Abstract
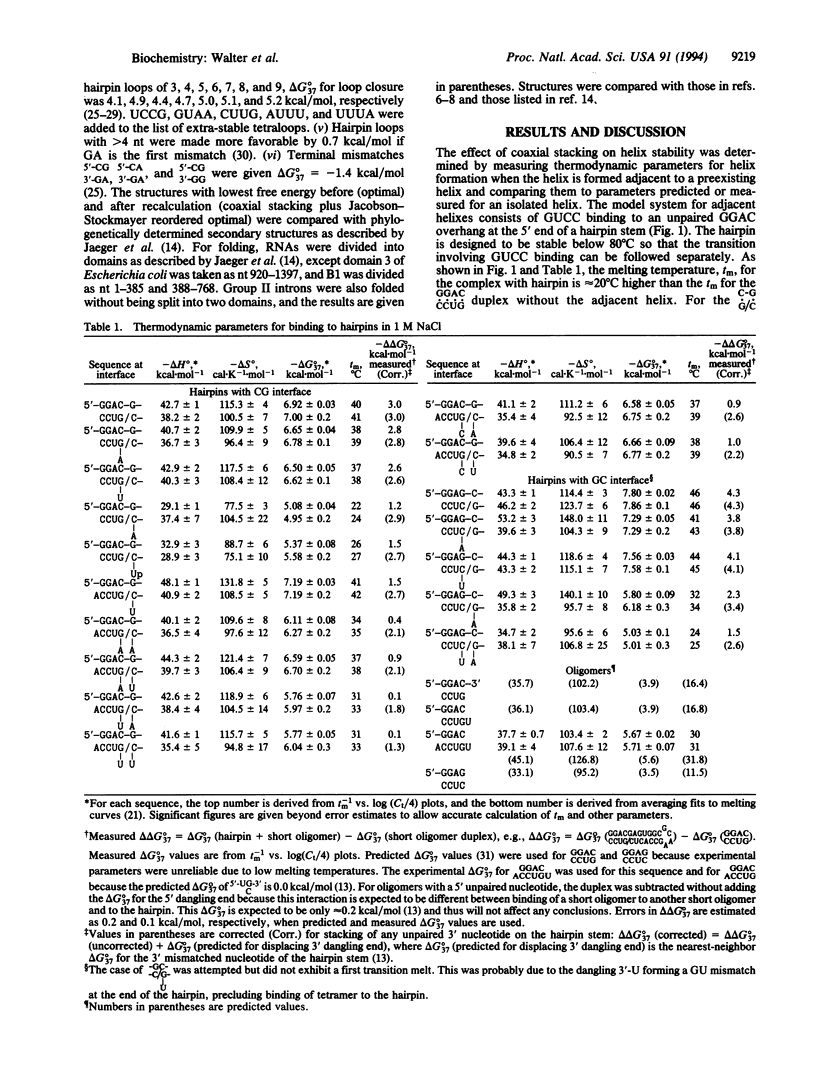
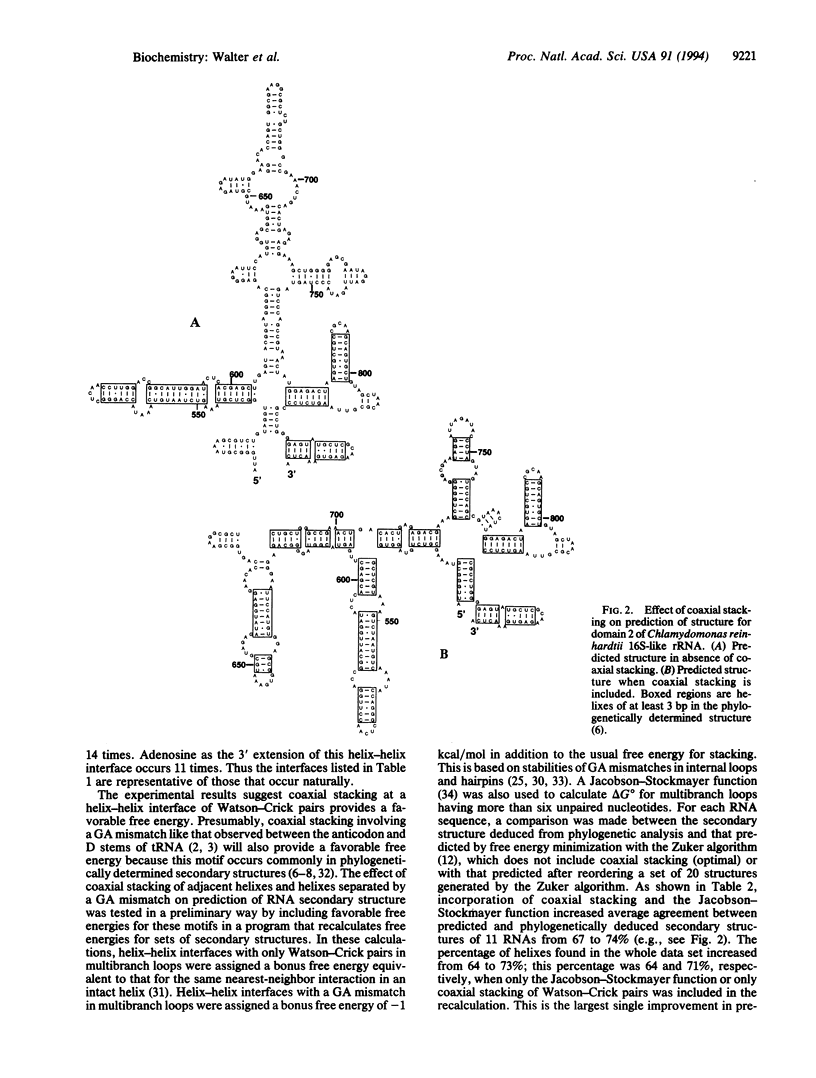
An RNA model system consisting of an oligomer binding to a 4-nt overhang at the 5' end of a hairpin stem provides thermodynamic parameters for helix-helix interfaces. In a sequence-dependent manner, oligomers bind up to 1000-fold more tightly adjacent to the hairpin stem than predicted for binding to a free tetramer at 37 degrees C. For the interface (/) in [formula: see text] additional free energy change, delta delta G 37 degrees, for binding is roughly the nearest-neighbor delta G 37 degrees for propagation of an uninterrupted helix of equivalent sequence, CGGC. When X and Z are omitted, the delta delta 37 degrees is even more favorable by approximately 1 kcal/mol (1 cal = 4.184J). On average, predictions of 11 RNA secondary structures improve from 67 to 74% accuracy by inclusion of similar stacking contributions.
Full text
PDF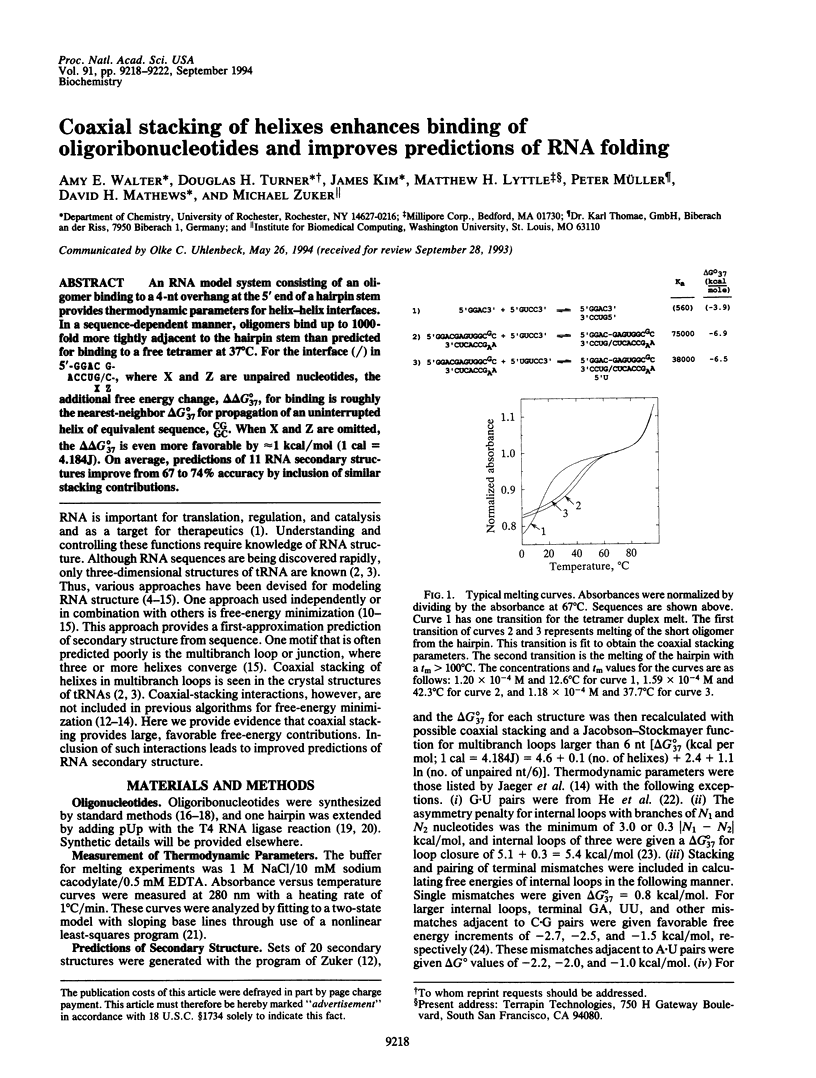


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bevilacqua P. C., Turner D. H. Comparison of binding of mixed ribose-deoxyribose analogues of CUCU to a ribozyme and to GGAGAA by equilibrium dialysis: evidence for ribozyme specific interactions with 2' OH groups. Biochemistry. 1991 Nov 5;30(44):10632–10640. doi: 10.1021/bi00108a005. [DOI] [PubMed] [Google Scholar]
- Coutts S. M., Gangloff J., Dirheimer G. Conformational transitions in tRNA Asp (brewer's yeast). Thermodynamic, kinetic, and enzymatic measurements on oligonucleotide fragments and the intact molecule. Biochemistry. 1974 Sep 10;13(19):3938–3948. doi: 10.1021/bi00716a019. [DOI] [PubMed] [Google Scholar]
- Delisi C., Crothers D. M. Prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 1971 Nov;68(11):2682–2685. doi: 10.1073/pnas.68.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker D. J., Vickers T. A., Bruice T. W., Freier S. M., Jenison R. D., Manoharan M., Zounes M. Pseudo--half-knot formation with RNA. Science. 1992 Aug 14;257(5072):958–961. doi: 10.1126/science.1502560. