D) Enriched transcription factor (TF) binding motifs with associated P-values for the 26% of active islet enhancers that are poised in PE versus the remaining 74% of active islet enhancers. (E) Percentage of enhancers, which are poised in PE and active in islets, versus random genomic regions bound by FOXA1 or FOXA2 in pancreatic endoderm. * P-value < 2.2e-16, chi-square test. (F) Enriched TF binding motifs with associated P-values for the 10% of poised enhancers in PE that become active in islets versus the remaining 89% of poised enhancers in PE. See also Figure S6 and Tables S5-S7.
To examine if TFs are also sequentially recruited to islet enhancers as observed during pancreatic and hepatic lineage induction, we analyzed binding motifs at enhancers that are poised in PE and active in islets. Specifically, to identify TFs with potential roles at poised enhancers, we compared motifs at active islet enhancers, which are poised at the PE stage, to motifs at active islet enhancers, which are not poised at the PE stage. This analysis revealed enrichment for FOXA motifs at poised islet enhancers (Fig. 6D). 39.3% (1348/3427; compared to an expected 2.5% by random chance) of these enhancers were occupied by either FOXA1 or FOXA2 in PE prior to endocrine differentiation (Fig. 6C,E), showing that FOXAs indeed associate with islet enhancers prior to their activation. When we compared the motifs at poised enhancers that become active in islets to motifs at poised PE enhancers that do not become active in islets, we found enrichment for RFX and FOXO1 motifs (Fig. 6F), indicating that these TFs could be important for activating islet enhancers during endocrine cell differentiation. In summary, these findings suggest a potential role for FOXA factors at poised islet enhancers in early embryonic intermediates and FOXO1 and RFX6 in the subsequent activation of islet enhancers during terminal differentiation. These predictions bear striking resemblance to our analysis identifying TFs regulating the stepwise activation of enhancers during pancreatic and hepatic lineage induction from the gut tube. Altogether, these observations support a model whereby FOXAs at poised enhancers play a critical role in establishing competence throughout development, including terminal differentiation into functional cell types.
Discussion
Acquisition of Developmental Competence through de novo Poising of Lineage-specific Enhancers
An unanswered question in developmental biology is which cell-intrinsic mechanisms enable developmental intermediates to specifically activate cell identity genes in response to extrinsic signaling cues. Here, we demonstrate that this cell-intrinsic property, referred to as developmental competence, is functionally linked to a poised chromatin state at cell type-specific enhancers. Our findings suggest that bookmarking cell identity genes at the level of enhancers endows cells with the ability to correctly interpret environmental differentiation cues. The annotation of the poised enhancer repertoire during developmental progression can thus provide prescient information about future cellular states.
A key characteristic of developmental competence is that it is not a passive state, but is actively acquired during differentiation. For example, endodermal cells activate pancreatic genes in response to co-culture with notochord. However, notochord is only capable of acting on endoderm after the endoderm has received prior instruction from mesoderm/ectoderm (Wells and Melton, 2000). Analogous to these findings in primary embryonic tissues, we observed that the competence to activate pancreas, lung and liver genes in response to extrinsic signaling cues is actively acquired at the transition from endoderm to gut tube during the in vitro differentiation of hESCs. Because the de novo poising of lineage-specific enhancers coincides with the acquisition of developmental competence for pancreas, lung and liver induction, our findings strongly suggest a functional link. We similarly find that enhancers associated with genes controlling endocrine cell function are poised in pancreatic progenitor cells, suggesting that poising cell type-specific enhancers is relevant at multiple developmental steps, including terminal differentiation. Certainly, other transcriptional priming mechanisms, such as a bivalent chromatin state at the level of promoters (Bernstein et al., 2006; Mikkelsen et al., 2007; Xie et al., 2013), may also be biologically important for a cell's developmental potential. However, in contrast to the poised enhancer state, which is acquired during development with precise timing, promoters of organ-specific genes often exhibit a bivalent state already in pluripotent stem cells. Thus, bivalent domains alone cannot explain the acquisition of developmental competence during lineage progression. Our findings help explain why developmental intermediates respond to signaling cues with high precision in time.
