Abstract
In most Gram-positive bacteria, including many clinically devastating pathogens from genera such as Bacillus, Clostridium, Listeria and Staphylococcus, T-box riboswitches sense and regulate intracellular availability of amino acids through a multipartite mRNA-tRNA interaction. The T-box mRNA leaders respond to nutrient starvation by specifically binding cognate tRNAs and sensing whether the bound tRNA is aminoacylated, as a proxy for amino acid availability. Based on this readout, T-boxes direct a transcriptional or translational switch to control the expression of downstream genes involved in various aspects of amino acid metabolism: biosynthesis, transport, aminoacylation, transamidation, etc. Two decades after its discovery, the structural and mechanistic underpinnings of the T-box riboswitch were recently elucidated, producing a wealth of insights into how two structured RNAs can recognize each other with robust affinity and exquisite selectivity. The T-box paradigm exemplifies how natural non-coding RNAs can interact not just through sequence complementarity, but can add molecular specificity by precisely juxtaposing RNA structural motifs, exploiting inherently flexible elements and the biophysical properties of post-transcriptional modifications, ultimately achieving a high degree of shape complementarity through mutually induced fit. The T-box also provides a proof-of-principle that compact RNA domains can recognize minute chemical changes (such as tRNA aminoacylation) on another RNA. The unveiling of the structure and mechanism of the T-box system thus expands our appreciation of the range of capabilities and modes of action of structured non-coding RNAs, and hints at the existence of networks of non-coding RNAs that communicate through both, structural and sequence specificity.
Introduction
Gram-positive bacteria are among the most ubiquitous microorganisms on Earth. This clade includes many harmless bacilli and cocci, but also a large number of clinically relevant pathogens from genera such as Bacillus, Clostridium, Listeria, and Streptococcus, as well as multi-drug resistant and extreme-drug resistant organisms such as methicilin-resistant Staphylococcos aureus (MRSA) and vancomycin-resistant Enterococcus (VRE).1 Thus, understanding the physiology and gene-regulatory circuits of Gram-positive bacteria not only can provide fundamental insights into the cellular and molecular mechanisms that enable microorganisms to adapt continuously to survive an ever-changing environment, but also can inform the design and development of new antibiotics that are urgently needed to battle the rising pandemic of antibiotic resistance.2
Compared to their Gram-negative counterparts, Gram-positive bacteria are characterized by larger operon size and complexity, gene-regulatory circuits with a bias toward transcriptional over translational control, and the widespread use of structured RNA devices such as riboswitches and ribozymes.3 Indeed, riboswitches, RNA genetic actuators that directly sense the intracellular concentration of metabolites and second messengers and regulate gene expression, are most abundant in Gram-positive bacteria. 3-7 This suggests a need to fine-tune the transcription of multi-gene operons whose expression consumes a large quantity of cellular resources.
The T-box riboswitches, discovered in the Henkin laboratory in 1993, are a class of gene-regulatory, non-coding RNA devices that are widely distributed in Gram-positive bacteria, in particular Firmicutes.8, 9 T-boxes sense and respond to amino acid starvation (Fig. 1). Rather than sensing free amino acids, as do other riboswitches10-13, T-boxes survey the amino acids immediately available to the translating ribosome by monitoring the aminoacylation levels of tRNAs.9 Since 1993, numerous studies of the T-box system have been undertaken using genetic, phylogenetic, biochemical, and biophysical approaches, producing important insights into their biological context and regulatory mechanism.14-19 Despite these important advances, three fundamental structural and mechanistic questions about the T-box had remained unanswered.8 First, how can an mRNA domain such as the T-box recognize a tRNA with high specificity and affinity outside of context of the ribosome? Second, how can the T-box determine the aminoacylation state of a bound tRNA? Third, how can an uncharged tRNA direct the outcome of a genetic switch? Here, we review studies reported by several groups in the past two years that have provided long-awaited answers to these questions.
Figure 1. T-box riboswitch senses and regulates intracellular amino acid availability.
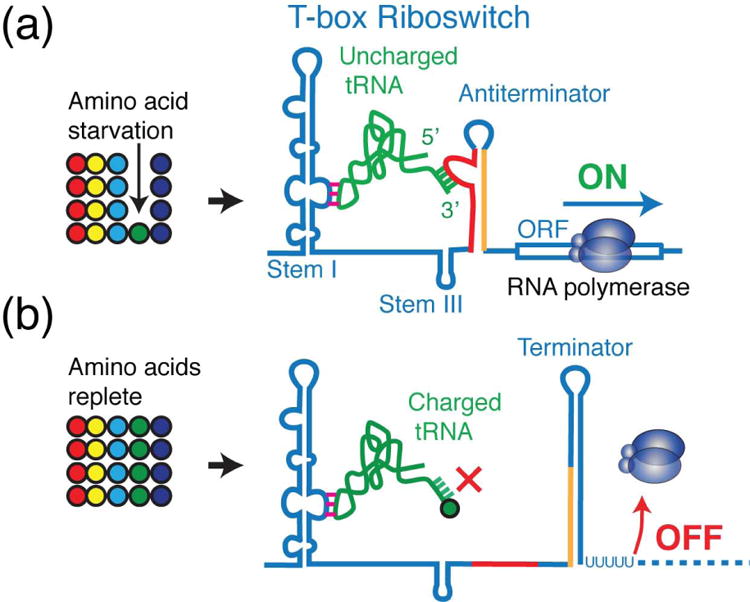
(a) Starvation for a particular amino acid (green circle) leads to reduced charging of its cognate tRNA (green). A cognate uncharged tRNA binds the T-box and stabilizes the otherwise unstable antiteriminator structure, allowing RNA polymerase to transcribe downstream genes. (b) The collective action of T-box downstream genes in (a) carries out amino acid biosynthesis, import, and tRNA charging, thus restoring cellular amino acid and tRNA charging levels (>95% charged). Charged tRNAs are rejected sterically, unable to stabilize the antiterminator. Thus, formation of the terminator prematurely terminates transcription.
Structural basis of specific tRNA recognition by the T-box riboswitches
T-box riboswitches are comprised of the highly conserved Stem I and antiterminator domains, connected by a variable linker (Fig. 2). Since 1993, genetic and phylogenetic analyses, in conjunction with biochemical structure probing, identified two key base-pairing interactions between the T-box and its cognate tRNA: one between the tRNA anticodon and the “specifier” trinucleotide in a loop near the base of the Stem I domain, the other between the single-stranded NCCA 3′ terminus of tRNA and the “antiterminator bulge” in the T-box antiterminator domain (Fig. 2).20, 21 However, it has remained unknown if additional contacts are formed between the T-box and the tRNA that confer structural selectivity beside this limited sequence complementarity. Recently, the first crystal structures of T-box riboswitch Stem I domains in complex with cognate tRNA have been determined, providing insight on the structural basis of this extra-ribosomal mRNA-tRNA interaction.22-24 The two reported structures share key commonalities and diverge in several informative aspects. Solution NMR analysis of a minimized complex consisting of the T-box specifier region and a tRNA anticodon stem-loop produced a structural model that is consistent with the co-crystal structures.25 Together, these studies indicate that the T-box riboswitches recognize their cognate tRNAs by closely tracking their three-dimensional architecture, recognizing defining tRNA features such as the anticodon and the elbow, accommodating and exploiting post-transcriptional tRNA modifications, and achieving a high degree of shape complementarity via mutually induced fit.24 The following sections explain how the various global and local structural features of the T-box Stem I-tRNA interaction contribute to attaining binding affinity and selectivity, thereby enabling tRNA-mediated metabolic surveillance and transcriptional response.
Figure 2. Two mutually exclusive secondary structures of a representative T-box riboswitch and its base-pairing interactions with its cognate tRNA.
