Abstract
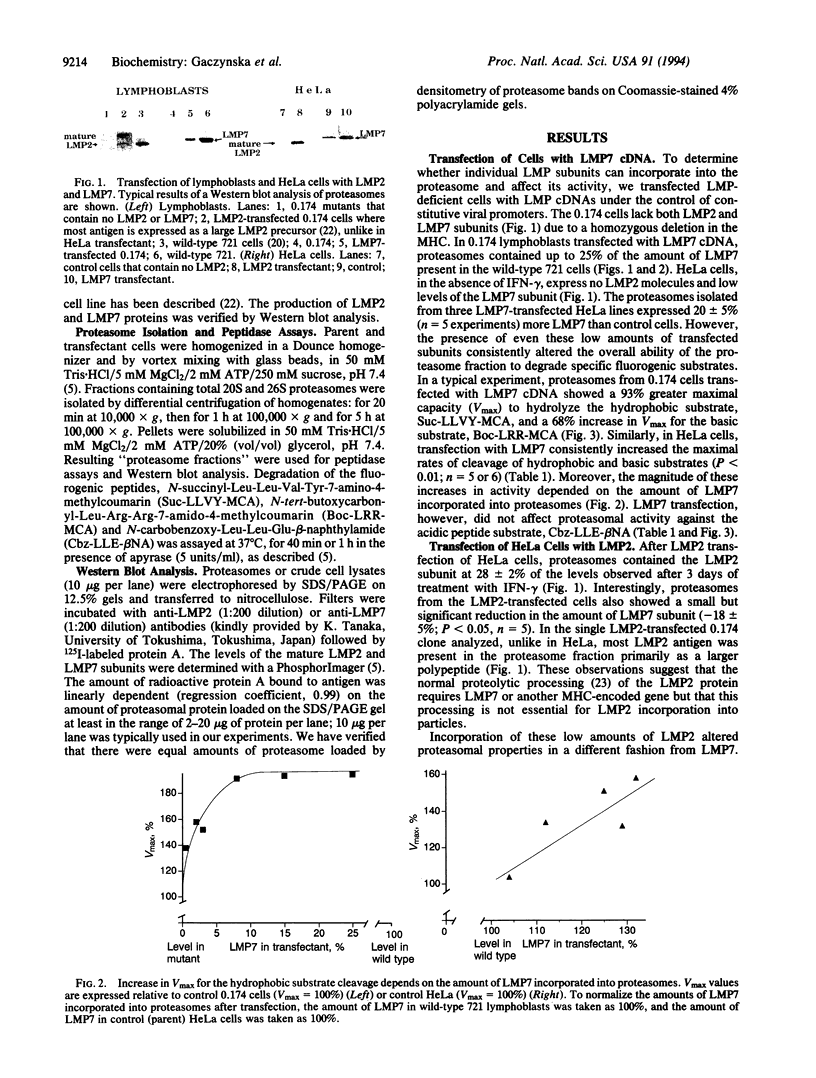
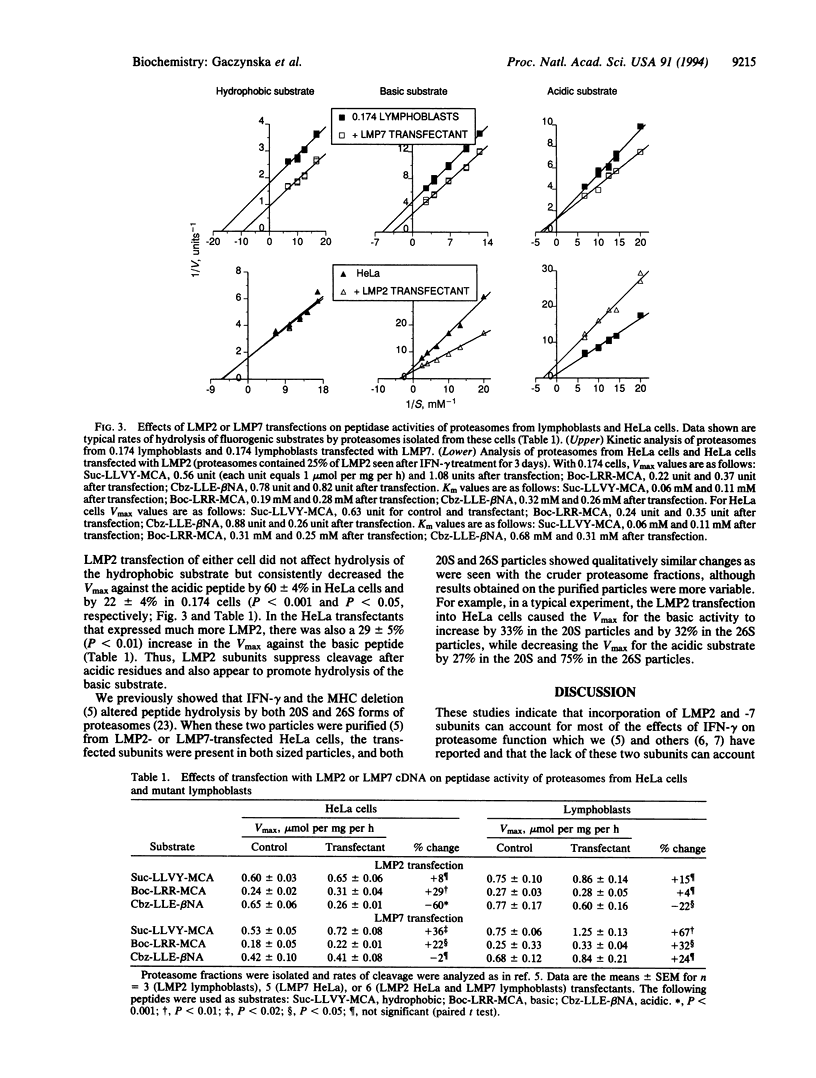
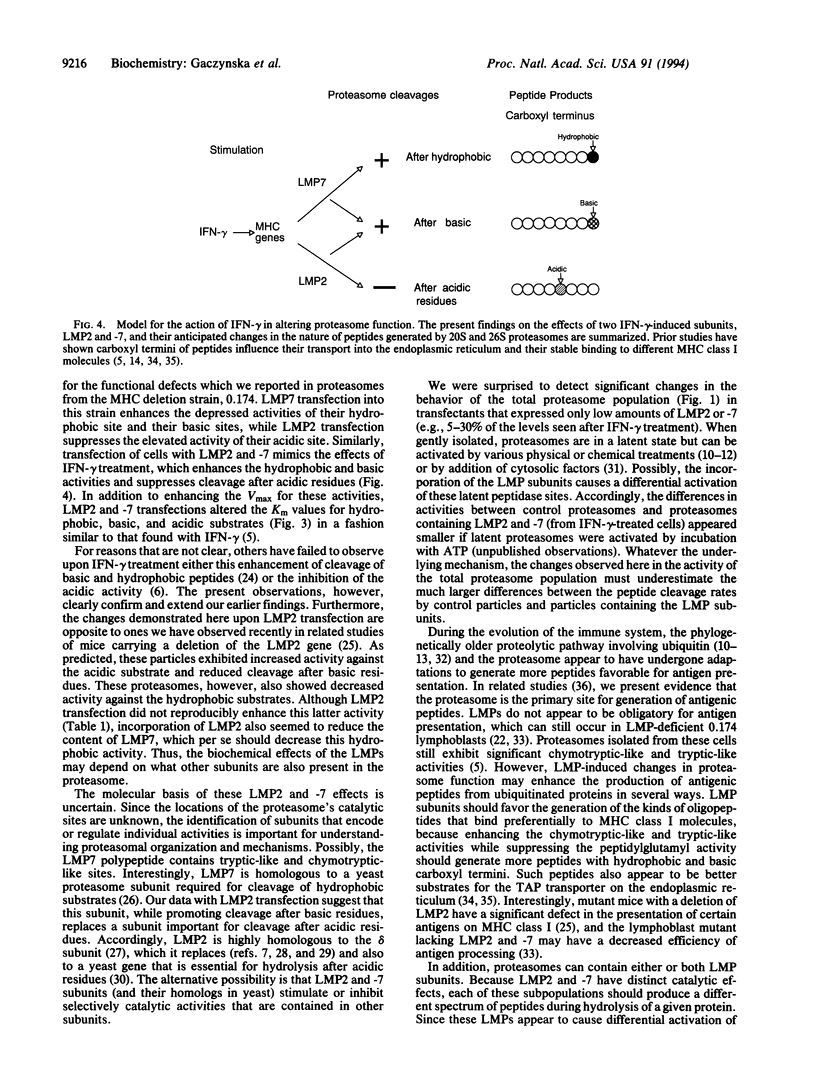
Recent studies have implicated proteasomes in the generation of the antigenic peptides that are presented on major histocompatibility complex class I molecules to T lymphocytes. Interferon gamma modifies the subunit composition of proteasomes and causes changes in their peptidase activities that should favor the production of peptides with hydrophobic or basic carboxyl termini (i.e., the types found on major histocompatibility complex class I molecules). It has been proposed that these changes in peptidase activity are due to incorporation into proteasomes of the major histocompatibility complex-encoded subunits LMP2 and -7, which are induced by interferon gamma. Here we show by gene transfection into lymphoblasts or HeLa cells that LMP7 increases the capacity (Vmax) of 20S and 26S proteasomes to cleave peptides after hydrophobic and basic residues without affecting hydrolysis after acidic residues. These changes depended on the amount of LMP7 subunits incorporated into proteasomes. Transfection of LMP2 reduced cleavage of peptides after acidic residues, increased hydrolysis after basic residues, and did not affect the hydrophobic activity. Since the activity of the total proteasome population changed after incorporation of only small amounts of LMP2 or -7, these subunits must cause major alterations in peptidase activity. Thus, their expression can account for the changes in proteasome activity induced by inteferon gamma, and these findings lend further support to the proposed roles of LMPs in altering the nature of the peptides generated for antigen presentation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aki M., Shimbara N., Takashina M., Akiyama K., Kagawa S., Tamura T., Tanahashi N., Yoshimura T., Tanaka K., Ichihara A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem. 1994 Feb;115(2):257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- Arnold D., Driscoll J., Androlewicz M., Hughes E., Cresswell P., Spies T. Proteasome subunits encoded in the MHC are not generally required for the processing of peptides bound by MHC class I molecules. Nature. 1992 Nov 12;360(6400):171–174. doi: 10.1038/360171a0. [DOI] [PubMed] [Google Scholar]