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978 May 30;17(11):2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Groebe D. R., Uhlenbeck O. C. Characterization of RNA hairpin loop stability. Nucleic Acids Res. 1988 Dec 23;16(24):11725–11735. doi: 10.1093/nar/16.24.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994 Mar;58(1):10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S-like) ribosomal RNA sequences presented in a secondary structure format. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2319–2330. doi: 10.1093/nar/18.suppl.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- He L., Kierzek R., SantaLucia J., Jr, Walter A. E., Turner D. H. Nearest-neighbor parameters for G.U mismatches: [formula; see text] is destabilizing in the contexts [formula; see text] and [formula; see text] but stabilizing in [formula; see text]. Biochemistry. 1991 Nov 19;30(46):11124–11132. doi: 10.1021/bi00110a015. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Pace N. R. Phylogenetic comparative analysis of RNA secondary structure. Methods Enzymol. 1989;180:227–239. doi: 10.1016/0076-6879(89)80104-1. [DOI] [PubMed] [Google Scholar]
- Kieleczawa J., Dunn J. J., Studier F. W. DNA sequencing by primer walking with strings of contiguous hexamers. Science. 1992 Dec 11;258(5089):1787–1791. doi: 10.1126/science.1465615. [DOI] [PubMed] [Google Scholar]
- Kierzek R., Caruthers M. H., Longfellow C. E., Swinton D., Turner D. H., Freier S. M. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986 Dec 2;25(24):7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Cech T. R. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Leontis N. B., Kwok W., Newman J. S. Stability and structure of three-way DNA junctions containing unpaired nucleotides. Nucleic Acids Res. 1991 Feb 25;19(4):759–766. doi: 10.1093/nar/19.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Guo Q., Marky L. A., Seeman N. C., Kallenbach N. R. Thermodynamics of DNA branching. J Mol Biol. 1992 Feb 5;223(3):781–789. doi: 10.1016/0022-2836(92)90989-w. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. Folding of circularly permuted transfer RNAs. Science. 1991 Nov 29;254(5036):1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Peritz A. E., Kierzek R., Sugimoto N., Turner D. H. Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry. 1991 Jul 2;30(26):6428–6436. doi: 10.1021/bi00240a013. [DOI] [PubMed] [Google Scholar]
