Abstract
Metallic nanoparticles provide versatile scaffolds for biosensing applications. In this review, we focus on the use of metallic nanoparticles for cell surface sensings. Examples of the use of both specific recognition and array-based “chemical nose” approaches to cell surface sensing will be discussed.
1. Introduction
Cell surface sensors for disease and detection of infection have direct access to the sensing target, in contrast to approached that detect intracellular proteins, nucleic acids, or other markers buried inside the cells. This ready access has the potential to provide rapid sensing with minimal processing. The rich environment presented by the cell exterior also gives cell surface sensors the capability to read out the phenotypes of cells, a property that is the final outcome of multiple factors including both genetic and epigenetic variations.1 For example, in the case of cancer, abnormal cells have been found to overexpress specific glycosylated proteins at their plasma membrane such as epithelial cell adhesion molecule (EpCAM) or carcinoembryonic antigen (CEA).2-4 Therefore, targeting cell surface phenotype provides a strategy for simple, rapid, and robust diagnostic pathways in diverse areas such as cancer and pathogenic bacteria.
Three integrated components are necessary to fabricate an effective sensor: (1) a recognition element to interact with a target analyte, (2) a signal transduction element to generate a measurable signal from an analyte-receptor binding event, and (3) a device that outputs a result. Metallic nanoparticles (NPs) can be easily engineered to provide scaffolds for recognition processes, with their physical properties facilitating the transduction process, making them excellent platforms for cell surface sensing.5,6
In this review, we will focus on the use of metallic NPs for the detection and quantification of cell properties, based on cell surface components. We will discuss examples of different engineered metallic NP systems 7, 8 that provide cell sensing through specific and selective interactions with the cell surfaces.
2. Cell surface and nanoparticle interactions
Enormous cell surface diversity exists among cells from plants, bacteria, and animals. The surface of a mammalian cell is composed of a complex structure featuring the lipid bilayer, proteins, nucleic acids as well as a range of polysaccharide structures that comprise the glycocalyx.9 This glycocalyx is composed of glycoproteins, proteoglycans and glycolipids.10 Phenotypically altered expression of each of these components provides diagnostic information for diseases such as cancer, Gaucher's, and Tay-Sachs diseases.11,12 Taken together, the complex array of biomolecules that comprise cell surfaces make them excellent targets for both specific biomarker sensing and selective “chemical nose” based methods.
The interaction between nanomaterials and cells is an important issue for designing systems not only for sensing, but also for imaging and delivery. In general, the following factors need to be taken into account: (1) specific receptors (biomarkers) on the cell membrane, (2) the size, shape, surface charge, roughness and hydrophobicity of nanoparticles and their role in selective interactions. While these topics are all central to the sensing described here, the in-depth discussion required for understanding this interaction is beyond the scope of this tutorial discussion. Nel and coworkers have provided a comprehensive review to help understand the biophysicochemical interactions at the nano-bio interface, which discussed cell-nanoparticle interactions in detail.13
3. Specific sensing
We will focus on spherical metallic nanoparticles in this review, as these systems have been widely employed for cell surface sensing. Metallic nanoparticles can be functionalized with small molecules 14 and biomacromolecules 15 to achieve the specific interactions with the biological targets. However, the vast majority of specific-based sensors have been using biomacromolecules to functionalize metallic nanoparticles, so we will focus on these bioconjugate systems. These platforms provide highly adaptable tools for rapid and/or point-of-care tools that provide alternatives to more complex and instrument-intensive techniques such as flow cytometry. 16
3.1. Antibody-based sensing
Antibodies are widely used as recognition elements in diagnostic and therapeutic applications.17 There are two key components of antibodies: the Fab (fragment, antigen-binding) region of an antibody that recognizes the antigen and the Fc (fragment, constant) that can be used for conjugation without disrupting the recognition process. Conjugation of either complete antibodies or Fab fragments to NPs provides an effective means of recognizing cell surface functionality. As described below, these binding events can be detected via various tools such as surface-enhanced Raman scattering (SERS) and electrochemistry.
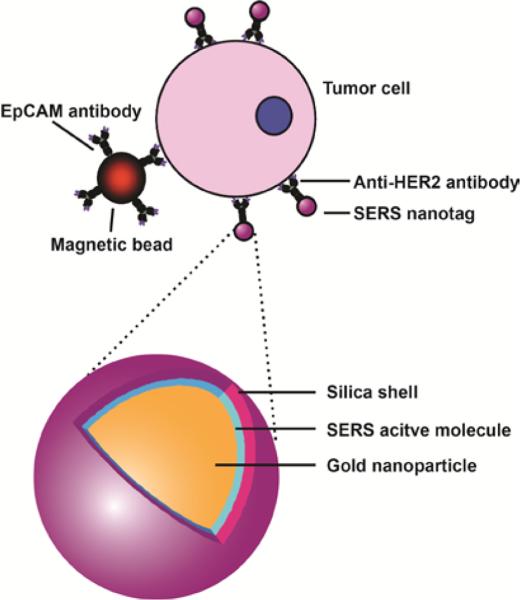
SERS is a technique in which the Raman signal can be dramatically amplified through surface plasmon resonance of metallic NPs.6 SERS-based techniques utilizing antibodies have been successfully applied to immunoassay-based methodologies. 18 In one example, Sha and coworkers detected cancer circulating cells (CTCs) by combining capturing capability of a magnetic bead and specific labeling of SERS nanotags.19 This bead was conjugated with anti-EpCAM antibody to capture SKBR3 cancer cells. These cells were then labeled for SERS detection by AuNPs functionalized with anti-HER2 antibody (human epidermal growth factor receptor-2). A silica shell was subsequently coated on this complex to enable the functionalization of antibody on the particle without interfering with the Raman response (Figure 1). In a similar study, SERS-based systems were further employed for in vivo tumor targeting. Poly(ethylene glycol)-capped AuNPs were used to stabilize Raman-active reporter molecules. The SERS-NPs were conjugated with antibodies specifically targeting the overexpressed epidermal growth factor receptor on tumor cells, resulting in highly specific in vivo tumor detection.20
Figure 1.
