Abstract
FAAP20 is a recently identified protein that associates with the Fanconi anemia (FA) core complex component, FANCA. FAAP20 contains a conserved ubiquitin-binding zinc-finger domain, and plays critical roles in the FA-BRCA pathway of DNA repair and genome maintenance. The function of FAAP20 in animals has not been explored. Here we report that deletion of Faap20 in mice led to a mild FA-like phenotype with defects in the reproductive and hematopoietic systems. Specifically, hematopoietic stem and progenitor cells (HSPCs) from Faap20−/− mice showed defects in long-term multi-lineage reconstitution in lethally irradiated recipient mice, with milder phenotype as compared to HSPCs from Fanca−/− or Fancc−/− mice. Faap20−/− mice are susceptible to mitomycin C (MMC)-induced pancytopenia. That is, acute MMC stress induced a significant progenitor loss especially the erythroid progenitors and megakaryocyte-erythrocyte progenitors (MEPs) in Faap20−/− mice. Furthermore, Faap20−/− HSPCs displayed aberrant cell cycle pattern during chronic MMC treatment. Finally, using Faap20−/− Fanca−/− double-knockout mice, we demonstrated a possible dominant effect of FANCA in the interaction between FAAP20 and FANCA. This novel Faap20 mouse model may be valuable in studying the regulation of the FA pathway during bone marrow failure progress in FA patients.
Introduction
Fanconi anemia (FA) is a genetic disorder associated with congenital developmental defects, bone marrow failure and predisposition to cancers particularly acute myelogenous leukemia.1-3 Complications of bone marrow failure and later myeloid malignancies are the major causes of the morbidity and mortality of FA patients.4,5 Loss of FA functions impairs cellular responses to genotoxic and cytotoxic stresses, leading to hematopoietic failure and bone marrow failure in the early stage of FA.6,7 As a genetically heterogeneous disease, eight FA core complex proteins (FANCA, -B,-C, -E, -F, -G, -L, and -M), five associated factors (FAAP100, FAAP24, HES1, MHF1, and MHF2) and two mono-ubiquitination dimer proteins (FANCD2-FANCI) have been identified.8-10 All these proteins coordinate together in the FA pathway, facilitate DNA cross-link repair and cell-cycle control, thereby maintain the genome stability.8,9
FAAP20 (20-kDa FANCA-associated Protein) was identified by several groups separately in the past two years as a novel integral subunit of the FA core complex. FAAP20 interacts with FANCA, an essential component of the FA core complex.11-14 By stabilizing FANCA, FAAP20 modulates the ubiquitin ligase activity of the FA core complex, which is required for monoubiquitination of FANCD2. FAAP20 contains a conserved ubiquitin-binding zinc-finger domain (UBZ), which binds to monoubiquitinated form of Rev1 and promotes the interaction of the FA core complex with PCNA/Rev1 DNA damage bypass complexes.14 FAAP20 also binds to the ubiquitin product of RNF8-UBC13, recruits FA core complex to DNA Interstrand Cross Link site (ICLs) and promotes cellular resistance to ICLs.11 Knockdown of FAAP20 in human somatic cells displays FA-like phenotypes.12,13 However, the function of FAAP20 in animals has not been explored.
In this report, we characterized a novel Faap20 mouse model. We showed that deletion of Faap20 in mice led to a mild FA-like phenotype compared to loss of the FA core complex. Faap20−/− mice show defects in the reproductive system and are susceptible to mitomycin C (MMC)-induced bone marrow failure resulting from defects in early hematopoietic progenitor compartments.
Materials and Methods
Mice
Heterozygous Faap20 mice in a C57BL/6 background were generated from the sperm purchased at the Knock-out Mice Program (KOMP) at the University of California Davis (Project ID# CSD23160). The IVF procedure was performed in Transgenic Animal and Genome Editing Core of Cincinnati Children’s Hospital Medical Center. Heterozygotes Faap20 mice were interbred to generate Faap20 null mice and littermate controls. Fanca+/− and Fancc+/− mice were provided by Dr. Madeleine Carreau (Laval University) and Dr. Manuel Buchwald (Hospital for Sick Children, University of Toronto),15,16 respectively. Mice were maintained on C57BL/B6 background in the animal barrier facility at Cincinnati Children’s Hospital Medical Center. Animals were kept in accordance with the Institutional Animal Care and Use Committees (2C06040).
LacZ Tissue Histology
Briefly, E10.5-E11.5 embryos were harvested and placed in PBS, followed by fixation in 0.2% glutaraldehyde overnight at 4°C. The embryos were then incubated overnight at room temperature in the β-Gal staining solution (GALS kit, Sigma). For frozen section, the harvested tissues were prefixed in 0.2% glutaraldehyde overnight in 4°C. All samples were transferred to a 30% sucrose PBS solution overnight. The samples were mounted in O.C.T mounting medium (Fisher Scientific), snap-frozen in isopentane/liquid nitrogen. Cryostat sectioning was performed and the 5mm thick sections were then stained with β-Gal solution (GALS kit, Sigma) for 4 hours at room temperature, and counterstained with Fast Red (N3020, Sigma).
Clonogenic survival assay
MEFs derived from different genotypes 11.5D embryos were treated with the indicated concentrations of MMC, then trypsinized and seeded in triplicate in 100-mm dishes at a density of 500 cells per dish. The cells were cultured in fresh medium for another 5-8 days. Colonies containing more than 50 cells were counted.
Flow cytometry
All the antibodies were obtained from eBioscience or BD Pharmingen unless otherwise indicated. Samples were examined on Fortessa or LSRII flow cytometry, and cytometry data were analyzed with BD FASC Diva or FlowJo software. For lineage cell analysis: Anti-CD71 and anti-Ter119 for erythroid, anti-Gr1 and anti-CD11b for granulocytic/monocytes, anti-CD45R (B220) for B-lymphoid, and anti-CD3e for T-lymphoid. For quantification of CMP, MEP, GMP, CLP, LK and LSK progenitors, mononucleated cell were separated by HISTOPAQUE 1083 (sigma) spin, then labeled with cocktail of anti-mouse lineage markers (CD3e, B220, Ter119, Mac1, and Gr1), anti-mouse CD117 (cKit), and anti-Ly-6A/E (Sca1), anti-CD34, anti-CD16/32, anti-CD150, anti-CD48 and anti-IL-7Ra. Common myeloid progenitors (CMPs) were defined as Lin-Sca1−cKit+CD34+ CD16/32low, and granulocyte-macrophage progenitors (GMPs) were defined as Lin-Sca1−cKit+CD34+CD16/32+. Megakaryocyte-erythroid progenitors (MEPs) were defined as Lin-Sca1−cKit+CD34−CD16/32−. Common lymphoid progenitors (CLPs) were defined as Lin-Sca1lowcKitlowIL-7Ra+, LK, LSK and SLAM cells were defined as Lin-Sca1−cKit+, Lin-Sca1+cKit+ and Lin-Sca1+cKit+CD48−CD150+, respectively (Fig. S4). Dead cells were excluded by 7-aminoactinomycin D staining (Sigma-Aldrich). FluoReporter lacZ Flow Cytometry Kit (F-1930, Life technologies) was used for the β-galactosidase activity flow cytometry quantification with fluorescein di-V-galactoside (FDG) in single cell. For cell cycle analysis, MEFs cells were fixed and permeabilized with Transcription Factor Buffer Set (BD Pharmingen), treated with 50ug/ml RNAase for 30 min at 37°C, then stained with Ki67 antibody (BD Pharmingen) and 10 mg/mL Propidium iodide (Sigma-Aldrich).
In vivo competitive repopulation assay
Briefly, bone marrow cells from different genotype mice were isolated and counted. Recipient CD45.1+ bone marrow cells were mixed with donor CD45.2+ bone marrow cells at a 1:1 ratio and then transplanted into lethally irradiated congenic C57BL/6 CD45.1+ recipient mice. Peripheral blood sample was collected through tail vein bleeding each month after transplantation. Mice were sacrificed 4 month later and subjected to bone marrow fraction analysis.
Hematopoietic colony assay
Bone marrow cells were isolated from femurs and tibiae of 6-8 weeks old mice, and 2×104 cells were put in 1 ml of Methocult M3434 media (StemCell Technologies) following standard protocols with or without MMC (sigma) treatment. Colonies were scored at day 10.
