Abstract
Processing of rRNA during ribosome assembly can proceed through alternative pathways but it is unclear whether this could affect the structure of the ribosome. Here, we demonstrate that shortage of a ribosomal protein can change pre-rRNA processing in a way that over time alters ribosome diversity in the cell. Reducing the amount of Rpl17 in mouse cells led to stalled 60S subunit maturation, causing degradation of most of the synthesized precursors. A fraction of pre-60S subunits, however, were able to complete maturation, but with a 5′-truncated 5.8S rRNA, which we named 5.8SC. The 5′ exoribonuclease Xrn2 is involved in the generation of both 5.8SC and the canonical long form of 5.8S rRNA. Ribosomes containing 5.8SC rRNA are present in various mouse and human cells and engage in translation. These findings uncover a previously undescribed form of mammalian 5.8S rRNA and demonstrate that perturbations in ribosome assembly can be a source of heterogeneity in mature ribosomes.
Keywords: ribosome, ribosomal protein, ribosome biogenesis, mammalian cells, RNA processing
INTRODUCTION
Eukaryotic ribosome biogenesis is a dynamic biosynthetic process in which 4 rRNAs and ∼80 ribosomal proteins (r-proteins) are assembled in a complex series of steps to generate small (40S) and large (60S) ribosomal subunits (Woolford and Baserga 2013; Henras et al. 2015). Incorporation of r-proteins into preribosomes in vivo follows a broadly hierarchical order, according to which these proteins can be divided into several earlier- and later-acting assembly groups (Auger-Buendia et al. 1979; Lastick 1980; Todorov et al. 1983; O'Donohue et al. 2010; Chen and Williamson 2013; Gamalinda et al. 2014). Early-binding r-proteins promote subsequent assembly steps by facilitating structural rearrangements in pre-rRNA and enabling binding of assembly factors (Ferreira-Cerca et al. 2007; Babiano and de la Cruz 2010; Jakob et al. 2012; Ohmayer et al. 2013). Studies in different species have demonstrated that lack of an essential r-protein stalls maturation of ribosomal precursors, which are then degraded by the nucleolar surveillance machinery (Moritz et al. 1990; Robledo et al. 2008; Pöll et al. 2009; Jakovljevic et al. 2012; Gamalinda et al. 2013).
Whereas a complete genetic ablation of individual r-proteins leads to severe growth defects and organismal lethality, with only a few known exceptions (Kirn-Safran et al. 2007; McIntosh et al. 2011; Babiano et al. 2012; Steffen et al. 2012; O'Leary et al. 2013), their suboptimal expression often results in complex pathological phenotypes. In humans, reduced expression or partial loss of function in a number of r-proteins are associated with a group of congenital disorders termed ribosomopathies, characterized by impaired proliferation and increased cell death in hematopoietic progenitors and certain other cell lineages (Ellis and Gleizes 2011; Raiser et al. 2014). On a subcellular level, defects in ribosome biogenesis lead to reduced protein synthesis capacity and misregulated translational controls (Horos et al. 2012; Teng et al. 2013; Ludwig et al. 2014). In addition, perturbations in ribosome biogenesis can activate the tumor suppressor p53, triggering lineage-specific cell cycle arrest or apoptosis (for review, see Bursac et al. 2014). Although these are fairly consistent molecular consequences of defects in ribosome biogenesis, the clinical manifestations of ribosomopathies are far from uniform (Armistead and Triggs-Raine 2014). The reasons for the phenotypical variability of r-protein deficiencies are only partially understood. Current hypotheses proposed to explain this phenomenon include extra-ribosomal functions of r-proteins, association of nonribosomal regulatory proteins with preribosomes, and the synthesis of specialized ribosomes with an altered r-protein composition (Kondrashov et al. 2011; Thapa et al. 2013).
Studies over several decades, particularly in animal cells, have shown that processing of pre-rRNA during eukaryotic ribosome maturation can occur through parallel and alternative pathways (Dudov et al. 1978; Bowman et al. 1981; Hadjiolova et al. 1993; Sloan et al. 2013; Henras et al. 2015). Both physiological stimuli and experimentally induced deficiencies in factors required for ribosome biogenesis can alter the kinetics of individual processing steps, changing the relative contribution of different pathways (Savino and Gerbi 1990; Lapik et al. 2004; Wang et al. 2014). An outstanding question raised by these observations is whether different routes in preribosome maturation always converge to yield a singular kind of ribosomes or may lead to ribosomes with nonidentical structural and functional properties. In the study presented here, we used mouse cells to analyze depletion of the r-protein L17 (also known as uL22) that has an essential role in ribosome architecture and function. L17/uL22 interacts with all six secondary structure domains in 28S rRNA and with 5.8S rRNA (Ben-Shem et al. 2011; Gamalinda et al. 2014), and is thought to stabilize long-range interactions important for establishing the structure of the polypeptide exit tunnel during 60S subunit assembly (Gamalinda et al. 2013). As shown below, Rpl17 deficiency in mouse cells resulted in marked changes in the way the 5′ end of the 32S pre-rRNA is generated, and caused instability of this precursor. Significantly, degradation of pre-rRNA intermediates was not the only outcome of the aberrant ribosome biosynthesis that we observed. A fraction of pre-60S subunits were able to successfully compete for the residual Rpl17 present in the cell and complete maturation. Such subunits, however, possessed a shortened form of 5.8S rRNA, which we termed 5.8SC. We further show that this rRNA exists as a minor 5.8S form in unstressed mouse and human cell lines and is present in translating polysomes. The structural implications of the newly discovered 5′ end heterogeneity in mammalian 5.8S rRNA are discussed.