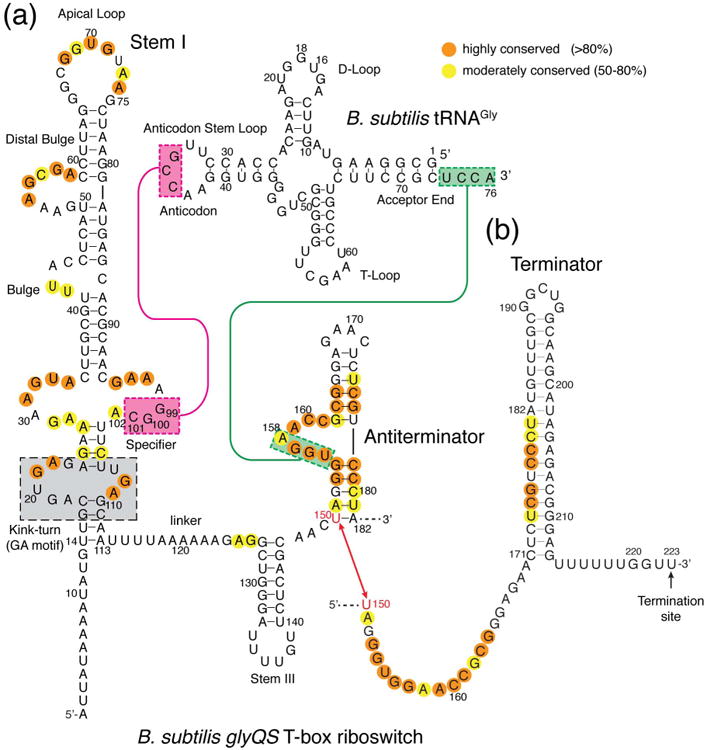
Unlike essentially all other T-boxes, tRNAGly-responsive T-boxes (such as the B. subtilis glyQS T-box depicted here and the closely related O. iheyensis glyQ T-box) do not contain the Stem II and Stem II A/B pseudoknot elements in the linker region. The mutually exclusive antiterminator and terminator conformations of the T-box riboswitch are shown in (a) and (b), respectively. Sequence conservation across known T-boxes is indicated in orange and yellow circles. Base pairing interactions between the Stem I specifier sequence in Stem I and the tRNA anticodon (magenta), and between the T-box antiterminator domain and the tRNA acceptor end (green), are indicated by colored lines and boxes. The gray box indicates the functionally important K-turn. The sequences of tRNAGly of B. subtilis and O. iheyensis are identical.
Global shape complementarity through flexible hinges and mutually induced fit
Higher order RNA structures are built by stitching together thermodynamically stable helices and structural motifs using a relatively modest set of tertiary and quaternary interaction strategies such as coaxial helical stacking, kissing loops, pseudoknots, tetraloop-tetraloop receptor interactions, A-minor interactions, etc.26-29 Among these, coaxial stacking is ubiquitous and to a large extent dictates the overall architecture of many complex RNAs.
The recent co-crystal structures of the T-box Stem I-tRNA complexes from Oceanobacillus iheyensis and Geobacillus kaustophilus revealed that T-boxes recognize their cognate tRNAs using a linear arrangement of discrete structural modules linked together by malleable hinges (Fig. 3).22-24, 30 This structurally flexible, segmented architecture gives the T-box the ability to recognize the bendable tRNA. All elongator tRNAs share a structural hinge near the t26•t44 pair (throughout, tRNA residue numbers are preceded by “t”), about which the anticodon stem loop and the rest of the tRNA can pivot by as much as 70° during the transit of tRNA through the ribosome (Fig. 3).31 Importantly, this motion changes the distance and orientation between two prominent structural features of the tRNA —the anticodon and the “elbow”, the latter characterized by the flat surface of the conserved tertiary base pair (tG19•tC56) linking the tRNA D- and T-loops.32, 33 In order to simultaneously engage both, the tRNA anticodon and elbow, the T-box has a hinged Stem I architecture that complements that of tRNA, and uses two discrete structural motifs for recognition (Fig. 3b). The flexibility of the Stem I hinge is evident in high crystallographic B-factors, and sequence and structural variation among even closely related species. The hinge of the O. iheyensis glyQS T-box is more compact than that from the G. kaustophilus GlyQ T-box. The latter features a C-loop motif.
Figure 3. Common and divergent features of the T-box Stem I-tRNA complex structures.
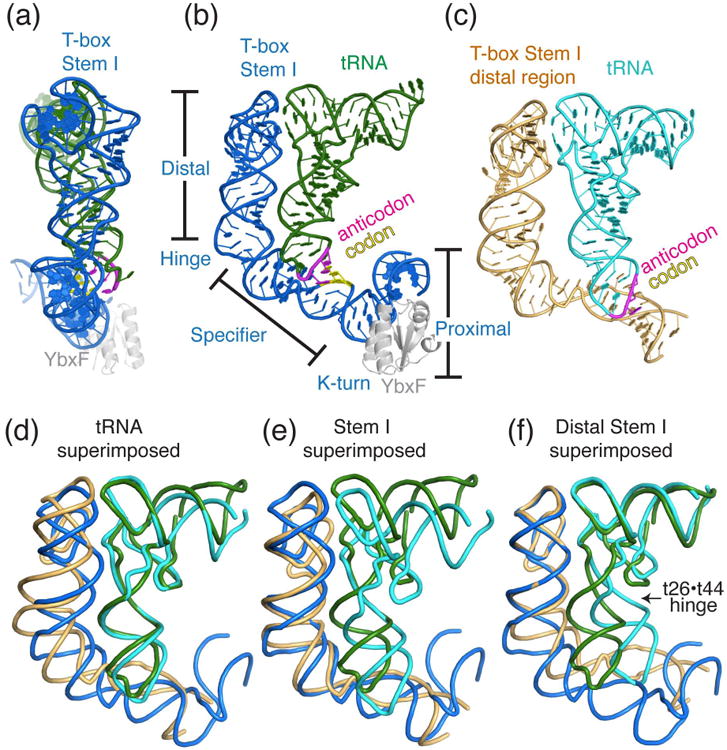
(a) and (b) Two orthogonal views of the overall structure of the O. iheyensis glyQ T-box–tRNAGly–B. subtilis YbxF ternary complex (PDB: 4LCK24), colored marine blue, forest green, and gray, respectively. The distal, specifier, and proximal regions of the Stem I are demarcated by the T-box hinge at T-box U30 and the K-turn. The tRNA anticodon and T-box specifier (codon) trinucleotides are colored magenta and yellow, respectively. (c) Overall structure of a truncated G. kaustophilus glyQ T-box Stem I in complex with tRNAGly (PDB: 4MGN22), colored light orange and cyan respectively. (d-f) Structural superpositions of the O. iheyensis and G. kaustophilus Stem I-tRNA complex structures made by least-squares alignment of the tRNAs (d), the Stem I (e), or the distal region of Stem I (f). It is evident that the tRNAs and the Stem Is cannot be superimposed simultaneously, due to structural differences between them. Superimposing the distal Stem I (f) automatically superimposes the tRNA elbow region, suggesting that the interaction between the T-box distal region and the tRNA elbow is structurally constrained. It is also clear that the degree of swivelling of the tRNA anticodon stem loop relative to the rest of the tRNA about the t19•t44 hinge (indicated by the arrowhead) varies considerably between the two T-box complex structures.