Hematology analysis
Peripheral blood was collected by tail vein bleeding. Measurements of the blood parameters were performed on a HEMAVET 950FS (Drew Scientific Group).
Western blot
FAAP20 protein expression was analyzed in homogenate tissue lysates by western blot using a polyclonal anti-C1orf86 isoform 2 (FAAP20) antibody (Sigma-Aldrich; HPA038829). Other antibody used in the experiments: anti-FANCD2 (Novus), anti-phospho-Histone H2A.X (Ser139) (Millipore), and anti-beta-actin (Sigma).
Statistical analysis
Student’s t-test, ANOVA test, the Pearson’s chi-square test were performed using Microsoft EXCEL software or Prism 6.0 software (GraphPad Software Inc). Error bars indicate SD. Differences were judged as significant if the P value was <0.05 (*), <0.01 (**) or <0.001(***).
Results
Generation and characterization of Faap20 mutant mice
To study the function of FAAP20, we generated Faap20 knockout mice using sperm purchased from KOMP at UCSD (Project ID# CSD23160) and the knock in strategy was illustrated in Figure S1A. The lacZ knock-in allele allowed us to assess FAAP20 protein expression based on β-galactosidase activity in Faap20+/− mice. We validated the knock-in and knock-out in the Faap20 allele by genotyping using four sets PCR primers (Fig. S1B,C) and by Western blot analyzing with lysates from testes (Fig.S1D).
The offspring (n>200) from Faap20+/− breeding followed predicted Mendelian frequencies, indicating no embryonic or perinatal lethality associated with Faap20 knockout (Fig. S2A). No gross phenotypic differences were noted between the wild-type and Faap20−/− offspring. The body weight of age- and sex-matched Faap20−/− mice was similar to that of wild type littermates (Fig. S2B). Hematological parameters including WBC (white blood count), RBC (red blood count), Hb (Hb level) and PLT (platelet count) were grossly normal in 2 month-old Faap20−/− mice comparing to wild type littermate controls (Fig. S2C).
Tissue specific FAAP20 protein expression in mice
We took advantage of Faap20 promoter-driven lacZ transcription and examined Faap20 expression in a developmental and tissue-specific manner. β-gal staining was observed in E11.5 of embryos development, with significantly higher expression in fetal liver, brain and limbs than other tissues (Fig. S3A, B). In addition, intensive β-galactosidase staining was observed in brain, intestine, testis, ovary, and uterus tissues of 2 month-old Faap20+/− mice (Fig. S3C). In contrast, Faap20 expression was low in muscle, heart, kidney, lung, liver and spleen (Fig. S3C). We next examined tissue-specific Faap20 expression by western blot. A strong 20-KDa band representing the mouse FAAP20 was detected from the lysates of testis, ovary, uterus, bone marrow and spleen of adult mice (Fig. S3D). This high level of Faap20 expression in the testis, ovary, spleen and bone marrow suggests an indispensable role of Faap20 in the reproductive and hematopoietic systems.
Reproductive defect in Faap20−/− mice
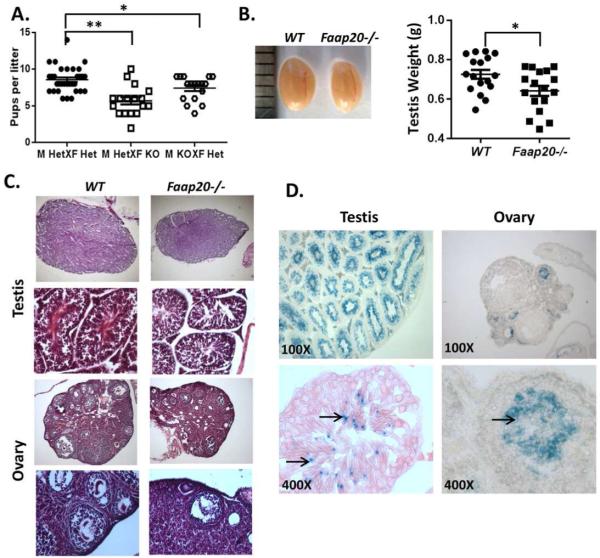
Because Faap20 was highly expressed in the testis and ovary, we evaluated reproductive performance of Faap20−/− mice by monitoring the number of pups born per litter over time. We found that Faap20−/− mice consistently produce small litters (Fig.1A). In contrast, Faap20 heterozygous mice did not show any defect in fertility (Fig.1A). The size of the testes of 2 month-old Faap20−/− mice was significantly smaller than those of wild-type littermate controls (Fig.1B). Consistent with a smaller size, Faap20−/− testes exhibited degenerated seminiferous tubules with decreased numbers of spermatocytes and spermatids (Fig. 1C). Histological analysis of ovaries from 2 month-old Faap20−/− females also showed reduced ovarian size and markedly degenerated follicles (Fig.1C).
Figure 1. Impaired fertility and gonadal development in Faap20−/− mice.
(A) Faap20−/− mice exhibit reduced reproduction ability. Reproductive performance of 4 pairs of mice per group was followed over time. Faap20−/− (KO) mice were crossed with heterozygote (Het) mice. Heterozygotes (Het) and heterozygotes (Het) crossing was used as controls. (B) The testes of Faap20−/− mice are smaller than their wild type littermates. Photograph show testis from wild type (left) male and Faap20−/− littermate (right). (C) H&E stained testis and ovary sections from 2 month-old Faap20−/− and wild type littermates show developmental defect in gonads. All stages of spermatogenesis are seen in the wild-type adult testis. Faap20−/− testis show seminiferous tubules with scattered sertoli cells and decreased number of spermatocytes and spermatids. Many follicles with oocytes at various stages of development can be seen in the wild type ovary. The Faap20−/− ovary is smaller in comparison to the wild type ovary with reduced number of follicles (Magnification 50X, 100X, 200X). (D) β-Gal staining of testis and ovary sections from 2 month-old Faap20+/− mice show cell type specific expression of FAAP20. There was no specific staining in epithelial and sertoli cells, but strong blue staining was seen in the spermatocytes and spermatids (arrows) in testis. In ovary section, the β-Gal signal was detected only in the follicles area (arrow). (Magnification 100X, 400X).
To identify specific germ cell types that Faap20 deletion likely affects the most, we examined Faap20 expression in different cell populations of testis and ovary. β-Gal staining on the 2 month-old Faap20+/− mice testis showed intensive β-galactosidase signal within the seminiferous tubules, but not on the Leydig cells or other interstitial tissue between tubules (Fig.1D). Inside the seminiferous tubules, β-galactosidase activity was detected specifically on spermatocytes and spermatids. No signal was detected on the supporting seminiferous epithelium sertoli cells (Fig.1D). On the other hand, Faap20 expression in ovaries of 2 month-old Faap20+/− mice was strictly observed in the small developmental primordial and primary follicles (Fig.1D). These results indicate that Faap20 expression is gametogenesis specific, mainly in the spermatocytes and spermatids or primordial and primary follicles, all of which are the major meiosis-stage haploid cells during spermatogenesis or oocytogenesis, respectively.
Loss of Faap20 leads to mild FA-like cellular phenotypes
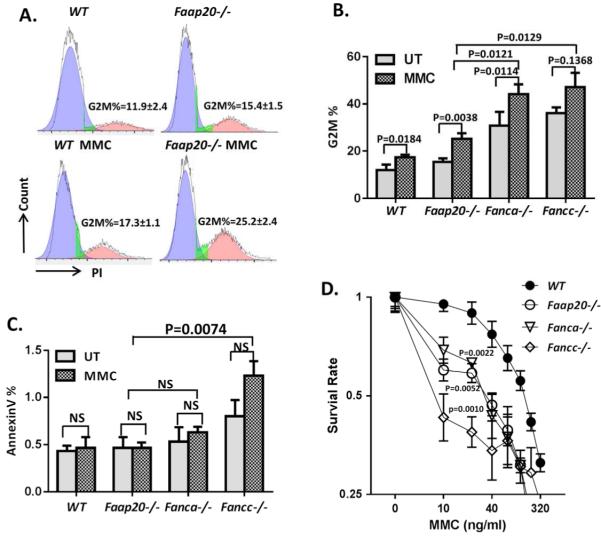
Since G2M arrest and mitomycin C (MMC) hypersensitivity are the cellular hallmarks of FA cells,17-19 we generated primary murine embryonic fibroblasts (MEFs) from E11.5 wild-type and Faap20−/− embryos and analyzed the cell cycle and survival in respond to MMC treatment. Cell cycle profiling revealed that there was a significant accumulation of G2M phase in Faap20−/− cells both at baseline and upon MMC treatment compared to wild-type MEFs (Fig. 2A). These data indicate that Faap20−/− cells undergo G2M arrest at both steady and stressed stages.