Tumor cell detection using anti-HER2 antibody-conjugated magnetic beads with SERS nanotags. Reprinted with permission from ref. 19. Copyright 2008 American Chemical Society.
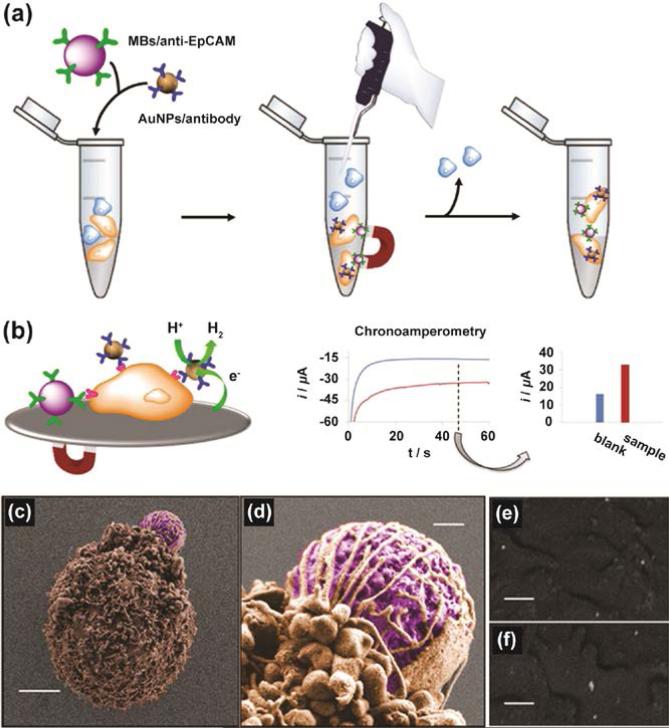
Electrochemical detection technique can utilize the electrocatalytic properties of AuNPs to provide rapid cell surface sensing. 21 This strategy has the advantages of simpler instrumentation coupled with the direct connection of sensor output with devices/computers. For example, Merkoci and coworkers employed antibody-conjugated magnetic beads and AuNPs for the detection of CTC (Figure 2).22 AuNPs fabricated with anti-EpCAM antibody were used for targeting EpCAM, an overexpressed transmembrane glycoprotein on human colon adenocarcinoma cells (Caco2 cells). The AuNP-antibody conjugates were used to generate an electrochemical signal through electrocatalytic hydrogen evolution. The signals generated from AuNP-antibody conjugates could detect 2.2 × 102 Caco2 cells in the presence of other interfering cells such as monocytes (THP-1).
Figure 2.
(a) Capture of Caco2 cells by magnetic beads conjugated to anti-EpCAM. Simultaneously, cells were labeled with AuNP-specific antibodies in the presence of interfering cells (THP-1). (b) Chronoamperometry of the hydrogen evolution reaction (HER) electrocatalyzed by AuNPs. (c), (d) False colors scanning electron microscopy (SEM) images of a Caco2 cell captured by magnetic beads (MBs)/anti-EpCAM. (e), (f) Backscattered images showing AuNPs distributed along the cell plasma membrane of Caco2 cells. Scale bars, 3 μm (c), 400 nm (d), and 200 nm (e, f). Reprinted with permission from ref. 22. Copyright 2012 American Chemical Society.
3.2. Lectin-based sensing
Lectins are proteins that exhibit strong and specific or selective binding towards carbohydrate moieties. Targeting carbohydrates has been the useful strategy for diagnosis because the alterations of carbohydrates found on plasma membrane have been correlated with disease, such as liver fibrosis, pancreatic cancer, and cervical cancers. 23 Thus, NPs functionalized with lectins can be a powerful tool for cell surface sensing.24
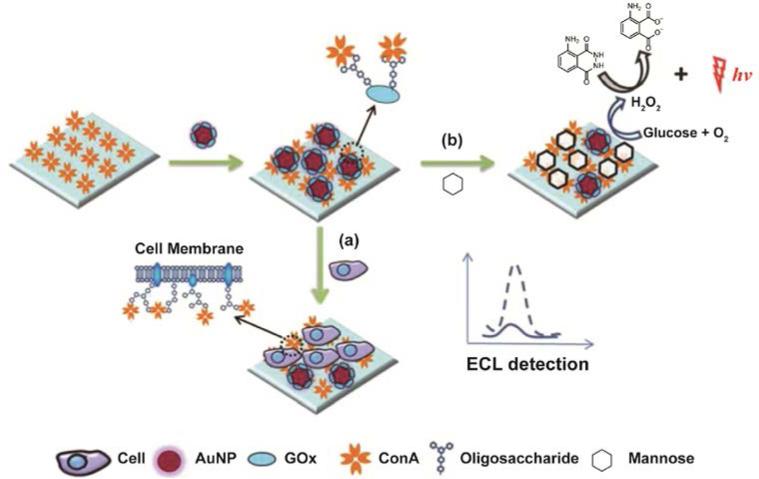
In a recent study, Liu and coworkers designed a sensitive electrochemiluminescence25 (ECL)-based biosensor using a displacement assay that relies on the interaction between NP-bound lectins and carbohydrates on the cell surface (Figure 3).26 In this system, a gold electrode immersed in luminol solution was coated with Concanavalin A (ConA), a lectin that recognizes mannose (a carbohydrate type found on the cell surface). These mannose moieties can also be found in glucose oxidase molecules (GOx), an enzyme that can catalyze the luminol ECL reaction. By coupling GOx with AuNPs (GOx-Au), they were able to fabricate a multifunctional probe. This GOx-Au probe can both compete with mannose moieties on the analyte cells for ConA-coated gold electrode and improve the ECL signal of luminol. In the presence of the target cells, the competition between GOx-Au and mannose-containing cells generates the alterations in ECL signal intensity, providing the ability to profile carbohydrate-lectin interaction and in situ cell surface carbohydrate expression.
