Abstract
Non-coding antisense RNAs regulate bacterial genes in response to nutrition or environmental stress, and can be engineered for artificial gene control. The RNA chaperone Hfq accelerates antisense pairing between non-coding RNAs and their mRNA targets, by a mechanism still unknown. We used a photocaged guanosine derivative in an RNA oligonucleotide to temporally control Hfq catalyzed annealing. Using a fluorescent molecular beacon as a reporter, we observed RNA duplex formation within 15 s following irradiation (3 s) of photocaged RNA complexed with Hfq. The results showed that the Hfq chaperone directly stabilizes the initiation of RNA base pairs, and suggests a strategy for light-activated control of gene expression by non-coding RNAs.
Keywords: Hfq, non-coding RNA, photocaged nucleotides, p-hydroxyphenacyl, RNA chaperones
In bacteria, hundreds of small non-coding RNAs (sRNAs) regulate the expression of genes involved in metabolism, stress response, and virulence.[1] Many bacterial sRNAs act by base pairing directly with an mRNA target, altering its translation or its half-life.[2] The association of two complementary RNAs depends on their sequences and secondary structures, and is typically inefficient at the low mRNA concentrations in the cell. The bacterial RNA chaperone Hfq increases the rate of base pairing with mRNA targets, and stabilizes sRNA-mRNA complexes.[3] Herein, we investigate the mechanism of Hfq-catalyzed annealing using a photocaged guanosine that provides rapid, light-dependent control of RNA base pairing.
Hfq forms a ring-shaped homo-hexamer that specifically binds sRNAs and mRNAs.[4] An arginine patch on the rim of the hexamer catalyzes RNA annealing and strand displacement.[5] In our working model (Figure 1A), Hfq forms a transient ternary complex with two RNA strands, increasing helix initiation 103 to 104 times above the uncatalyzed rate.[5, 6] The remaining base pairs zipper, releasing double-stranded RNA. Although previous experiments suggested Hfq helps nucleate base pairing between RNA strands,[5, 6] how it does so is not understood.
Figure 1.
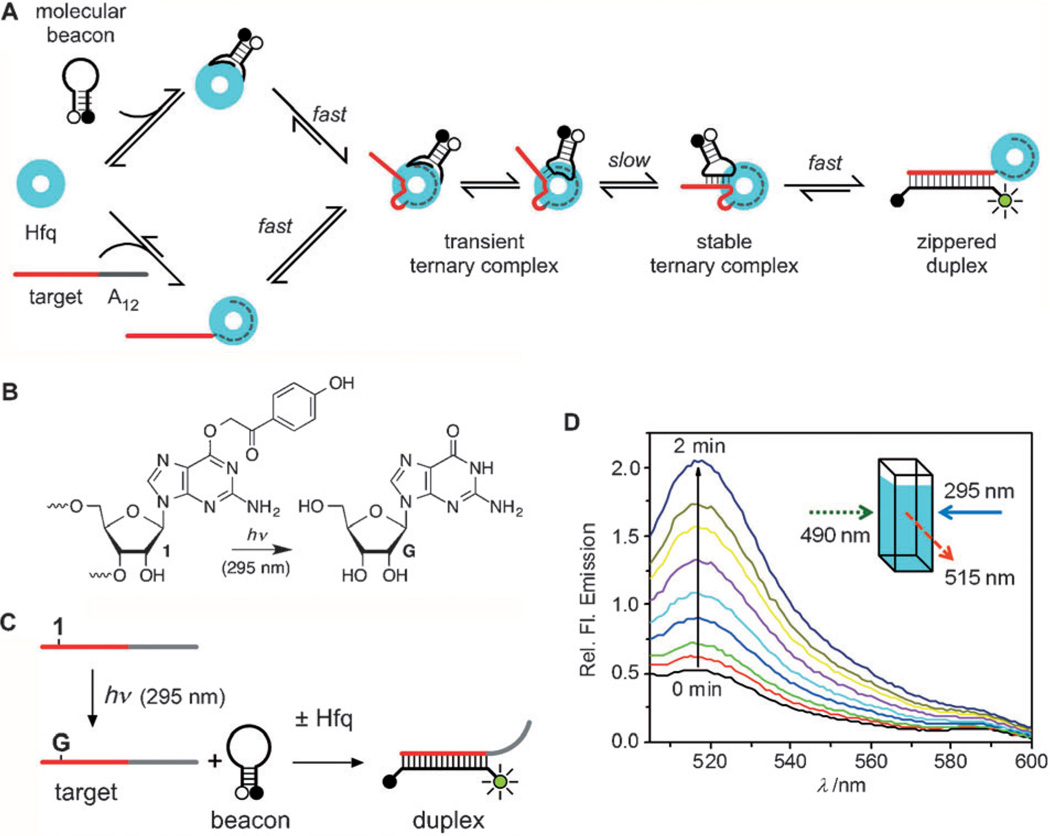
Photocaged control of RNA annealing. A) A working model for Hfq-catalyzed RNA annealing. This work shows Hfq directly stabilizes helix initiation complexes. B) Conversion of photocaged guanosine (1) to guanosine (G) by UV irradiation. C) Target RNA containing 1 does not anneal with a complementary molecular beacon until after photochemical uncaging. The fluorescence intensity of the FAM-labeled beacon reports the extent of annealing. D) Emission spectra of the beacon–target complexes after irradiation with 295 nm UV diodes.