RESULTS AND DISCUSSION
Rpl17 is required for pre-rRNA processing in two internal transcribed spacers
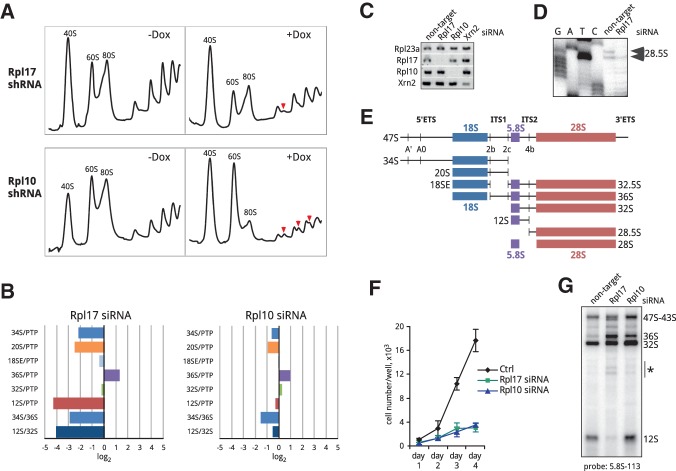
Previous biochemical studies in yeast indicated that the essential r-protein L17 belongs to a “middle-acting” group of r-proteins necessary for processing of 5.8S/25S rRNA and recruitment of ribosome assembly factors required for late stages of 60S subunit maturation (Gamalinda et al. 2013, 2014). To address the role of Rpl17 in pre-rRNA processing in a mammalian system, we established stable 3T3 cell lines carrying doxycycline-inducible Rpl17 shRNA. Consistent with a role in 60S biogenesis, depletion of mouse Rpl17 led to diminished 60S subunit levels and a reduction in translating polysomes (Fig. 1A, top). To identify the stages in 60S subunit maturation affected by Rpl17 depletion, we examined relative levels of various rRNA processing intermediates using the ratio analysis of multiple precursors (RAMP) (Wang et al. 2014) in cells transfected with gene-specific siRNAs (Fig. 1C). To generate a RAMP profile for Rpl17 knockdown, we normalized pre-rRNA ratios in cells transfected with siRNA targeting Rpl17 to the corresponding ratios in cells transfected with nontargeting siRNA. The observed alterations in pre-rRNA ratios (Fig. 1B, left) indicated impaired pre-rRNA processing in internal transcribed spacers 1 and 2 (ITS1 and ITS2). First, ratios of pre-rRNAs to PTP (“primary transcript plus,” the combined early 47S-45S pre-rRNAs) (Wang et al. 2014) showed lowered levels of 34S and 20S pre-rRNAs, which are generated through cleavage at site 2c in ITS1 (Fig. 1E), whereas 18SE and 36S pre-rRNAs, which require site 2b cleavage in ITS1, were only weakly affected or increased (Fig. 1B). This indicated that Rpl17 deficiency inhibited ITS1 cleavage at the 5.8S-proximal site 2c, forcing the splitting of the pre-rRNA transcript through site 2b, similar to other previously studied 60S synthesis factors (Wang et al. 2014). Second, the strongly reduced 12S/PTP and 12S/32S ratios (Fig. 1B) suggested that Rpl17 deficiency also inhibited cleavage at site 4b within ITS2, the spacer that separates 5.8S and 28S rRNAs. Primer extension analysis corroborated this result by showing that the 3′ product of site 4b cleavage, 28.5S pre-rRNA, was decreased in Rpl17-depleted cells (Fig. 1D).
FIGURE 1.
Effects of Rpl17 and Rpl10 knockdowns on translation and ribosome biogenesis. (A) Depletion of Rpl17 and Rpl10 results in distinct alterations in polysome profiles. Cells stably transfected with doxycycline-inducible shRNAs against Rpl17 and Rpl10 were treated with doxycycline for 48 h or remained untreated (+Dox, −Dox). Cell lysates were centrifuged through 10%–45% (w/w) sucrose gradients and fractionated with the continuous measurement of absorbance at 254 nm. Halfmers are marked with red arrowheads. (B) RAMP profiles of Rpl17 and Rpl10 knockdowns showing changes in steady-state levels of pre-rRNA intermediates 48 h after siRNA transfections. Total cellular RNA was extracted and analyzed by northern hybridizations, ratios of various rRNA intermediates in knockdowns were normalized to the corresponding ratios in control cells transfected with nontargeting siRNA (i.e., log2 = 0 indicates no change relative to control cells; see Wang et al. [2014] for further details on this assay). (PTP) Primary transcript plus, early 47–45S pre-rRNAs. (C) RT-PCR analysis of Rpl17, Rpl10, and Xrn2 knockdowns; Rpl23a was used as a reference gene. (D) Primer extension to examine levels of 28.5S pre-rRNA, a short-lived product of ITS2 cleavage at site 4b. (E) Schematic representation of the mouse 47S pre-rRNA transcript with cleavage sites and major pre-rRNA processing intermediates indicated. (ETS) External transcribed spacer; (ITS) internal transcribed spacer. (F) Knockdown of Rpl17 or Rpl10 severely inhibits cell proliferation. Cells were transfected with nontargeting siRNA (Ctrl), or siRNAs against Rpl17 and Rpl10 siRNA. Cell numbers were determined in triplicate transfections; error bars indicate SD. (G) Depletion of Rpl17 and Rpl10 results in altered 36S and 12S pre-rRNA levels and accumulation of pre-rRNA decay products (asterisk). Total RNA was analyzed by a northern hybridization at 48 h after transfection with the indicated siRNAs.
Defects in pre-rRNA processing caused by deficiency in an r-protein may result not only from the protein's direct involvement in ribosome assembly, but also from translational inhibition of the synthesis of other ribosomal components and/or slower recycling of ribosome assembly factors. To address this possibility, we analyzed depletion of another essential r-protein, Rpl10 (uL16). Unlike Rpl17, Rpl10 is incorporated into nascent 60S subunits after their cytoplasmic export, as shown by previous studies in both mammalian cells (Auger-Buendia et al. 1979; Lastick 1980) and yeast (Saveanu et al. 2003; West et al. 2005; Ohmayer et al. 2013). Depletion of Rpl10 severely inhibited cell growth, as did depletion of Rpl17 (Fig. 1F); however, 60S subunit levels were not significantly reduced in Rpl10-depleted cells (Fig. 1A). Instead, the gradient analysis of cytoplasmic ribosomes revealed an increase in free subunits, while the amount of translating ribosomes was greatly reduced. In addition, prominent halfmer peaks, indicative of polyribosomes containing an extra 40S subunit were observed (Fig. 1A, bottom), consistent with an essential role of Rpl10 in subunit joining during translation (Eisinger et al. 1997). RAMP profiling of the Rpl10 knockdown showed changes in rRNA precursors that were much less pronounced in comparison with Rpl17 knockdown (Fig. 1B), but nevertheless they also pointed to a slightly reduced frequency of pre-rRNA splitting at site 2c. Because Rpl10 is recruited at a late cytoplasmic step of ribosome maturation and not expected to play a direct role in pre-rRNA processing, the observed mild effects on ITS1 cleavage may be due to negative feedback from late stages of ribosome maturation and/or translation. In contrast to Rpl17, Rpl10 depletion did not alter significantly the 12S/PTP and 12S/32S ratios, indicating normal ITS2 processing (Fig. 1B).