Stepwise Enhancer Assembly during Lineage Progression
Our data shows that the stepwise developmental transition at lineage-specific enhancers from 1. unmarked chromatin, to 2. poised chromatin, to 3. histone H3K27 acetylation is associated with the sequential assembly of distinct classes of TFs at these enhancers. Specifically, our findings suggest that pioneer TFs, in particular FOXAs, play a role at poised enhancers, while lineage-specifying TFs promote the transition from a poised to an active enhancer state. Whereas our work experimentally demonstrates a role for PDX1 in activating poised pancreatic enhancers, motifs overrepresented at hepatic and islet enhancers indicate similar functions for HNF4A and RFX factors during hepatic and islet cell differentiation, respectively. Such notion is consistent with reported phenotypes of Hnf4a and Rfx6 knockout mice (Li et al., 2000; Smith et al., 2010). In contrast to PDX1 knockdown, which prevented H3K27ac deposition at pancreatic enhancers, FOXA1 knockdown did not affect H3K4me1 levels. As reported in cell lines (Lupien et al., 2008), it appears that although necessary for target gene activation, FOXA1 activity is not required for H3K4 methylation in the context of endoderm development. Given the known property of FOXAs to displace nucleosomes (Li et al., 2012), a likely mechanism by which FOXAs regulate enhancer activity is by establishing a transcriptionally permissive enhancer chromatin state.
This raises the question as to which, if any, TFs are responsible for the deposition of H3K4me1 at lineage-specific enhancers. Although our data suggests that FOXA1 is not required, it is possible that other FOXA TFs compensate for FOXA1 and mediate H3K4me1 deposition in FOXA1-deficient cells. Furthermore, other TFs not studied here could likewise play this role. Also unclear is the functional role of H3K4me1 in priming enhancers for future activation. While our data suggest that H3K4me1 deposition alone is not sufficient for future gene activation in a FOXA1-depleted state, it remains to be investigated whether enhancer activation requires H3K4 methylation and how TFs and epigenetic modifications cooperatively shape a transcriptionally permissive enhancer landscape prior to gene activation. Recent studies in the context of adipogenesis suggest that H3K4me1 deposition is indeed necessary for enhancer activation (Lee et al., 2013).
Implications for Cellular Reprogramming and Stem Cell Differentiation
The transcriptional priming of lineage-specific enhancers in early lineage intermediates prior to gene activation helps explain the highly context-dependent activity of lineage-specific TFs in cellular programming. Our findings suggest that effective cellular reprogramming of somatic cells requires a combination of both pioneer and lineage-specific TFs. Recent studies show that the reprogramming of fibroblasts into liver, neurons or pluripotent cells indeed requires the inclusion of pioneer factors (Huang et al., 2011; Soufi et al., 2012; Wapinski et al., 2013). Similarly, pioneer and lineage-specific TFs cooperatively regulate macrophage and B cell gene transcription at the levels of enhancers (Heinz et al., 2010; Heinz et al., 2013). We speculate that this regulatory logic is pervasive throughout development and relevant for reprogramming lineages from all germ layers.
As stem cell differentiation protocols to derive various terminal differentiated cell types continue to be developed, scaled and optimized, assessing the acquisition of competence may be an important consideration. For example, the design of large-scale screens to identify molecules or factors which promote a specific differentiation step may be ineffective if the responder cell population has not yet acquired the transcriptional competence to appropriately respond. In addition, one emerging strategy for cell replacement therapy is the transplantation of stem cell-derived lineage-specific progenitor cells. Inquiring if these progenitors have acquired the competency or poised chromatin to form the desired therapeutic cell type may be beneficial in the assessment of suitability for transplantation.
Experimental Procedures
hESC Culture
CyT49 hESCs were maintained and differentiated as previously described with minor modifications (Kroon et al., 2008; Schulz et al., 2012). hESC research was approved by the University of California San Diego Institutional Review Board and Embryonic Stem Cell Research Oversight Committee. For further details, see Supplemental Experimental Procedures.
ChIP-seq and Data Analysis
ChIP-seq was performed as previously described with minor modifications (Hawkins et al., 2010). All the sequencing experiments were performed using Illumina Hi-Seq 2000 instruments. Each read was aligned to the human genome build hg18 with Bowtie (Langmead et al., 2009). We used the first 36 bp for the alignment and only kept reads with up to two mismatches. Duplicated reads from the same library were removed. Data sets from highly correlated biological replicates were pooled for subsequent analysis. MACS (Zhang et al., 2008) was used for peak calling. Peaks were further filtered as described (Shen et al., 2012). For further details, see Supplemental Experimental Procedures.
Enhancer Predictions
Enhancers were predicted as described, using H3K4me1, H3K4me3 and H3K27ac (Rajagopal et al., 2013). We first divided the human genome into 100 bp bins and counted the number of reads that fell within each bin. Then the tag counts in each bin were normalized against the total number of reads and input as described (Shen et al., 2012). The normalized signals for each mark were merged as one input file for the enhancer prediction pipeline. To compute the FDR, we first shuffled the rows and columns of the input data. Second, we ran the enhancer prediction pipeline on this simulated data. The FDR was computed as the ratio of the number of predicted enhancers from simulated data over the real data. We required that predicted enhancers have an FDR of < 2% and are at least 3 kb away from a known transcriptional start site.