- Boes B., Hengel H., Ruppert T., Multhaup G., Koszinowski U. H., Kloetzel P. M. Interferon gamma stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994 Mar 1;179(3):901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. G., Driscoll J., Monaco J. J. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991 Sep 26;353(6342):355–357. doi: 10.1038/353355a0. [DOI] [PubMed] [Google Scholar]
- Catterall W. A. A Nobel Prize for single channel recording. New Biol. 1992 Jan;4(1):1–3. [PubMed] [Google Scholar]
- DeMars R., Rudersdorf R., Chang C., Petersen J., Strandtmann J., Korn N., Sidwell B., Orr H. T. Mutations that impair a posttranscriptional step in expression of HLA-A and -B antigens. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8183–8187. doi: 10.1073/pnas.82.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino G. N., Orth K., McCullough M. L., Lee L. W., Munn T. Z., Moomaw C. R., Dawson P. A., Slaughter C. A. The primary structures of four subunits of the human, high-molecular-weight proteinase, macropain (proteasome), are distinct but homologous. Biochim Biophys Acta. 1991 Aug 9;1079(1):29–38. doi: 10.1016/0167-4838(91)90020-z. [DOI] [PubMed] [Google Scholar]
- Driscoll J., Brown M. G., Finley D., Monaco J. J. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993 Sep 16;365(6443):262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- Driscoll J., Goldberg A. L. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J Biol Chem. 1990 Mar 25;265(9):4789–4792. [PubMed] [Google Scholar]
- Falk K., Rötzschke O. Consensus motifs and peptide ligands of MHC class I molecules. Semin Immunol. 1993 Apr;5(2):81–94. doi: 10.1006/smim.1993.1012. [DOI] [PubMed] [Google Scholar]
- Früh K., Yang Y., Arnold D., Chambers J., Wu L., Waters J. B., Spies T., Peterson P. A. Alternative exon usage and processing of the major histocompatibility complex-encoded proteasome subunits. J Biol Chem. 1992 Nov 5;267(31):22131–22140. [PubMed] [Google Scholar]