The combined data from the Rpl17 and Rpl10 knockdowns lead us to conclude that Rpl17 is required for processing of the 32S pre-rRNA to 28S and 5.8S precursors during pre-60S subunit assembly in mouse cells. This function of mammalian Rpl17 in the context of assembly parallels the function of yeast L17 (Gamalinda et al. 2013). The mouse 32S pre-rRNA, however, did not significantly accumulate in Rpl17-depleted cells, as evidenced by the lack of increase in the 32S/PTP ratio (Fig. 1B). Northern hybridizations revealed an elevated amount of pre-rRNA degradation products after Rpl17 knockdown (Fig. 1G), indicating active turnover of 32S pre-rRNA in pre-60S complexes. In addition to the abortive maturation of 32S pre-rRNA, earlier cleavage at site 2c was affected to a higher extent than what can be attributed to translational inhibition, indicating that Rpl17 assembly also promotes the splitting of the pre-rRNA transcript through the 5.8S-proximal site 2c in ITS1.
Rpl17 deficiency alters 5′ end processing of 5.8S rRNA precursors
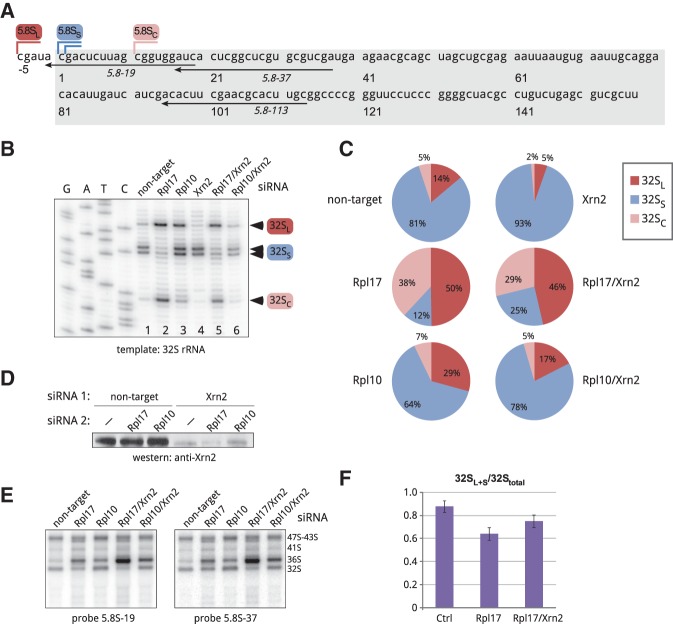
In previously studied eukaryotic species, 5.8S rRNA was found to exist in two forms, long and short, which in yeast result from the use of alternative processing pathways (Henry et al. 1994). The major short form of mammalian 5.8S rRNA, 5.8SS, exhibits a one-nucleotide heterogeneity of its 5′ ends, as illustrated in Figure 2A, while the less abundant long species 5.8SL has a 5–6-nucleotide 5′ extension. As the above results show, 32S pre-rRNA is the last 5.8S/28S precursor that can be formed in Rpl17-depleted mouse cells. Our attempts to examine 32S pre-rRNA by primer extensions were initially hampered by a failure of reverse transcriptases to extend ITS2-specific primers beyond a strong GC-rich hairpin formed in 5.8S rRNA, whereas primers hybridizing upstream of this structure primarily extended the abundant mature 5.8S rRNA. To circumvent this problem, we separated large pre-rRNAs from 5.8S rRNA by electrophoresis in guanidine thiocyanate-containing agarose gels (Goda and Minton 1995). Guanidine thiocyanate efficiently denatures RNA without introducing chemical modifications into RNA molecules and allows recovery of RNA suitable for primer extensions. Primer extension with 32S pre-rRNA (Fig. 2B, lane 1) revealed a doublet of stops that matched the 5′ ends of the major short form of mammalian 5.8S rRNA (Fig. 2A) and a 5–6-nt extended species; the corresponding pre-rRNAs were designated 32SS and 32SL, respectively. Depletion of Rpl17 greatly reduced 32SS pre-rRNA and increased 32SL pre-rRNA (Fig. 2B, lane 2). Thus, the major short 5′ ends of 5.8S rRNA could not be formed and/or 32SS pre-rRNA was rapidly degraded when Rpl17 was limiting, while the long precursors continued to be generated. Strikingly, a prominent stop located 9–10 nt downstream from the 5′ end of 32SS rRNA was observed after Rpl17 depletion (Fig. 2B, lane 2); for consistency with other truncated mammalian processing intermediates (Carron et al. 2011; Preti et al. 2013), this RNA species was designated 32SC (“cropped”). The 32SC pre-rRNA was also detectable at low levels in cells transfected with nontargeting siRNA (Fig. 2B, lane 1). Quantification of primer extension stops (Fig. 2C) showed that under normal conditions, ∼80% of 32S pre-rRNA was comprised of the 32SS form, whereas in Rpl17-depleted cells 32SS was greatly decreased and the 32SL and 32SC forms together accounted for >80% 32S pre-rRNA. A significant accumulation of the 32SC form was observed in Rpl17-depleted cells, but not after Rpl10 depletion (Fig. 2B, lane 3; Fig. 2C). Interestingly, the 32SL fraction increased moderately in Rpl10-depleted cells relative to cells transfected with nontargeting siRNA (Fig. 2C), suggesting that functional defects in ribosomes caused by lack of this late assembling protein can feed back to affect earlier processing steps at the 5′ end of 5.8S rRNA to some extent.
FIGURE 2.
