Abstract
Background
Of recent interest is the finding that certain CSF biomarkers traditionally linked to Alzheimer’s disease (AD), specifically amyloid beta protein (Aβ), are abnormal in PD CSF. The aim of this exploratory investigation was to determine if genetic variation within the amyloid precursor protein (APP) processing pathway genes, correlate with cerebrospinal fluid (CSF) Aβ42 levels in Parkinson’s disease (PD).
Method
PD (n=86) and control (n=161) DNA were genotyped for 19 regulatory region tagging single nucleotide polymorphisms (SNPs) within nine genes (APP, ADAM10, BACE1, BACE2, PSEN1, PSEN2, PEN2, NCSTN and APH1B) involved in the cleavage of APP. SNP genotypes were tested for their association with CSF biomarkers and PD risk while adjusting for age, gender, and APOE ε4 status.
Results
Significant correlation with CSF Aβ42 levels in PD was observed for two SNPs, (APP rs466448 and APH1B rs2068143). Conversely, significant correlation with CSF Aβ42 levels in controls was observed for three SNPs (APP rs214484 and rs2040273 and PSEN1 rs362344).
Conclusion
The results of this exploratory investigation suggest that an APP SNP and an APH1B SNP are marginally associated with PD CSF Aβ42 levels in APOE ε4 non-carriers. Further hypotheses generated include that decreased CSF Aβ42 levels are in part driven by genetic variation in APP processing genes. Additional investigation into the relationship between these findings and clinical characteristics of PD, including cognitive impairment, compared to other neurodegenerative diseases, such as AD, are warranted.
Keywords: APP, ADAM10, BACE1, BACE2, PSEN1, PSEN2, PEN2, NCSTN, APH1B, Parkinson’s disease, Cerebrospinal fluid
Introduction
Amyloid precursor protein (APP) is an integral membrane protein that is best known as the protein whose processing generates amyloid beta (Aβ), a peptide that is the primary component of the amyloid plaques characteristic of Alzheimer’s disease (AD). Aβ peptides of varying sizes are found in cerebrospinal fluid (CSF). Low CSF Aβ42 levels are a promising biomarker for AD1 and have been described in Parkinson’s disease (PD)2.
Many proteins are involved in the post-translational cleavage of APP into Aβ. An α-secretase, ADAM10, cleaves APP in one processing pathway that does not form an amyloidgenic Aβ peptide3–7. In another processing pathway, APP cleavage by β-secretases BACE1 or BACE2 produces a peptide, which subsequently can undergo γ-secretase cleavage to produce an amyloidgenic Aβ peptide8–12. The γ-secretase protein complex is composed of several subunits including presenilin (either PSEN1 or PSEN2), nicastrin (NCSTN), APH1B and PEN213–15, and is uniquely involved in APP cleavage according to tissue specificity, or the stability of the protein complex16–20.
Rare autosomal-dominant forms of early-onset familial AD, are caused by mutations in the PSEN1, PSEN2 or APP21. The majority of the rare coding mutations in APP alter processing of APP so that the relative levels of Aβ42 are increased22–24. Triplication of the APP gene, due to chromosome 21 trisomy in Down’s Syndrome, is associated with increased APP expression and early amyloid plaque formation25–28. In addition, APP promoter polymorphisms have been associated with AD29,30.
Aβ peptides of varying sizes are normally present in both the brain and CSF. Low CSF Aβ42 levels are associated with increased Aβ deposition in the brain31–33, age and APOE ε4 genotype in cognitively normal adults34, and in AD and mild cognitive impairment35–37.
Some studies, but not all, report reduced CSF Aβ42 associated with PD or cognitive decline in PD38–40. In addition, the APOE ε4 allele appears to be a risk factor across the Lewy body disease (LBD) spectrum, including PD41.
Our group has reported an association between CSF biomarker levels and APP processing genes in AD42. However, to the best of our knowledge, the genetic influences of APP processing related genes on AD-associated CSF biomarkers have not been studied in PD. Therefore, we hypothesized that genetic variation within regulatory regions of APP processing genes, would correlate with CSF Aβ42 levels in PD according to APOE ε4 status. Specifically, the aim of this investigation was to determine if genetic variation in common transcriptional regulatory regions of APP, ADAM10, BACE1, BACE2, PSEN1, PSEN2, PEN2, NCSTN and APH1B correlate with PD CSF Aβ42 levels. A total of 19 single nucleotide polymorphisms (SNPs) were analyzed while taking into account age, gender, and APOE ε4 status.
Methods
Subjects
All procedures were approved by the institutional review boards of the participating institutions. Following informed consent, all PD subjects (n=86) underwent evaluation that consisted of medical history, family history, physical and neurologic examinations, and laboratory tests. All PD subjects fulfilled criteria for a diagnosis of PD43.
All control subjects (n=161) underwent thorough clinical and neuropsychological assessment as prescribed by the Alzheimer Centers’ uniform data set44. All control subjects had a Clinical Dementia Rating (CDR) scale score of 0, and underwent consensus conference review.
Cerebrospinal Fluid
All CSF samples were collected in the morning after participants fasted overnight. CSF samples were collected as previously described34,45. Results reported here are from assays run from comparable lumbar puncture fractions to limit variability from rostrocaudal concentration gradients. Concentrations of Aβ42 in the 5th to 10th ml of collected CSF Aβ42 were measured using the INNO-BIA AlzBio3 kit obtained from Innogenetics (Gent, Belgium) following the manufacturer’s instructions except that the CSF samples were diluted 1:4 before performing the assay. CSF Aβ42 was measured using multiplexed Luminex reagents from InnoGenetics according to manufacturer’s instructions and as previously described46. All CSF samples were analyzed using a LiquiChip Luminex 200TM Workstation (Qiagen, Valencia, CA). Intra-assay coefficient of variation was <10 % for all assays. Assays were performed blind to clinical diagnosis.
Genes and SNP selection
The nine studied genes were chosen for their biologically characterized role in encoding proteins that are involved in the processing of APP. SNPs were chosen within these genes according to the following criteria; (1) the SNP was located within a known or putative regulatory region of the gene. Tagging SNPs were chosen to capture regulatory regions when the actual regulatory region SNP was not available; (2) the SNP had a minor allele frequency (MAF) of ≥ 0.1 in HapMap Northern European population (CEU) and a minor genotype frequency in our study sample of ≥ 0.01; and (3) the SNP genotyping assay was commercially available. Based on these criteria, a total of 19 SNPs were selected (Table 2). Additionally, SNPs rs429358 and rs7412 were genotyped to determine APOE ε4 status.