In addition to the hinge between the distal and specifier regions, the T-box Stem I contains a second hinge formed by a kink-turn (K-turn) or “GA” motif. Originally recognized because of its recurrence in ribosomal RNA, the K-turn is a ubiquitous, dimorphic RNA structural motif that can create a sharp, 120° bend in the RNA helical trajectory. 34-38 Canonical K-turns, such as those frequently found in the proximal region of the T-box Stem I, feature a trinucleotide bulge at the sharp bend between a 5′ Watson-Crick helix and a 3′ helix characterized by tandem sheared A•G and G•A pairs (thus the “GA motif”). Comparing the NMR structure of the Stem I proximal region in isolation to the co-crystal structure of the complete Stem I bound to tRNA reveals major differences in Stem I trajectories.24, 39, 40 The K-turn has an extended conformation in the NMR structure whereas the co-crystal structure shows it in a canonical, kinked conformation. This conformation, further stabilized by the bound YbxF protein (a member of the L7Ae-family of K-turn binding proteins41-44) dramatically shortens the distance between the 3′ terminus of Stem I and acceptor end of the tRNA, and may help bring the antiterminator domain of the T-box closer to the site of aminoacylation on the tRNA, thereby facilitating sensing of tRNA charge state and genetic switching.24 The functional importance of the YbxF protein or its close paralog YlxQ to the T-box regulatory system remains unclear.8
Decoding the tRNA anticodon: structural specificity augments sequence specificity
As predicted from genetic experiments, the tRNA anticodon is recognized by the T-box “specifier” trinucleotide using three canonical Watson-Crick base pairs (Fig. 4a-b). Previous phylogenetic and structural probing analyses proposed a possible fourth base pair between the conserved tRNA tU33 and T-box A90 residues.16 The co-crystal structures as well as the NMR and calorimetric analyses establish that a fourth base pair does not occur. Instead, A90 stacks underneath the 3 bp codon-anticodon duplex and provides stabilization energy (Fig. 4a-b). Substitutions of A90 with all other nucleosides reveal a clear preference for a purine at this position, as the A90U substitution abolishes tRNA binding, and A90C causes a 4-fold drop in binding affinity, whereas A90G produced no defect.24 While purines are known to be superior to pyrimidines in base-stacking interactions, the large difference between uridine and cytidine is not immediately understood. Reciprocally, a conserved tRNA purine, t37, stacks atop the codon-anticodon duplex, thus completing a geometry where the codon-anticodon duplex is reinforced on both flanks by stacked purines. This geometry is remarkably similar to that used for tRNA decoding in the ribosomal P site (Fig. 4c). Curiously, in place of the conserved A/G90 purine, the ribosomal P site uses an invariant rC1400 residue (ribosomal RNAs numbers are preceded by an “r”) to stack underneath the codon-anticodon duplex.45, 46 The ribosome uses a combination of mRNA and ribosomal RNA to achieve the same geometry that the T-box attains using a single mRNA segment. In both instances, effective decoding of the tRNA anticodon is accomplished by supplementing the limited sequence specificity from three base pairs with additional structural specificity from the flanking stacked purines (Fig. 4d). Such a strategy not only supplies thermodynamic stability, but may also grant necessary specificity against non-tRNA structures that may fortuitously contain the same anticodon trinucleotide sequence.
Figure 4. A conserved geometric arrangement decodes tRNA anticodon using both structural and sequence determinants.
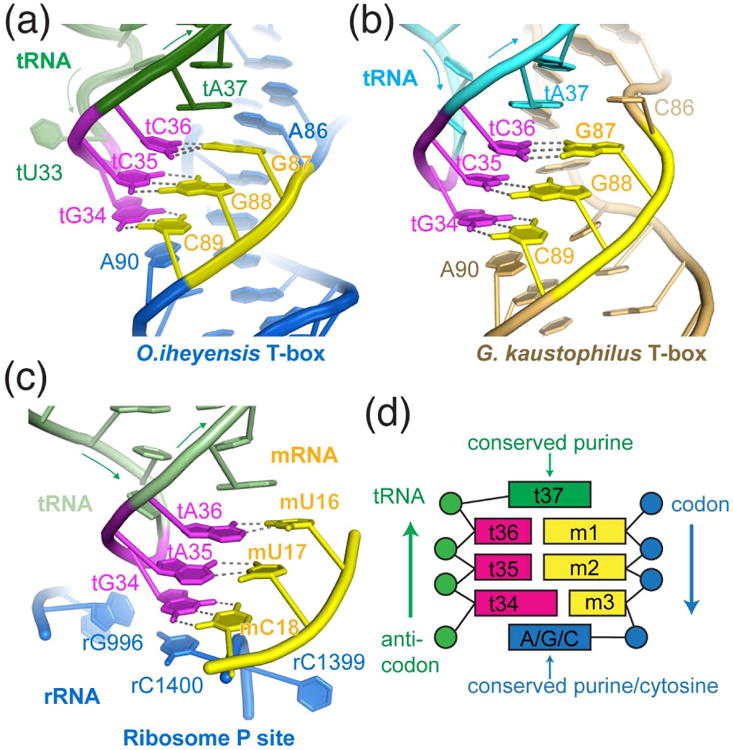
(a-b) Structural arrangements of tRNA anticodon (magenta) decoding by the T-box specifier (yellow) and neighbouring residues observed in the O. iheyensis (a, marine blue) and G. kaustophilus (b, light orange) T-boxes. (c) Structural arrangement that decodes the anticodon of the P site tRNA (PDB: 4V5D46). The tRNA, mRNA, and rRNA are shown in light green, yellow, and marine blue, respectively. (d) Cartoon illustration of the geometric arrangement responsible for tRNA anticodon decoding by the T-box riboswitch and the ribosome. Codon and anticodon are colored as in Fig. 3.
Accommodation of tRNA anticodon posttranscriptional modifications
In addition to recognizing both structural and sequence features of the tRNA anticodon, the T-box system also has to accommodate idiosyncratic tRNA chemical features in the form of post-transcriptional modifications.47, 48 In particular, the tRNA t37 nucleobase is frequently modified into bulky hypermodified bases such as wybutosine, cyclic N6-threonyl-carbamoyladenosine (ct6A), 2-methylthio-N6-Isopentenyladenosine (ms2i6A), etc.49, 50 To make room for these bulky modifications that could occur at either the Hoogsteen, Watson-Crick, or sugar edge, the T-box Stem I backbone bends away from the t37 nucleobase, forming an overwound loop E motif (also known as the bulged-G motif or S turn; Fig. 5). The specific geometry of the loop E motif creates a pocket that would accommodate even the largest known tRNA anticodon loop post-transcriptional modifications. Since all reported T-box co-crystal structures have been determined employing unmodified tRNA, the extent to which posttranscriptional tRNA modifications contribute to the affinity and specificity of T-box-tRNA interaction remains to be clarified. In addition, in light of the recent discovery of widespread mRNA posttranscriptional modifications, it is conceivable that the T-box RNA itself might be chemically modified to provide added affinity or tRNA selectivity.
Figure 5. Variability in the T-box-tRNA interface adjacent to the codon-anticodon duplex.