Analysis of the 5′ end formation in mouse 32S pre-rRNA. (A) Nucleotide sequence of the mature short 5.8SS rRNA in the mouse (boxed) and positions of 5′ ends of the long, short, and cropped forms (5.8SL, 5.8SS, and 5.8SC). Positions of oligonucleotides used in hybridizations and primer extensions are indicated. (B) Primer extension analysis of the 5′ ends of gel-purified 32S pre-rRNA using primer 5.8S-113. Cells were analyzed 48 h after transfections with the indicated siRNAs. (C) Intensities of primer extension stops corresponding to 32SL, 32SS, and 32SC pre-rRNAs shown in B were quantified by phosphorimaging analysis and expressed as a percentage of the total (32SL+S+C). (D) Immunoblotting analysis of Xrn2 levels in cells transfected with the indicated siRNA combinations. (E) Northern hybridizations of the RNA samples used for primer extensions in B. RNA was separated on a formaldehyde–agarose gel, blotted and hybridized with the indicated probes. Probe 5.8S-19 overlaps with site C (see A) and does not hybridize with 32SC, while probe 5.8S-37 detects all three 32S forms (32SL, 32SS, and 32SC). (F) Quantification of 32SL+S forms relative to total 32S pre-rRNA from hybridizations with two 5.8S probes, as shown in E. Data are mean values from three independent transfections with the indicated siRNAs. Error bars, SEM; see Materials and Methods for details.
The results obtained through primer extensions revealed both similarities and differences between mouse and yeast cells with respect to the formation of 5.8S rRNA precursors after L17 depletion. A 5′-truncated species analogous to 32SC was observed in L17-depleted yeast (Sahasranaman et al. 2011; Gamalinda et al. 2013). Levels of 27SB precursors, equivalent to mammalian 32S pre-rRNAs, were reported to increase in yeast after L17 depletion, with 27SBS increasing more than 27SBL pre-rRNA (Sahasranaman et al. 2011). In contrast, levels of the mouse 32SS pre-rRNA showed a strong reduction after Rpl17 depletion, suggesting that either the formation of the major short ends of 5.8S rRNA in mouse cells cannot be accomplished without Rpl17 assembly or the 32SS pre-rRNA is selectively destabilized.
Role of Xrn2 in the formation of the 5′ ends of 5.8S precursors
The exonuclease Xrn2 mediates trimming and proofreading of 5′ extensions resulting from cleavages in the spacers of the mammalian pre-rRNA transcript (Wang and Pestov 2011; Preti et al. 2013; Sloan et al. 2013). To understand the role that Xrn2 plays in the formation of the different 5′ ends of 5.8S precursors, we transfected cells with Xrn2 siRNA (Fig. 2D) and examined gel-purified 32S pre-rRNA by primer extensions (Fig. 2B, lanes 4–6). Quantification of stop intensities (Fig. 2C) showed that Xrn2 depletion increased the ratio of 32SS pre-rRNA to 32SL and 32SC pre-rRNAs; this trend also held for Xrn2 knockdowns in cells depleted of Rpl17 and Rpl10. Thus, the long and cropped forms of 32S pre-rRNA are more sensitive to cellular Xrn2 levels than the short form. Interestingly, inactivation of the Xrn2 homolog Rat1 in yeast cells affects short 5.8S precursors to a larger extent than the long forms (Henry et al. 1994; El Hage et al. 2008). While formation of short 5.8S precursors in yeast requires an exonuclease, long forms are thought to be generated by a direct endonucleolytic cleavage at the B1L site (Faber et al. 2006). Rat1 also promotes turnover of pre-rRNA in L17-depleted yeast cells by removing ∼10 nt from the 5′ end of the 27SB precursor, suggesting that L17 may act as a steric inhibitor limiting 5′ exonuclease progression (Sahasranaman et al. 2011). In agreement with this model, our primer extensions showed a decrease in the 32SC fraction when Xrn2 was depleted in addition to Rpl17 (Fig. 2C). Therefore, the activity of Xrn2 in the mammalian complexes that lack Rpl17 may be similar to that of Rat1 in yeast. To corroborate primer extension data, we estimated the combined amount of the 32SL+S forms in the total 32S pre-rRNA using northern hybridizations with different 5.8S probes (Fig. 2E). Quantification of band intensities showed that Rpl17 depletion decreased the fraction contributed by the long and short forms, and Xrn2 codepletion partially counteracted this effect (Fig. 2F).
Collectively, these data indicate that Xrn2 promotes the accumulation of the 32SL and 32SC pre-rRNAs in mouse cells. It is currently unclear if the mammalian short 32SS form may be generated through a distinct endonucleolytic mechanism, as postulated for the long form of yeast 27SB pre-rRNA. An endonucleolytic cleavage generating 32SS pre-rRNA would yield a 5′ product with an ITS1 fragment extending all the way to the 5.8S sequence. Such species were reported in early studies of mammalian pre-rRNA processing (Bowman et al. 1983), but are not readily detectable in 3T3 cells in our hands. We cannot exclude that these intermediates have a very short life span owing to their rapid 3′ trimming by exonucleases. Alternatively, all three 5′ ends in 32S pre-rRNA might be generated through a 5′ exonucleolytic mechanism, in which the precise position where the nuclease terminates is determined by the state of the local structure in pre-60S complexes at the time of nuclease action.
Cropped 32S precursors are processed into functional 5.8SC rRNA
The unexpected complexity of the 5′ end formation in 32S pre-rRNA led us to investigate 5′ ends in mature 5.8S rRNA. Primer extensions with gel-purified 5.8S rRNA revealed a stop that matched the 5′ end of 32SC pre-rRNA (Fig. 3A). Although the existence of two 5.8S forms (5.8SS and 5.8SL) in eukaryotes has been long recognized (Rubin 1974; Smith et al. 1984), this result suggested that a fraction of mature 5.8S rRNA might be derived from 32SC pre-rRNA. To verify that the stop corresponding to the 5′ end of 5.8SC rRNA was not an artefact of primer extension, we performed northern hybridizations (Fig. 3B) with probes complementary to different parts of 5.8S rRNA (Fig. 2A). Indeed, a band of a 5′-truncated 5.8S species was detectable (Fig. 3B) with probes hybridizing downstream from the 5′ end of 5.8SC rRNA, but not with probe 5.8S-19 that overlaps the 5′ end of this rRNA. We also reasoned that if the 5.8SC rRNA is generated from 32SC pre-rRNA, increasing the frequency of 32SC formation should increase the fraction of 5.8SC rRNA in the cell. Because of the long half-life of rRNA present in mature ribosomes, changes in cellular 5.8S rRNA may not become evident after short-term r-protein depletion. To allow sufficient time for the pre-existing 60S subunits to turn over, we depleted Rpl17 for 7 d using doxycycline-inducible shRNA. Hybridization analysis showed a pronounced accumulation of the 5.8SC rRNA after long Rpl17 depletion, but not in Rpl10 depletion that does not significantly increase 32SC pre-rRNA (Fig. 3C). To further rule out that 5.8SC rRNA is an aberrant species destined for degradation, we separated cytoplasmic extracts on a sucrose gradient. Hybridization analysis revealed the presence of 5.8SC rRNA in polysomal fractions (Fig. 3D), indicating that 60S subunits containing this 5′-truncated 5.8S form are functional in translation. Finally, we asked whether cells other than the 3T3s analyzed above contain 5.8SC rRNA. Hybridizations showed that the 5.8SC species was present in all tested mouse and human cell lines (Fig. 3E).