We synthesized a target RNA containing a photocaged guanosine (1) that affords temporal control of the annealing reaction on the Hfq chaperone. Photocaged compounds have found numerous applications in diverse fields of chemistry and biology due to their ability to act as “ON/OFF” switches regulated by a specific wavelength of light.[7–11] To be useful in kinetic experiments, the uncaging reaction should be much faster than the molecular process under investigation.
In the present work, the photocaged guanosine utilizes the p-hydroxyphenacyl (pHP) photosolvolysis reaction (Figure 1B).[12] In contrast to the often used o-nitrobenzyl photoredox reaction, which proceeds through an intermediate that can exist for seconds to a minute, pHP photosolvolysis typically liberates its contents far more rapidly following excitation. The deprotection rate of pHP correlates inversely with the pKa of the conjugate acid of the leaving group. The rate constant for release of phenolate (phenol pKa≈10) is 108 s−1. Although the rate constant for guanine (pKa≈9) release is unknown, the similarity in pKa values between it and phenol suggested that a pHP caged guanosine would provide suitable temporal resolution for studying the effects of Hfq on RNA hybridization. We anticipated that the altered H-bonding pattern of the caged guanosine containing a pHP group at the O6 position, combined with the steric bulk of pHP group, would prevent RNA annealing (“OFF” state).
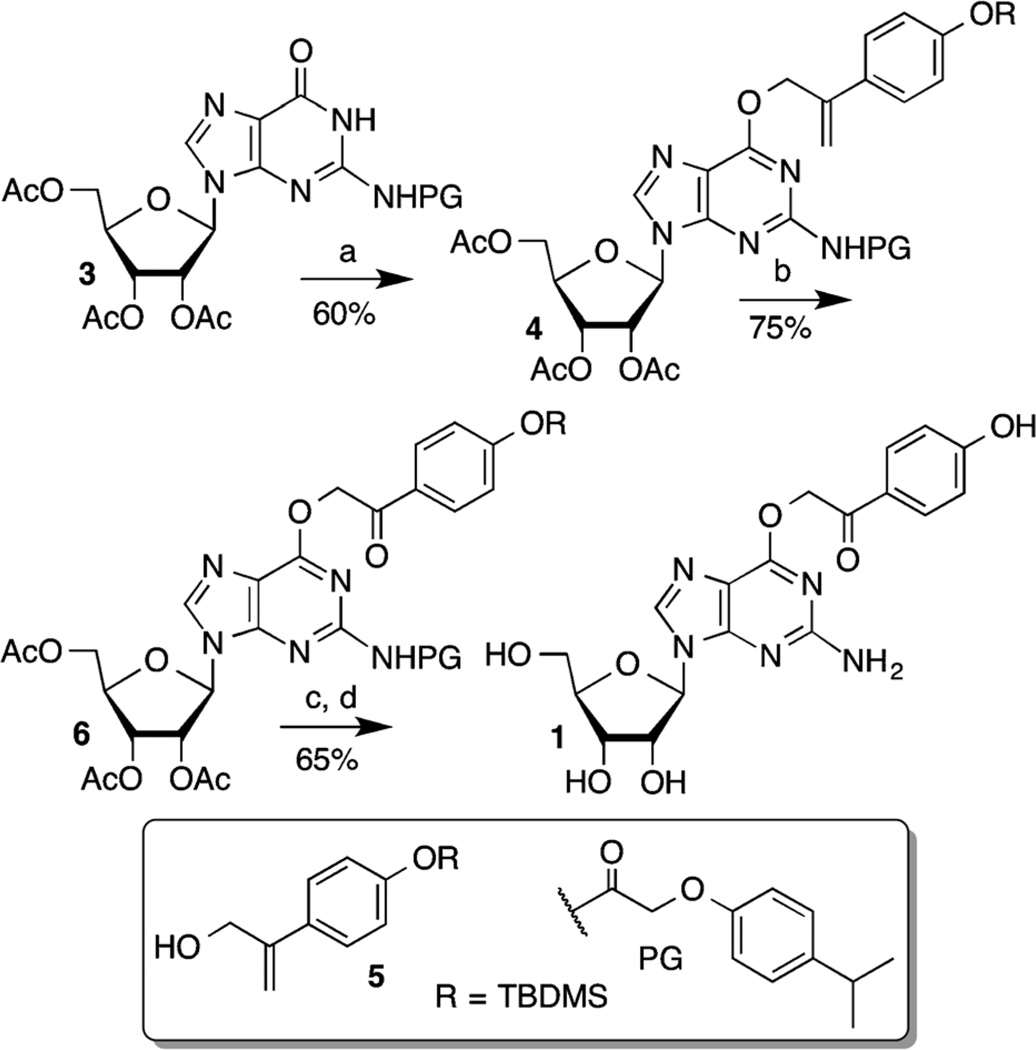
The syntheses of the photocaged guanosine nucleoside (1, Scheme 1) and corresponding phosphoramidite (2, Supporting information) began from 3. Various methods involving coupling the corresponding α-hydroxyacetophenone with 3 were unsuccessful. Ultimately, the p-hydroxyphenacyl group was introduced indirectly via a Mitsunobu reaction between 3 and allyl alcohol 5.