Figure 3.
Schematic illustration of the lectin-based sensing strategy for (a) carbohydrate-ConA interaction analysis and (b) cell surface carbohydrate expression. Reprinted with permission from ref. 26. Copyright 2013 Royal Society of Chemistry.
3.3. Aptamer-based sensing
Short, single-stranded oligonucleotides (ssDNA or ssRNA), known as aptamers, provide an emerging strategy for biorecognition. Aptamers are produced from an in vitro method known as SELEX (systematic evolution of ligands by exponential enrichment). In this process, SELEX uses polymerase chain reaction (PCR) to specifically amplify the sequence that has high affinity and selectivity to the target analyte. The iterative process generates aptamers that often fold into unique three-dimensional conformations. Aptamers can bind to target molecules ranging from small organic molecules to biomacromolecules,27 making them promising candidates to serve as recognition elements in biosensors.
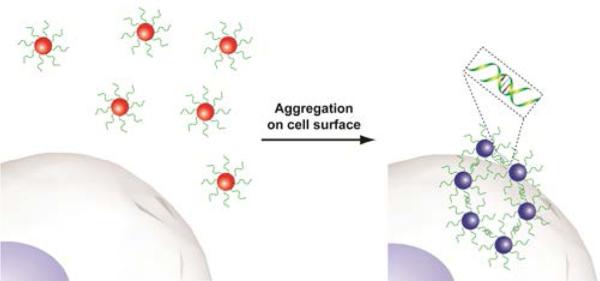
The recognition capabilities of aptamers can be combined with the spectroscopic advantages of AuNPs for cell detection applications. AuNPs possess strong distance dependent optical properties due to surface plasmon resonance.28 The aggregation of aptamer-conjugated AuNPs causes a shift in their absorption spectra, resulting in a change in their scattering profile and color from red to blue/purple.29 This colorimetric sensing method has been applied using aptamers for the detection of cancer cells. For instance, Tan and coworkers successfully applied the aggregation-based colorimetric sensing platform to detect cancer cells using AuNPs functionalized with the aptamers of interest.30 The specific interaction between AuNP-aptamer conjugates and the target cells (CCRF-CEM acute leukemia cell) induced a distinct color change (Figure 4).
Figure 4.
Aptamer-conjugated gold nanoparticles used in colorimetric sensing of cancer cells.
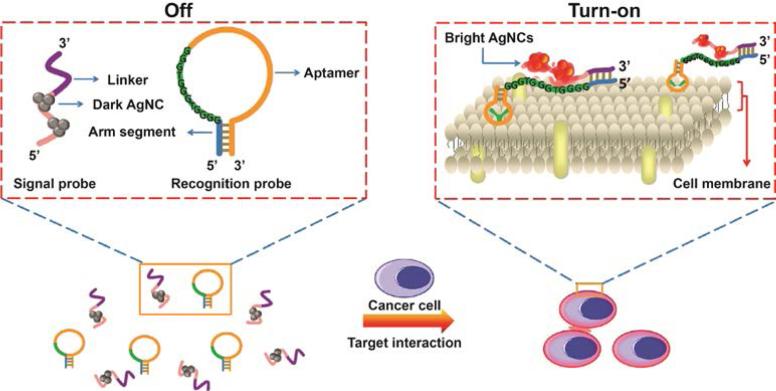
Recently, DNA-templated silver nanoclusters (DNA-AgNCs) were used as a signal transduction element for use with aptamers. The fluorescence of DNA-AgNCs can be significantly amplified in proximity of guanine-rich DNA sequences, 31 a phenomena Wang and coworkers have applied in cell surface-based sensing.32 They designed a “turn-on” system for cancer cell detection, utilizing the fluorescence enhancement of DNA-AgNCs and the recognition capability of aptamers. Two separate DNA-based probes were involved in this system, denoted as the recognition probe and signal probe. The recognition probe was designed as a hairpin-shaped structure that contains a CCRF-CEM cancer cell specific aptamer sequence, a guanine-rich DNA sequence and an arm segment. The signal probe contains a sequence for AgNC-templated synthesis and a link sequence that is complementary to the arm segment of the recognition probe. Once the aptamer sequence from the recognition probe recognizes and binds to CCRF-CEM cells, the recognition probe undergoes a conformational alteration. This conformational alternation then initiates the hybridization of the two probes and consequently brings DNA-AgNCs close to the guanine-rich DNA sequence, resulting in an enhanced fluorescence readout (Figure 5).