Table 2.
Genotype description and frequency distribution.
| Gene ID | Chromosome Location |
SNP | NCBI Gene Location |
Alleles | PD Genotype Frequency |
Control Genotype Frequency |
PD vs Controls | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted Genotype |
Adjusted Genotype |
Unadjusted Collapsed Genotype |
Adjusted Collapsed Genotype |
|||||||||||
| EE | EF | FF | EE | EF | FF | p-value | p-value | p-value | p-value | |||||
| Amyloid Precusor Protein | ||||||||||||||
| APP | 21q21.3 | rs466448 | 5' region | G/a | 0.19 | 0.52 | 0.29 | 0.25 | 0.49 | 0.26 | 0.324 | 0.122 | 0.578 | 0.136 |
| rs2830101 | intron 1 | G/a | 0.08 | 0.46 | 0.46 | 0.13 | 0.41 | 0.46 | 0.601 | 0.394 | 0.990 | 0.728 | ||
| rs214484 | intron 17 | C/g | 0.07 | 0.47 | 0.46 | 0.11 | 0.42 | 0.47 | 0.697 | 0.909 | 0.917 | 0.987 | ||
| rs2040273 | 3' region | A/g | 0.13 | 0.51 | 0.36 | 0.16 | 0.43 | 0.41 | 0.888 | 0.661 | 0.491 | 0.545 | ||
| α-Secretase | ||||||||||||||
| ADAM10 | 15q22.1 | rs514049 | 5' region | C/a | 0.13 | 0.46 | 0.41 | 0.22 | 0.45 | 0.34 | 0.089 | 0.191 | 0.237 | 0.298 |
| rs2305421 | Intron 13 | A/g | 0.04 | 0.21 | 0.75 | 0.04 | 0.27 | 0.70 | 0.414 | 0.778 | 0.345 | 0.933 | ||
| β-Secretase | ||||||||||||||
| BACE1 | 11q23-24 | rs573801 | 5' region | G/a | 0.12 | 0.28 | 0.60 | 0.03 | 0.42 | 0.55 | 0.631 | 0.466 | 0.478 | 0.695 |
| rs11601511 | intron 0 | G/c | 0.01 | 0.22 | 0.76 | 0.02 | 0.24 | 0.74 | 0.556 | 0.463 | 0.661 | 0.697 | ||
| rs638405 | exon 5 | C/g | 0.14 | 0.53 | 0.33 | 0.16 | 0.50 | 0.34 | 0.929 | 0.449 | 0.848 | 0.948 | ||
| BACE2 | 21q22.3 | rs734757 | intron 1 | C/t | 0.08 | 0.49 | 0.42 | 0.19 | 0.44 | 0.37 | 0.097 | 0.102 | 0.438 | 0.434 |
| rs12149 | exon 9 | C/t | 0.22 | 0.47 | 0.31 | 0.25 | 0.47 | 0.28 | 0.557 | 0.252 | 0.665 | 0.417 | ||
| γ-Secretase | ||||||||||||||
| PSEN1 | 14q24.3 | rs362344 | 3' region | C/t | 0.05 | 0.35 | 0.60 | 0.05 | 0.35 | 0.60 | 0.999 | 0.900 | 0.970 | 0.956 |
| rs362408 | 3' region | G/a | 0.04 | 0.18 | 0.79 | 0.02 | 0.23 | 0.75 | 0.762 | 0.736 | 0.520 | 0.608 | ||
| PSEN2 | 1q42.2 | rs1295652 | 5' region | A/g | 0.05 | 0.33 | 0.62 | 0.07 | 0.37 | 0.56 | 0.295 | 0.194 | 0.330 | 0.224 |
| rs2802268 | 3' region | T/g | 0.04 | 0.28 | 0.68 | 0.06 | 0.40 | 0.55 | 0.048 | 0.083 | 0.040 | 0.078 | ||
| PEN2 (PSENEN) | 19q13.1 | rs2293688 | intron 2 | C/g | 0.11 | 0.47 | 0.42 | 0.12 | 0.48 | 0.39 | 0.570 | 0.964 | 0.625 | 0.925 |
| NCSTN | 1q22-q23 | rs7540865 | intron 5 | G/t | 0.07 | 0.48 | 0.45 | 0.11 | 0.41 | 0.48 | 0.978 | 0.414 | 0.577 | 0.223 |
| APH1B | 15q22.2 | rs35408871 | Intron 4 | G/a | 0.01 | 0.25 | 0.74 | 0.02 | 0.24 | 0.73 | 0.748 | 0.984 | 0.889 | 0.820 |
| rs2068143 | intron 4 | G/a | 0.16 | 0.35 | 0.48 | 0.07 | 0.34 | 0.60 | 0.020 | 0.081 | 0.088 | 0.152 | ||
Adjusted p-values are based on a logistic regression model with diagnosis as the dependent variable and SNP as a predictor variable along with the covariates gender, age and APOE ε4 status.
P-values < 0.05 are in bold but do not remain significant after adjustment for multiple comparisons.
SNP Genotyping
Genomic DNA (10 ng) was genotyped using TaqMan allelic discrimination detection on 384 well plates as previously described47. Briefly, for each reaction, SNP TaqMan Assay (Applied Biosystems), TaqMan Universal PCR Master Mix (Applied Biosystems) and DNA were pipetted into each well. PCR was carried out using a 9700 Gene Amp PCR System (Applied Biosystems). Plates were then subjected to an end-point read on a 7900 Real-Time PCR System (Applied Biosystems). The results were first evaluated by cluster variations; the allele calls were then assigned automatically before being integrated into the genotype database.
Seven PD patients with an age-at-onset of younger than 40 years were screened for Parkin (PARK2) mutations using a method previously described48. One patient was found to be a compound heterozygote and was excluded from the analysis leaving 85 PD subjects for analysis (Table 1).
Table 1.