[13] Nucleoside 1 was obtained from 6 via exhaustive deprotection following transformation of the terminal alkene (4) into the ketone (6) via a one-pot osmylation/periodate oxidation.[14] Photolysis of monomeric 1 produced guanosine in 60% yield. Oligonucleotides containing 1 were prepared via standard methods with the exception that 2 was coupled manually.
Scheme 1.
Synthesis of the photocaged guanosine nucleoside 1. a) DEAD, PPh3, 5, THF, −10 to 0°C; b) OsO4, NaIO4, 2,6-lutidine, dioxane/H2O, 25 °C; c) TABF, THF, 0°C; d) NH3, MeOH then NaOMe, MeOH, 0 to 25°C.
To measure the RNA annealing kinetics, we used a FAM-labeled molecular beacon and a complementary 16 nt target sequence (Figure 1C and Experimental Section).[15] A 3’-A12 extension of the target binds the distal face of Hfq (KD ≈ 0.1 nm).[16] As shown previously,[6] Hfq protein accelerated annealing of beacon and target RNAs from 0.06 s−1 to 2.1 s−1 (Supporting Information, Figure S12). To design a caged RNA target, we tested three different G to A substitutions (Supporting Information, Table S1) and found that a mismatch at position 4 reduced the extent of base pairing to 5%, with or without Hfq (Supporting Information, Figure S12). As anticipated, placement of 1 at position 4 of the target RNA also blocked duplex formation, judging from the small increase in the fluorescence signal of the beacon when the two RNAs were mixed (Figure 1D). Irradiation at 295 nm sharply increased the fluorescence emission, owing to base pairing with the uncaged target. Although the half-life for uncaging was 20 s (Supporting Information, Figure S13), subsequent experiments used a 3 s UV pulse, which provided sufficient (1 nm) sensitivity and time resolution.