Figure 5.
Schematic representation of cancer cell detection based on DNA-templated silver nanoclusters (AgNCs). Reprinted with permission from ref. 32. Copyright 2013 American Chemical Society.
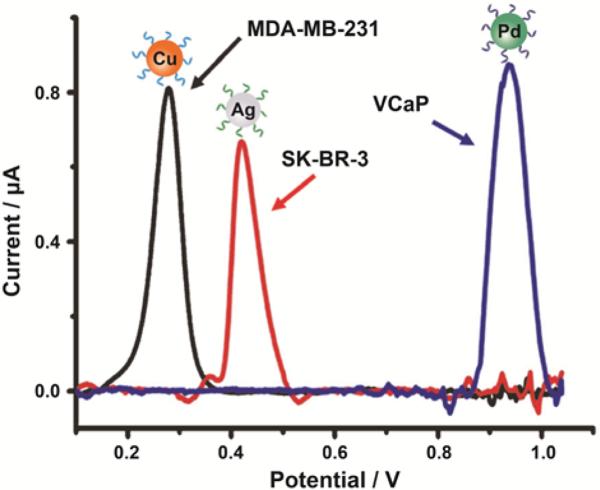
Aptamer-based specific sensing can also be used to detect different cancer cells using an electrochemical approach. By combining multiple metallic NPs with electrochemical analysis, Kelley and coworkers designed a chip-based strategy for the analysis of cancer cells associated with different tumor phenotypes.33 Pd, Ag, and Cu NPs were chosen as signal reporters since they have well-separated potentials as redox-active probes. Biomarker-specific aptamers were conjugated with these three types of metallic NPs to form Pd-anti-PSMA, Ag-anti-HER2, and Cu-anti-MUC1 NPs. A mixture of these three NPs were successfully used for the specific detection of different prostate and breast cancer cell lines such as VCaP, SK-BR-3, and MDAMB-231 (Figure 6).
Figure 6.
Linear-sweep voltammetry of specific cancer cell detection with a mixture of Pd-anti-PSMA, Cu-anti-MUC1, and Ag-anti-HER2 nanoparticles: VCaP (blue), MDA-MB-231 (black) and SK-BR-3 (red) cells. Reprinted with permission from ref. 33. Copyright 2014 WILEY-VCH KGaA, Weinheim.
3.4. DNAzyme-based sensing
Deoxyribozymes, known as DNAzymes or catalytic DNAs, provide an alternative approach to biosensing. DNAzymes are selected from random DNA sequences through combinatorial screening techniques for catalytic and ligand-binding activities. 34 DNAzyme-based sensing relies on the optical property of AuNPs for target recognition role for analytes such as as metal ions and small organic molecules. Using such DNAzyme-functionalized AuNPs, the DNAzyme-catalyzed cleavage or ligation of the nucleic acid substrates affects the assembly of AuNPs, resulting in a colorimetric readout for the cofactors.34
DNAzymes can also be used for signal amplification by behaving as peroxidase mimics. For example, it has been found that when one certain DNA sequence binds with hemin, DNAzyme can be formed with G-quadruplex motifs. This type of DNAzyme can catalyze the generation and enhancement of chemiluminescence (CL) signals in the presence of luminol and H2O2. In this process, AuNPs are employed as carriers for these horseradish peroxidase (HRP)-mimicking DNAzymes. 35 In a recent study, Zhang and coworkers applied HRP-mimicking DNAzyme-functionalized NPs to cancer cell detection through the amplified CL signals.36
4. Selective sensing
Specific recognition-based sensors require pre-identification of the biomarkers, and face certain limitations when used in systems containing multiple analytes. For example, cancer cells present multiple biomarkers on the cell surface. The level of biomarkers may vary among cell populations. In addition, subtle changes in the biomarker levels may be indicative of dramatic phenotypic differences. As an alternative, sensors using selectivity-based modality do not require the knowledge of a specific biomarker. On the contrary, selectivity-based approaches capture the responses from complex analytes to generate a signature for each sample. Such selective sensing approach can be utilized to detect non-specific analytes with either a single recognition element or more commonly an array of recognition elements. In a typical array-based sensor, a set of recognition elements interacts with a number of different analytes or classes of analytes, providing a process reminiscent of mammalian olfaction.37 This mechanistic similarity is why array-based sensors are often denoted as chemical “noses” or “tongues”.