Population description.
| PD | Controls | χ2 p-value | |
|---|---|---|---|
| n=85 | n=161 | ||
| % Female | 24 | 58 | < 0.0001 |
| % White | 98 | 92 | 0.170 |
| % APOE ε4+ | 25 | 37 | 0.060 |
| Mean Age (Minimum-Maximum) | 67 (48–83) | 68 (52–88) | 0.454 |
| Mean Age-at-motor symptom onset (Minimum-Maximum) | 57 (28–76) | ||
| MMSE (Minimum-Maximum) | 27.6 (10–30) |
Statistical Analysis
Logistic regression was used to compare SNP genotype (FF major homozygotes, EF heterozygotes, EE minor homozygotes) or collapsed genotype (EF, EE compared to FF; minor allele positive compared to minor allele negative) between PD and controls both with and without adjusting for age, gender and APOE ε4 status. SNP genotype frequencies were tested for Hardy-Weinberg equilibrium (HWE) using the chi-squared test with one degree of freedom49 (Table 2). All subsequent analyses involving SNPs were based on the collapsed genotype group.
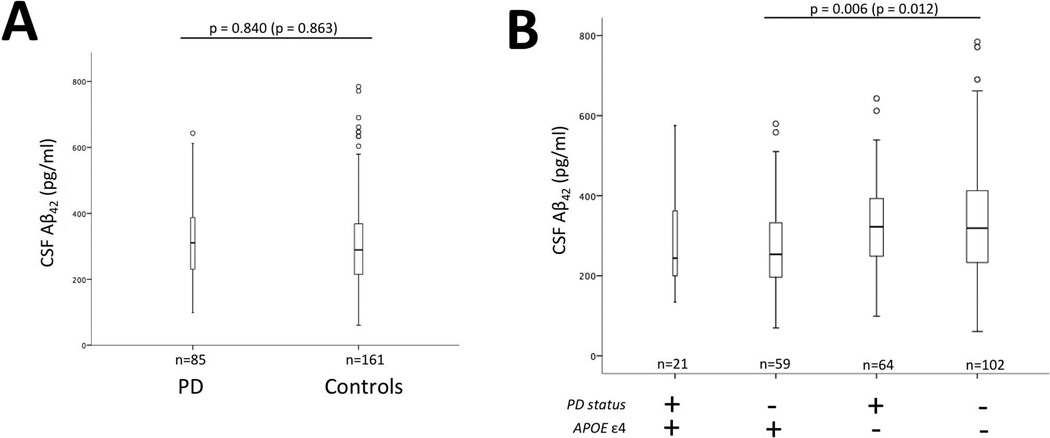
Linear regression models were used to compare PD and control CSF Aβ42 protein levels with and without adjusting for gender, age, and APOE ε4 (Figure 1, Panel A) and to evaluate the relationship between APOE ε4 status and CSF Aβ42 levels while taking into account gender and age (Figure 1; Panels B).
Figure 1.
Cerebrospinal fluid (CSF) levels stratified by disease status and APOE status. There is not a significant difference in CSF Aβ42 levels between Parkinson’s disease (PD) and cognitively normal control subjects (Controls) (Panel A). There is a significant difference in CSF Aβ42 levels in controls between APOE ε4+ and APOE ε4− but not PD (P-values are Bonferroni corrected for multiple comparisons: Panel B). Bars represent interquartile range for CSF Aβ42 levels. P-values are adjusted for covariates gender and age. P-values in parentheses are not adjusted for covariates and bar graphs are not adjusted for covariates. The horizontal line within the quartiles represents the median. The vertical lines represent minimum and maximum CSF Aβ42 levels. Circles represent outliers.
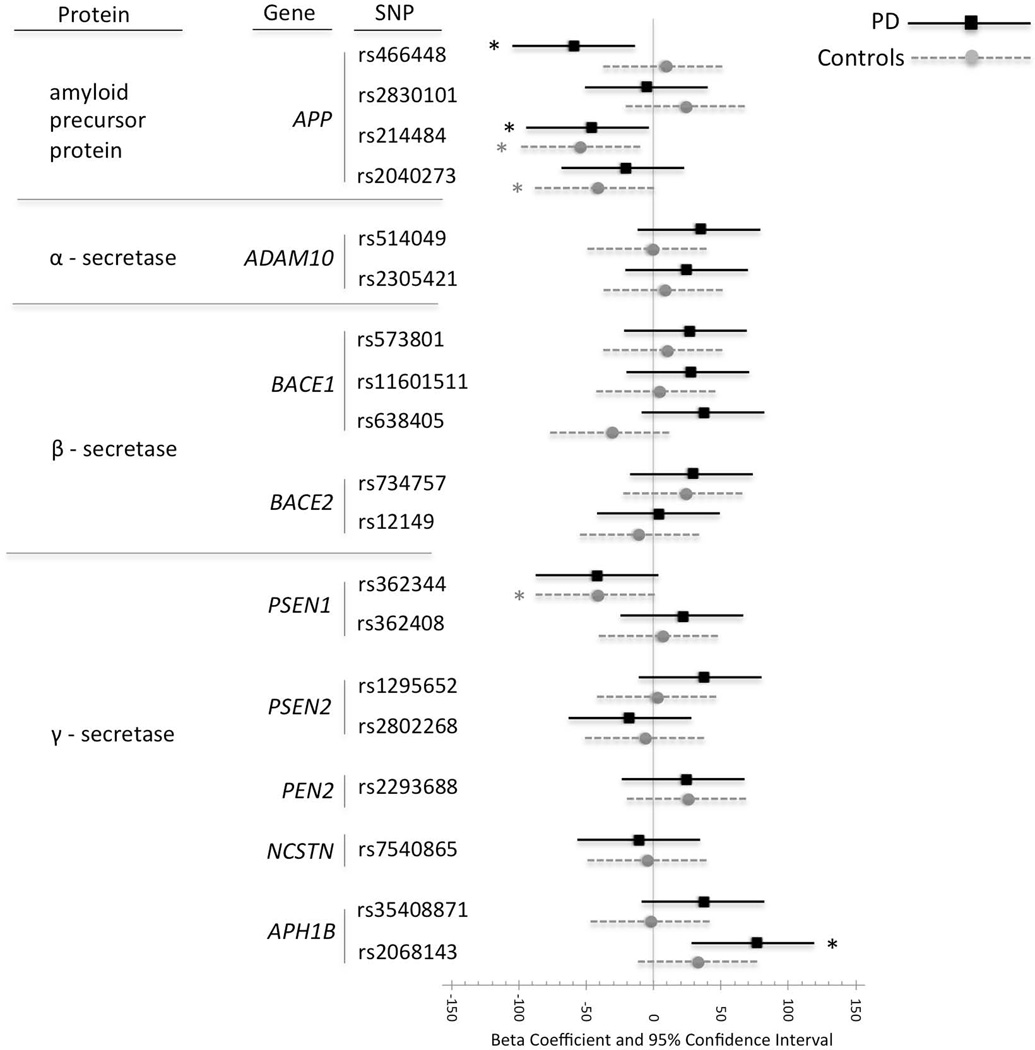
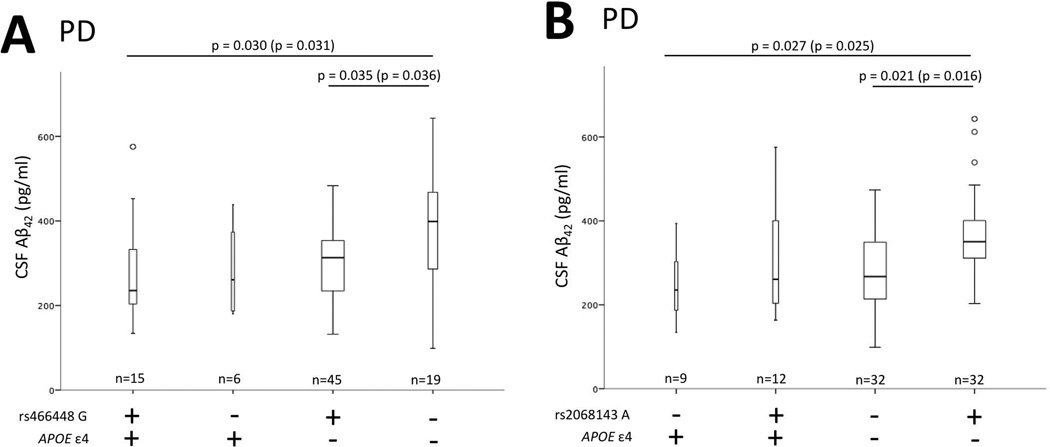
Linear regression main effect models both with a single SNP or all SNPs were used to examine the relationship between SNP (for collapsed genotype) and CSF Aβ42 levels with and without adjusting for gender, age, and APOE ε4 status (Figure 2). Additional regression models included a SNP-by-APOE ε4 status interaction term or a term for group (1) ε4+ and SNP allele +, (2) ε4+ and SNP allele −, (3) ε4− SNP and allele +, and (4) ε4− and SNP allele −, both with and without adjusting for age and gender (Figure 3).
Figure 2.
Linear regression beta-coefficients for cerebrospinal fluid (CSF) Aβ42 levels in cognitively normal control subjects (Controls: grey circle) or Parkinson’s disease patients (PD: black square) for each collapsed genotype. A confidence interval that does not cross the vertical line at zero indicates that the difference in genotype is significant (p < 0.05) before adjusting for multiple comparisons as indicated by asterisk (*). A beta coefficient to the left of the vertical line indicates lower CSF Aβ42 levels for the collapsed genotype group (EE, EF) than for the major homozygote genotype (FF). Beta coefficients, 95% confidence intervals and p-values in parentheses are not adjusted for covariates.
Figure 3.