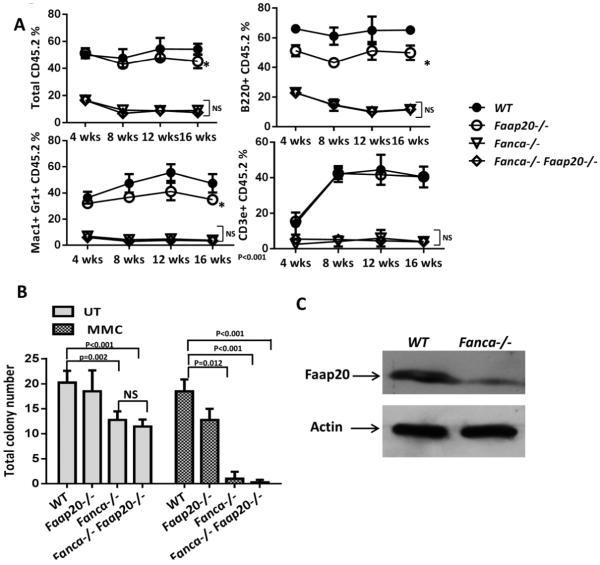
Figure 2. Faap20−/− MEFs and bone marrow cells show typical FA features, but with milder phenotypes in comparison to Fanca−/− or Fancc−/− cells.
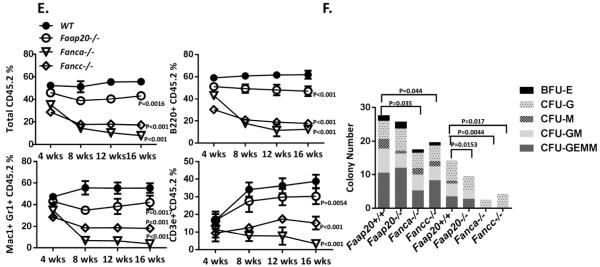
(A) Faap20−/− MEFs exhibit increased G2M arrest before and after MMC treatment. Wild type and Faap20−/− MEFs cells cultured with or without MMC (100 nM) for 24 hours were analyzed by propidium iodide (PI) DNA content staining. The G2M phase percentage was quantified by cell cycle tools in FlowJo. (B) Faap20−/− , Fancc−/− and Fanca−/− MEFs show increased G2M phase than wild type MEFs before and after MMC treatment. The G2M arrest in Fancc−/− and Fanca−/− MEFs was more severe than Faap20−/− MEFs. Different genotypes of MEFs were stained for PI, and analyzed with FACS 24 hours after MMC treatment. Results were expressed as the percentages of G2M phase cells treated with (MMC) or without (UT) MMC. (C) Cell apoptosis rate was increased in Fancc−/− MEFs, but not in Faap20−/− and Fanca−/− MEFs. Different genotype of MEFs were stained for Annexin V, and analyzed with FACS 24 hours after MMC treatment. (D) Faap20−/− , Fancc−/− and Fanca−/− MEFs show reduced survival rate than wild type MEFs after MMC treatment. Fancc−/− MEFs is more sensitive than the Faap20−/− and Fanca−/− MEFs. The MEFs were plated after treatment with or without MMC for 24 hours and colonies were counted after 8 days. Data represent percentage survival, comparing each dose to untreated cells. (E) Faap20−/− bone marrow show reduced long-term repopulating activity in vivo, but not as severe as the Fanca−/− or Fancc−/− bone marrow. Irradiated CD45.1+ recipient mice were competitively reconstituted with 1×106 CD45.2+ bone marrow cells from wild type, Faap20−/−, Fanca−/− or Fancc−/− along with 1×106 CD45.1+ recipient bone marrow cells. Donor-derived total (CD45.2+), B cell (B220+), myeloid cell (Mac1+ Gr1+), and T cell (CD3e+) engraftment in peripheral blood of transplanted recipients were analyzed each month for 4 months. Each line represents average donor cell reconstitution levels from 3 independent experiments with 4-6 recipients/experiment. (F) Decreased colony-forming activity of Faap20−/− bone marrow progenitors under genotoxic condition. Total colony numbers formed by wild type, Faap20−/−, Fanca−/− or Fancc−/− BM cells treated with (MMC) or without (UT) MMC are shown. BFU-E: Burst-Forming Unit-Erythroid; CFU-GM: Colony-Forming Unit-Granulocyte/Macrophage; CFU-G: Colony Forming Unit-Granulocyte; CFU-M: Colony-Forming Unit-Macrophage; CFU-GEMM: Colony-Forming Unit-Granulocyte/Erythrocyte/Macrophage/Megakaryocyte. Data represent mean ± SD of 3 experiments with four replicates each.
Because FAAP20 interacts with FANCA and regulates the FA core complex,11-14 we compared MMC sensitivity of primary MEFs from Faap20−/− mice to those from Fanca−/− or Fancc−/− mice. Cell cycle analysis of cells subjected to100 nM MMC treatment for 24 h showed that Faap20−/− cells underwent less severe G2M arrest than Fanca−/− or Fancc−/− cells (Fig. 2B). Consistently, Faap20−/− cells exhibited less apoptosis (Fig. 2C) and a better survival rate (Fig. 2D) than Fancc−/− cells in response to MMC treatment. Taken together, these results indicate that deletion of Faap20 leads to a milder cellular phenotype compared to loss of the FA core complex in the context of MMC sensitivity.
It has been shown that FA-deficient mouse HSC and progenitor cells are defective in hematopoietic repopulation.20-26 To assess the effect of Faap20 loss on HSC function, we performed competitive long-term reconstitution assays using bone marrow cells from Faap20−/−, Fanca−/− or Fancc−/− mice and their littermate controls. Although less robust than wild-type controls, Faap20−/− HSCs showed milder defect in long-term multi-lineage reconstitution for myeloid, B, and T cells in lethally irradiated recipient mice, as compared to bone marrow HSCs from Fanca−/− or Fancc−/− mice (Fig. 2E). We also performed colony-forming assays to evaluate the effect of Faap20 deficiency on the proliferation of hematopoietic progenitor cells in the absence of functional bone marrow niche and under condition of DNA damage by MMC. Again, loss of Faap20 resulted in a milder defect than deficiency in the FA core complex (i.e. Fanca−/− or Fancc−/−) in the colony forming capacity of hematopoietic precursors in response to MMC (Fig. 2F). Collectively, these results provide in vivo evidence that deletion of the murine Faap20 gene induces mild FA-like cellular phenotypes.