FIGURE 3.
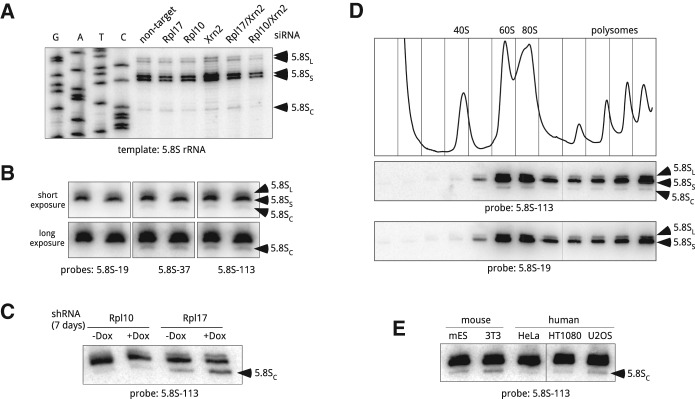
Heterogeneity of 5.8S rRNA in ribosomes from mouse and human cells. (A) Primer extension analysis of the 5′ ends of gel-purified 5.8S rRNA using primer 5.8S-113. Cells were analyzed 48 h after transfections with the indicated siRNAs. (B) A shortened form of 5.8S rRNA is detectable by northern hybridizations. Duplicate samples of RNA from growing 3T3 cells were resolved on a polyacrylamide/urea gel, transferred to a nylon membrane and hybridized with the indicated probes. Probes 5.8S-37 and 5.8S-113 detect all three forms of 5.8S, whereas 5.8S-19 does not hybridize with 5.8SC; see Figure 2A for probe positions. (C) Prolonged depletion of Rpl17, but not Rpl10, increases the amount of 5.8SC rRNA in mouse cells. Cells harboring doxycycline-inducible shRNAs were incubated with doxycycline for 7 d or remained untreated (+Dox, −Dox). RNA from these cells was separated by polyacrylamide gel electrophoresis and analyzed by Northern blotting. (D) 5.8SC rRNA is incorporated into ribosomes that are translationally competent. Growing 3T3 cells were lysed and cytoplasmic ribosomes were separated by centrifugation through 10%–45% (w/w) sucrose gradients. The absorbance profile of the gradient at 254 nm is shown at the top. RNA isolated from each gradient fraction was analyzed by polyacrylamide gel electrophoresis and northern hybridizations with probes that either detect (5.8S-113) or do not detect (5.8S-19) the 5.8SC isoform. (E) 5.8SC rRNA is formed in different mouse and human cell lines. RNA was extracted from the indicated mouse and human cell lines, and analyzed as in C. (mES) Mouse embryonic stem cells.
Together, these findings provide a strong indication that 32SC pre-rRNA can be processed to 5.8SC rRNA, which gives rise to translationally competent ribosomes. Although the generation of 5.8SC is greatly increased when Rpl17 is limiting, the presence of the 5.8SC rRNA in normal cells indicates that this species is continuously generated in a fraction of 60S maturation events. It seems plausible that stochastic variations during assembly of individual subunits may involve delayed loading of Rpl17, leading to occasional Xrn2 overruns when this enzymes trims the 5′ ends of 5.8S precursors. Notably, northern hybridizations of yeast 5.8S rRNA do not show detectable “cropped” forms (data not shown). The 5′ heterogeneity of mammalian 5.8S rRNA might thus reflect a less rigid assembly order and/or more relaxed surveillance mechanisms, leading to a greater structural diversity of ribosomes in these species.
Interactions of Rpl17 with full-length and cropped 5.8S rRNA in mature ribosomes
The 5′ end truncation in 5.8SC rRNA removes a large portion of the evolutionarily conserved helix 2 formed by 5.8S and domain I of 28/25S rRNA (Fig. 4A). To obtain insight into possible consequences of this disruption, we looked at the local ribosome environment in a high-resolution structure of the mammalian ribosome-Sec61 complex (Voorhees et al. 2014). Helix 2 in a mature 60S subunit is positioned in close proximity to Rpl17, which interacts with 28S and 5.8S rRNAs at several sites (Fig. 4B,C). Rpl17 is comprised of two distinct structural elements: a compact globular domain and an extended β-hairpin (Zhang et al. 2013). The globular domain extensively interacts with domain I of the 60S subunit (Fig. 4C). Specific interactions involve two Rpl17 residues, R62 and Y63, engaged in hydrogen bonding with helix 2 (Fig. 4D), and two asparagine residues (N97, N101) forming hydrogen bonds with nucleotide residues of helix 24 (Fig. 4E). The β-hairpin extends deep inside the ribosome so that its tip faces the exit tunnel and forms additional hydrophobic/electrostatic contacts with a hairpin loop in domain III of 28S rRNA (Fig. 4F; Voorhees et al. 2014). The absence of ten 5′-terminal nucleotides in 5.8SC rRNA destabilizes the interaction of the globular domain of Rpl17 with helix 2 (Fig. 4G,H), likely affecting conformational rearrangements in this r-protein at the subunit surface in the vicinity of the exit tunnel. Interestingly, the globular domain of yeast Rpl17 has been previously shown to be in proximity with Sec61 translocon components; moreover, the presence of a transmembrane segment in the nascent polypeptide chain inside the exit tunnel is sensed by Rpl17 to recruit the small translocon-associated protein RAMP4 (Pool 2009). An altered conformational flexibility of mammalian Rpl17 in 60S subunits harboring 5.8SC rRNA may thus impact the interface between the ribosome and the translocon, and potentially other cellular factors interacting with nascent polypeptide chains. Experiments are in progress to address the functional consequences of these changes.