PD CSF levels stratified by APOE and SNP. There is a significant difference in CSF Aβ42 levels between APOE ε4+ rs466448 G+ and APOE ε4− rs466448 G− (p=0.030) as well as between APOE ε4− rs466448 G+ and APOE ε4− rs466448 G− (p=0.035) (Panel A). There is a significant difference in CSF Aβ42 levels between APOE ε4+ rs2068143 A− and APOE ε4− rs2068143 A+ (p=0.027) as well as between APOE ε4− rs2068143 A− and APOE ε4− rs2068143 A+ (p=0.021) (Panel B). Bars represent interquartile range for CSF Aβ42 levels and width of bars a adjusted for number of subjects. P-values are adjusted for covariates gender and age. P-values in parentheses are not adjusted for covariates and bar graphs are not adjusted for covariates. The horizontal line within the quartiles represents the median. The vertical lines represent minimum and maximum CSF Aβ42 levels. Circles represent outliers. All p-values are Bonferroni corrected for multiple comparisons.
Diagnostic plots of model residuals were inspected to assess any major departures from normality or homoscedasticity. Statistical analyses were performed in R (version 3.0.2; R Core Team, 2013; http://www.R-project.org.) or SPSS (Version 22). When correcting for multiple comparisons, the Holm (1979) was used.
Results
Sample Population
Subjects were 161 healthy cognitively normal control subjects with a mean age of 68 years, and 85 PD patients with a mean age of 67 years and a mean age-of-motor-onset symptom of 57 years (Table 1). All normal control subjects were reviewed by a consensus panel and designated as neurologically and cognitively normal based on cognitive testing and examination. All PD subjects fulfilled criteria for PD, based on history and examination43. All but two PD cases had a mini-mental status examination (MMSE), with a mean MMSE score of 27.6 (+/− 3.5) for these cases. Only thirteen cases had a MMSE below 26.50 The percentage of female subjects was significantly higher in control subjects compared to PD subjects (Table 1). Out of 86 PD patients, there were 7 PD patients with an age-at-onset of younger than 40 years (Table 1). These 7 patients were screened for Parkin (PARK2) mutations. One patient was found to be a compound heterozygote. This patient was excluded from the analysis leaving 85 PD patients for the analyses.
SNP Genotype Frequency
All 19 APP processing gene SNPs passed the HWE test after correcting for multiple comparisons. PSEN2 rs2802268 and APH1B rs2068143 frequency was significantly different between PD and controls. Significance did not remain after Holm multiple comparison correction (Table 2).
CSF Aβ42 Levels by Disease Group and APOE ε4 Status
CSF Aβ42 levels were not significantly different between PD and controls (Figure 1; Panel A). In the control sample, but not in PD, APOE ε4 carriers had significantly lower CSF Aβ42 levels compared to non-carriers both with and without adjusting for gender and age (p = 0.006; p=0.012, respectively) (Figure 1; Panel B).
SNP Effect on CSF Aβ42 Levels
The effect of SNP (collapsed genotypes) on CSF Aβ42 levels was analyzed for each group (controls or PD) both with and without adjusting for the covariates; gender, age and APOE ε4-status. The results are presented graphically as beta coefficients and 95% confidence intervals without adjusting for covariates.
Within the PD group, Aβ42 levels were significantly lower for minor allele carriers of the APP rs466468 (p = 0.014 adjusted, p = 0.015 unadjusted), the rs214484 (p = 0.032 adjusted, p = 0.036 unadjusted),and significantly higher for minor allele carriers of the APH1B SNP rs2068143 (p = 0.002 adjusted, p = 0.003 unadjusted). The rs2068143 adjusted value remained significant after Holm correction for multiple comparisons (p = 0.040). The rs466468 and rs2068143, but not rs214484, remained significant in main effect models containing all SNPs and covariates (p = 0.049, p = 0.011, p = 0.106, respectively).
