Abstract
We describe the nearly complete genome of a highly divergent parvovirus, we tentatively name Sesavirus, from the feces of a California sea lion pup (Zalophus californianus) suffering from malnutrition and pneumonia. The 5,049-base-long genome contained two major ORFs encoding a 553-aa nonstructural protein and a 965-aa structural protein which shared closest amino acid identities of 25 and 28 %, respectively, with members of the copiparvovirus genus known to infect pigs and cows. Given the low degree of similarity, Sesavirus might be considered as prototype for a new genus with a proposed name of Marinoparvovirus in the subfamily Parvovirinae.
Keywords: Parvovirus, Sea lion, Feces, Genus, Metagenomics
The study
Parvoviruses are small, non-enveloped, single-stranded DNA viruses. The family Parvoviridae currently consists of two subfamilies: Densovirinae and Parvovirinae. While the members of the subfamily Densovirinae infect invertebrates, those of the subfamily Parvovirinae infect vertebrates [1]. The subfamily Parvovirinae is currently classified into eight genera that have been reported in a wide range of hosts including mammalian and bird species [1].
Here we used metagenomics deep sequencing to analyze five rectal swabs collected from wild sea lions from Northern California. Fecal suspensions were vortexed and then clarified by 15,000×g centrifugation for 10 min. The supernatants were pooled and filtered through a 0.45-μm filter (Millipore) to remove bacterium-sized particles. The filtrate was treated with a mixture of enzymes to digest unprotected nucleic acids, and then viral nucleic acids were extracted [2]. Random RT-PCR was then used to amplify RNA and DNA, and a library was constructed for Illumina sequencing (MiSeq 2 × 250 bases) using Nextera™ XT Sample Preparation Kit. Translated sequence reads showing sequence similarity to viral sequences with E score <10−5 were identified using BLASTx.
Out of 18,098 sequence reads, 187 reads encoded parvovirus-related proteins. Forty-three reads and 31 reads encoded picornavirus- and anelloviruses-related proteins. Picornavirus has been previously reported in feces of sea lions [3] and was not analyzed further. The majority of anellovirus reads mapped closest to a previously described sea lion anellovirus [4]. Anelloviruses are highly prevalent infections in many mammals and generally considered to cause commensal infections [5]. The nearly complete genome of the parvovirus, named Sesavirus for sea lion stool associated parvovirus, was then determined by filling gaps in the metagenomics sequence reads by PCR and Sanger-sequencing the amplicons. The individual swab specimen within the pool containing this viral genome was then identified by PCR. Primers Sesa-F1 (5′-AGA TTC GCT GCC AAC TAC GGA ACT GT-3′) and Sesa-R1 (5′-GTC CTT GCC TTT TTC GTG TCT GGT TC-3′) were used for the first round of PCR, and primers Sesa-F2 (5′-GGT TGG TGG GAC GAA GGT ATG ATG ACA-3′) and Sesa-R2 (5′-AGG ACT TGG AAT GGT ATG GGT CTG TG-3′) for the second round of PCR. The PCR conditions were 95 °C for 5 min, 35 cycles 95 °C for 30 s, 55 °C (for the first or second round) for 30 s, and 72 °C for 1 min, a final extension at 72 °C for 10 min, resulting in an amplicon of 368-bp. The positive rectal swab came from a 31.5-kg, 106-cm-long female pup that was sampled due to severe malnutrition and pneumonia. The animal (CSL-10538) was recovered and was subsequently released. Because the distribution of the picornaviruses and anelloviruses in the feces of the five animals studied was not determined, their presence in the sesavirus infected sea lion and their possible role in its symptoms are unknown.