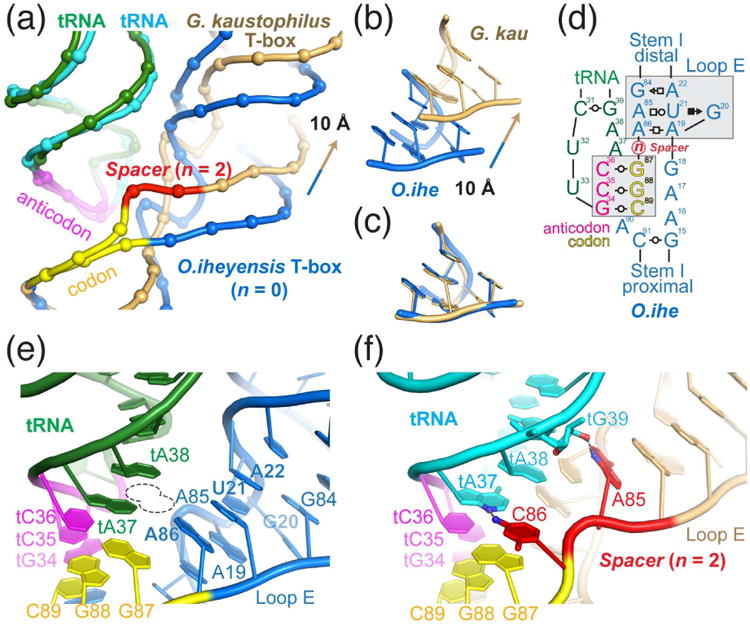
(a) Overlay of the O. iheyensis (marine blue) and G. kaustophilus (light orange) T-box-tRNA complex structures by superimposing the tRNA anticodon trinucleotides (magenta). This leads to superimposition of the T-box specifier codons (yellow) and reveals differences between the two structures in the region immediately distal to the codon-anticodon duplex. Backbone phosphates are shown in spheres and the spacer nucleotides in G. kaustophilus T-box are shown in red. Nucleobases and riboses are omitted for clarity. (b) Compared to the O. iheyensis structure, the Loop E motif (or bulged-G or S-turn motif) in the G. kaustophilus complex is translated more distal by 10 Å. (c) Upon translation, the two loop E motifs are superimposable. (d) Cartoon illustration of the T-box-tRNA interaction adjacent to the codon-anticodon duplex. “n” denotes the variable number of nucleotides that are inserted in between the codon trinucleotide and the loop E motif. n = 0 and 2 for O. iheyensis (e) and G. kaustophilus (f) T-boxes respectively. (e) In the O. iheyensis structure, the formation of the loop E motif immediately adjacent to the codon trinucleotide bends the Stem I backbone away from tA37, thus easily accommodating bulky hypermodifications at that position (dotted line). (f) In contrast, in the G. kaustophilus structure, where n = 2, the two inserted nucleotides, A85 and C86, shown in red, each makes a single hydrogen bond to the tRNA tG39 and tA37. Compared to the O. iheyensis structure, the G. kaustophilus T-box is in closer proximity to the tRNA in the anticodon region (compare e and f), possibly offsetting its reduced shape complementarity and increased distance to the tRNA t26•t44 hinge region (compare Figs. 3b and 3c). Further, the inserted nucleotides in the specifier loop may permit presentation and utilization of alternative codons, thus allowing a single T-box riboswitch to respond to two or more tRNAs.
Dual- and multi-specificity T-boxes that respond to more than one tRNA
Despite a high degree of sequence and structural conservation of the codon-anticodon duplex and the loop E motif, a number of T-boxes contain additional nucleotides inserted in between these two structural modules, leading to architectural diversification and functional elaboration in tRNA anticodon decoding (Fig. 5d). Recently, a T-box from Clostridium acetobutylicum was shown to respond to not one, but two tRNAs (bearing different anticodon sequences) by exploiting a guanosine inserted between the loop E motif (which ends with a GAA triplet) and the AAC codon.51 This T-box can present two codons depending on the reading frame: GAA for glutamate and AAC for asparagine, thus conferring dual specificity for tRNAGlu or tRNAAsn and allowing this single T-box to monitor starvation for two amino acids. Intriguingly, and consistent with the metabolic connections between glutamate and asparagine biosynthetic pathways, this T-box controls the expression of four genes that comprise the tRNA-dependent transamidation pathway, which catalyzes the initial misacylation (with Glu or Asp) and subsequent transamidation to produce Gln-tRNAGln or Asn-tRNAAsn.51 This study further demonstrated that the specifier loop tolerates shifts of the reading frame of the codon sequence and proposed that many other T-boxes could potentially respond to more than two tRNA species through a longer specifier loop and the resulting codon ambiguity.
Comparison of the sequences of three glycine-responsive T-boxes from Bacillus subtilis, G. kaustophilus, and O. iheyensis shows that the spacer (“n”, Fig. 5d) between the loop E motif and the codon varies between 0 and 2 (Table 1). This suggests that O. iheyensis glyQS T-box (n = 0) is most likely a single-tRNA riboswitch while the other two T-boxes can potentially respond to other tRNAs such as tRNAArg and tRNAThr. In the case of G. kaustophilus T-box, the insertion of a 2-nucleotide spacer (Fig. 5a, red nucleotides) effectively shifts the entire top half of the Stem I, including the loop E motif, more distal by ∼10 Å (Fig. 5b and 5c). Interestingly, this structural shift did not impact a separate interaction between the tRNA elbow and the distal loops of the Stem I because the structural shift above the anticodon-codon duplex is compensated by the tRNA assuming a more bent structure in the G. kaustophilus complex (Fig. 3d-f).
Table 1. Length variation of the specifier loops in three glycine-responsive T-boxes.
| n | Specifier sequence | Species | Gene | Codon | Possible substrate tRNAs |
|---|---|---|---|---|---|
| 0 | GGC | O. iheyensis | glyQ | GGC | tRNAGly |
| 1 | A GGC | B. subtilis | glyQS | GGC, AGG | tRNAGly, tRNAArg |
| 2 | AC GGC | G. kaustophilus | glyQ | GGC, CGG, ACG | tRNAGly, tRNAArg, tRNAThr |
n: the number of nucleotides between the bulged-G motif and the codon trinucleotide.
It remains unknown how codon ambiguity and binding of more than one tRNA is structurally accommodated by such T-boxes. It is noteworthy that such codon slippage changes the identities of the stacking nucleotides on both flanks of the codon-anticodon duplex. Sequence and structural constraints, such as the finding that at least the nucleotide stacking underneath the duplex cannot be a uridine in the O. iheyensis glyQS T-box (A90U abolishes tRNA binding 24), may restrict the extent to which codon slippage and alternative tRNA pairing can occur. In addition, as both spacer nucleotides in G. kaustophilus T-box (A85 and C86) make a single hydrogen bond to the tRNA (Fig. 5f), using the spacer nucleotides as alternative codons will break these interactions and might also create new contacts. It remains unknown how such interface remodelling impacts the affinity and selectivity of anticodon decoding. Importantly, it remains uncertain whether the dual- or multi-specificity T-boxes actually respond to limitations of more than one amino acid in vivo, as might be predicted from the in vitro and in silico studies. Nonetheless, it is a fascinating development that T-boxes can utilize codon ambiguity and reading-frame slippage to link and coordinate two or more related amino acid metabolic pathways. Structurally speaking, this finding also provides an interesting parallel to tRNASufJ–mediated suppression of ribosomal frameshifting, during which an elongated tRNASufJ anticodon loop causes recoding through tRNA distortion.52
Recognition of the tRNA elbow: convergent use of interdigitated T-loops
While it is clear that codon-anticodon pairing is the chief tRNA specificity determinant for T-boxes, switching the codon (or specifier trinucleotide) does not always result in the switching of tRNA and amino acid specificity and the recoded T-boxes do not achieve 100% efficiency.8 This is presumably due to structural and chemical differences among different tRNAs. This and several other lines of evidence pointed early on to the existence of additional tRNA-T-box contacts beyond the genetically predicted base-pairing interactions to the tRNA anticodon and single-stranded 3′-NCCA terminus.53 Strong phylogenetic conservation of the two distal loops of T-box Stem I has long been recognized but their structure and function remained elusive (Fig. 2a). The recognition of the conservation pattern as conforming to two T-loops (a recurring pentanucleotide RNA loop that bends the backbone by 180° and leaves a stacking gap between its fourth and fifth nucleotide, thus poised for intercalation from an external nucleobase 54, 55, Fig. 6) and its striking sequence similarity to the ribosome L1 stalk, allowed Lehmann et al. to boldly propose a structural model in which the two non-adjacent T-loops interdigitate with each other to form a “head” domain that produces flat, stackable platforms on both faces, in a manner analogous to the RNAse P ribozyme and the ribosome L1 stalk.56 By computing the frequency of occurrences of purines, which are strongly preferred over pyrimidines for stacking interactions, Lehmann et al., correctly identified which platform is used to recognize tRNA elbow.56
Figure 6. Recognition of tRNA elbow through two interdigitated T-loops.