- Gaczynska M., Rock K. L., Goldberg A. L. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993 Sep 16;365(6443):264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., Rock K. L. Proteolysis, proteasomes and antigen presentation. Nature. 1992 Jun 4;357(6377):375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Heemels M. T., Schumacher T. N., Wonigeit K., Ploegh H. L. Peptide translocation by variants of the transporter associated with antigen processing. Science. 1993 Dec 24;262(5142):2059–2063. doi: 10.1126/science.8266106. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W., Gruhler A., Möhrle V., Mahé Y., Wolf D. H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993 Mar 5;268(7):5115–5120. [PubMed] [Google Scholar]
- Hilt W., Enenkel C., Gruhler A., Singer T., Wolf D. H. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J Biol Chem. 1993 Feb 15;268(5):3479–3486. [PubMed] [Google Scholar]
- Howard J. C., Seelig A. Antigen processing. Peptides and the proteasome. Nature. 1993 Sep 16;365(6443):211–212. doi: 10.1038/365211a0. [DOI] [PubMed] [Google Scholar]
- Kelly A., Powis S. H., Glynne R., Radley E., Beck S., Trowsdale J. Second proteasome-related gene in the human MHC class II region. Nature. 1991 Oct 17;353(6345):667–668. doi: 10.1038/353667a0. [DOI] [PubMed] [Google Scholar]
- Ma C. P., Slaughter C. A., DeMartino G. N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J Biol Chem. 1992 May 25;267(15):10515–10523. [PubMed] [Google Scholar]
- Martinez C. K., Monaco J. J. Homology of proteasome subunits to a major histocompatibility complex-linked LMP gene. Nature. 1991 Oct 17;353(6345):664–667. doi: 10.1038/353664a0. [DOI] [PubMed] [Google Scholar]
- Michalek M. T., Grant E. P., Gramm C., Goldberg A. L., Rock K. L. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature. 1993 Jun 10;363(6429):552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- Momburg F., Ortiz-Navarrete V., Neefjes J., Goulmy E., van de Wal Y., Spits H., Powis S. J., Butcher G. W., Howard J. C., Walden P. Proteasome subunits encoded by the major histocompatibility complex are not essential for antigen presentation. Nature. 1992 Nov 12;360(6400):174–177. doi: 10.1038/360174a0. [DOI] [PubMed] [Google Scholar]