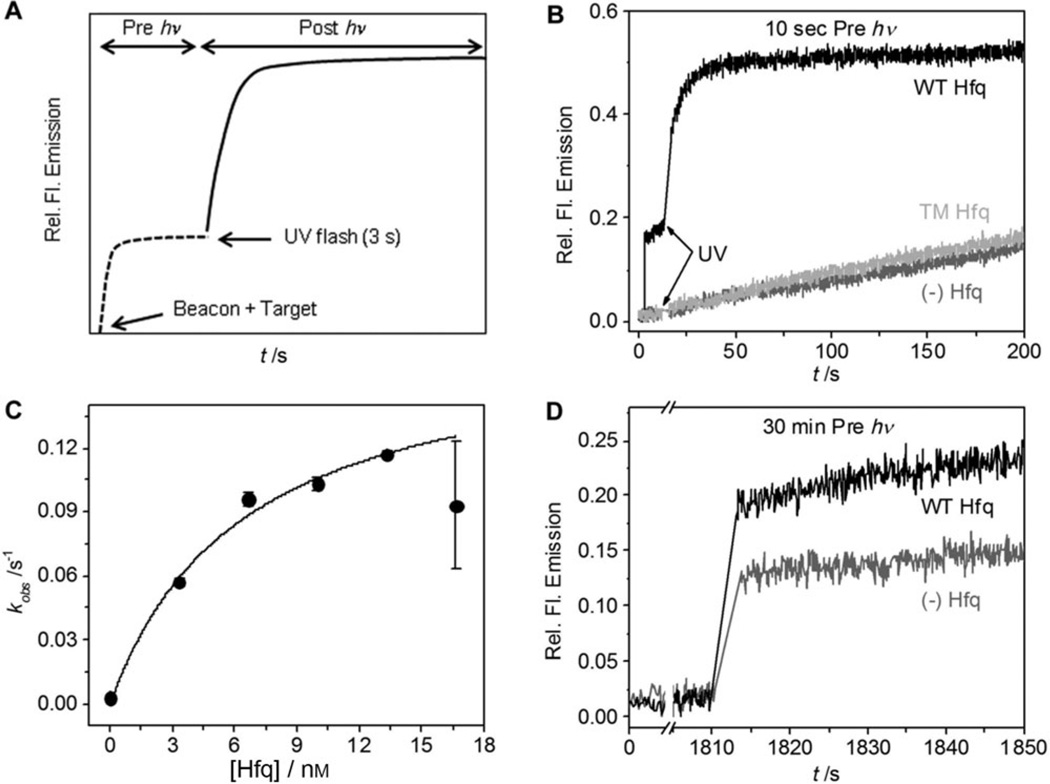
To determine whether 1 blocks annealing by the Hfq chaperone, we combined beacon (10 nm) and photocaged target RNA (10 nm) with or without 10 nm Hfq hexamer and recorded the fluorescence emission at 515 nm. After 10 s, samples were flashed with UV light for 3 s, and the growth in fluorescence recorded for a further 200 s (Figure 2 A). The uncaged target RNA annealed with the molecular beacon 35 times faster in the presence of Hfq (kobs=0.1 s−1; Figure 2B) than without the chaperone (kobs=0.003 s−1; Figure 2B). A small increase in fluorescence intensity before the flash (black, Figure 2B) indicated that the molecular beacon and target RNA bound Hfq during the 10 s pre-incubation, as expected.[6]
Figure 2.
Hfq catalyzed annealing. A) Representation of the experimental procedure. 10 nm molecular beacon and 10 nmcaged RNA were incubated (Pre hv) with or without Hfq before 3 s UV flash (Post hv). B) Samples were irradiated 10 s after mixing with no Hfq (gray), 10 nm WT Hfq (black), and 10 nm inactive TM Hfq (light gray). C) Rate constants for RNA annealing (Post hv) as in B versus [Hfq] hexamer. The data points were fitted into a two-state binding equation with an apparent Kd=6.5 nm. D) Kinetic traces of samples irradiated after 30 min of mixing; no Hfq, gray; 10 nm Hfq, black.
Hfq increased the RNA annealing rate and stabilized the beacon-target duplex, as manifested by higher reaction endpoints (Figure 2B). This enhancement depended on the specific chaperone activity of Hfq, as no burst was observed in experiments performed with an inactive triple mutant (TM) lacking the arginine patch that catalyzes annealing (Figure 2B). The rate of RNA annealing after uncaging increased with Hfq concentration, and reached saturation when the concentration of Hfq equaled the concentration of target RNA (Figure 2C; Supporting Information, Figure S14) in agreement with previous measurements of the annealing kinetics.[6] This required recruitment of Hfq to the RNA, as the maximum annealing rate was three times lower when the target RNA lacked the A12 binding site (Supporting Information, Figure S15).
Surprisingly, when the target and beacon were equilibrated for 30 min before the UV flash, base pairing occurred within a few seconds after UV irradiation (Figure 2D). This rapid annealing was observed with or without Hfq, although Hfq increased the yield of double-stranded RNA as before (Figure 2D; Supporting Information, Figure S14). Although helix initiation is energetically unfavorable, we deduced the beacon and caged target slowly form partially base-paired intermediates that are poised to anneal as soon as the caged RNA is photolyzed. This partial base pairing does not unfold the beacon, as the fluorescence remained low until the target was uncaged (Supporting Information, Figure S16).