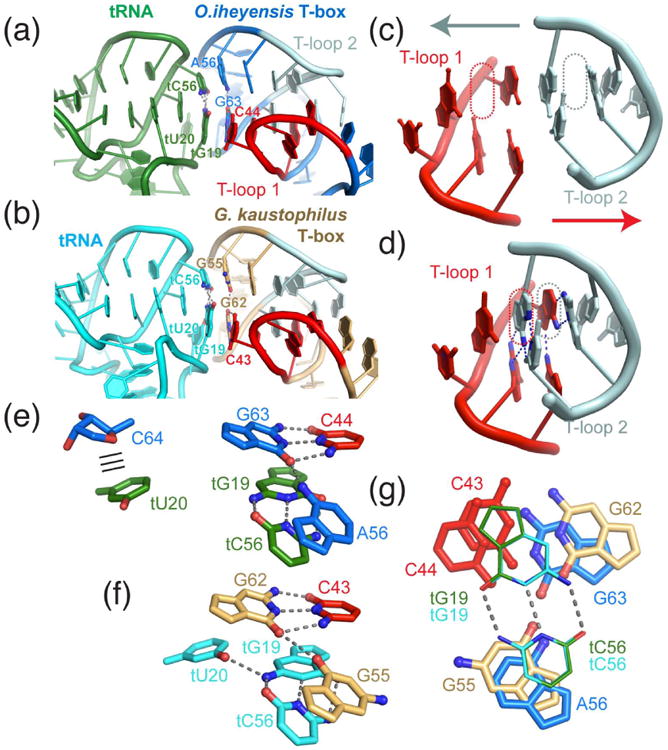
(a-b) Recognition of tRNA elbow by the O. iheyensis (marine blue, a) and G. kaustophilus (light orange, b) T-box Stem I distal region, the core of which is formed by the interdigitation of two T-loops (shown in red and light cyan). Interfacial nucleotides are labelled. (c-d) Depiction of how the isolated T-loops approach each other to fill each other's stacking gap (dotted lines), arriving in an extensively paired (two base triples in the center; hydrogen bonds on each base triple are indicated by blue dashes) and heavily stacked structure, as shown in (d). (e-f) Detailed interaction between the T-box base triple (C44•G63•A56) and the tRNA tertiary base pair (tG19•tC56) seen in the O. iheyensis (e) and G. kaustophilus (f) structures. The position and interaction of tU20 residue, albeit extrahelically flipped in both instances, differ in the two structures. The nucleobase of tU20 is seen forming a packing interaction (parallel lines) with the ribose of T-box C64 in the O. iheyensis structure, whereas the same residue in the G. kaustophilus structure is shifted closer to and forms a base triple with the tRNA tG19•tC56 pair and appears to stack against T-box G62. (g) Superimposition of the tRNA tG19•tC56 pairs (thin lines) of the two structures reveals appreciable variability in the precise location of the T-box base triples.
The co-crystal structures of the T-box Stem I-tRNA complex revealed the precise structural basis of tRNA elbow recognition by the T-box distal element (Fig. 6a-b).22, 24 Overall, the structures of the two crystallized T-box complexes are similar but also diverge in several important features. Interdigitation of the two distal T-loops in a head-to-tail arrangement allows them to fill each other's stacking gaps, forming a central core consisting of two stacked base triples, which are further reinforced by two base pairs on each flank (Fig. 6c-d). This heavily stacked structure enables the presentation of a purine-rich base triple against the tertiary base pair (tG19•tC56) formed between the tRNA D- and T-loops (Fig. 6e-g). Curiously, tRNA tU20 from the highly conserved D-loop assumes very different locations in the O. iheyensis and G. kaustophilus T-box structures. In the O. iheyensis structure, the nucleobase of an extrahelically flipped tU20 is seen packed against the ribose of T-box C64 (Fig. 6e). In the G. kaustophilus structure, the similarly extrahelically flipped tU20 is appreciably closer to tG19 and makes a single hydrogen bond to the latter nucleobase, thus forming a base triple with the tG19•tC56 pair that stacks with the T-box surface base triple (C43•G62•G55; Fig. 6f). In either case, extrahelical flipping of tU20 of the D-loop is necessary for it to expand the RNA-RNA interface. Previous biophysical analyses suggest that tU20 flipping is stimulated by its canonical post-transcriptional modification to dihydrouridine (the highly conserved modification after which the D-loop is named).57
The precise position of the T-box base-triple relative to the tRNA tG19•tC56 pair and its base composition are both variable, providing considerable flexibility and sequence diversity while maintaining the essential structural configuration (Fig. 6g). Extensive mutagenesis and calorimetry suggest that purines are strongly preferred in the T-box base triple (C44•G63•A56) but Watson-Crick pairing between C44 and G63 is not required.24 For instance, C44A and even C44A∷G63A substitutions are both well tolerated, producing a mere 2-3 fold reduction in affinity. This suggests that the presentation of the base triple is largely dictated by the overall domain structure and backbone trajectory directed by the dual-T-loop core, rather than being determined by the chemical compatibility of the three nucleobases to form a triple. This finding further exemplifies how unique RNA structural features can sometimes override sequence specificity, making sequence-based structural prediction more challenging. In contrast, substitutions that destabilize the T-loop architecture reduce binding by more than three orders of magnitude, or outright abolish tRNA binding. Consistent with the bioinformatic analyses and the structures, destabilizing T-loop 1, which directly contacts the tRNA elbow, is much more detrimental to binding than equivalent substitutions that disrupt T-loop 2.24
Elucidation of the T-box riboswitch structure and identification of the interdigitated T-loops motif as an essential recognition determinant for the tRNA elbow revealed interesting parallels with two other large RNA machines whose functions depend on their recognition of overall tRNA architecture. The universal ribozyme RNase P, responsible for endonucleolytic maturation of pre-tRNA 5′ ends, measures the distance from the pre-tRNA elbow to determine the appropriate site of pre-tRNA cleavage, using essentially the same structure, formed in this case by the interdigitation of two T-loops from the J11/12-J12/J11 internal loops of the RNase P RNA.58 During translation, the ribosome L1 stalk, a mobile RNA structure, engages the E-site tRNA elbow using a similar pair of interdigitated T-loops to escort and facilitate tRNA release from the ribosome.59, 60 As there are currently no known evolutionary connections between the ribosome, the RNase P, and the T-box riboswitches, the recurring use of the interdigitated T-loops in these three instances to recognize the tRNA elbow appears to be a product of convergent evolution. Alternatively, it is conceivable that T-boxes might have evolved from a fragment of the primordial ribosomal RNA, given the striking structural resemblances in how the two RNA machines decode the tRNA antidodon and recognize the tRNA elbow structure. It is unknown whether there are other naturally occurring structured RNAs that also interact with tRNAs besides these three, and if so whether they too use interdigitated T-loops to recognize the tRNA elbow. For instance, a ribozyme that carries out tRNA aminoacylation may have existed, as in vitro selected ribozymes such as the flexizyme can aminoacylate the tRNA 3′ end using chemically activated amino acids.61, 62 Such a ribozyme would have been a key component of a protein synthesis pathway made entirely of RNA, where ribozymes would have catalyzed amino acid activation, tRNA aminoacylation, and peptidyl transfer.63
Mechanisms for tRNA aminoacylation readout and genetic switching by the T-box
The recent co-crystal structures and NMR model have elucidated how T-box riboswitch Stem I domains recognize their cognate tRNAs with both sequence and structural specificity. However, the crystal structures do not contain the antiterminator domain, and thus do not address how the tRNA 3′ end, and in particular, the tRNA aminoacylation status, is recognized by the T-box, and further how this readout is used to dictate the genetic outcome of the switch (Fig. 1). In fact, until 2014, it was not even established whether the T-box riboswitch is capable of directly sensing ther tRNA aminoacylation state without the aid of protein factors such as EF-Tu.