- Momburg F., Roelse J., Howard J. C., Butcher G. W., Hämmerling G. J., Neefjes J. J. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature. 1994 Feb 17;367(6464):648–651. doi: 10.1038/367648a0. [DOI] [PubMed] [Google Scholar]
- Monaco J. J. A molecular model of MHC class-I-restricted antigen processing. Immunol Today. 1992 May;13(5):173–179. doi: 10.1016/0167-5699(92)90122-N. [DOI] [PubMed] [Google Scholar]
- Monaco J. J. Structure and function of genes in the MHC class II region. Curr Opin Immunol. 1993 Feb;5(1):17–20. doi: 10.1016/0952-7915(93)90075-4. [DOI] [PubMed] [Google Scholar]
- Morrison L. A., Lukacher A. E., Braciale V. L., Fan D. P., Braciale T. J. Differences in antigen presentation to MHC class I-and class II-restricted influenza virus-specific cytolytic T lymphocyte clones. J Exp Med. 1986 Apr 1;163(4):903–921. doi: 10.1084/jem.163.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Navarrete V., Seelig A., Gernold M., Frentzel S., Kloetzel P. M., Hämmerling G. J. Subunit of the '20S' proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature. 1991 Oct 17;353(6345):662–664. doi: 10.1038/353662a0. [DOI] [PubMed] [Google Scholar]
- Rivett A. J. Proteasomes: multicatalytic proteinase complexes. Biochem J. 1993 Apr 1;291(Pt 1):1–10. doi: 10.1042/bj2910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnelle C., DeMars R., Long E. O. DO beta: a new beta chain gene in HLA-D with a distinct regulation of expression. EMBO J. 1985 Nov;4(11):2839–2847. doi: 10.1002/j.1460-2075.1985.tb04012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend A., Bodmer H. Antigen recognition by class I-restricted T lymphocytes. Annu Rev Immunol. 1989;7:601–624. doi: 10.1146/annurev.iy.07.040189.003125. [DOI] [PubMed] [Google Scholar]
- Yang Y., Waters J. B., Früh K., Peterson P. A. Proteasomes are regulated by interferon gamma: implications for antigen processing. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4928–4932. doi: 10.1073/pnas.89.11.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]