Within the control group, Aβ42 levels were significantly lower for minor allele carriers of the APP rs214484 (p = 0.002 adjusted, p = 0.007 unadjusted) and rs2040273 (p = 0.017 adjusted, p = 0.039 unadjusted), and for the PSEN1 rs362344 (p = 0.023 adjusted, p = 0.040 unadjusted). The APP SNP rs214484 adjusted value within controls remained significant after Holm correction (p = 0.040) and after including all the SNPs and covariates in the model (p = 0.044) (Figure 2). When a SNP-by-APOE ε4 status interaction term was included in the model it was not significant within controls and not significant within PD patients, while APOE ε4 and SNP remained significant, suggesting an additive effect.
CSF Aβ42 Levels by APOE ε4 Status and SNP status
The nature of this additive effect for 3 SNPs positive in the control group was evaluated further using linear regression models described in the methods section. For the one SNP that remained significant after correcting for multiple comparisons there was a significant difference in CSF Aβ42 levels between APOE ε4 carriers with the rs214484 G allele and APOE ε4 non-carriers without the rs214484 G allele with (p = 0.0002) and without adjusting for gender and age (p = 0.0001). There was a significant difference between APOE ε4 carriers with the rs2040273 G allele and APOE ε4 non-carriers without the rs2040273 G allele with (p = 0.002) and without adjusting for gender and age (p =0.001). There was a significant difference in CSF Aβ42 levels between APOE ε4 carriers with the rs362344 T allele and APOE ε4 non-carriers without the rs362344 T allele with (p =0.001) and without adjusting for gender and age (p =0.001). There was also a significant difference in CSF Aβ42 levels between APOE ε4 carriers with the rs362344 T allele and APOE ε4 non-carriers with the rs362344 T allele with (p = 0.027) and without adjusting for gender and age (p = 0.020) (data not shown).
The nature of this additive effect for 2 SNPs positive in the PD group was evaluated further using the linear regression models described in the methods section. There was a significant difference in CSF Aβ42 levels between APOE ε4 carriers with the rs466448 G allele and APOE ε4 non-carriers without the rs466448 G allele with (p = 0.030) and without adjusting for gender and age (p = 0.031) (Figure 3; Panel A). There was also a significant difference in CSF Aβ42 levels between APOE ε4 non-carriers with the rs466448 G allele and APOE ε4 non-carriers without the rs466448 G allele with (p = 0.035) and without adjusting for gender and age (p = 0.036) (Figure 3; Panel A). There was a significant difference in CSF Aβ42 levels between APOE ε4 carriers with the rs2068143 A allele carriers and APOE ε4 non-carriers without the rs2068143 A allele with (p = 0.027) and without adjusting for gender and age (p = 0.025) (Figure 3; Panel A). There was also a significant difference in CSF Aβ42 levels between APOE ε4 non-carriers with the rs2068143 A allele and APOE ε4 non-carriers without the rs2068143 A allele with (p = 0.021) and without adjusting for gender and age (p = 0.016) (Figure 3; Panel A).
Discussion
This exploratory investigation demonstrates that genetic variation within regulatory regions of APP processing pathway genes correlate with CSF Aβ42 levels in PD and normal controls.
There was not a significant difference in CSF Aβ42 levels between PD and controls (Figure 1, Panel A). Some, but not all, previous studies describe PD CSF Aβ42 levels as lower than in controls51–57. A more consistent finding is an association between lower CSF Aβ42 levels and cognitive impairment in PD38,56,57. Upon stratification by APOE ε4, control, but not PD, APOE ε4 carriers had significantly lower CSF Aβ42 levels, both with and without age adjustment, compared to APOE ε4 non-carriers (Figure 1, Panel B). These results are supported by previous reports that show cognitively normal subject CSF Aβ42 levels decrease with increasing age in APOE ε4 carriers34. In addition, a lack of association between PD CSF Aβ42 levels and APOE ε4 has been described58,59.
One APP rs214484 was significantly associated with CSF Aβ42 levels in cognitively normal controls after correcting for multiple comparisons and after adjusting for gender, age and APOE ε4 (Figure 2). APP rs2040273 and PSEN1 rs362344 nominally correlated with CSF Aβ42 levels in controls and APP rs466448 and APH1B rs2068143, nominally correlated with CSF Aβ42 levels in PD (Figure 2). To our knowledge this is the first investigation to find an association between SNPs in the APP processing pathway and CSF Aβ42 levels in PD. However, in support of these findings, a recent report suggests that disturbed APP processing has an effect on the variable rate of motor and functional decline in PD60. Coding mutations in the APP gene that influence the processing of APP and increase the production of Aβ cause a rare early onset form of AD and emphasize the importance of the APP processing pathway in AD cognitive decline29,6121. Our group previously reported an association between soluble CSF biomarkers and APP processing genes in AD42. Specifically, a SNP allele located at the α-secretase ADAM10 locus was associated with CSF APPα levels in AD and was significantly different between AD and controls. However, there was no association between APP processing genes and CSF Aβ42 levels in AD42. Taken together, a lack of association between APP processing genes and CSF Aβ42 levels in AD in the previous study, but an association in PD in the present study, suggests that a different biological factor may play a role in the pathobiology of amyloid pathology in PD. Furthermore, the association between an APP 3’ region SNP in controls (rs214484) and an APP 5’ region SNP in PD (rs466448), generates further hypotheses regarding CSF Aβ42 levels and APP genetic variants as therapeutic targets or markers of disease specific neurodegeneration.
To determine if these 5 APP pathway SNPs in combination with APOE ε4 have an additive effect on CSF Aβ42 levels, all 5 SNPs were grouped by APOE ε4 in regression models. All 5 SNPs exhibited an additive effect where the carriers of both APOE ε4 and one of the APP processing SNP alleles had significantly lower CSF Aβ42 levels compared to APOE ε4 non-carriers and APP processing SNP allele non-carriers. Interestingly, rs466448 and rs2068143 were the only SNPs that showed significantly lower PD CSF Aβ42 levels within the APOE ε4 non-carriers, suggesting that these SNPs may influence PD CSF Aβ42 levels, but not control levels, regardless of APOE ε4 status (Figure 3). Rs466448 is located in the 5’ region at position -1023 upstream of TSS of the APP promoter62 and is within an ENCODE described promoter associated histone mark (H3K4Me3)63 suggesting that allele specific variation may play a mechanistic role in APP expression. In addition, rs214484 is located in the 3’ region of APP within an ENCODE described H3K27Ac mark often found near active regulatory elements63 further suggesting that APP expression level may be influenced by genetic variation at these locations.