Evaluation of tRNA aminoacylation status by the T-box
EF-Tu is among the most abundant proteins in the bacterial cell and exhibits extraordinarily high selectivity and affinity (KD ∼ 10-10 M) toward charged (aminoacylated) tRNAs.64 It follows that most charged tRNAs would be associated with EF-Tu in vivo, and that the tRNA pools encountered by T-box riboswitches could be either uncharged free tRNAs, or charged and EF-Tu-bound tRNAs. Thus it may be possible for the T-box to lack an inherent ability to sense tRNA charging state directly, and to use EF-Tu as a proxy to distinguish charged from uncharged tRNAs.23 The true selectivity of the T-box riboswitch, (or T-box ribonucleoprotein switch if it requires EF-Tu to function) has remained unresolved due to several technical challenges. This has not been assayed in vivo because EF-Tu is essential for life. In vitro, it was demonstrated early on that uncharged cognate tRNA strongly promotes readthrough transcription of the T-box terminator region into the coding region.18, 65 However, it has remained unknown how the T-box would respond to aminoacylated tRNA, as such experiments require a homogeneous preparation of aminoacylated tRNA that is essentially free of contaminating uncharged tRNAs. tRNA aminoacylation reactions using cognate aminoacyl-tRNA synthetases, like many enzymatic reactions, do not go to completion, producing a mixture of charged and uncharged tRNAs. Further, it has remained difficult to separate the two forms of tRNAs, in particular, those tRNAs charged with small amino acids (such as glycine) from their uncharged congeners. A third technical difficulty lies in the hydrolytic instability of the aminoacyl bond under even slightly alkaline conditions.17, 66 Due to these difficulties, the effect of charged tRNA has been generally approximated using a tRNA variant that carries an extra cytosine (tRNAEX1C) on its 3′ terminus, which indeed is unable to induce transcription readthrough.17, 67 However, tRNAEX1C does not truly mimic aminoacylated tRNA because the extra 3′ cytosine (a) is six times as large as an esterified glycine, (b) occupies a different spatial location than the esterified amino acid due to its ability to stack on the nucleobase of the now penultimate adenosine, and (c) directly base pairs with a near-invariant guanosine (G154) at the trailing edge of the antiterminator helix A1 (Fig. 7a) and might thus strand-invade helix A1 and actively destabilize the antiterminator structure. Formation of this additional intermolecular C-G pair was confirmed by calorimetric measurements and explains why tRNAEX1C is particularly ineffective in inducing transcription read through, compared to other tRNAs that carry 3′ extra nucleosides that do not pair with G154.67
Figure 7. Mechanisms of sensing tRNA aminoacylation state and genetic switching by the T-box riboswitch.
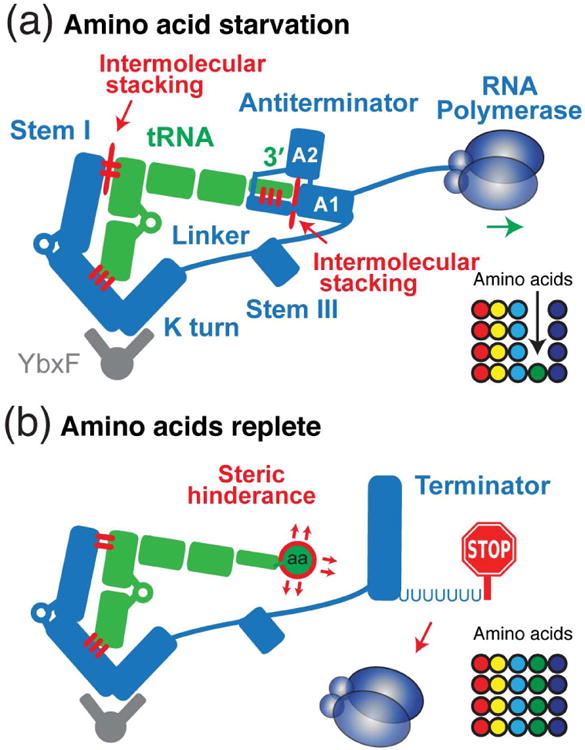
(a) Starvation of a particular amino acid (green sphere) leads to deacylation of its carrier tRNA (green). The uncharged tRNA 3′ NCCA terminus base pairs with the antiterminator bulge, forming a 4-bp intermolecular helix. This helix further coaxially stacks against the helix A1 of the antiterminator, thereby forming an extended continuous helical stack (29 layers) and stabilizing the anititerminator structure and permitting the RNA polymerase to proceed through into the downstream coding region. Thus, the uncharged tRNA is incorporated into the antiterminator assembly and dictates the outcome of the genetic switch. (b) When amino acids are replete, an aminoacylated tRNA 3′ terminus destabilizes the antiterminator structure by steric hindrance of the amino acid, causing the thermodynamically more stable transcription terminator (strong RNA hairpin followed by a track of uridines) to form. Transcription is therefore terminated and the downstream coding genes are shut off, completing a negative feedback loop that senses and maintains steady-level supplies of amino acids for the ribosome in the form of aminoacylated tRNAs.
A definitive analysis of the true tRNA selectivity of the T-box requires homogeneous aminoacylated tRNAs, which are traditionally prepared by aminoacylation using cognate aminoacyl-tRNA synthetases, followed by isolation of aminoacylated tRNAs exploiting a specific interaction between the 3′ terminal ribose cis-diol and immobilized boronate68, or taking advantage of the selectivity for aminoacylated tRNAs of the activated EF-Tu•GTP complex.69 However, neither method is efficient or scalable. Both are subject to on-column tRNA deacylation and other issues such as the very low (∼5%) efficiency of EF-Tu activation70 using a GTP-GDP regeneration and exchange system. To provide a robust, flexible, and scalable alternative, a new method was developed to produce preparative quantities of aminoacylated tRNAs combining ribozyme-mediated tRNA aminoacylation71 (thus conferring superior flexibility in both tRNA and the amino acid to be charged) and a reversible chemical protection approach.72 First, tRNA was aminoacylated using flexizyme, a 46-nt in vitro selected ribozyme capable of aminoacylating virtually any single-stranded RNA 3′ terminus with any natural amino acid, many unnatural amino acids, D-amino acids, or even hydroxy acids.62 The resulting mixture of charged and uncharged tRNA was immediately reacted with N-pentenoyl succinimide, which appends a pentenoyl protecting group to the α-amine of the 3′-esterified amino acid. The pentenoyl group not only stabilizes the aminoacyl bond against hydrolysis, but also serves as hydrophobic purification handle, permitting isolation of N-protected charged tRNAs using reversed-phase high-performance liquid chromatography (RP-HPLC).66, 73 Immediately before use, treatment with aqueous iodine under very mild conditions readily regenerates highly purified aminoacylated tRNAs (>95%).
Using highly purfied aminoacylated tRNA obtained using the new method, it became clear that the T-box riboswitches, exemplified by the B. subtilis glyQS T-box, strongly reject tRNAs charged with even the smallest amino acid, glycine, to the fullest extent, i.e., glycyl-tRNAGly, was unable to induce any transcription readthrough.66 Further, using a panel of tRNA variants carrying a range of 3′ chemical groups that vary in molecular volume, it was demonstrated that as the van der Waals volume of its 3′ substituent increases, tRNA is progressively less able to induce transcription read through. In other words, the size of the tRNA 3′ group is an excellent inverse predictor of its ability to stabilize the antiterminator conformation of the T-box riboswitch. Thus the T-box gauges the molecular volume of tRNA 3′ substituents and the aminoacylated tRNA is rejected by structural destabilization due to steric hindrance. Importantly, a 3′-esterified glycine already elicits the full effect, and making the substituent larger does not further reduce transcription readthrough.66 This means that the T-box steric sieve has a threshold size that is of the order of that of glycine, the smallest amino acid. This allows a common antiterminator architecture to function in all the T-boxes that sense and respond to any of the amino acids. Consistent with this, phylogenetic analyses show that the antiterminator sequence and secondary structure do not vary based on the amino acid specificity of the T-box of which they are part.20, 21 Moreover, switching the Stem I specifier sequence allows at least partial switching of the tRNA and amino acid specificity of a T-box.