FIGURE 4.
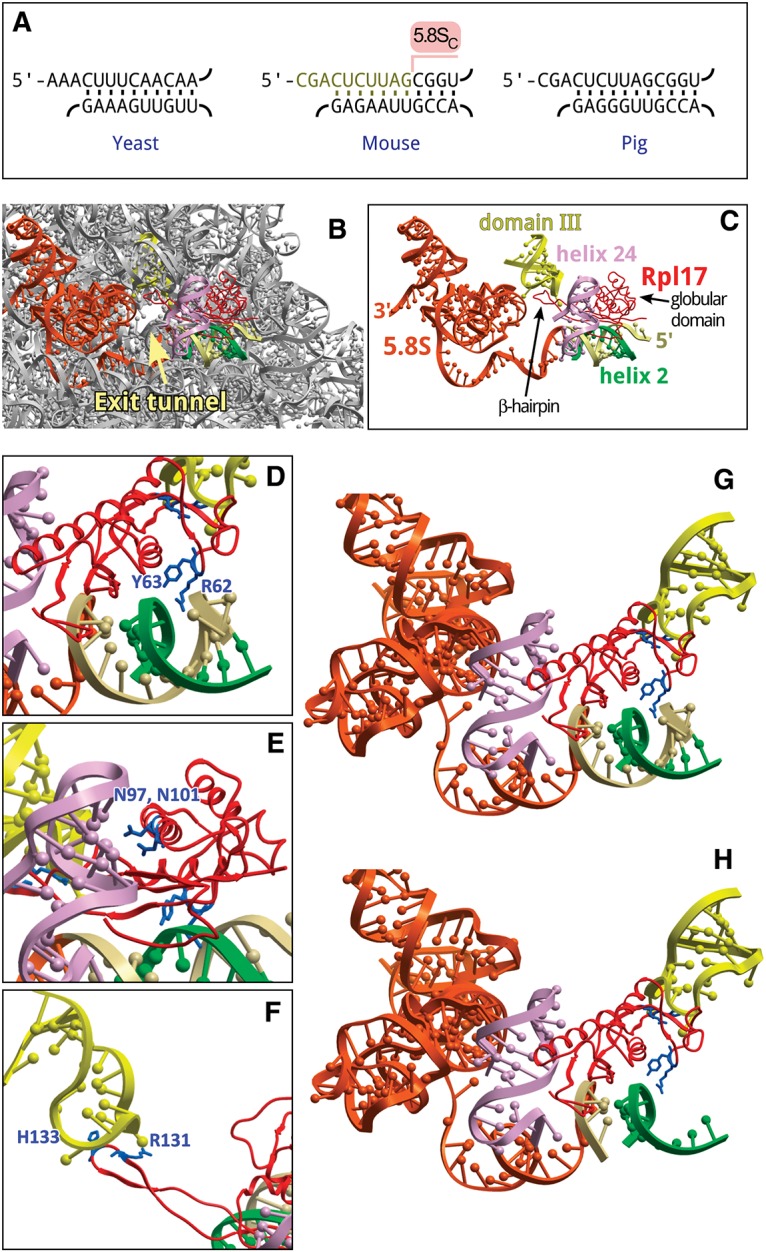
Structural implications of the 5′ truncation in 5.8S rRNA. (A) Conservation of helix 2 in the large subunit rRNA of Saccharomyces cerevisiae, Mus musculus, and Sus scrofa. Top strand, 5.8SS rRNA; bottom strand, 25/28S rRNA. Ten nucleotides missing in 5.8SC rRNA are indicated in the mouse sequence. (B) Position of 5.8S rRNA, helix 2 and Rpl17/uL22 relative to the exit tunnel in the 60S subunit of S. scrofa (PDB: 3J71). (C) The same view showing only structural elements that surround the exit tunnel and interact with Rpl17 (red). Orange, 5.8S rRNA, the first 14 nucleotides at the 5′ terminus shown in beige. Green, helix 2, domain I (28S rRNA nucleotides 419–430, 5′-accguuaagag-3′); light purple, helix 24, domain I (28S rRNA nucleotides 388–410, 5′-aacuuugaagagagaguucaaga-3′); yellow, domain III hairpin loop (28S rRNA nucleotides 1587–1606, 5′-guccugacgugcaaaucggu-3′). (D–F) Rpl17 residues that contact helix 2, helix 24, and the loop in domain III, respectively. (G,H) Residues of the Rpl17 globular domain facing intact or cropped helix 2, respectively. Images were generated using ICM-Browser (Molsoft).
Concluding remarks
In this study, we analyzed effects of depletion of Rpl17, an integral r-protein of the large ribosomal subunit, in mouse cells. Our results revealed that like its previously studied yeast ortholog, mouse Rpl17 is required for pre-rRNA processing in ITS2. Depletion of Rpl17 also strongly affected processing efficiency at the 5.8S-proximal ITS1 site 2c, similar to several other factors involved in 60S subunit maturation in mouse cells (Wang et al. 2014). Interestingly, formation of the short 5.8S precursor 32SS in Rpl17-depleted cells was affected to a much greater extent than the long form 32SL. Furthermore, depletion of the 5′ exoribonuclease Xrn2, whose yeast homolog Rat1 was implicated in the generation of short 5′ ends of 5.8S rRNA, preferentially affected long 5.8S precursors in mouse cells. These differences between yeast and mouse cells might reflect distinct kinetics of ITS1 processing or in fact indicate that generation of the long rather than short 5.8S species requires a 5′-exonucleolytic activity in mouse cells.