4.1. Single recognition element system
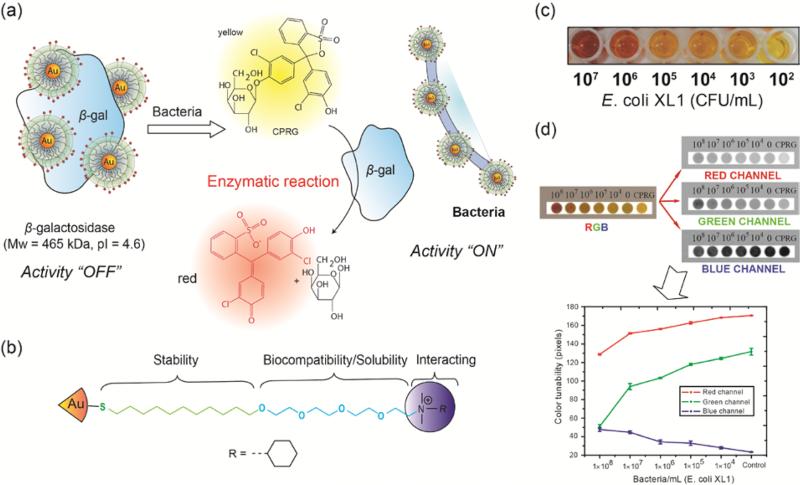
Rotello and coworkers have developed an enzyme amplification sensor using cationic AuNP to inhibit the activity of β-galactosidase (β-gal) based on electrostatic interaction.38 Such enzyme catalysis can amplify the weak signals generated by the system. Bacterial cell surfaces are negatively charged which can disrupt the AuNP-β-gal conjugates. During the sensing process, bacteria cells replace β-gal from the NP-β-gal conjugates, restoring the activity of β-gal towards the chromogenic substrate. Finally, the enzymatic reaction on the substrate gives the corresponding readout to quantify the analytes (Figure 7).39 The enzyme-amplified colorimetric readout was able to detect 102 CFU/mL of Escherichia coli (E. coli) in solution. Furthermore, the performance of this methodology was tested on a paper strip format against concentrations of bacteria ranging from 104~108 CFU/mL. The designed bacteria test strips demonstrate the potential for field applications such as a test of drinking water safety. However, this strategy displays limitations in sensitivity and multiple analyte detection capability due to insufficient interactions between the single recognition element and the analytes.
Figure 7.
(a) Schematic demonstration of enzyme amplified sensing of bacteria using gold nanoparticles. (b) The structure of quaternary amine functionalized gold nanoparticles. (c) The colorimetric sensing of Escherichia coli (E. coli) in solution. (d) Schematic illustration of the RGB analysis for monitoring color changes on test strips for different concentrations of E. coli. Reprinted with permission from ref. 39. Copyright 2011 American Chemical Society.
4.2. Array based sensing systems
Multiple recognition elements can maximize the variation in interactions between sensors and analytes. This array-based strategy combines responses from many individual sensors and analytes to generate a distinct pattern (fingerprint) for each analyte, either based on specific or selective interactions. Since multiple responses can be obtained from array-based sensors, these data matrices are generally analyzed using a variety of multivariate analyses such as principal component analysis (PCA) or linear discriminant analysis (LDA).40
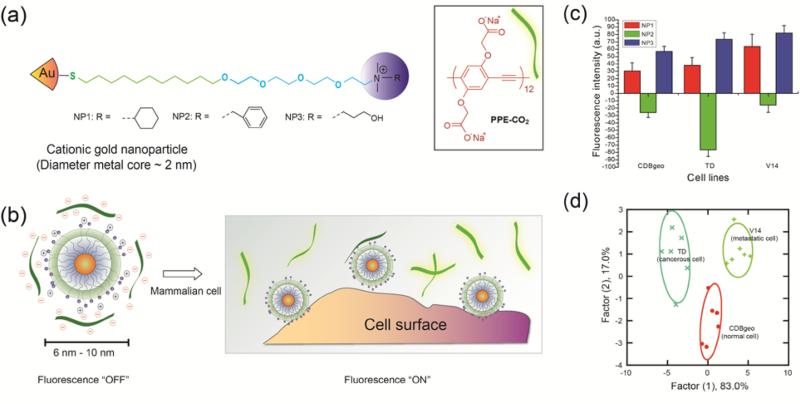
NPs can be readily functionalized with ligands to generate diverse sensor elements. These head groups exhibit differential affinity towards various analytes, leading to variations in the sensor that can be correlated to cellular signatures. Rotello and coworkers have designed several small molecule based sensor arrays that used the phenomenon of fluorescence quenching.41 Fluorescence quenching is reduction in the fluorescence quantum yield due to energy transfer from the photo-excited fluorophore to the AuNPs. In one approach, gold nanoparticles (AuNPs) with a fluorescent polymer [carboxylate poly(para-phenyleneethynylene) (PPE-CO2)] were used to discriminate a series of cell lines (Figure 8).42 The sensor was comprised of three NP types with different quaternary amine functional head groups. An array of AuNPs was used to quench the intensity of the fluorescent polymers via electrostatic interaction. The subsequent binding of cells disrupted the AuNP-polymer complex, thereby generating different fluorescence response patterns for each cell lines. This AuNP-polymer complex was able to identify human cancerous (MCF-7), metastatic (MDA-MB231) and normal (MCF10A) breast cell lines. Since these cell lines came from different individuals, their differentiation might be originating from genetic variation. To avoid this possibility, isogenic cell lines [CDBgeo (normal), TD (cancerous) and V14 (metastatic)] derived from BALB/c mice were used to validate the sensor. Fluorescent proteins can also be used as a transducer in an array based sensor. Cell differentiation using AuNP-green fluorescent protein (GFP) conjugates resulted in four-fold enhancement in the sensitivity of “chemical nose”-based sensor.43 Moreover, in a subsequent study, AuNP-GFP constructs were used to discriminate site specific metastases and healthy state using cell lysates as well as tissue lysates, providing a promising strategy for medical diagnosis.44
Figure 8.
(a) Cationic gold nanoparticles (NP1-NP3) and the fluorescent polymer, carboxylate poly(para-phenyleneethynylene) (PPE-CO2). (b) Fluorescence quenching of the polymers and the restoration of fluorescence after AuNP-polymer complex disrupted by the incubation with cells (dark green strips, fluorescence off; light green strips, fluorescence on). (c) Detection of three isogenic mammalian cell lines (CDBgeo, TD cell and V14) determined by fluorescence change using nanoparticle-polymer supramolecular complexes. (d) Canonical score plot using linear discrimination analysis (LDA) for the first two factors of simplified fluorescence response patterns obtained with NP-polymer assembly arrays against isogenic cell types. Reprinted with permission from ref. 42. Copyright 2009 National Academy of Sciences, USA.