Mechanism of uncharged tRNA-directed genetic switching
A central mechanistic mystery of the T-box system is how an uncharged tRNA molecule can dictate the outcome of a transcriptional switch. Is the genetically predicted base-pairing interaction between the tRNA 3′ NCCA terminus and a 7-nt bulge that separates the two antiterminator helices14, 15, 74 (helices A1 and A2, Fig. 7a) sufficient to stabilize the antiterminator conformation against the formation of a much more stable terminator (ΔΔG ∼ 16.5 kcal mol-1)? Quantitative analyses of both the fluorescence intensity and lifetimes of fluorescent nucleoside analogues (2-aminopurine and pyrrolo-cytosine) strategically placed at the tRNA-T-box interface, complemented by calorimetric analyses, reveal that the tRNA 3′ NCCA terminus not only base pairs to the antiterminator bulge, but also forms an intermolecular coaxial stack with helix A1, thereby stabilizing it against terminator formation (Fig. 7a).66 Thus, the two conserved elements of the T-box, the Stem I and antiterminator domains, sandwich the top half of the uncharged tRNA (coaxially stacked acceptor stem and T-stem), assembling into a long (a remarkable 29 layers) coaxial stack that comprises two instances of intermolecular stacking, and in doing so, stabilize the otherwise unstable antiterminator (in particular, helix A1, Fig. 7a). The collective thermodynamic contributions from these multi-partite T-box-tRNA base-pairing and stacking interactions, in conjunction with the kinetic advantage of being transcribed and folding first, allow the antiterminator conformation to be populated and to persist over the terminator, overcoming the otherwise very unfavorable difference in thermodynamic stability. The T-box thus leverages the structural stability of tRNA to direct the co-transcriptional folding pathways of the antiterminator/terminator region to ultimately gain control of the genetic switch. All of this regulation is coupled to the direct sensing of small chemical changes to the tRNA, in this case aminoacylation, using steric hindrance by the amino acid itself (Fig. 7b), in a compact and versatile design consisting of just over thirty nucleotides.
Conclusion
Recent genetic, structural, and mechanistic analyses have produced a wealth of insight into the multi-faceted T-box gene-regulatory system, which is also a paradigm for high-affinity, high-specificity interactions between two natural non-coding RNAs. The modular nature of the T-box RNA allows the initial docking of tRNAs with its Stem I domain, irrespective of their aminoacylation status.17 This interaction is selective for both sequence and structural features of the anticodon region of the tRNA and the receptor specifier (codon) region of the T-box Stem I. This interaction appears to be the first committed contact that occurs between the two structured RNAs because disruption of the codon-anticodon pairing completely abolishes binding whereas disruption of other interactions (such as the tRNA elbow-distal Stem I interaction) preserves weak binding.24 An unexpected elaboration on the specifier-anticodon interaction is that the structurally characterized specificity can somehow be reconciled with flexibility of reading frame and alternative codon selection by the single-stranded nucleotides of the specifier loop.51 Initial engagement of the anticodon-specifier interaction then induces the formation of secondary contacts, in particular, the interaction between the flat surfaces of the tRNA elbow and the distal region of the Stem I. This latter interaction appears to be sequence-nonspecific and is instead primarily driven by structural complementarity from juxtaposed, stackable base planes from both RNAs. As cognate tRNA productively docks with Stem I, its K-turn (possibly aided by L7Ae-superfamily proteins such as YbxF) sharply bends the trajectory of the T-box RNA toward the tRNA 3′ terminus, thereby setting the stage for the antiterminator domain to sense the tRNA aminoacylation status. The T-box antiterminator domain wraps around the tRNA 3′ terminus, making four bases pairs with the tRNA 3′ NCCA terminus and coaxially stacks this inter-molecular helix on both flanks, one side with the antiterminator helix A1 and on the other side with the rest of the tRNA. This structural configuration yields a strikingly long continuous helical stack that traverses both T-box domains as well as the entire top half of tRNA, lending crucial stabilization to the antiterminator to prevent the formation of the competing, strong terminator. The intimate interaction of the tRNA 3′ terminus with the antiterminator is apparently destabilized by steric hindrance from an esterified amino acid, allowing the stabilizing interaction to be selective for uncharged tRNAs.
Despite the insights from two decades of research on the T-box system and the recent elucidation of much of its structural and mechanistic underpinnings, a number of important questions remain. What is the precise structure of the steric sieve created by the antiterminator that is capable of sensing minute chemical changes to the tRNA 3′-terminus? What functional roles does the linker between the Stem I and antiterminator domains play? T-boxes with specificities other than glycine contain elaborate and structurally conserved Stem II and Stem II A/B pseudoknot elements in the linker. Why do glycine T-boxes not require such a structured linker? Do these complex linker regions contact other parts of tRNA to provide added affinity or specificity? What is the precise timeline of formation of the various T-box-tRNA contacts, and how do tRNAs release from the tight T-box grip? To what extent is the T-box riboswitch under kinetic or thermodynamic control? 75 To what extent can T-boxes bind more than one tRNA and respond to starvation of more than one amino acid? How do structurally divergent T-boxes function? Curious examples include the ileS T-boxes from Actinomyces that are truncated at the top of Stem I so that they no long contain the interdigitated T-loops.76 How do translationally acting T-boxes differ in structure and mechanism from their transcriptionally acting, better-understood counterparts? Further, can the T-box riboswitch be engineered to function as RNA devices to sense, track, or modulate the distribution and dynamics of specific tRNAs in eukaryotes from which T-boxes are absent? Finally, are there structurally complex eukaryotic or viral RNAs such as long non-coding RNAs (lncRNAs) that utilize RNA-RNA recognition strategies similar to the T-box system to interact with cellular tRNAs or other tRNA-like elements to achieve cellular or viral gene regulation? With the structural and mechanistic framework established, we anticipate an accelerated pace in addressing these open questions and engineering challenges.
Acknowledgments
We thank N. Baird, M. Lau, J. Lehmann, C. Stathopoulos, N. Walter, W. Winkler, and E. Westhof for discussions. J.Z. is a recipient of a NHLBI Career Transition Award (K22) from the National Heart, Lung and Blood Institute. This work was supported in part by the intramural program of the NHLBI, NIH.
Contributor Information
Jinwei Zhang, National Heart, Lung, and Blood Institute.
Adrian R. Ferré-D'Amaré, Email: adrian.ferre@nih.gov, National Heart, Lung, and Blood Institute.