Faap20−/− mice are susceptible to MMC-induced pancytopenia
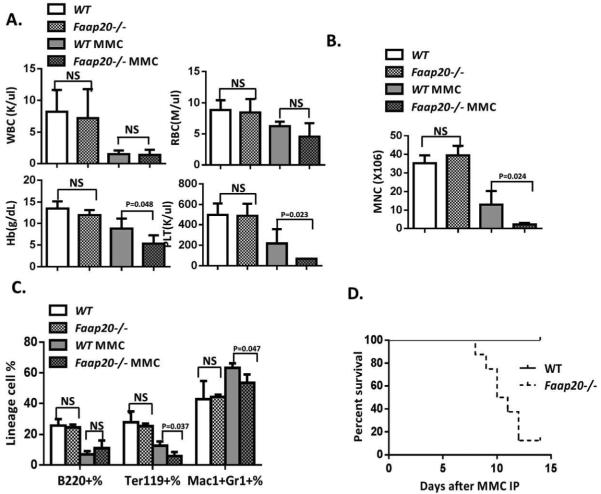
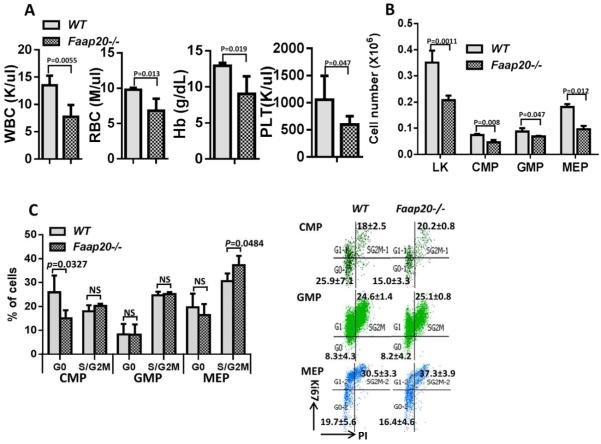
BM failure does not occur spontaneously but can be induced by DNA damage or other stresses in some FA mouse models.20,25 We next examined whether MMC could induce BM failure in Faap20−/− mice. No grossly difference was observed between Faap20−/− mice and wild-type littermates in hematological parameters in the peripheral blood (Fig. 3A& S2C), BM cellularity (Fig. 3B), and lineage differentiation (Fig. 3C) at the steady stage. However, two weeks after acute exposure to one single injection of 8 mg/kg MMC, Faap20−/− mice showed severe pancytopenia, with significantly reduced BM cellularity, Hb, PLT (Fig. 3A-B). In the MMC-treated cohorts, we observed more severe reduction in erythroid lineage than myeloid/lymphoid lineage between Faap20−/− and wild-type mice (Fig. 3C). Consequently, all MMC-treated Faap20−/− mice died within 15 days, mostly due to BM failure (Fig. 3D). Thus, these results indicate that like FA deficiency, loss of Faap20 renders mice susceptible to MMC-induced pancytopenia.
Figure 3. Faap20−/− mice are vulnerable to MMC-induced pancytopenia.
Faap20−/− mice and their wild type littermates at 6-8 weeks of age were injected with 8mg/kg MMC. PBS injected Faap20 −/− or wild type mice were used as controls. Mice were analyzed 8 days after injection. (A) Hematological parameters of peripheral blood cells show bone marrow failure in Faap20−/− mice after MMC injection. Data represent mean ± SD of two to four independent experiments with more than three mice for each group per experiment. (B) Reduced bone marrow cellularity in Faap20−/− mice after MMC treatment. Total bone marrow mononuclear cells from two femurs plus two tibias of individual animals (n>4) of indicated genotypes and treatment were isolated and counted. (C) Bone marrow cells were divided into individual lineage compartments according to cell surface markers using flow cytometric analysis. Numbers represent percentages of the B cells (B220+), erythroid (Ter119+) or myeloid (Mac1+Gr1+) lineage of bone marrow cells. (D) Survival of recipients (n>10) was monitored by Kaplan-Meier curve method. Note that Faap20−/− mice died at around 8-14 days after MMC injection.
MMC induces specific defect in early erythroid progenitor and MEP compartments in Faap20−/− mice
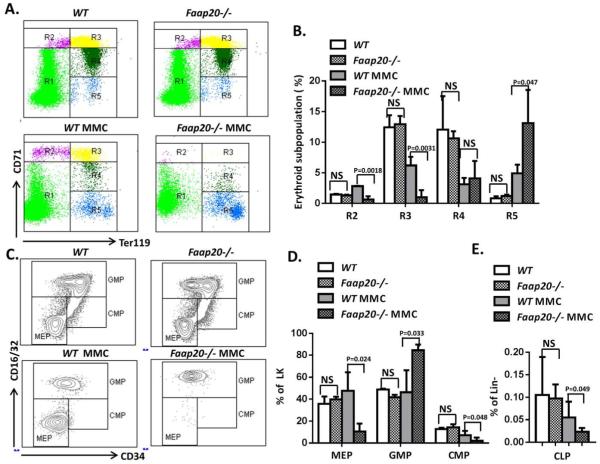
The observation that MMC treatment dramatically reduced RBC and erythroid lineage prompted us to analyze the erythroid differentiation program in Faap20−/− mice, using the erythroid progenitor markers CD71 and TER119 to identify distinct cell populations at specific stages of erythrocyte differentiation (Fig. 4A). We found that MMC induced significant reduction of basophilic erythroblasts (R2, 77.6%, p<0.01) and late basophilic and chromatophilic erythroblasts (R3, 78.5%, p<0.001), but not orthochromatophilic erythroblasts (R4) in Faap20−/− mice compared to wild-type littermate controls (Fig. 4A, B). In contrast, we observed a compensative enrichment in the more mature reticulocyte (R5) population in MMC-treated Faap20−/− mice (Fig. 4A, B).
Figure 4. Loss of Faap20 induces early erythroid progenitors and MEPs ablation after MMC treatment.
Faap20−/− or wild type mice at 6-8 weeks of age were injected with one dose 8mg/kg MMC to induce the bone marrow failure. PBS injected Faap20−/− or wild type mice were used as controls. Mice were analyzed 8 days after injection. (A) The early erythroid progenitors in Faap20−/− mice were depleted after MMC exposure. Freshly dissociated bone marrow cells were labeled with CD71 and Ter119 mAb, and subjected to flow cytometric analysis. The five regions (R1 to R5) within a flow cytometry scatter plot contain distinct cell types found in the different stages of erythrocyte differentiation were selected as indicated. There are predominantly proerythroblasts in region R1, basophilic erythroblasts in region R2, late basophilic and chromatophilic erythroblasts in region R3, orthochromatophilic erythroblasts in region R4, and reticulocytes and erythrocytes in R5. (B) The percentages of R2-R5 cells in the bone marrow were quantitated from the data shown in (A). (C) Flow cytometry analysis show early progenitor depletion in Faap20−/− mice treated with MMC. (D) Quantitation of CMPs, MEPs and GMPs in Lin-cKit+ (LK) compartment. Data represent summary of more than three mice of each genotype from two independent experiments. (E) Quantitation of CLPs. Data represent summary of more than three mice of each genotype from two independent experiments.
We next examined the effect of MMC on the upstream progenitor compartments in Faap20−/− mice. We found that MMC induced a significant percentage decrease in common myeloid progenitors (CMPs) and megakaryocyte-erythrocyte progenitors (MEPs) but a compensatory increase in granulocyte–macrophage progenitors (GMPs) in the LK (Lin-cKit+) population in Faap20−/− mice compared to WT littermate controls (Fig. 4C, D). A significant reduction of common lymphoid progenitors (CLPs) was also observed in MMC-treated Faap20−/− mice (Fig. 4E). Thus, it appears that Faap20 deficiency affects CMPs and MEPs more than GMPs in the context of MMC sensitivity. In agreement with a differential functional role of Faap20 in lineage progenitors, we found that Faap20 expression was significantly higher in CMPs and MEPs than in GMPs (Fig. S5).
Faap20−/− HSPCs display aberrant cell cycle pattern under chronic MMC stress
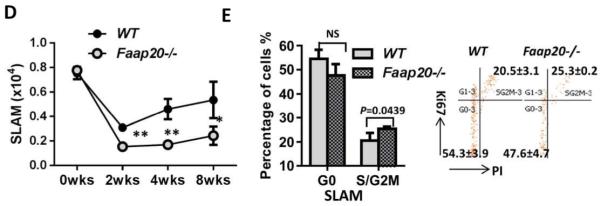
To understand the mechanism underlying genotoxic stress-induced pancytopenia in Faap20−/− mice, we performed cell cycle analysis on the HSPCs in Faap20−/− mice. Because acute high dose MMC treatment on the Faap20−/− mice caused almost complete bone marrow HSPCs depletion with not enough cells for FACS analysis and chronic MMC stress may reflect more physiologic challenges from the environment, the Faap20−/− and WT control mice were subject to weekly injection with low dosage MMC (0.8mg/kg) in accordance to the published protocol for FA mice 20,27. The pancytopenia phenotype was observed in Faap20−/− mice around 6-8 weeks, as evidenced by a significant reduction in the number of WBC, RBC, platelets counts, and Hb level in Faap20−/− mice in comparison with WT mice (Fig. 5A). Again, MMC induced a significant decrease in the LK (Lin-cKit+) compartment, particularly MEPs of Faap20−/− mice, as compared with those of WT mice (Fig. 5B). In addition, we observed a progressive reduction of phenotypic BM HSCs (LSK CD150+ CD48−; SLAM) in MMC-treated Faap20−/− mice (Fig. 5D). Cell cycle analysis with PI and Ki67 staining showed aberrant cell cycle pattern characterized by decreased quiescent (G0) or increased cycling (S/G2M) cells in CMPs, MEPs and SLAMs populations of Faap20−/− mice after 8 weeks of MMC treatment (Fig. 5C, E). Thus, these results suggest that low level of chronic genotoxic stress may cause replicative exhaustion of Faap20−/− HSPCs, which may be one major mechanism responsible for HSPC depletion and pancytopenia phenotype in MMC-treated Faap20−/− mice.