In an analogous study, a swallowtail substituted carboxylate PPE (Sw-CO2) was used as a transducer for AuNP-polymer sensor. 45 This array-based system comprised of amine functionalized hydrophobic NPs that served as recognition units for microorganisms such as bacteria. Bacterial cell walls are negatively charged and furnish a polyvalent environment to interact selectively with AuNP-polymer complex. For example, the Gram-positive microorganisms are highly negatively charged due to the presence of techoic acid residue, whereas E. coli bacteria possess pili (rich in lectins) emanating from the surface. This array-based sensor enables the detection of bacteria cells within minutes. The AuNP-polymer complex was disrupted via competitive binding of different bacteria strains with the AuNP. Twelve different bacteria strains including Gram-positive strains such as Bacillus subtilis, Amycolatopsis azurea and Gram-negative bacteria such as E. coli, and Pseudomonas putida were identified.
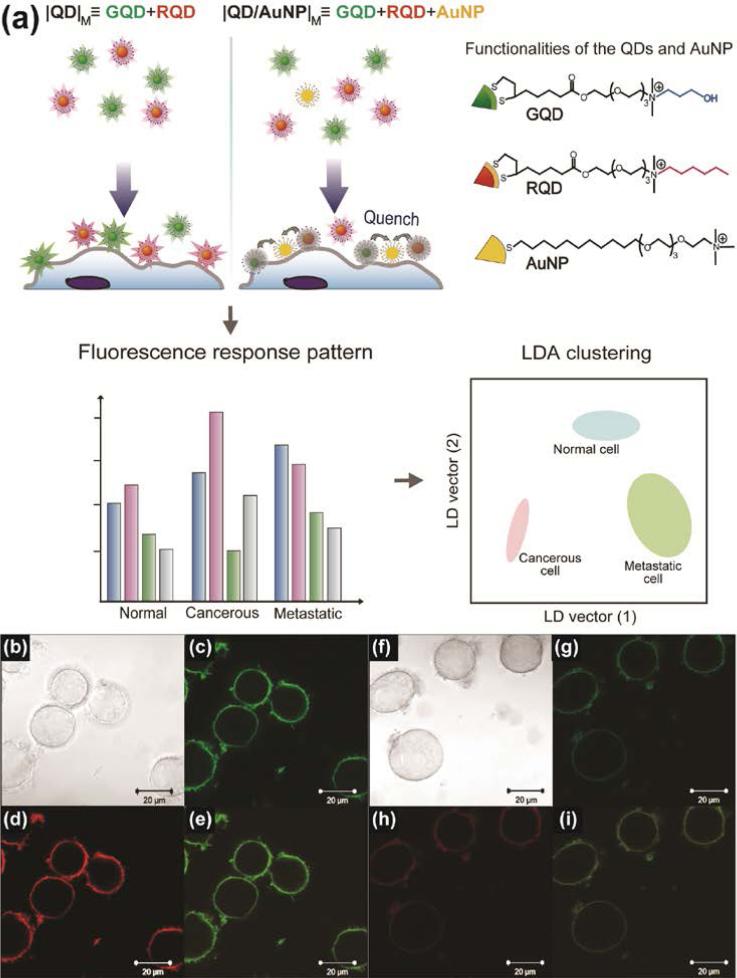
Recently, quantum dots (QDs) were used in an array-based sensor as a recognition element alongside AuNPs. When added together to the cells, co-localization of AuNPs and QDs resulted in quenching, generating different patterns based on cell type/state. This sensor was used to differentiate four different types of cancer cells as well as isogenic normal, cancer and metastatic cells (Figure 9).46 The dual channel fluorescence response obtained from the QDAuNP sensor array could identify 30 unknown samples with 100% accuracy. Besides pairing with QDs, AuNPs can also be combined with upconversion nanoparticles (UCNPs) as a fluorescence resonance energy transfer (FRET) couple to design biosensors.47
Figure 9.
(a) Schematic illustration of the interaction between the nanoparticles and cell surface. The sensing system generated differential quenching and provided distinct patterns to discern different types/states of cells. Two arrays (|QD|M and |QD/AuNP|M) were used in the system and placed in separated wells, with each array providing two fluorescence responses. |QD|M, the mixture of GQD and RQD; |QD/AuNP|M, the mixture of GQD, RQD, and AuNP. (b)-(i) Confocal microscopy images of (b)-(e) |QD|M and (f)-(i) |QD/AuNP|M after the incubation with HeLa cells for 15 min: (b), (f) bright field; (c), (g) green channel; (d), (h) red channel; (e), (i) merged images. Reprinted with permission from ref. 46. Copyright 2013 Elsevier.
Functionalization of biomolecules such as aptamers on NPs can also be used for selective identification of target analytes. Aptamers with selective binding properties towards were coated on citrate capped AuNPs. 48 Upon addition of the target cells, aptamer-protected AuNPs displayed different aggregation, generating different color patterns. Human cancer cells (Jurkat, Reh, Raji) and normal human cells (WIL2-S) were distinguished with the array-based approach by using one human immunoglobulin E aptamer (HIgE-1) and two thrombin aptamers (Tro-1 and Tro-2).