The rs2068143 is located in the 3’ region of APH1B in intron 4 in an ENCODE DNase I hypersensitivity cluster as well as an enhancer associated histone mark (H3K4Me1)63 suggesting that allele specific variation may play a mechanistic role in APH1B expression. Inactivation of the Aph1B subunit of γ-secretase in a mouse model of AD has been described. In this model inactivation of Aph1B led to improvements in AD relevant phenotypic features64. Given that the genetic variation of this gene has not been extensively investigated, and given that only 2 SNPs were analyzed in the present investigation, further analysis is needed to determine if rs2068143 is a surrogate marker for another genetic variant at the APH1B locus. Interestingly, there is a modest effect by all these described SNPs after multiple comparison adjustment. However, the information provided by this exploratory investigation may be important as it suggests that APOE ε4 non-carriers have a different APP pathway related association with CSF Aβ42 compared to APOE ε4 carriers. This information suggests that it may be important, in future PD therapeutic strategies related to amyloid treatment of cognitive decline, to take into account APOE ε4 status.
An advantage of this investigation, in contrast to genome wide association studies, is that a small number of SNPs (n=19), within the context of a specific biological pathway were analyzed, and therefore increased the power to test the hypothesis that the APP processing pathway genes influence PD CSF Aβ42 levels. A limitation of this investigation’s approach was that only a few APP processing genes were tested. Thus, many other contributors to APP processing may have been missed, such as genes related to the clearance or deposition of Aβ in the brain. In addition, a limited number of SNPs were used to capture putative regulatory genetic variation within and surrounding the genes of interest. Therefore, these results must be approached with caution since many important SNPs may have been missed. Another limitation of this investigation is that sAPPα and sAPPβ were not measured. Previously, our group described an association between CSF sAPPα and ADAM10 in AD42. Others have described CSF sAPPα, sAPPβ and various CSF Aβ42 fragments in PD, Lewy body disease, and AD60,65,66. Unfortunately, we did not have sAPPα and sAPPβ data available for this PD cohort.
Others have reported correlation between cognitive decline in PD and CSF Aβ4266. In the present investigation the relationship between cognition (MMSE) and SNP was evaluated (data not shown) and there was not a significant association for any of the SNPs or CSF Aβ42 levels. However, we did not have cognitive data available for this cohort beyond MMSE, which has been shown to have low sensitivity to cognitive impairment in PD. Thus, additional cognitive testing would be necessary to adequately assess this issue. Given the results described here, future investigation of the relationship between CSF Aβ42, APP pathway genes and cognition in PD may be warranted.
In conclusion, the main novel findings are that an APP SNP and an APH1B SNP are marginally associated with PD CSF Aβ42 levels in APOE ε4 non-carriers. Further hypotheses generated by this exploratory investigation, include that decreased CSF Aβ42 levels are in part driven by genetic variation in APP processing genes. This information may have important implications for therapeutic strategies related to amyloid treatment of cognitive decline that take into account APOE ε4 status. Investigation into the relationship between these genes and clinical characteristics of PD, such as cognitive impairment, compared to other neurodegenerative diseases, are warranted. Characterization of the functional influence of APP and APH1B genetic variation on expression may lead to a better understanding of PD, help identify novel PD biomarkers and help identify PD therapeutic targets.
Acknowledgements
This work is supported in part by the U.S. Department of Veterans Affairs, Office of Research and Development Clinical Research and Development Program, the Biomedical Laboratory Research Program and NIH Grants P50AG005136, P50NS062684, K99AG034214/4R00AG034214. Additional support includes University of Washington Alzheimer’s Disease Research Center NIH P50-AB005136 and the Jane and Lee Seidman Fund, Joseph Hahn MD Endowed Chair.
Footnotes
Disclosure Statement for Authors: James B. Leverenz; consultant Boehringer-Ingelheim, Citibank, Navidea Biopharmaceuticals, Piramal Healthcare.
References
- 1.Zetterberg H, Lunn MP, Herukka SK. Clinical use of cerebrospinal fluid biomarkers in Alzheimer's disease. Biomark Med. 2012;6:371–376. doi: 10.2217/bmm.12.47. [DOI] [PubMed] [Google Scholar]
- 2.Parnetti L, et al. Cerebrospinal fluid biomarkers in Parkinson disease. Nat Rev Neurol. 2013;9:131–140. doi: 10.1038/nrneurol.2013.10. [DOI] [PubMed] [Google Scholar]
- 3.Postina R. A closer look at alpha-secretase. Curr Alzheimer Res. 2008;5:179–186. doi: 10.2174/156720508783954668. [DOI] [PubMed] [Google Scholar]
- 4.Deuss M, Reiss K, Hartmann D. Part-time alpha-secretases: the functional biology of ADAM 9, 10 and 17. Curr Alzheimer Res. 2008;5:187–201. doi: 10.2174/156720508783954686. [DOI] [PubMed] [Google Scholar]
- 5.Postina R, et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. 2004;113:1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colciaghi F, et al. [alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients. Mol Med. 2002;8:67–74. [PMC free article] [PubMed] [Google Scholar]
- 7.Qin W, Ho L, Wang J, Peskind E, Pasinetti GM. S100A7, a novel Alzheimer's disease biomarker with non-amyloidogenic alpha-secretase activity acts via selective promotion of ADAM-10. PLoS ONE. 2009;4:e4183. doi: 10.1371/journal.pone.0004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zetterberg H, et al. Elevated cerebrospinal fluid BACE1 activity in incipient Alzheimer disease. Arch Neurol. 2008;65:1102–1107. doi: 10.1001/archneur.65.8.1102. [DOI] [PubMed] [Google Scholar]