Figure 5. Aberrant cell cycle pattern of HSPCs in Faap20−/− mice after chronic MMC stress.
Faap20−/− mice and their WT littermates at 6-8 weeks of age were injected with 0.8mg/kg MMC weekly. The mice were sacrificed and analyzed after 8 weeks of IP administration. (A) WBC, RBC,PLT counts and the Hb level in Faap20−/− were significantly decreased compared to WT mice after 8 weeks of MMC treatment. (B) Decreased number of LKs and MPPs subpopulation in Faap20−/− mice after MMC stress. (C) The CMPs and MEPs, but not GMPs population show aberrant cell cycle pattern in MMC-treated Faap20−/− mice. The representative FACS graphs are shown on right. (D) The number of SLAM cells was progressively declined during long-term MMC stress in Faap20−/− mice. (E) Increased cycling (S/G2M) cells in the SLAM population of Faap20−/− mice after 8 weeks of MMC injection. The representative FACS graphs are shown on right.
Possible dominant effect of FANCA protein in the interaction between FAAP20 and FANCA in response to MMC
To study the relationship between Faap20 and Fanca, we generated Faap20−/−Fanca−/− double knockout (dKO) mice. Analysis of donor-derived total or lineage-specific repopulation in the peripheral blood show no significant difference between the Fanca−/− and Faap20−/−Fanca−/− recipient cohorts (Fig. 6A). The repopulating capacity of Fanca−/− BM HSCs in lethally irradiated recipient mice was not further reduced by a simultaneous Faap20 deficiency. Also, colony-forming assays showed that loss of Faap20 did not aggravate the extent of MMC effect on Fanca−/− BM HSPC proliferation in vitro (Fig. 6B). Consistent with previous reports in human cell lines,12,13 we observed a lower level of FAAP20 protein in Fanca−/− MEFs than in wild-type cells (Fig. 6C), supporting the notion that FANCA stabilizes FAAP20. These data imply a possible dominant effect of FANCA over FAAP20.
Figure 6. Loss of Faap20 does not aggravate the bone marrow phenotype in Fanca−/− mice.
(A) Faap20 deficiency does not further compromise long-term repopulating activity of Fanca−/− bone marrow HSPCs. Irradiated CD45.1+ recipient mice were competitively reconstituted with 1×106 CD45.2+ bone marrow cells from wild type, Faap20−/−, Fanca−/− or Fanca−/−Faap20−/− along with 1×106 CD45.1+ recipient bone marrow cells. Donor-derived total engraftment, B cell (B220+), myeloid cell (Mac1+ Gr1+), and T cell (CD3e+) engraftment in the peripheral blood of recipients were analyzed each month for 4 months. Each line represents average donor cell reconstitution levels from two independent experiments with 4-6 recipients per experiment. (B) Colony forming activity of WT, single KO (Fanca−/− or Faap20−/−), double KO (Fanca−/−Faap20−/−) bone marrow cells treated with (MMC) or without (UT) MMC. The double KO (Fanca−/−Faap20−/−) bone marrow cells did not exhibit reduced colony forming ability comparing to single Fanca−/− KO bone marrow cells. Data represent mean ± SD of two independent experiments with 4 replicates each genotype per experiment. (C) Immunoblotting show reduced expression levels of FAAP20 in Fanca−/− MEFs cells.
To further understand the mechanistic role of FAAP20 in the FA pathway, we performed western blot and immunostaining on the MEFs and lineage-negative cells derived from WT, Faap20−/−, Fanca−/−, or Fancd2−/− mice, with or without MMC treatment (Fig.S6 & S7). We observed partial FANCD2 monoubiquitination in the Faap20−/− MEFs in comparison to non-FANCD2 ubiquitination in Fanca−/− MEFs. In addition, less intensive phospho-Histone H2A.X (Ser139) signal was detected in the Faap20−/− cells in comparison to Fanca−/− or Fancd2−/− cells, indicating less DNA damage. These data suggest that the FA complex is not completely disrupted in the absence of FAAP20.
Discussion
Multiple Fanconi Anemia (FA) mouse models have been generated to study the role of FA proteins in growth and development.15,16,27-32 These FA mice generally exhibit decreased fertility with abnormal germ cell development and increased DNA cross-linker sensitivity.16,27,30 The FA mice also show hematopoietic stem cell (HSPC) dysfunction upon transplantation and develop bone marrow failure after MMC injection.20,24,26 To explore the in vivo function of the newly discovered FAAP20 protein, we have characterized the Faap20−/− mice with the exon1c and exon2 of the Faap20 gene replaced by a β-galactosidase gene. The Faap20−/− mice display similar features of other FA knockout mouse models. For example, similar to Fanca−/−, Fancc−/−, Fancg−/− and Fancd2−/− mice16,27,30,31, Faap20−/− mice show defects in reproductive organs, hematopoietic repopulation and cellular resistance to DNA cross-linking agent MMC. Interestingly, Faap20−/− mice exhibit milder FA-like phenotypes than mice deficient for the FA core complex components such as FANCA or FANCC. For instance, although inter-cross of Faap20−/− mice exhibited significantly reduced litter sizes, the Fanca−/−, Fancc−/− and Fancd2−/− mice are completely infertile. In addition, BM HSC and progenitor cells from Fanca−/− or Fancc−/− mice are more sensitive to replicative stress induced by transplantation or genotoxic stress resulting from DNA cross-links than those from Faap20−/− mice. Our results suggest that loss of Faap20 may not completely disrupt the formation of the FA core complex. Thus, partial monoubiquitination of FANCD2 and less DNA damage in the Faap20−/− cells may be responsible for the mild FA-like phenotypes we observed in the Faap20−/− mice (Fig.S6 & S7). In supporting this notion, previously study indicates that residual monoubiquitination of FANCD2 was detected in FAAP20-deficient human cell lines.12
Another interesting finding of the current study is the observation that the expression of mouse Faap20 is regulated in developmental and tissue-specific manner. We observed high levels of Faap20 expression in stages of E11.5 embryos, suggesting that a role of Faap20 during the early development. Consistent with this, it has been reported that FANCA is also expressed in E7.5 and E11.5 embryos,15 and that 60–75% of FA patients were born with congenital defects, prenatal lethality and growth retardation.4,33,34 Interestingly, Faap20 is specifically expressed in the hippocampus and cortex region in the brain (Fig. S3C). Relevant to this, it is noteworthy that some FA patients display central nervous system symptoms, including increased fluid in the center of the brain (hydrocephalus) or an unusually small head size (microcephaly).35-37 The function of Faap20 in hippocampus and cortex of the brain remains to be investigated. Also, the Faap20 expression was observed in the cyst of intestine tissue and part of the uterus.
The highest level of Faap20 expression was detected in the testes and ovaries of adult mice. This is consistent with the reproductive defects observed in Faap20−/− mice. Similarly, FANCA and FANCD2 are also highly expressed in the testes and Fanca or Fancd2 deficiency causes reproductive defects in mice.15,23 Furthermore, we observed intensive β-galactosidase signal specifically in spermatocytes and spermatids of testes and in the small primordial and primary follicles of ovaries (Fig. 2D). This is the stage during gametogenesis when meiotic recombination results in formation and repair of DNA double-stranded breaks. In addition, it has been shown that increased FANCA expression is restricted to mid- to late-pachytene spermatocytes.15 Collectively, these observations suggested that FAAP20 may function in meiotic recombination during spermatogenesis and oocytogenesis.