4.3. Multiplexed output sensing system
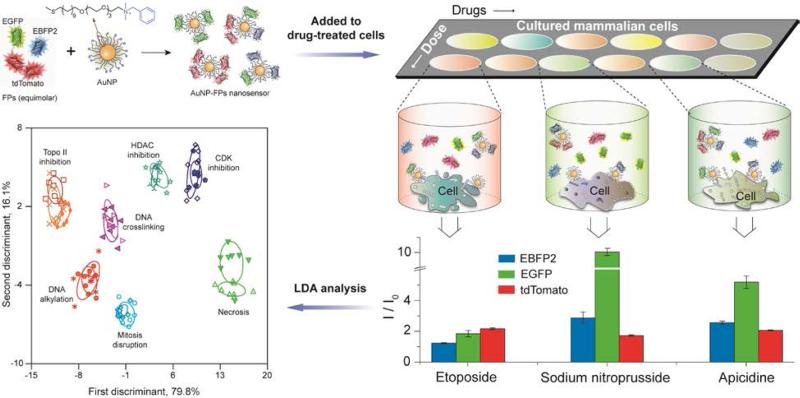
The previous array-based sensing examples use separate recognition elements to generate the multiple sensor outputs required for identification of analytes. An alternative strategy to generating information-rich data would be to use a single recognition element with multiple outputs. Very recently, Rotello and coworkers have developed a high-throughput multi-channel sensor that classifies the mechanism of chemotherapeutic drugs in minutes. 49 This sensor consists of a single AuNP complexed with three different fluorescent proteins (FPs) that is used to sense drug-induced physicochemical changes on cell surfaces. In the presence of cells, differential displacement of the fluorophores with concomitant fluorogenesis provide a ratiometric output that is measurable from a single readout (Figure 10). This result demonstrates the ability of cell surface sensing to be used for high throughput screening of therapeutics, and suggests the utility of these sensors for applications in toxicology and related fields.
Figure 10.
Multi-channel sensor fabricated by incubating AuNP to an equimolar mixture of three fluorescent proteins (FPs): tdTomato (red), EBFP2 (blue) and EGFP (green). Different drug-treated cells result in distinct cell surface phenotypes, leading to different FP displacement patterns as schematically shown for the three wells. The bar plot shows differential fluorescence responses for three representative drugs. These fluorescence responses were further analyzed by linear discriminant analysis (LDA) to generate different clusters corresponding to different categories of drug mechanisms. Each ellipse represents each drug in that mechanism category. Adapted with permission from ref. 49. Copyright 2014 by Nature Publishing Group.
5. Conclusions and prospects
Metallic NPs present a versatile platform for the creation of recognition elements for analyzing the biological targets. NPs can be fabricated with different recognition elements to provide specific or selective interactions with the target analytes. Moreover, physiochemical properties of the NPs such as fluorescence quenching or enhancement, surface enhanced Raman scattering and electrochemical activity can be harnessed to signal the transduction of the binding events. Hence, inclusion of NPs can simplify the system design as well as increase the sensitivity of the biosensors.
Incorporation of metallic nanoparticles in diagnostic techniques has opened promising avenues for a wide range of sensing strategies that feature combinations of simplicity, rapid output, low cost platforms and multiplexing. As we develop better strategies for particle functionalization and signal transduction, a wide range of platforms ranging from microfluidic sensors through inexpensive paper test strips will be enabled, provide solutions to address health issues worldwide.
Key learning points.
Both cell surface biomarkers (such as carbohydrates and proteins) and the overall cell surface signatures provide crucial information for identifying cell types.
Metallic nanoparticles provide multiple modes of signal transduction for biosensing applications.
Surface functionalization determines how nanoparticles interact with cell surfaces.
The specific recognition capabilities of biomacromolecules such as antibodies, lectins, aptamers, and DNAzymes can be coupled with nanoparticle transduction processes to design cell sensing strategies.
Nanoparticle surface can be functionalized with a variety of small molecule ligands to provide the selective recognition required for array-based sensing.
Acknowledgment
The authors thank Rubul Mout (University of Massachusetts--Amherst) and Rui Tang (University of Massachusetts--Amherst) for their help in preparing the manuscript. This work was supported by The NIH (GM077173) and the NSF (Center for Hierarchical Manufacturing, CMMI-1025020).
Footnotes
The authors declare no competing financial interest.