- 9.Wu G, et al. Decrease in age-adjusted cerebrospinal fluid beta-secretase activity in Alzheimer's subjects. Clin Biochem. 2008;41:986–996. doi: 10.1016/j.clinbiochem.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Ewers M, et al. Increased CSF-BACE 1 activity is associated with ApoE-epsilon 4 genotype in subjects with mild cognitive impairment and Alzheimer's disease. Brain. 2008;131:1252–1258. doi: 10.1093/brain/awn034. [DOI] [PubMed] [Google Scholar]
- 11.Sun X, et al. Distinct transcriptional regulation and function of the human BACE2 and BACE1 genes. FASEB J. 2005;19:739–749. doi: 10.1096/fj.04-3426com. [DOI] [PubMed] [Google Scholar]
- 12.Stockley JH, O'Neill C. The proteins BACE1 and BACE2 and beta-secretase activity in normal and Alzheimer's disease brain. Biochem Soc Trans. 2007;35:574–576. doi: 10.1042/BST0350574. [DOI] [PubMed] [Google Scholar]
- 13.Francis R, et al. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 2002;3:85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 14.Baulac S, et al. Functional gamma-secretase complex assembly in Golgi/trans-Golgi network: interactions among presenilin, nicastrin, Aph1, Pen-2, and gamma-secretase substrates. Neurobiol Dis. 2003;14:194–204. doi: 10.1016/s0969-9961(03)00123-2. [DOI] [PubMed] [Google Scholar]
- 15.Kimberly WT, et al. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci U S A. 2003;100:6382–6387. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirotani K, Edbauer D, Prokop S, Haass C, Steiner H. Identification of distinct gamma-secretase complexes with different APH-1 variants. J Biol Chem. 2004;279:41340–41345. doi: 10.1074/jbc.M405768200. [DOI] [PubMed] [Google Scholar]
- 17.Lee SF, et al. A conserved GXXXG motif in APH-1 is critical for assembly and activity of the gamma-secretase complex. J Biol Chem. 2004;279:4144–4152. doi: 10.1074/jbc.M309745200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YW, et al. Nicastrin is critical for stability and trafficking but not association of other presenilin/gamma-secretase components. J Biol Chem. 2005;280:17020–17026. doi: 10.1074/jbc.M409467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe N, et al. Pen-2 is incorporated into the gamma-secretase complex through binding to transmembrane domain 4 of presenilin 1. J Biol Chem. 2005;280:41967–41975. doi: 10.1074/jbc.M509066200. [DOI] [PubMed] [Google Scholar]
- 20.Prokop S, Shirotani K, Edbauer D, Haass C, Steiner H. Requirement of PEN-2 for stabilization of the presenilin N-/C-terminal fragment heterodimer within the gamma-secretase complex. J Biol Chem. 2004;279:23255–23261. doi: 10.1074/jbc.M401789200. [DOI] [PubMed] [Google Scholar]
- 21.Goedert M, Spillantini MG. A century of Alzheimer's disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 22.Goedert M, Klug A, Crowther RA. Tau protein, the paired helical filament and Alzheimer's disease. J Alzheimers Dis. 2006;9:195–207. doi: 10.3233/jad-2006-9s323. [DOI] [PubMed] [Google Scholar]
- 23.Scheuner D, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 24.Walker LC, et al. Emerging prospects for the disease-modifying treatment of Alzheimer's disease. Biochem Pharmacol. 2005;69:1001–1008. doi: 10.1016/j.bcp.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Giaccone G, et al. Down patients: extracellular preamyloid deposits precede neuritic degeneration and senile plaques. Neurosci Lett. 1989;97:232–238. doi: 10.1016/0304-3940(89)90169-9. [DOI] [PubMed] [Google Scholar]
- 26.Lemere CA, et al. Sequence of deposition of heterogeneous amyloid beta-peptides and APO E in Down syndrome: implications for initial events in amyloid plaque formation. Neurobiol Dis. 1996;3:16–32. doi: 10.1006/nbdi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 27.Englund H, et al. Increase in beta-amyloid levels in cerebrospinal fluid of children with Down syndrome. Dement Geriatr Cogn Disord. 2007;24:369–374. doi: 10.1159/000109215. [DOI] [PubMed] [Google Scholar]
- 28.Leverenz JB, Raskind MA. Early amyloid deposition in the medial temporal lobe of young Down syndrome patients: a regional quantitative analysis. Exp Neurol. 1998;150:296–304. doi: 10.1006/exnr.1997.6777. [DOI] [PubMed] [Google Scholar]
- 29.Guyant-Marechal L, et al. Variations in the APP gene promoter region and risk of Alzheimer disease. Neurology. 2007;68:684–687. doi: 10.1212/01.wnl.0000255938.33739.46. [DOI] [PubMed] [Google Scholar]
- 30.Lv H, Jia L, Jia J. Promoter polymorphisms which modulate APP expression may increase susceptibility to Alzheimer's disease. Neurobiol Aging. 2008;29:194–202. doi: 10.1016/j.neurobiolaging.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Fagan AM, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 32.Visser PJ, et al. Prevalence and prognostic value of CSF markers of Alzheimer's disease pathology in patients with subjective cognitive impairment or mild cognitive impairment in the DESCRIPA study: a prospective cohort study. Lancet Neurol. 2009;8:619–627. doi: 10.1016/S1474-4422(09)70139-5. [DOI] [PubMed] [Google Scholar]
- 33.Tapiola T, et al. Cerebrospinal fluid {beta}-amyloid 42 and tau proteins as biomarkers of Alzheimer-type pathologic changes in the brain. Arch Neurol. 2009;66:382–389. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- 34.Peskind ER, et al. Age and apolipoprotein E*4 allele effects on cerebrospinal fluid beta-amyloid 42 in adults with normal cognition. Arch Neurol. 2006;63:936–939. doi: 10.1001/archneur.63.7.936. [DOI] [PubMed] [Google Scholar]
- 35.Motter R, et al. Reduction of beta-amyloid peptide42 in the cerebrospinal fluid of patients with Alzheimer's disease. Ann Neurol. 1995;38:643–648. doi: 10.1002/ana.410380413. [DOI] [PubMed] [Google Scholar]
- 36.Galasko D, et al. High cerebrospinal fluid tau and low amyloid beta42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype. Arch Neurol. 1998;55:937–945. doi: 10.1001/archneur.55.7.937. [DOI] [PubMed] [Google Scholar]
- 37.Sunderland T, et al. Decreased beta-amyloid1–42 and increased tau levels in cerebrospinal fluid of patients with Alzheimer disease. Jama. 2003;289:2094–2103. doi: 10.1001/jama.289.16.2094. [DOI] [PubMed] [Google Scholar]