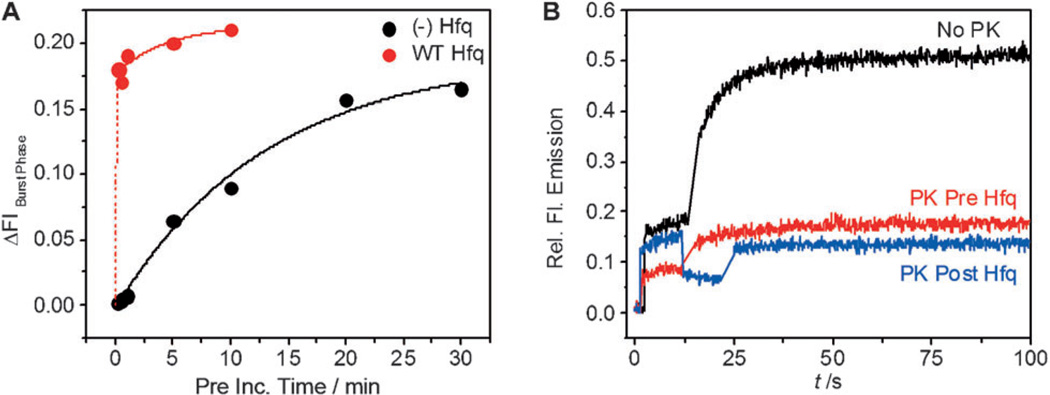
Hfq may lower the transition state free energy for RNA annealing by creating stable helix initiation complexes. To test this, we varied the pre-incubation time of the components before the target RNA was photolyzed. Without Hfq, a burst of annealing appeared as the pre-flash incubation was lengthened (Supporting Information, Figure S17), and the amplitude of the burst increased with kobs=0.07 min−1 with respect to pre-incubation time (black; Figure 3A). This is equal within error to kobs for annealing uncaged target RNA (Supporting Information, Figure S12A). In the presence of Hfq, the amplitude of the burst phase rose dramatically after just 10 s pre-incubation (red; Figure 3A and Supporting Information, Figure S18), indicating that most of the RNA-Hfq complexes were competent to base pair once the target was uncaged. Since 30 min was needed to reach a similar fraction of reactive intermediate without Hfq, this corresponds to a 180-fold increase in the effective rate of helix initiation.
Figure 3.
Hfq stabilizes helix initiation. A) Burst of annealing following uncaging, after different pre-incubation times, was fit to an exponential rate equation with kobs=0.07 min−1, no Hfq; >10 min−1, 10 nm Hfq. B) Samples treated with proteinase K (PK) before uncaging; no treatment, black; after 10 s Pre hv, blue; before adding Hfq, red.
To ask whether Hfq is still needed to facilitate base pairing after the helix initiation takes place, we removed Hfq with proteinase K (PK) at different stages of the annealing reaction. The target and beacon RNAs were pre-incubated with Hfq for 10 s as usual to form the helix initiation complexes (Figure 3B). When proteinase K was added for 10 s before photolysis, the fluorescence intensity dropped to the no protein background, suggesting that Hfq was no longer bound to the RNAs (blue; Figure 3B). After the UV flash, only a small increase in fluorescence was observed, showing that the protease destroyed the helix initiation complexes. Proteinase K had no effect on the RNA in the absence of Hfq (Supporting Information, Figure S19A). A control reaction in which proteinase K was added to the RNAs before Hfq also yielded little RNA duplex (red; Figure 3B). Semi-native PAGE confirmed that Hfq was digested under these conditions (Supporting Information, Figure S19B). Thus, Hfq must remain bound to the target RNA at the time of photolysis, to convert the helix initiation complexes into duplex product.
RNA binding proteins such as Hfq have been proposed to passively chaperone RNA interactions by neutralizing the negative charge of the RNA or by simultaneously binding more than one RNA strand.[17] By trapping helix initiation complexes with photocaged RNA, we show that Hfq directly overcomes the energetic barrier for nucleating the RNA helix, explaining its potent activity as an RNA chaperone (Figure 1A). That the complexes become more stable over time, and that Hfq is required at the moment of uncaging, suggest that Hfq also facilitates zippering of additional base pairs, in contrast to previous models.[2, 18] Future experiments will test if the extent of zippering depends on the location of the caged base. Bacterial sRNAs have modular structures that can be targeted to new genes,[19, 20] suggesting an approach for activating Hfq-dependent sRNAs with light. Our results demonstrate that photosolvolysis of pHP enables light-activated control of rapid biopolymer transitions such as base pairing, providing new insight into the mechanism of biological regulation.