Supplementary Material
Acknowledgments
We thank J. Wysocka for the GFP enhancer-reporter construct, C. Wright for anti-Pdx1 antibody, and members of the Sander and Ren laboratories for helpful discussions and critical reading of the manuscript. We are grateful to Viacyte Inc. for CyT49 hESCs and advice on hESC differentiation. This work was supported by National Institutes of Health grants U01-DK089567 to M.S., U01-DK072473 to M.S. and B.R., U01-ES017166 to B.R., U01-DK089540 to D.A.S, California Institute for Regenerative Medicine (CIRM) grant RB5-07236 and Helmsley Charitable Trust grant 2012PG-T1D074 to M.S., JDRF-3-2012-177 postdoctoral fellowship and T32 DK7494-27 training grant to A.W., CIRM-TG2-01154 training grant to R.X. and N.A.P., and the CIRM Bridges to Stem Cells Program (T.H., K.M., and J.P.).
Footnotes
Accession number: All ChIP-seq and GRO-seq data sets have been deposited into Gene Expression Omnibus under accession number GSE54471.
Author contributions: AW and MS conceived the project. AW, FY, YL, BR and MS designed the experiments and data analysis. AW, YL, RX, TH, NP, KM, JP, JW, DL and JR performed the experiments. FY, AW and YQ performed the data analysis. AW, FY and MS wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boj SF, Petrov D, Ferrer J. Epistasis of transcriptomes reveals synergism between transcriptional activators Hnf1alpha and Hnf4alpha. PLoS genetics. 2010;6:e1000970. doi: 10.1371/journal.pgen.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends in genetics : TIG. 2012;28:276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A, Biggin MD. A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. The EMBO journal. 1999;18:1598–1608. doi: 10.1093/emboj/18.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Huang SX, Islam MN, O'Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jin F, Li Y, Ren B, Natarajan R. PU.1 and C/EBP(alpha) synergistically program distinct response to NF-kappaB activation through establishing monocyte specific enhancers. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5290–5295. doi: 10.1073/pnas.1017214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Flanagan SE, Shaw-Smith C, De Franco E, Akerman I, Caswell R, Ferrer J, Hattersley AT, Ellard S. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nature genetics. 2012;44:20–22. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker C, Stern CD. Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development. 2004;131:5671–5681. doi: 10.1242/dev.01445. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Molecular cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N, Xie W, Li Y, Wagner U, Wang W, Stamatoyannopoulos J, Ernst J, Kellis M, Ren B. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS computational biology. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TC, Young HY, Agulnick AD, Babin MJ, Baetge EE, Bang AG, Bhoumik A, Cepa I, Cesario RM, Haakmeester C, et al. A Scalable System for Production of Functional Pancreatic Progenitors from Human Embryonic Stem Cells. PLoS ONE. 2012;7:e37004. doi: 10.1371/journal.pone.0037004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Wang A, Sander M. Pancreas organogenesis: from lineage determination to morphogenesis. Annual review of cell and developmental biology. 2013;29:81–105. doi: 10.1146/annurev-cellbio-101512-122405. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Smith SB, Qu HQ, Taleb N, Kishimoto NY, Scheel DW, Lu Y, Patch AM, Grabs R, Wang J, Lynn FC, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nature genetics. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Storey KG, Crossley JM, De Robertis EM, Norris WE, Stern CD. Neural induction and regionalisation in the chick embryo. Development. 1992;114:729–741. doi: 10.1242/dev.114.3.729. [DOI] [PubMed] [Google Scholar]
- van Hoek M, Dehghan A, Witteman JC, van Duijn CM, Uitterlinden AG, Oostra BA, Hofman A, Sijbrands EJ, Janssens AC. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes. 2008;57:3122–3128. doi: 10.2337/db08-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Organisers & genes. Cambridge Eng.: The University Press; 1940. [Google Scholar]
- Wang A, Sander M. Generating cells of the gastrointestinal system: current approaches and applications for the differentiation of human pluripotent stem cells. J Mol Med (Berl) 2012;90:763–771. doi: 10.1007/s00109-012-0923-y. [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Zhao R, Li J, Duncan SA. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC developmental biology. 2007;7:37. doi: 10.1186/1471-213X-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Cebola I, Patch AM, Flanagan SE, De Franco E, Caswell R, Rodriguez-Segui SA, Shaw-Smith C, Cho CH, Lango Allen H, et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nature genetics. 2014;46:61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–1572. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- Xie R, Everett Logan J, Lim HW, Patel Nisha A, Schug J, Kroon E, Kelly Olivia G, Wang A, D'Amour Kevin A, Robins Allan J, et al. Dynamic Chromatin Remodeling Mediated by Polycomb Proteins Orchestrates Pancreatic Differentiation of Human Embryonic Stem Cells. Cell Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Molecular cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.