References
- 1.Earl AM, Losick R, Kolter R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008;16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 3.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 4.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 5.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 6.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 7.Peselis A, Serganov A. Themes and variations in riboswitch structure and function. Biochim Biophys Acta. 2014;1839:908–918. doi: 10.1016/j.bbagrm.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 10.Serganov A, Patel DJ. Amino acid recognition and gene regulation by riboswitches. Biochim Biophys Acta. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ames TD, Breaker RR. Bacterial aptamers that selectively bind glutamine. RNA Biology. 2011;8 doi: 10.4161/rna.8.1.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 14.Means JA, Simson CM, Zhou S, Rachford AA, Rack JJ, Hines JV. Fluorescence probing of T box antiterminator RNA: insights into riboswitch discernment of the tRNA discriminator base. Biochem Biophys Res Commun. 2009;389:616–621. doi: 10.1016/j.bbrc.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauzi H, Agyeman A, Hines JV. T box transcription antitermination riboswitch: influence of nucleotide sequence and orientation on tRNA binding by the antiterminator element. Biochim Biophys Acta. 2009;1789:185–191. doi: 10.1016/j.bbagrm.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 17.Grundy FJ, Yousef MR, Henkin TM. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J Mol Biol. 2005;346:73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 18.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci U S A. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson AR, Henkin TM, Agris PF. tRNA regulation of gene expression: interactions of an mRNA 5′-UTR with a regulatory tRNA. RNA. 2006;12:1254–1261. doi: 10.1261/rna.29906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigg JC, Ke A. Structural Determinants for Geometry and Information Decoding of tRNA by T Box Leader RNA. Structure. 2013;21:2025–2032. doi: 10.1016/j.str.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Ferré-D'Amaré AR. Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration. Structure. 2014;22:1363–1371. doi: 10.1016/j.str.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Ferré-D'Amaré AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500:363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang AT, Nikonowicz EP. Solution NMR determination of hydrogen bonding and base pairing between the glyQS T box riboswitch Specifier domain and the anticodon loop of tRNA(Gly) FEBS Lett. 2013;587:3495–3499. doi: 10.1016/j.febslet.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Jones CP, Ferré-D'Amaré AR. Global analysis of riboswitches by small-angle X-ray scattering and calorimetry. Biochim Biophys Acta. 2014;1839:1020–1029. doi: 10.1016/j.bbagrm.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferré-D'Amaré AR, Doudna JA. RNA folds: insights from recent crystal structures. Annu Rev Biophys Biomolec Struct. 1999;28:57–73. doi: 10.1146/annurev.biophys.28.1.57. [DOI] [PubMed] [Google Scholar]
- 28.Hermann T, Patel DJ. Stitching together RNA tertiary architectures. J Mol Biol. 1999;294:829–849. doi: 10.1006/jmbi.1999.3312. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Ferré-D'Amaré AR. New molecular engineering approaches for crystallographic studies of large RNAs. Curr Opin Struct Biol. 2014;26C:9–15. doi: 10.1016/j.sbi.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigg JC, Chen Y, Grundy FJ, Henkin TM, Pollack L, Ke A. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci U S A. 2013;110:7240–7245. doi: 10.1073/pnas.1222214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JHD. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AHJ, Seeman NC, Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 33.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 34.Strobel SA, Adams PL, Stahley MR, Wang J. RNA kink turns to the left and to the right. RNA. 2004;10:1852–1854. doi: 10.1261/rna.7141504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McPhee SA, Huang L, Lilley DM. A critical base pair in k-turns that confers folding characteristics and correlates with biological function. Nat Commun. 2014;5:5127. doi: 10.1038/ncomms6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lilley DM. The K-turn motif in riboswitches and other RNA species. Biochim Biophys Acta. 2014;1839:995–1004. doi: 10.1016/j.bbagrm.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Henkin TM, Nikonowicz EP. NMR structure and dynamics of the specifier loop domain from the Bacillus subtilis tyrS T box leader RNA. Nucleic Acids Res. 2010;38:3388–3398. doi: 10.1093/nar/gkq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Nikonowicz EP. Solution structure of the K-turn and specifier loop domains from the Bacillus subtilis tyrS T-Box leader RNA. J Mol Biol. 2011;408:99–117. doi: 10.1016/j.jmb.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamma T, Ferré-D'Amaré AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 43.Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DMJ. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baird NJ, Zhang J, Hamma T, Ferré-D'Amaré AR. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA. 2012;18:759–770. doi: 10.1261/rna.031518.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 46.Korostelev A, Trakhanov S, Laurberg M, Noller H. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 47.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giege R, Juhling F, Putz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip Rev RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 49.Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 50.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013;41:D262–267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saad NY, Stamatopoulou V, Braye M, Drainas D, Stathopoulos C, Becker HD. Two-codon T-box riboswitch binding two tRNAs. Proc Natl Acad Sci U S A. 2013;110:12756–12761. doi: 10.1073/pnas.1304307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fagan CE, Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA. 2014;20:1944–1954. doi: 10.1261/rna.046953.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van de Guchte M, Ehrlich SD, Chopin A. Identity elements in tRNA-mediated transcription antitermination: implication of tRNA D- and T-arms in mRNA recognition. Microbiology. 2001;147:1223–1233. doi: 10.1099/00221287-147-5-1223. [DOI] [PubMed] [Google Scholar]
- 54.Chan CW, Chetnani B, Mondragón A. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip Rev RNA. 2013 doi: 10.1002/wrna.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krasilnikov AS, Mondragón A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lehmann J, Jossinet F, Gautheret D. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNase P and T-box leaders. Nucleic Acids Res. 2013;41:5494–5502. doi: 10.1093/nar/gkt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 1996;24:1073–1079. doi: 10.1093/nar/24.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikulin A, Eliseikina I, Tishchenko S, Nevskaya N, Davydova N, Platonova O, Piendl W, Selmer M, Liljas A, Drygin D, et al. Structure of the L1 protuberance in the ribosome. Nature Struct Biol. 2003;10:104–108. doi: 10.1038/nsb886. [DOI] [PubMed] [Google Scholar]
- 60.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 61.Xiao H, Murakami H, Suga H, Ferré-D'Amaré AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- 62.Murakami H, Ohta A, Ashiai H, Suga H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nature Methods. 2006;3:357–359. doi: 10.1038/nmeth877. [DOI] [PubMed] [Google Scholar]
- 63.Ferré-D'Amaré AR. Use of a coenzyme by the glmS ribozyme-riboswitch suggests primordial expansion of RNA chemistry by small molecules. Phil Trans Royal Soc B. 2011;366:2942–2948. doi: 10.1098/rstb.2011.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Louie A, Ribeiro NS, Reid BR, Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem. 1984;259:5010–5016. [PubMed] [Google Scholar]
- 65.Putzer H, Condon C, Brechemier-Baey D, Brito R, Grunberg-Manago M. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 2002;30:3026–3033. doi: 10.1093/nar/gkf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Ferré-D'Amaré AR. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol Cell. 2014;55:148–155. doi: 10.1016/j.molcel.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yousef MR, Grundy FJ, Henkin TM. tRNA requirements for glyQS antitermination: a new twist on tRNA. RNA. 2003;9:1148–1156. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCutchan TF, Gilham PT, Soll D. An improved method for the purification of tRNA by chromatography on dihydroxyboryl substituted cellulose. Nucleic Acids Res. 1975;2:853–864. doi: 10.1093/nar/2.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohtsuki T, Yamamoto H, Doi Y, Sisido M. Use of EF-Tu mutants for determining and improving aminoacylation efficiency and for purifying aminoacyl tRNAs with non-natural amino acids. J Biochem. 2010;148:239–246. doi: 10.1093/jb/mvq053. [DOI] [PubMed] [Google Scholar]
- 70.Asahara H, Uhlenbeck OC. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu. GTP Biochemistry. 2005;44:11254–11261. doi: 10.1021/bi050204y. [DOI] [PubMed] [Google Scholar]
- 71.Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nature Protocols. 2011;6:779–790. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- 72.Lodder M, Wang B, Hecht SM. The N-pentenoyl protecting group for aminoacyl-tRNAs. Methods. 2005;36:245–251. doi: 10.1016/j.ymeth.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Ferré-D'Amaré AR. A Flexible, Scalable Method for Preparation of Homogeneous Aminoacylated tRNAs. Methods Enzymol. 2014;549:105–113. doi: 10.1016/B978-0-12-801122-5.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerdeman MS, Henkin TM, Hines JV. Solution structure of the Bacillus subtilis T-box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility. J Mol Biol. 2003;326:189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J, Lau MW, Ferré-D'Amaré AR. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sherwood AV, Grundy FJ, Henkin TM. T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions. Proc Natl Acad Sci U S A. 2015;112:1113–1118. doi: 10.1073/pnas.1424175112. [DOI] [PMC free article] [PubMed] [Google Scholar]