Previously studies suggested that FAAP20 might also possess DNA repair function outside of the FA pathway, as the ubiquitin-binding activity elicited by the UBZ domain of FAAP20 is independent of the FAAP20-FANCA interaction.13 Based on this information, we hypothesized that deletion of Faap20 in Fanca−/− mice might exhibit a more severe phenotype than Fanca−/− mice. However, our results show that loss of Faap20 did not aggravate Fanca−/− phenotype and FAAP20 protein level was almost negligible in Fanca null MEFs by western blot (Fig. 6). In addition, previously studies indicate that FAAP20 could bind to ubiquitinated Rev1 or histone H2A then trigger the recruitment of FA core complex to the ICLs site, suggesting that FAAP20 may function downstream of the FA core complex.11,14 In this context, we propose a possible dominant effect of the FANCA protein in the interaction between FANCA and FAAP20 during ICL repair.
Interestingly, we observed differential effects of MMC-induced genotoxic stress on specific lineages of Faap20−/− BM progenitors. Both high-dose acute and low-dose chronic exposure of Faap20−/− mice to MMC induced severe pancytopenia, most likely resulting from injuries to HSPCs especially the erythroid progenitors and their upstream MEPs (Figs. 4, 5). MMC-induced BM failure has been reported in Fancg−/− and Fancc−/− mice.20,27 However, the cellular mechanism responsible for this hallmark phenotype was not further investigated. In this study, we showed that the LKs and SLAM populations in MMC-treated Faap20−/− mice displayed an aberrant cell cycle pattern featuring decreased quiescent (G0) and increased cycling (S/G2M) cells (Fig. 5), typical of replicative exhaustion in these HSPCs. It is possible that the increased MMC injury observed in erythroid progenitors and MEPs over GMPs cells was due to the fact that erythroid progenitors and MEPs cycle more rapidly than myeloid and GMPs do.38 Consistently, we detected higher levels of Faap20 expression in the erythroid progenitors, CMPs and MEPs compared with mature myeloid (Mac1+Gr1+) cells and GMPs (Fig. S5). A recent clinical study of FA patients identifies dyserythropoiesis and dysmegakaryopoiesis as the most common morphologic abnormalities,5 further corroborating the notion that the erythroid cells and their precursor MEPs may be more vulnerable to DNA damage stress than other lineages in FA. Our results are consistent with the role of FAAP20 in cell survival following MMC-induced DNA damage and replicative stress. In this context, the Faap20 mouse model may be a useful tool for studying mechanisms underlying the pathophysiology of FA BM failure.
Supplementary Material
Supplementary Materials
Supplementary Figure 1. Generation of Faap20 /LacZ knock-in mice. (A) Schematic tm1(KOMP)Mbp diagram of Faap20 (KOMP, 2610002J02Rik) mice knock-in strategy. The Faap20/LacZ knock-in allele is shown to illustrate the Faap20 gene in which the exon1c and exon2 is replaced by the LacZ-neomycin expression cassette, resulting in a Faap20 null allele. This cassette contains the splice acceptor of mouse engrailed 2 exon 2 (En2 SA), an internal ribosome entry sequence (IRES) to initiate LacZ translation, and polyadenylation (pA) to terminate transcription after the LacZ gene. The neo gene is under human beta actin promoter (hBactP) and contains its own polyA. The exons are indicated by black boxes. (B, C) Genotyping by PCR. Four sets PCR primer were used to validate the Faap20/LacZ knock-in allele. For both 5′ and 3′ set, one end of the primer set was designed outside the homology arm and the other end was within the LacZ/neo sequence. The 5′ set primer produced a predicted 6.7 kb amplicon from correct recombinants. The 3′ set produced a predicted 5.7 kb amplicon from correct recombinants. The genotyping internal primer set produced a 383 kb amplicon from the wild-type allele and 775 kb amplicon from the knockout allele. (D) Cell lysates derived from wild-type (WT) and Faap20 heterozygous (Faap20+/−) and homozygous (Faap20−/−) testes were analyzed by immunoblotting with FAAP20 antibodies. Similar amount of FAAP20 protein was detected in the testis tissue from the Faap20+/+ and Faap20+/− mouse. FAAP20 protein expression was abrogated in Faap20−/− mice.
Supplementary Figure 2. No gross phenotypic difference was noted between the wild-type and Faap20−/− offspring. (A) The genotype of offspring from crossing Faap20+/− mice followed a predicted Mendelian frequency, indicating that no embryonic or perinatal lethality was associated with homozygous Faap20 knockout.(B) No significant difference was observed in the male and female body weight between wild-type and Faap20−/− mice (n>6 per group). (C) No significant difference was observed in peripheral blood WBC counts , RBC counts, Hb level and PLT level between wild-type and Faap20−/− mice (n>10 per group).
Supplementary Figure 3. Tissue specific expression of FAAP20 protein.(A) Whole mount staining was performed on 11.5 day Faap20+/− embryos and their wild type (WT) littermates. The photograph illustrates the broad β-Gal staining throughout the Faap20+/− embryo especially in fetal liver, brain and limbs. No β-galactosidase activity was detected in WT littermates. (B) Western blot of FAAP20 on mouse embryonic fibroblast cells derived from E11.5 embryos indicated the expression of FAAP20 in embryonic stage. (C) Representative micrographs were shown to illustrate specific β-Gal staining of tissue sections from 2 month old Faap20+/− mice and wild type littermates. Note intensive staining in brain, intestine, testes, ovary, and uterus tissues. In contrast, there was no or little staining in muscle, heart, liver, kidney, lung and spleen tissues from Faap20+/− mice, or any tissues from the WT littermates.(D) Western blot of FAAP20 in different tissue of wild type mice show tissue specific expression of FAAP20. The FAAP20 protein was highly expressed in the testis, ovary, uterus, bone marrow and spleen.
Supplementary Figure 4. Cytometric gates strategy for lineages and early progenitors in bone marrow.(A) For bone marrow cell lineage analysis, the following antibodies were used: anti-CD45R (B220) for B-lymphoid, anti-CD3e for T-lymphoid, anti-Gr1 and anti-CD11b for granulocytic/monocytes, anti-CD71 and anti-Ter119 for erythroid (R1-R5 gates indicated specific development stages of erythroid cells). (B) LK, LSK and SLAM cells were gated as Lin-Sca1−cKit+, Lin-Sca1+cKit+ and Lin-Sca1+cKit+CD48−CD150+, respectively. (C) Common myeloid progenitors (CMPs) were gated as Lin-Sca1−cKit+CD34+ CD16/32low, and granulocyte-macrophage progenitors (GMPs) were gated as Lin-Sca1−cKit+CD34+CD16/32+. Megakaryocyte-erythroid progenitors (MEPs) were gated as Lin-Sca1−cKit+CD34−CD16/32−. Common lymphoid progenitors (CLPs) were gated as Lin-Sca1lowcKitlowIL-7Ra+.
Supplementary Figure 5. FAAP20 expression in different population of bone marrow cells by FDG staining. (A) FDG staining for β-galactosidase activity in bone marrow cells from 2 month old Faap20+/− mice indicates that about 10.6% of total bone marrow cells expressed FAAP20. 0.3% background staining was observed in wild type (WT) control bone marrow cells. (B) The FDG+ percentage is higher in the erythroid and lymphoid lineages compared to myeloid cells. LK, CMPs, CLPs and MEPs show higher percentage of FDG positive cells compared to LSK, SLAM and GMPs, with the highest FAAP20 expression in the MEPs population. (C) FDG positive cells in each lineage and progenitor population are presented in cytometric plots.
Supplementary Figure 6: Less severe DNA damage in Faap20−/− than in Fanca−/− or Fancd2−/− MEFs. (A) Western blot was performed on the MEFs cells derived from WT, Faap20−/−, Fanca−/−, or Fancd2−/− mice, with or without the MMC (100nM for 16h) treatment. The blot using anti-FANCD2 (Novus) antibody show partial FANCD2 monoubiquitination in the Faap20−/− MEFs in comparison to non-FANCD2 ubiquitination in Fanca−/− MEFs and complete depletion of FANCD2 protein in the Fancd2−/− MEFs. The blot using anti-phospho-Histone H2A.X (Ser139) antibody (Millipore) indicates less phospho-Histone H2A.X (Ser139) in the Faap20−/− MEFs in comparison to Fanca−/− or Fancd2−/− MEFs. (B) The relative abundance of phospho-Histone H2A.X (Ser139) to Actin in (A) was analyzed through quantification of the western blot with ImageJ software. (C, D) DNA damage immunofluoresence staining with anti-phospho-Histone H2A.X (Ser139) antibody (Millipore) was performed on MEFs cells derived from WT, Faap20−/−, Fanca−/−, or Fancd2−/− mice, with or without the MMC (100nM for 16 h) treatment. Less DNA damage staining was detected in the Faap20−/− MEFs compared to Fanca−/−, Fancd2−/− cells. Scale bar, 10 µm. (D) The intensity of phospho-Histone H2A.X (Ser139) staining in (C) was quantified using ImageJ software.
