Abstract
Existing methods for detecting RNA intermediates resulting from exonuclease degradation are low-throughput and laborious. In addition, mapping the 3′ ends of RNA molecules to the genome after high-throughput sequencing is challenging, particularly if the 3′ ends contain post-transcriptional modifications. To address these problems, we developed EnD-Seq, a high-throughput sequencing protocol that preserves the 3′ end of RNA molecules, and AppEnD, a computational method for analyzing high-throughput sequencing data. Together these allow determination of the 3′ ends of RNA molecules, including nontemplated additions. Applying EnD-Seq and AppEnD to histone mRNAs revealed that a significant fraction of cytoplasmic histone mRNAs end in one or two uridines, which have replaced the 1–2 nt at the 3′ end of mature histone mRNA maintaining the length of the histone transcripts. Histone mRNAs in fly embryos and ovaries show the same pattern, but with different tail nucleotide compositions. We increase the sensitivity of EnD-Seq by using cDNA priming to specifically enrich low-abundance tails of known sequence composition allowing identification of degradation intermediates. In addition, we show the broad applicability of our computational approach by using AppEnD to gain insight into 3′ additions from diverse types of sequencing data, including data from small capped RNA sequencing and some alternative polyadenylation protocols.
Keywords: bioinformatics, high-throughput sequencing, histone mRNA, mRNA
INTRODUCTION
The synthesis, processing, and degradation of RNA are complex processes, with every stage of an RNA's lifetime, from transcription initiation to degradation, requiring careful control. Much attention has been focused on regulation of transcription and pre-mRNA processing, but the detailed pathways of mRNA degradation remain poorly understood. During exonucleolytic degradation of RNA some portions of the molecule are more difficult to degrade than others, resulting in accumulation of intermediates in regions that are degraded more slowly. Eukaryotic mRNAs can be degraded in either 5′–3′ or 3′–5′ directions, or in some cases in both directions (Mullen and Marzluff 2008). Critical to understanding the pathway of degradation or modification of the mRNA is a method for determining the precise termini of RNA molecules. Here we describe a method to determine the 3′ end of RNA molecules, which can be applied to mapping degradation intermediates generated during 3′–5′ degradation. The presence of RNA binding proteins and secondary structure motifs may block the progress of 3′–5′ degradation resulting in a spectrum of partly degraded transcripts that differ only at the 3′ end (Fig. 1A). Additionally, the 3′ ends of RNAs are often modified by the addition of short, nontemplated 3′ tails, and we are just starting to appreciate the broad range of these modifications (Chang et al. 2014). For example, during degradation of mammalian histone mRNAs, there is oligouridylation of mature mRNA to initiate degradation (Mullen and Marzluff 2008; Hoefig et al. 2013; Su et al. 2013) as well as uridylation of a large variety of degradation intermediates (Slevin et al. 2014).
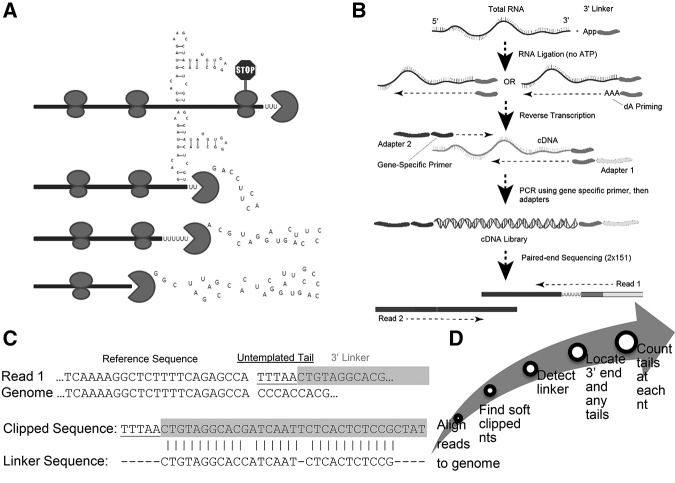
FIGURE 1.
EnD-Seq and AppEnD strategy. (A) Schematic of the 3′ end of a hypothetical RNA molecule, indicating potential intermediates in 3′–5′ degradation resulting from bound proteins or RNA secondary structure that might slow 3′–5′ exonuclease degradation. (B) EnD-seq sequencing strategy. We ligate a preadenylated 3′ linker onto the 3′ end to preserve the information at the 3′ end of RNAs in total cell RNA. We then prime cDNA synthesis using a primer antisense to the linker. Alternatively, we can prime cDNA with the antisense primer extended with a short sequence (e.g., three A's to enrich uridylated RNAs). Gene-specific primers can be used to enrich for transcripts of interest. (C) Example of sequence containing an untemplated tail and one containing a single U-tail obtained from the EnD-Seq data. Note that the linker sequence can be identified even if it contains sequencing errors. (D) Flow chart for the analysis of sequencing data by App-EnD.
Existing methods for studying RNA degradation intermediates or RNAs with nontemplated nucleotides are low-throughput and laborious, requiring cloning of individual degradation intermediates, limiting our ability to probe intermediates in mRNA degradation. In general, these studies have identified a small number of cloned and sequenced intermediates (Shen and Goodman 2004; Ibrahim et al. 2006; Eberle et al. 2009; Rissland and Norbury 2009; Hoefig et al. 2013; Mullen and Marzluff 2008; Sement et al. 2013). Conventional RNA-seq techniques do not yield precise 3′ ends of RNA molecules, since the sequences are generated using cDNA priming. As a result the first nucleotides identified are located internal to the 3′ end of the molecule. A number of methods for locating alternative polyadenylation sites have been developed (Mayr and Bartel 2009; Shepard et al. 2011; Lianoglou et al. 2013; Hoque et al. 2014; Masamha et al. 2014), only some of which rely on sequencing the junction between the nontemplated poly(A) tail and the cleavage site to identify the precise nucleotide where poly(A) is added (Martin et al. 2012; Hoque et al. 2014; Yao and Shi 2014).