- 38.Siderowf A, et al. CSF amyloid {beta} 1–42 predicts cognitive decline in Parkinson disease. Neurology. 2010;75:1055–1061. doi: 10.1212/WNL.0b013e3181f39a78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang JH, et al. Association of Cerebrospinal Fluid beta-Amyloid 1–42, T-tau, P-tau181, and alpha-Synuclein Levels With Clinical Features of Drug-Naive Patients With Early Parkinson Disease. JAMA Neurol. 2013 doi: 10.1001/jamaneurol.2013.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montine TJ, et al. CSF Abeta(42) and tau in Parkinson's disease with cognitive impairment. Mov Disord. 2010;25:2682–2685. doi: 10.1002/mds.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuang D, et al. APOE epsilon4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 2013;70:223–228. doi: 10.1001/jamaneurol.2013.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bekris LM, et al. Amyloid precursor protein (APP) processing genes and cerebrospinal fluid APP cleavage product levels in Alzheimer's disease. Neurobiol Aging. 2011;32:556, 13–23. doi: 10.1016/j.neurobiolaging.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibb WR, Lees AJ. A comparison of clinical and pathological features of young- and old-onset Parkinson's disease. Neurology. 1988;38:1402–1406. doi: 10.1212/wnl.38.9.1402. [DOI] [PubMed] [Google Scholar]
- 44.Morris JC, et al. The Uniform Data Set (UDS): clinical and cognitive variables and descriptive data from Alzheimer Disease Centers. Alzheimer Dis Assoc Disord. 2006;20:210–216. doi: 10.1097/01.wad.0000213865.09806.92. [DOI] [PubMed] [Google Scholar]
- 45.Peskind ER, et al. Safety and acceptability of the research lumbar puncture. Alzheimer Dis Assoc Disord. 2005;19:220–225. doi: 10.1097/01.wad.0000194014.43575.fd. [DOI] [PubMed] [Google Scholar]
- 46.Shi M, et al. Cerebrospinal fluid biomarkers for Parkinson disease diagnosis and progression. Ann Neurol. 2011;69:570–580. doi: 10.1002/ana.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bekris LM, et al. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J Alzheimers Dis. 2008;13:255–266. doi: 10.3233/jad-2008-13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kay DM, et al. A comprehensive analysis of deletions, multiplications, and copy number variations in PARK2. Neurology. 2010;75:1189–1194. doi: 10.1212/WNL.0b013e3181f4d832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis CM, Knight J. Introduction to genetic association studies. Cold Spring Harb Protoc. 2012;2012:297–306. doi: 10.1101/pdb.top068163. [DOI] [PubMed] [Google Scholar]
- 50.Dubois B, et al. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol. 2014;13:614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 51.Holmberg B, Johnels B, Blennow K, Rosengren L. Cerebrospinal fluid Abeta42 is reduced in multiple system atrophy but normal in Parkinson's disease and progressive supranuclear palsy. Mov Disord. 2003;18:186–190. doi: 10.1002/mds.10321. [DOI] [PubMed] [Google Scholar]
- 52.Bibl M, et al. CSF amyloid-beta-peptides in Alzheimer's disease, dementia with Lewy bodies and Parkinson's disease dementia. Brain. 2006;129:1177–1187. doi: 10.1093/brain/awl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mollenhauer B, et al. Beta-amlyoid 1–42 and tau-protein in cerebrospinal fluid of patients with Parkinson's disease dementia. Dement Geriatr Cogn Disord. 2006;22:200–208. doi: 10.1159/000094871. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, et al. CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am J Clin Pathol. 2008;129:526–529. doi: 10.1309/W01Y0B808EMEH12L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parnetti L, et al. Cerebrospinal fluid biomarkers in Parkinson's disease with dementia and dementia with Lewy bodies. Biol Psychiatry. 2008;64:850–855. doi: 10.1016/j.biopsych.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 56.Alves G, et al. CSF amyloid-beta and tau proteins, and cognitive performance, in early and untreated Parkinson's disease: the Norwegian ParkWest study. J Neurol Neurosurg Psychiatry. 2010;81:1080–1086. doi: 10.1136/jnnp.2009.199950. [DOI] [PubMed] [Google Scholar]
- 57.Andersson M, Zetterberg H, Minthon L, Blennow K, Londos E. The cognitive profile and CSF biomarkers in dementia with Lewy bodies and Parkinson's disease dementia. Int J Geriatr Psychiatry. 2011;26:100–105. doi: 10.1002/gps.2496. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, et al. Longitudinal assessment of tau and amyloid beta in cerebrospinal fluid of Parkinson disease. Acta Neuropathol. 2013;126:671–682. doi: 10.1007/s00401-013-1121-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beyer MK, et al. Cerebrospinal fluid Abeta levels correlate with structural brain changes in Parkinson's disease. Mov Disord. 2013;28:302–310. doi: 10.1002/mds.25282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alves G, et al. Cerebrospinal fluid amyloid-beta and phenotypic heterogeneity in de novo Parkinson's disease. J Neurol Neurosurg Psychiatry. 2013;84:537–543. doi: 10.1136/jnnp-2012-303808. [DOI] [PubMed] [Google Scholar]
- 61.Miar A, et al. Amyloid precursor protein gene (APP) variation in late-onset Alzheimer's disease. J Mol Neurosci. 2011;45:5–9. doi: 10.1007/s12031-011-9510-x. [DOI] [PubMed] [Google Scholar]
- 62.Lahiri DK, Ge YW, Maloney B. Characterization of the APP proximal promoter and 5'-untranslated regions: identification of cell type-specific domains and implications in APP gene expression and Alzheimer's disease. Faseb J. 2005;19:653–655. doi: 10.1096/fj.04-2900fje. Epub 2005 Feb 9. [DOI] [PubMed] [Google Scholar]
- 63.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Serneels L, et al. gamma-Secretase heterogeneity in the Aph1 subunit: relevance for Alzheimer's disease. Science. 2009;324:639–642. doi: 10.1126/science.1171176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mulugeta E, et al. Cerebrospinal Fluid Levels of sAPPalpha and sAPPbeta in Lewy Body and Alzheimer's Disease: Clinical and Neurochemical Correlates. Int J Alzheimers Dis. 2011;2011:495025. doi: 10.4061/2011/495025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mollenhauer B, Rochester L, Chen-Plotkin A, Brooks D. What can biomarkers tell us about cognition in Parkinson's disease? Mov Disord. 2014;29:622–633. doi: 10.1002/mds.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]