Supplementary Figure 7: Less severe DNA damage induced by MMC in bone marrow HSPCs from Faap20−/− mice than Fanca−/− or Fancd2−/− mice. Bone marrow Lin-cells from WT, Faap20−/−, Fanca−/−, Fancd2−/− mice were treated with or without MMC (100nM for 16 h) and used for phospho-Histone H2A.X (Ser139) western blot (A, B) and immuno-staining (C, D). Scale bar, 5 µm. (B) The relative abundance of phospho-Histone H2A.X (Ser139) to Actin in (A) was analyzed through quantification of the western blot with ImageJ software. (D) The intensity of phospho-Histone H2A.X (Ser139) staining in (C) was quantified using ImageJ software. Faap20−/− cells show less severe DNA damage compared to Fanca−/− or Fancd2−/− cells.
Key Points.
Deletion of Faap20 in mice causes a mild FA-like phenotype. Faap20−/− mice show reproductive defect and are susceptible to genotoxic stress induced bone marrow failure resulting from depletion of early hematopoietic progenitors.
Acknowledgements
We thank Dr. Madeleine Carreau (Laval University) for the Fanca+/− mice, Dr. Manuel Buchwald (Hospital for Sick Children, University of Toronto) for the Fancc+/−mice, the Transgenic Animal and Genome Editing Core (Cincinnati Children’s Hospital Medical Center) for IVF service, and the Comprehensive Mouse and Cancer Core of the Cincinnati Children's Research Foundation (Cincinnati Children’s Hospital Medical Center) for bone marrow transplantation service. This investigation was supported by NIH grants R01 HL076712, R01 CA157537 and T32 HL091805. Q.P. is supported by a Leukemia and Lymphoma Scholar award.
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 2.Bagby GC., Jr Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10(1):68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Auerbach AD, Allen RG. Leukemia and preleukemia in Fanconi anemia patients. A review of the literature and report of the International Fanconi Anemia Registry. Cancer Genet Cytogenet. 1991;51(1):1–12. doi: 10.1016/0165-4608(91)90002-c. [DOI] [PubMed] [Google Scholar]
- 3.Tischkowitz M, Dokal I. Fanconi anaemia and leukaemia - clinical and molecular aspects. Br J Haematol. 2004;126(2):176–191. doi: 10.1111/j.1365-2141.2004.05023.x. [DOI] [PubMed] [Google Scholar]
- 4.Kutler DI, Singh B, Satagopan J, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101(4):1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 5.Cioc AM, Wagner JE, MacMillan ML, DeFor T, Hirsch B. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133(1):92–100. doi: 10.1309/AJCP7W9VMJENZOVG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller LU, Williams DA. Finding the needle in the hay stack: hematopoietic stem cells in Fanconi anemia. Mutat Res. 2009;668(1-2):141–149. doi: 10.1016/j.mrfmmm.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123(1):26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26(13):1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol. 2012;226(2):326–337. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- 10.Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24(16):1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Z, Guo R, Paramasivam M, et al. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol Cell. 2012;47(1):61–75. doi: 10.1016/j.molcel.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung JW, Wang Y, Fong KW, Huen MS, Li L, Chen J. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci U S A. 2012;109(12):4491–4496. doi: 10.1073/pnas.1118720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali AM, Pradhan A, Singh TR, et al. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood. 2012;119(14):3285–3294. doi: 10.1182/blood-2011-10-385963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Yang K, Dejsuphong D, D'Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19(2):164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12(16):2063–2076. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 16.Chen M, Tomkins DJ, Auerbach W, et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996;12(4):448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 17.Seyschab H, Friedl R, Sun Y, et al. Comparative evaluation of diepoxybutane sensitivity and cell cycle blockage in the diagnosis of Fanconi anemia. Blood. 1995;85(8):2233–2237. [PubMed] [Google Scholar]
- 18.Heinrich MC, Hoatlin ME, Zigler AJ, et al. DNA cross-linker-induced G2/M arrest in group C Fanconi anemia lymphoblasts reflects normal checkpoint function. Blood. 1998;91(1):275–287. [PubMed] [Google Scholar]
- 19.Akkari YM, Bateman RL, Reifsteck CA, D'Andrea AD, Olson SB, Grompe M. The 4N cell cycle delay in Fanconi anemia reflects growth arrest in late S phase. Mol Genet Metab. 2001;74(4):403–412. doi: 10.1006/mgme.2001.3259. [DOI] [PubMed] [Google Scholar]
- 20.Carreau M, Gan OI, Liu L, et al. Bone marrow failure in the Fanconi anemia group C mouse model after DNA damage. Blood. 1998;91(8):2737–2744. [PubMed] [Google Scholar]
- 21.Haneline LS, Gobbett TA, Ramani R, et al. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94(1):1–8. [PubMed] [Google Scholar]
- 22.Haneline LS, Li X, Ciccone SL, et al. Retroviral-mediated expression of recombinant Fancc enhances the repopulating ability of Fancc−/− hematopoietic stem cells and decreases the risk of clonal evolution. Blood. 2003;101(4):1299–1307. doi: 10.1182/blood-2002-08-2404. [DOI] [PubMed] [Google Scholar]
- 23.Parmar K, Kim J, Sykes SM, et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010;28(7):1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Le Beau MM, Ciccone S, et al. Ex vivo culture of Fancc−/− stem/progenitor cells predisposes cells to undergo apoptosis, and surviving stem/progenitor cells display cytogenetic abnormalities and an increased risk of malignancy. Blood. 2005;105(9):3465–3471. doi: 10.1182/blood-2004-06-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milsom MD, Lee AW, Zheng Y, Cancelas JA. Fanca−/− hematopoietic stem cells demonstrate a mobilization defect which can be overcome by administration of the Rac inhibitor NSC23766. Haematologica. 2009;94(7):1011–1015. doi: 10.3324/haematol.2008.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro S, Meza NW, Quintana-Bustamante O, et al. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14(4):525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Koomen M, Cheng NC, van de Vrugt HJ, et al. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum Mol Genet. 2002;11(3):273–281. doi: 10.1093/hmg/11.3.273. [DOI] [PubMed] [Google Scholar]
- 28.Bakker ST, de Winter JP, te Riele H. Learning from a paradox: recent insights into Fanconi anaemia through studying mouse models. Dis Model Mech. 2013;6(1):40–47. doi: 10.1242/dmm.009795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parmar K, D'Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668(1-2):133–140. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng NC, van de Vrugt HJ, van der Valk MA, et al. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet. 2000;9(12):1805–1811. doi: 10.1093/hmg/9.12.1805. [DOI] [PubMed] [Google Scholar]
- 31.Houghtaling S, Timmers C, Noll M, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17(16):2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakker ST, van de Vrugt HJ, Rooimans MA, et al. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet. 2009;18(18):3484–3495. doi: 10.1093/hmg/ddp297. [DOI] [PubMed] [Google Scholar]
- 33.Antonio Casado J, Callen E, Jacome A, et al. A comprehensive strategy for the subtyping of patients with Fanconi anaemia: conclusions from the Spanish Fanconi Anemia Research Network. J Med Genet. 2007;44(4):241–249. doi: 10.1136/jmg.2006.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ameziane N, Errami A, Leveille F, et al. Genetic subtyping of Fanconi anemia by comprehensive mutation screening. Hum Mutat. 2008;29(1):159–166. doi: 10.1002/humu.20625. [DOI] [PubMed] [Google Scholar]