We also found that depletion of mouse Rpl17 promoted the formation of “cropped” 32SC pre-rRNA. Similar 5′-truncated 28SB forms were observed in yeast as an intermediate in Rat1-mediated turnover of pre-rRNA in stalled pre-60S subunits (Sahasranaman et al. 2011; Gamalinda et al. 2013). Surprisingly, mammalian 32SC pre-rRNA gives rise to functional ribosomes in a fraction of maturation events. These results indicate that deficiency in assembly components does not always end up with the elimination of ribosomal precursors; moreover, altered ribosome maturation can lead to structural changes in the newly synthesized ribosomes, thus impacting ribosome diversity in the cell. Importantly, changes in mature ribosomes were only observed through experimentally induced long-term r-protein depletion, which allowed sufficient time for turnover of the pre-existing ribosomes. Because many ribosomopathies result from a chronic reduction of r-protein expression in the organism, it is tempting to speculate that slowly occurring changes in ribosome diversity may underlie some of the protein-specific phenotypes observed in these disorders. Heterogeneity of cytoplasmic ribosomes is emerging as an important, yet still poorly understood, mechanism of regulation in gene expression (Xue and Barna 2012). Understanding the functional differences between structurally divergent ribosomes will be an important direction for the future studies. It would also be interesting to see if imbalanced expression of other r-proteins may translate into distinct alterations of rRNA in mature ribosomes.
MATERIALS AND METHODS
Cell culture and transfections
Cells were maintained and transfected with siGENOME SMARTpool siRNAs (Dharmacon) as described previously (Shcherbik et al. 2010). Knockdown efficiency of r-proteins was assessed by regular and quantitative RT-PCR (Wang et al. 2014). Transfections that reduced Rpl17 and Rpl10 mRNA levels to <5% relative to cells transfected with nontargeting siRNA were used for quantitative pre-rRNA assays. Xrn2 knockdowns were monitored using immunoblotting with antibodies sc-99237 (Santa Cruz) and estimated to provide 60%–80% reduction of Xrn2 protein levels. For shRNA-mediated knockdowns, Rpl17- and Rpl10-specific cassettes were transferred from pGIPZ constructs (Dharmacon) into the doxycycline-inducible pIVRE vector (Wang et al. 2014), the resulting plasmids were transfected into NIH 3T3 cells and stable clones were selected with puromycin. Expression of shRNA was induced with 2 µg/mL doxycycline. Because of a slight leakiness of the pIVRE-shRNA constructs, siRNA transfections were used for all quantitative assays. Cell numbers were determined using the CyQUANT cell proliferation assay (Life Technologies).
RNA analysis
RNA was extracted from cells using RNazol RT (Molecular Research Center, Inc.). Long RNA species were separated using modified formaldehyde–agarose gels (Mansour and Pestov 2013) and hybridized with 32P-labeled oligonucleotide probes (Pestov et al. 2008). Steady-state levels of rRNA processing intermediates were determined by phosphorimager analysis, and their ratios analyzed as described (Wang et al. 2014). Primer extensions were carried out according to our previously published protocol (Wang and Pestov 2011). Cytoplasmic ribosomes were fractionated using the protocol from Strezoska et al. (2000).
To estimate the fraction of 32SL+S pre-rRNAs in the 32S band (Fig. 2E,F), we hybridized the same membrane sequentially with probes 5.8S-19 (detects 32SL + S only) and 5.8S-37 (detects 32SL + S + C). After each hybridization, the membrane was scanned using a phosphorimager. The total pixel volume V36S in bands containing 36S pre-rRNA, which has the same number of binding sites for both probes, was used to correct for differences between probe labeling and hybridization efficiencies. The normalization coefficient Knorm was defined as the mean of the individual ratios k = V36S, 19/V36S, 37 for each 36S band on the blot; the coefficient of variation between k values in our experimental set was <3%. The fraction of 32SL + S pre-rRNA in each sample was then calculated as V32S, 19/(Knorm × V32S, 37), where V32S, 19 and V32S, 37 are total pixel volumes obtained for the 32S pre-rRNA band with probes 5.8S-19 and 5.8S-37.
To analyze 5.8S rRNA, 0.25 μg total cellular RNA was mixed with loading buffer (95% formamide, 1 mM EDTA, 0.02% bromophenol blue and 0.03% xylene cyanol), heated at 70°C for 5 min and loaded on 0.75 mm, 10 × 10 cm 6% polyacrylamide gels containing 8 M urea and 0.5× TBE that were prerun for 15 min at 250 V. Gels were run at the same voltage in 0.5× TBE. RNA was transferred to nylon membranes in 0.5× TBE for 30 min at 10 V using a Trans-blot SD semi-dry apparatus (Bio-Rad).
Gel purification of RNA
To separate long pre-rRNAs from mature rRNAs, we modified the analytical gel electrophoresis method of Goda and Minton (1995) as follows. A total of 3 μg of RNA dissolved in formamide was mixed with an equal volume of 2× loading buffer (80 mM guanidine thiocyanate, 2× HT buffer (60 mM HEPES, 60 mM triethanolamine Mansour and Pestov 2013), 2 mM EDTA and 0.08% bromophenol blue), incubated at 65°C for 3 min and cooled to room temperature. RNA samples were loaded onto 1.2% (w/v) agarose gels that were both prepared and run in 1× HT buffer containing 40 mM guanidine thiocyanate and 0.2 μg/mL ethidium bromide. Separated RNA was visualized under long-wave UV light and the desired bands were cut out from the gel. RNA was isolated from gel slices with the Zymoclean Gel RNA Recovery Kit (Zymo Research) according to the manufacturer's recommendations.
Oligonucleotides
The sequences of oligonucleotides used in this study are as follows: ITS2-pEx1: GGGGTGGAGGATCTTACTCA; 5.8-19: 5′-GATCCACCGCTAAGAGTCGT; 5.8-37: 5′-TCGACGCACGAGCCGAGTGAT; 5.8-113: 5′-GCAAGTGCGTTCGAAGTGT. PCR primer sequences are available upon request.
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) grant GM074091 to D.G.P. and the Foundation of UMDNJ grant to N.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.051169.115.