References
- 1.Shen H, Laird PW. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 3.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr. J. Clin. Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 4.Trzpis M, McLaughlin PMJ, de Leij LMFH, Harmsen MC. Am. J. Pathol. 2007;171:386–395. doi: 10.2353/ajpath.2007.070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain PK, Huang XH, El-Sayed IH, El-Sayed MA. Acc. Chem. Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 6.Sperling RA, Rivera Gil P, Zhang F, Zanella M, Parak WJ. Chem. Soc. Rev. 2008;37:1896–1908. doi: 10.1039/b712170a. [DOI] [PubMed] [Google Scholar]
- 7.Katz E, Willner I. Angew. Chem. Int. Ed. 2004;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- 8.Mout R, Moyano DF, Rana S, Rotello VM. Chem. Soc. Rev. 2012;41:2539–2544. doi: 10.1039/c2cs15294k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edidin M. Nat. Rev. Mol. Cell Biol. 2003;4:414–418. doi: 10.1038/nrm1102. [DOI] [PubMed] [Google Scholar]
- 10.Mager MD, LaPointe V, Stevens MM. Nat. Chem. 2011;3:582–589. doi: 10.1038/nchem.1090. [DOI] [PubMed] [Google Scholar]
- 11.Dube DH, Bertozzi CR. Nat. Rev. Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 12.Ohtsubo K, Marth JD. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Nel AE, Maedler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 14.Sudimack J, Lee RJ. Adv. Drug Del. Rev. 2000;41:147–162. doi: 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 15.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 16.Rifai N, Gillette MA, Carr SA. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 17.Holliger P, Hudson PJ. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 18.Porter MD, Lipert RJ, Siperko LM, Wang G, Narayanana R. Chem. Soc. Rev. 2008;37:1001–1011. doi: 10.1039/b708461g. [DOI] [PubMed] [Google Scholar]
- 19.Sha MY, Xu H, Natan MJ, Cromer R. J. Am. Chem. Soc. 2008;130:17214–17215. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian X, Peng X-H, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 21.Katz E, Willner I, Wang J. Electroanalysis. 2004;16:19–44. [Google Scholar]
- 22.Maltez-da Costa M, de la Escosura-Muniz A, Nogues C, Barrios L, Ibanez E, Merkoci A. Nano Lett. 2012;12:4164–4171. doi: 10.1021/nl301726g. [DOI] [PubMed] [Google Scholar]
- 23.Jelinek R, Kolusheva S. Chem. Rev. 2004;104:5987–6016. doi: 10.1021/cr0300284. [DOI] [PubMed] [Google Scholar]
- 24.Ding L, Cheng W, Wang X, Ding S, Ju H. J. Am. Chem. Soc. 2008;130:7224–7225. doi: 10.1021/ja801468b. [DOI] [PubMed] [Google Scholar]
- 25.Hu L, Xu G. Chem. Soc. Rev. 2010;39:3275–3304. doi: 10.1039/b923679c. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Chen Z, Liu Y, Li J. Nanoscale. 2013;5:7349–7355. doi: 10.1039/c3nr01598j. [DOI] [PubMed] [Google Scholar]
- 27.Fang X, Tan W. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava S, Frankamp BL, Rotello VM. Chem. Mater. 2005;17:487–490. [Google Scholar]
- 29.Liu J, Lu Y. Nat. Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 30.Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Anal. Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 31.Yeh H-C, Sharma J, Han JJ, Martinez JS, Werner JH. Nano Lett. 2010;10:3106–3110. doi: 10.1021/nl101773c. [DOI] [PubMed] [Google Scholar]
- 32.Yin J, He X, Wang K, Xu F, Shangguan J, He D, Shi H. Anal. Chem. 2013;85:12011–12019. doi: 10.1021/ac402989u. [DOI] [PubMed] [Google Scholar]
- 33.Wan Y, Zhou Y-G, Poudineh M, Safaei TS, Mohamadi RM, Sargent EH, Kelley SO. Angew. Chem. Int. Ed. 2014;53:13145–13149. doi: 10.1002/anie.201407982. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niazov T, Pavlov V, Xiao Y, Gill R, Willner I. Nano Lett. 2004;4:1683–1687. [Google Scholar]
- 36.Bi S, Zhang J, Zhang S. Chem. Commun. 2010;46:5509–5511. doi: 10.1039/c0cc00127a. [DOI] [PubMed] [Google Scholar]
- 37.Le NDB, Yazdani M, Rotello VM. Nanomedicine. 2014;9:1487–1498. doi: 10.2217/nnm.14.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miranda OR, Chen H-T, You C-C, Mortenson DE, Yang X-C, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miranda OR, Li X, Garcia-Gonzalez L, Zhu Z-J, Yan B, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2011;133:9650–9653. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez AM, Kak AC. IEEE Trans. Pattern Anal. 2001;23:228–233. [Google Scholar]
- 41.Bunz UHF, Rotello VM. Angew. Chem. Int. Ed. 2010;49:3268–3279. doi: 10.1002/anie.200906928. [DOI] [PubMed] [Google Scholar]
- 42.Bajaj A, Miranda OR, Kim I-B, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Chem. Sci. 2010;1:134–138. [Google Scholar]
- 44.Rana S, Singla AK, Bajaj A, Elci SG, Miranda OR, Mout R, Yan B, Jirik FR, Rotello VM. ACS Nano. 2012;6:8233–8240. doi: 10.1021/nn302917e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips RL, Miranda OR, You C-C, Rotello VM, Bunz UHF. Angew. Chem. Int. Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 46.Liu Q, Yeh Y-C, Rana S, Jiang Y, Guo L, Rotello VM. Cancer Lett. 2013;334:196–201. doi: 10.1016/j.canlet.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang LY, Yan RX, Hao ZY, Wang L, Zeng JH, Bao J, Wang X, Peng Q, Li YD. Angew. Chem. Int. Ed. 2005;44:6054–6057. doi: 10.1002/anie.200501907. [DOI] [PubMed] [Google Scholar]
- 48.Lu Y, Liu Y, Zhang S, Wang S, Zhang S, Zhang X. Anal. Chem. 2013;85:6571–6574. doi: 10.1021/ac4014594. [DOI] [PubMed] [Google Scholar]
- 49.Rana S, Le NDB, Mout R, Saha K, Tonga GY, Bain RES, Miranda OR, Rotello CM, Rotello VM. Nat. Nanotechnol. 2015;10:65–69. doi: 10.1038/nnano.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]