- Petersheim M., Turner D. H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983 Jan 18;22(2):256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- Riesner D., Maass G., Thiebe R., Philippsen P., Zachau H. G. The conformational transitions in yeast tRNAPhe as studied with tRNAPhe fragments. Eur J Biochem. 1973 Jul 2;36(1):76–88. doi: 10.1111/j.1432-1033.1973.tb02887.x. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992 Apr 10;256(5054):217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Stabilities of consecutive A.C, C.C, G.G, U.C, and U.U mismatches in RNA internal loops: Evidence for stable hydrogen-bonded U.U and C.C.+ pairs. Biochemistry. 1991 Aug 20;30(33):8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- Serra M. J., Lyttle M. H., Axenson T. J., Schadt C. A., Turner D. H. RNA hairpin loop stability depends on closing base pair. Nucleic Acids Res. 1993 Aug 11;21(16):3845–3849. doi: 10.1093/nar/21.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Cameron V. Equimolar addition of oligoribonucleotides with T4 RNA ligase. Nucleic Acids Res. 1977 Jan;4(1):85–98. doi: 10.1093/nar/4.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Y., Lyttle M. H., Borer P. N. Enzymatic and NMR analysis of oligoribonucleotides synthesized with 2'-tert-butyldimethylsilyl protected cyanoethylphosphoramidite monomers. Nucleic Acids Res. 1990 Jun 11;18(11):3347–3352. doi: 10.1093/nar/18.11.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Ogilvie K. K., Pon R. T. Prevention of chain cleavage in the chemical synthesis of 2'-silylated oligoribonucleotides. Nucleic Acids Res. 1989 May 11;17(9):3501–3517. doi: 10.1093/nar/17.9.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J. R., Puglisi J. D., Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J Mol Biol. 1990 Jul 20;214(2):455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- Zuker M., Jaeger J. A., Turner D. H. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991 May 25;19(10):2707–2714. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]