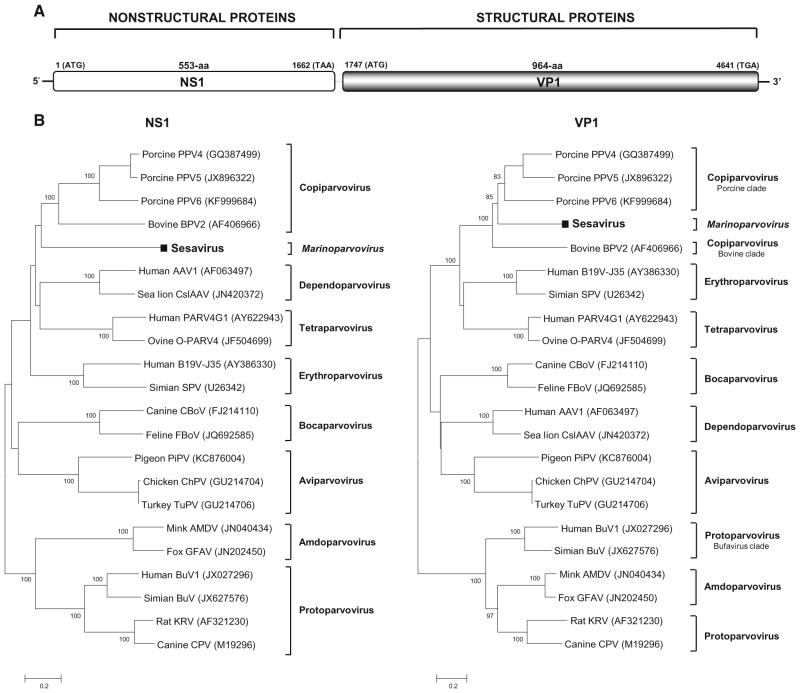
The nearly complete genome (5,049 bases) of the sea lion parvovirus, including a partial 5′ UTR (313-bp), complete nonstructural protein (NS1), complete structural protein (VP1), and a partial 3′ UTR (95-bp), was generated (Fig. 1a) (GenBank KM035804). NS1 encoded a 553-aa long protein which contained the ATP or GTP binding Walker loop motif 318GPPTTGKS325 [GXXXXGK(T/S)] [6]. It also contained two conserved replication initiator motifs RWHIHYAYE and INNYLLPKQY (where conserved amino acids are in boldface type) [7]. Pair-wise amino acid sequence analysis showed that sesavirus NS1 shared its highest level of identity (25 %) to the NS1 of porcine parvovirus 5 (PPV5 GenBank NC_023020) [8]. Similar to PPV5 and other members of the genus Copiparvovirus [1], Sesavirus also possessed the phospholipase A2 (PLA2) motif in the VP1 N-termini with expected calcium-binding site and catalytic residues. Its VP1 protein showed the highest identity of 28 % to porcine parvovirus 5. Figure 1b shows the Sesavirus clade with a very deep branch relative to other genera. According to a recent report from the International Committee on Taxonomy of Viruses (ICTV), members of a parvovirus genus should share >30 % identity in NS1 [1]. Based on genetic distance calculation, Sesavirus is therefore proposed as prototype of a novel genus with a proposed name of Marinoparvovirus (for its origin in a sea lion temporarily housed at The Marine Mammal Center in Marin County, California) in the subfamily Parvovirinae pending ICTV approval.
Fig. 1.
New parvovirus genome and phylogeny. a Organization of the Sesavirus genome. b Phylogenetic trees generated with NS1 and VP1 of Sesavirus and the representatives of eight genera in the subfamily Parvovirinae. The scale indicated amino acid substitutions per position. Bootstrap values (based on 100 replicates) for each node are given if >70
Acknowledgments
We acknowledge NHLBI Grant R01 HL105770 to E.L.D and the Blood Systems Research Institute for support.
Footnotes
Nucleotide sequence accession number
The nearly complete genome sequence of Sesavirus is available in GenBank under Accession Number KM035804.
Contributor Information
Tung Gia Phan, Blood Systems Research Institute, San Francisco, CA 94118, USA. Department of Laboratory, Medicine University of California, San Francisco, CA 94118, USA.
Frances Gulland, The Marine Mammal Center, Sausalito, CA 94965, USA.
Claire Simeone, The Marine Mammal Center, Sausalito, CA 94965, USA.
Xutao Deng, Blood Systems Research Institute, San Francisco, CA 94118, USA.
Eric Delwart, Email: delwarte@medicine.ucsf.edu, Blood Systems Research Institute, San Francisco, CA 94118, USA. Department of Laboratory, Medicine University of California, San Francisco, CA 94118, USA.
References
- 1.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. Arch Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, Shan T, Wang C, Cote C, Kolman J, Onions D, Gulland FM, Delwart E. J Virol. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng TF, Suedmeyer WK, Wheeler E, Gulland F, Breitbart M. J Gen Virol. 2009;90:1256–1261. doi: 10.1099/vir.0.008987-0. [DOI] [PubMed] [Google Scholar]
- 5.Nishiyama S, Dutia BM, Stewart JP, Meredith AL, Shaw DJ, Simmonds P, Sharp CP. J Gen Virol. 2014;95:1544–1553. doi: 10.1099/vir.0.065219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker JE, Saraste M, Runswick MJ, Gay NJ. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilyina TV, Koonin EV. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao CT, Gimenez-Lirola LG, Jiang YH, Halbur PG, Opriessnig T. PLoS ONE. 2013;8:e65312. doi: 10.1371/journal.pone.0065312. [DOI] [PMC free article] [PubMed] [Google Scholar]