- 35.Porteous ME, Cross I, Burn J. VACTERL with hydrocephalus: one end of the Fanconi anemia spectrum of anomalies? Am J Med Genet. 1992;43(6):1032–1034. doi: 10.1002/ajmg.1320430624. [DOI] [PubMed] [Google Scholar]
- 36.Alter BP. Hydrocephalus in Fanconi anemia. Am J Med Genet. 1993;45(6):785. doi: 10.1002/ajmg.1320450627. [DOI] [PubMed] [Google Scholar]
- 37.Cox PM, Gibson RA, Morgan N, Brueton LA. VACTERL with hydrocephalus in twins due to Fanconi anemia (FA): mutation in the FAC gene. Am J Med Genet. 1997;68(1):86–90. [PubMed] [Google Scholar]
- 38.Matarraz S, Teodosio C, Fernandez C, et al. The proliferation index of specific bone marrow cell compartments from myelodysplastic syndromes is associated with the diagnostic and patient outcome. PLoS One. 2012;7(8):e44321. doi: 10.1371/journal.pone.0044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Supplementary Figure 1. Generation of Faap20 /LacZ knock-in mice. (A) Schematic tm1(KOMP)Mbp diagram of Faap20 (KOMP, 2610002J02Rik) mice knock-in strategy. The Faap20/LacZ knock-in allele is shown to illustrate the Faap20 gene in which the exon1c and exon2 is replaced by the LacZ-neomycin expression cassette, resulting in a Faap20 null allele. This cassette contains the splice acceptor of mouse engrailed 2 exon 2 (En2 SA), an internal ribosome entry sequence (IRES) to initiate LacZ translation, and polyadenylation (pA) to terminate transcription after the LacZ gene. The neo gene is under human beta actin promoter (hBactP) and contains its own polyA. The exons are indicated by black boxes. (B, C) Genotyping by PCR. Four sets PCR primer were used to validate the Faap20/LacZ knock-in allele. For both 5′ and 3′ set, one end of the primer set was designed outside the homology arm and the other end was within the LacZ/neo sequence. The 5′ set primer produced a predicted 6.7 kb amplicon from correct recombinants. The 3′ set produced a predicted 5.7 kb amplicon from correct recombinants. The genotyping internal primer set produced a 383 kb amplicon from the wild-type allele and 775 kb amplicon from the knockout allele. (D) Cell lysates derived from wild-type (WT) and Faap20 heterozygous (Faap20+/−) and homozygous (Faap20−/−) testes were analyzed by immunoblotting with FAAP20 antibodies. Similar amount of FAAP20 protein was detected in the testis tissue from the Faap20+/+ and Faap20+/− mouse. FAAP20 protein expression was abrogated in Faap20−/− mice.
Supplementary Figure 2. No gross phenotypic difference was noted between the wild-type and Faap20−/− offspring. (A) The genotype of offspring from crossing Faap20+/− mice followed a predicted Mendelian frequency, indicating that no embryonic or perinatal lethality was associated with homozygous Faap20 knockout.(B) No significant difference was observed in the male and female body weight between wild-type and Faap20−/− mice (n>6 per group). (C) No significant difference was observed in peripheral blood WBC counts , RBC counts, Hb level and PLT level between wild-type and Faap20−/− mice (n>10 per group).
Supplementary Figure 3. Tissue specific expression of FAAP20 protein.(A) Whole mount staining was performed on 11.5 day Faap20+/− embryos and their wild type (WT) littermates. The photograph illustrates the broad β-Gal staining throughout the Faap20+/− embryo especially in fetal liver, brain and limbs. No β-galactosidase activity was detected in WT littermates. (B) Western blot of FAAP20 on mouse embryonic fibroblast cells derived from E11.5 embryos indicated the expression of FAAP20 in embryonic stage. (C) Representative micrographs were shown to illustrate specific β-Gal staining of tissue sections from 2 month old Faap20+/− mice and wild type littermates. Note intensive staining in brain, intestine, testes, ovary, and uterus tissues. In contrast, there was no or little staining in muscle, heart, liver, kidney, lung and spleen tissues from Faap20+/− mice, or any tissues from the WT littermates.(D) Western blot of FAAP20 in different tissue of wild type mice show tissue specific expression of FAAP20. The FAAP20 protein was highly expressed in the testis, ovary, uterus, bone marrow and spleen.
Supplementary Figure 4. Cytometric gates strategy for lineages and early progenitors in bone marrow.(A) For bone marrow cell lineage analysis, the following antibodies were used: anti-CD45R (B220) for B-lymphoid, anti-CD3e for T-lymphoid, anti-Gr1 and anti-CD11b for granulocytic/monocytes, anti-CD71 and anti-Ter119 for erythroid (R1-R5 gates indicated specific development stages of erythroid cells). (B) LK, LSK and SLAM cells were gated as Lin-Sca1−cKit+, Lin-Sca1+cKit+ and Lin-Sca1+cKit+CD48−CD150+, respectively. (C) Common myeloid progenitors (CMPs) were gated as Lin-Sca1−cKit+CD34+ CD16/32low, and granulocyte-macrophage progenitors (GMPs) were gated as Lin-Sca1−cKit+CD34+CD16/32+. Megakaryocyte-erythroid progenitors (MEPs) were gated as Lin-Sca1−cKit+CD34−CD16/32−. Common lymphoid progenitors (CLPs) were gated as Lin-Sca1lowcKitlowIL-7Ra+.
Supplementary Figure 5. FAAP20 expression in different population of bone marrow cells by FDG staining. (A) FDG staining for β-galactosidase activity in bone marrow cells from 2 month old Faap20+/− mice indicates that about 10.6% of total bone marrow cells expressed FAAP20. 0.3% background staining was observed in wild type (WT) control bone marrow cells. (B) The FDG+ percentage is higher in the erythroid and lymphoid lineages compared to myeloid cells. LK, CMPs, CLPs and MEPs show higher percentage of FDG positive cells compared to LSK, SLAM and GMPs, with the highest FAAP20 expression in the MEPs population. (C) FDG positive cells in each lineage and progenitor population are presented in cytometric plots.
Supplementary Figure 6: Less severe DNA damage in Faap20−/− than in Fanca−/− or Fancd2−/− MEFs. (A) Western blot was performed on the MEFs cells derived from WT, Faap20−/−, Fanca−/−, or Fancd2−/− mice, with or without the MMC (100nM for 16h) treatment. The blot using anti-FANCD2 (Novus) antibody show partial FANCD2 monoubiquitination in the Faap20−/− MEFs in comparison to non-FANCD2 ubiquitination in Fanca−/− MEFs and complete depletion of FANCD2 protein in the Fancd2−/− MEFs. The blot using anti-phospho-Histone H2A.X (Ser139) antibody (Millipore) indicates less phospho-Histone H2A.X (Ser139) in the Faap20−/− MEFs in comparison to Fanca−/− or Fancd2−/− MEFs. (B) The relative abundance of phospho-Histone H2A.X (Ser139) to Actin in (A) was analyzed through quantification of the western blot with ImageJ software. (C, D) DNA damage immunofluoresence staining with anti-phospho-Histone H2A.X (Ser139) antibody (Millipore) was performed on MEFs cells derived from WT, Faap20−/−, Fanca−/−, or Fancd2−/− mice, with or without the MMC (100nM for 16 h) treatment. Less DNA damage staining was detected in the Faap20−/− MEFs compared to Fanca−/−, Fancd2−/− cells. Scale bar, 10 µm. (D) The intensity of phospho-Histone H2A.X (Ser139) staining in (C) was quantified using ImageJ software.
Supplementary Figure 7: Less severe DNA damage induced by MMC in bone marrow HSPCs from Faap20−/− mice than Fanca−/− or Fancd2−/− mice. Bone marrow Lin-cells from WT, Faap20−/−, Fanca−/−, Fancd2−/− mice were treated with or without MMC (100nM for 16 h) and used for phospho-Histone H2A.X (Ser139) western blot (A, B) and immuno-staining (C, D). Scale bar, 5 µm. (B) The relative abundance of phospho-Histone H2A.X (Ser139) to Actin in (A) was analyzed through quantification of the western blot with ImageJ software. (D) The intensity of phospho-Histone H2A.X (Ser139) staining in (C) was quantified using ImageJ software. Faap20−/− cells show less severe DNA damage compared to Fanca−/− or Fancd2−/− cells.