Experimental Section
Compound 1 and caged RNA, 5’-GUG1UCAGUCGAGUGGA12, were synthesized and characterized as described in Supporting Information. The molecular beacon was 5’-6-FAM-GGUCCCCCACUCGACUCACCACCGGACC-DABCYL (Trilink). Light-triggered RNA annealing was performed in a Fluorolog-3 (Horiba) spectrofluorometer modified as described in the Supporting Information. Molecular beacon and caged target RNA (10 nm each) were combined in 500 µL 10mm Tris-HCl pH 7.5, 50 mm NaCl, 50 mm KCl (TNK) at room temperature before uncaging with 3 s irradiation at 295 nm (35 mW). FAM emission was recorded at 515 nm (see the Supporting Information) and normalized to the maximum change after 1 min irradiation. The time origin was defined by the moment of mixing. For protease treatment, 5 µL (4 U) proteinase K was added at the times indicated. Hfq concentrations are given per hexamer.
Supplementary Material
Footnotes
This work was supported by the NIH (R01 GM46686 to S.W. and R01 GM54996 to M.M.G.)
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201501658.
Contributor Information
Dr. Subrata Panja, T.C. Jenkins Department of Biophysics, Johns Hopkins University 3400 N. Charles St., Baltimore MD 21218 (USA)
Dr. Rakesh Paul, Department of Chemistry, Johns Hopkins University 3400 N. Charles St., Baltimore MD 21218 (USA)
Prof. Marc M. Greenberg, Email: mgreenberg@jhu.edu, Department of Chemistry, Johns Hopkins University 3400 N. Charles St., Baltimore MD 21218 (USA).
Prof. Sarah A. Woodson, Email: swoodson@jhu.edu, T.C. Jenkins Department of Biophysics, Johns Hopkins University 3400 N. Charles St., Baltimore MD 21218 (USA).
References
- 1.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Cold Spring Harbor Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G, Opdyke JA, Zhang A. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Vogel J, Luisi BF. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan RG, Link TM. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Panja S, Schu DJ, Woodson SA. Nucleic Acids Res. 2013;41:7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopkins JF, Panja S, Woodson SA. Nucleic Acids Res. 2011;39:5193–5202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schäfer F, Wagner J, Knau A, Dimmeler S, Heckel A. Angew. Chem. Int. Ed. 2013;52:13558–13561. doi: 10.1002/anie.201307502. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013;125:13801–13805. [Google Scholar]
- 8.Govan JM, Uprety R, Thomas M, Lusic H, Lively MO, Deiters A. ACS Chem. Biol. 2013;8:2272–2282. doi: 10.1021/cb400293e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höbartner C, Silverman SK. Angew. Chem. Int. Ed. 2005;44:7305–7309. doi: 10.1002/anie.200502928. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2005;117:7471–7475. [Google Scholar]
- 10.Tang X, Dmochowski IJ. Angew. Chem. Int. Ed. 2006;45:3523–3526. doi: 10.1002/anie.200600954. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2006;118:3603–3606. [Google Scholar]
- 11.Resendiz MJE, Schön A, Freire E, Greenberg MM. J. Am. Chem. Soc. 2012;134:12478–12481. doi: 10.1021/ja306304w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klán P, Solomek T, Bochet CG, Blanc A, Givens R, Rubina M, Popik V, Kostikov A, Wirz J. Chem. Rev. 2013;113:119–191. doi: 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong W, Liu Y, Xue J, Xie Z, Li Y. Chem. Lett. 2012;41:1597–1599. [Google Scholar]
- 14.Yu W, Mei Y, Kang Y, Hua Z, Jin Z. Org. Lett. 2004;6:3217–3219. doi: 10.1021/ol0400342. [DOI] [PubMed] [Google Scholar]
- 15.Panja S, A.Woodson S. Nucleic Acids Res. 2012;40:8690–8697. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Haseth PL, Uhlenbeck OC. Biochemistry. 1980;19:6138–6146. doi: 10.1021/bi00567a029. [DOI] [PubMed] [Google Scholar]
- 17.Woodson SA. RNA Biol. 2010;7:677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauer E. RNA Biol. 2013;10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beisel CL, Storz G. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee T, Feig AL. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.