REFERENCES
- Armistead J, Triggs-Raine B. 2014. Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett 588: 1491–1500. [DOI] [PubMed] [Google Scholar]
- Auger-Buendia MA, Longuet M, Tavitian A. 1979. Kinetic studies on ribosomal proteins assembly in preribosomal particles and ribosomal subunits of mammalian cells. Biochim Biophys Acta 563: 113–128. [DOI] [PubMed] [Google Scholar]
- Babiano R, de la Cruz J. 2010. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 38: 5177–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiano R, Gamalinda M, Woolford JL Jr, de la Cruz J. 2012. Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol Cell Biol 32: 3228–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bowman LH, Rabin B, Schlessinger D. 1981. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res 9: 4951–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman LH, Goldman WE, Goldberg GI, Hebert MB, Schlessinger D. 1983. Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol 3: 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac S, Brdovcak MC, Donati G, Volarevic S. 2014. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim Biophys Acta 1842: 817–830. [DOI] [PubMed] [Google Scholar]
- Carron C, O'Donohue M-F, Choesmel V, Faubladier M, Gleizes P-E. 2011. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res 39: 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Williamson JR. 2013. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol 425: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov KP, Dabeva MD, Hadjiolov AA, Todorov BN. 1978. Processing and migration of ribosomal ribonucleic acids in the nucleolus and nucleoplasm of rat liver nuclei. Biochem J 171: 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger DP, Dick FA, Trumpower BL. 1997. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol 17: 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D. 2008. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SR, Gleizes P-E. 2011. Diamond Blackfan anemia: ribosomal proteins going rogue. Semin Hematol 48: 89–96. [DOI] [PubMed] [Google Scholar]
- Faber AW, Vos HR, Vos JC, Raué HA. 2006. 5′-end formation of yeast 5.8SL rRNA is an endonucleolytic event. Biochem Biophys Res Commun 345: 796–802. [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Pöll G, Kühn H, Neueder A, Jakob S, Tschochner H, Milkereit P. 2007. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell 28: 446–457. [DOI] [PubMed] [Google Scholar]
- Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL. 2013. Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res 41: 1965–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL. 2014. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 28: 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda SK, Minton NP. 1995. A simple procedure for gel electrophoresis and Northern blotting of RNA. Nucleic Acids Res 23: 3357–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie J-P. 1993. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem 212: 211–215. [DOI] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O'Donohue M-F, Chakraborty A, Gleizes P-E. 2015. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J 13: 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horos R, IJspeert H, Pospisilova D, Sendtner R, Andrieu-Soler C, Taskesen E, Nieradka A, Cmejla R, Sendtner M, Touw IP, et al. 2012. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood 119: 262–272. [DOI] [PubMed] [Google Scholar]
- Jakob S, Ohmayer U, Neueder A, Hierlmeier T, Perez-Fernandez J, Hochmuth E, Deutzmann R, Griesenbeck J, Tschochner H, Milkereit P. 2012. Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes. PLoS One 7: e32552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL. 2012. Ribosomal proteins L7 and L8 function in concert with six A3 assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn-Safran CB, Oristian DS, Focht RJ, Parker SG, Vivian JL, Carson DD. 2007. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn 236: 447–460. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapik YR, Fernandes CJ, Lau LF, Pestov DG. 2004. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell 15: 17–29. [DOI] [PubMed] [Google Scholar]
- Lastick SM. 1980. The assembly of ribosomes in HeLa cell nucleoli. Eur J Biochem FEBS 113: 175–182. [DOI] [PubMed] [Google Scholar]
- Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, et al. 2014. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med 20: 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour FH, Pestov DG. 2013. Separation of long RNA by agarose-formaldehyde gel electrophoresis. Anal Biochem 441: 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh KB, Bhattacharya A, Willis IM, Warner JR. 2011. Eukaryotic cells producing ribosomes deficient in Rpl1 are hypersensitive to defects in the ubiquitin-proteasome system. PLoS One 6: e23579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Paulovich AG, Tsay YF, Woolford JL. 1990. Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol 111: 2261–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue M-F, Choesmel V, Faubladier M, Fichant G, Gleizes P-E. 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Pöll G, Linnemann J, Hierlmeier T, Perez-Fernandez J, Kumcuoglu B, Leger-Silvestre I, et al. 2013. Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae. PLoS One 8: e68412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary MN, Schreiber KH, Zhang Y, Duc A-CE, Rao S, Hale JS, Academia EC, Shah SR, Morton JF, Holstein CA, et al. 2013. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet 9: e1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov DG, Lapik YR, Lau LF. 2008. Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol 39: 22.11.1–22.11.16. [DOI] [PubMed] [Google Scholar]
- Pöll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL, Tschochner H, Milkereit P. 2009. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One 4: e8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR. 2009. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol 185: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M, O'Donohue M-F, Montel-Lehry N, Bortolin-Cavaille M-L, Choesmel V, Gleizes P-E. 2013. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res 41: 4709–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiser DM, Narla A, Ebert BL. 2014. The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leuk Lymphoma 55: 491–500. [DOI] [PubMed] [Google Scholar]
- Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. 2008. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14: 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM. 1974. Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem FEBS 41: 197–202. [DOI] [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL. 2011. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J 30: 4020–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Namane A, Gleizes P-E, Lebreton A, Rousselle J-C, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 23: 4449–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Gerbi SA. 1990. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J 9: 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep 11: 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJM, Watkins NJ. 2013. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J Cell Biol 200: 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Banerjee N, Sitz TO. 1984. Gene heterogeneity: a basis for alternative 5.8S rRNA processing. Biochemistry 23: 3648–3652. [DOI] [PubMed] [Google Scholar]
- Steffen KK, McCormick MA, Pham KM, MacKay VL, Delaney JR, Murakami CJ, Kaeberlein M, Kennedy BK. 2012. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol Cell Biol 20: 5516–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Mercer CA, Hexley P, Thomas G, Fumagalli S. 2013. Loss of tumor suppressor RPL5/RPL11 does not induce cell cycle arrest but impedes proliferation due to reduced ribosome content and translation capacity. Mol Cell Biol 33: 4660–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa M, Bommakanti A, Shamsuzzaman M, Gregory B, Samsel L, Zengel JM, Lindahl L. 2013. Repressed synthesis of ribosomal proteins generates protein-specific cell cycle and morphological phenotypes. Mol Biol Cell 24: 3620–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov IT, Noll F, Hadjiolov AA. 1983. The sequential addition of ribosomal proteins during the formation of the small ribosomal subunit in friend erythroleukemia cells. Eur J Biochem 131: 271–275. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Fernández IS, Scheres SHW, Hegde RS. 2014. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157: 1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Pestov DG. 2011. 5′-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res 39: 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Anikin L, Pestov DG. 2014. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res 42: 11180–11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Hedges JB, Chen A, Johnson AW. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol 25: 3802–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wölfle T, Rospert S. 2013. Interaction of nascent chains with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 of Saccharomyces cerevisiae. J Biol Chem 288: 33697–33707. [DOI] [PMC free article] [PubMed] [Google Scholar]