The computational analysis involved in detecting nontemplated tails from sequencing data is not trivial. A common approach to this problem is to strip homopolymers from raw reads before genomic read alignment (Henriques et al. 2013; Yao and Shi 2014). Such a prealignment read stripping approach is less than ideal, making restrictive assumptions about the length and nucleotide composition of the nontemplated additions.
We developed EnD-Seq (Exonuclease Degradation sequencing) and AppEnD (Application for mapping EnD-Seq data), a customized high-throughput sequencing strategy and computational method for identifying 3′ ends of RNA molecules, including any nontemplated additions, with no assumptions about sequence composition. Here we demonstrate the utilization of EnD-Seq and AppEnD to identify nontemplated nucleotides as short as 1 nt, allowing us to define an unanticipated modification of the 3′ end of histone mRNA after processing. We also use AppEnD to gain insight into 3′ nontemplated additions from diverse types of sequencing data, including small capped RNA sequencing data and PAS-SEQ and A-SEQ polyadenylation data.
RESULTS
Overview of approach
Our EnD-Seq strategy is designed to identify the 3′ end of nonpolyadenylated RNA molecules, including degradation intermediates of polyadenylated mRNAs after deadenylation which leaves oligo(A) tails. Starting with total cell RNA we ligate on a preadenylated 3′ linker to preserve the information on the 3′ end of RNAs. We then prime cDNA synthesis using a primer antisense to the linker. We also primed cDNA synthesis with an antisense primer whose 3′ end was appended with a short sequence designed to identify specific nontemplated modifications (e.g., three A's to enrich uridylated RNAs) (Fig. 1B). A number of strategies can then be used to amplify cDNA of appropriate size for paired-end sequencing. Following ligation of the 3′ linker, the RNA can be cleaved either chemically (Yao and Shi 2014) or enzymatically (Chang et al. 2014) followed by ligation of a linker to the 5′ end. To amplify a specific set of cDNAs, a set of 5′ primers can be used to amplify the desired cDNAs (Slevin et al. 2014; Newman et al. 2011). Subsequent paired-end sequencing produces two reads: Read one contains the transcript 3′ end and read two contains indices for multiplexing and aids in properly aligning the first read.
To obtain information about the position of the transcript end and any nontemplated tails, we examined the first read which begins with the linker sequence, followed by a nontemplated tail or the 3′ end with no tail. Our computational method, AppEnD, aligns the paired-end reads to the genome using an RNA-seq aligner, e.g., bowtie2 (Langmead and Salzberg 2012) for unspliced RNAs or MapSplice (Wang et al. 2010) for spliced RNAs. Since read 1 contains the linker sequence and any nontemplated 3′ additions, the end of the read sequence that diverges from the genome is soft-clipped. We identify the linker sequence within the soft-clipped portion of the read using the Needleman–Wunsch algorithm (Needleman and Wunsch 1970). Any nontemplated 3′ additions are identified as nucleotides after the end of the linker in the soft-clipped portion of the read (Fig. 1C). After identifying the 3′ ends at single nucleotide resolution, we plot the abundance of transcripts ending at each nucleotide. This gives the positional distribution of the last templated nucleotides, and the pattern of nontemplated additions, if any are present.
Human histone mRNAs have modified 3′ ends containing nontemplated uridines
We applied EnD-Seq to study the metabolism of the nonpolyadenylated replication-dependent histone mRNAs in HeLa cells. Histone mRNAs end in a conserved stem–loop structure at the 3′ end (Marzluff et al. 2008), which is formed by endonucleolytic cleavage 5 nt after the stem–loop (Scharl and Steitz 1994). As previously reported, the cytoplasmic histone mRNAs end only 2–3 nt after the stem–loop (Mullen and Marzluff 2008; Hoefig et al. 2013). The histone 3′ end is trimmed by Eri1 (3′hExo) following the initial cleavage 5 nt after the stem–loop (Scharl and Steitz 1994) leaving a 2–3 nt overhang (Dominski et al. 2003; Yang et al. 2006; Hoefig et al. 2013). We analyzed the 3′ ends of histone mRNAs using the primer antisense to the linker to prime cDNA in S-phase cells, when histone mRNAs are not rapidly degraded, and histone-specific primers fused with Illumina linkers were used for second-strand synthesis to amplify the histone cDNAs specifically for sequencing (Slevin et al. 2014).
Surprisingly, 30%–50% of the histone mRNA molecules contained nontemplated nucleotides at the 3′ end. These were found on all the histone mRNAs and were added to molecules that had previously been trimmed by an additional 2–3 nt, presumably by 3′hExo, restoring the cytoplasmic histone mRNA to its normal length (Fig. 2B–F), with a 2- to 3-nt extension after the stem. The tails are short, 1–2 nt long, and are almost exclusively uridines (Fig. 2E). These tailed molecules are likely not decay intermediates, since they persist during degradation, but may be the result of a uridylation reaction that restores the normal length of histone mRNA on slightly shortened histone messages (see Discussion).
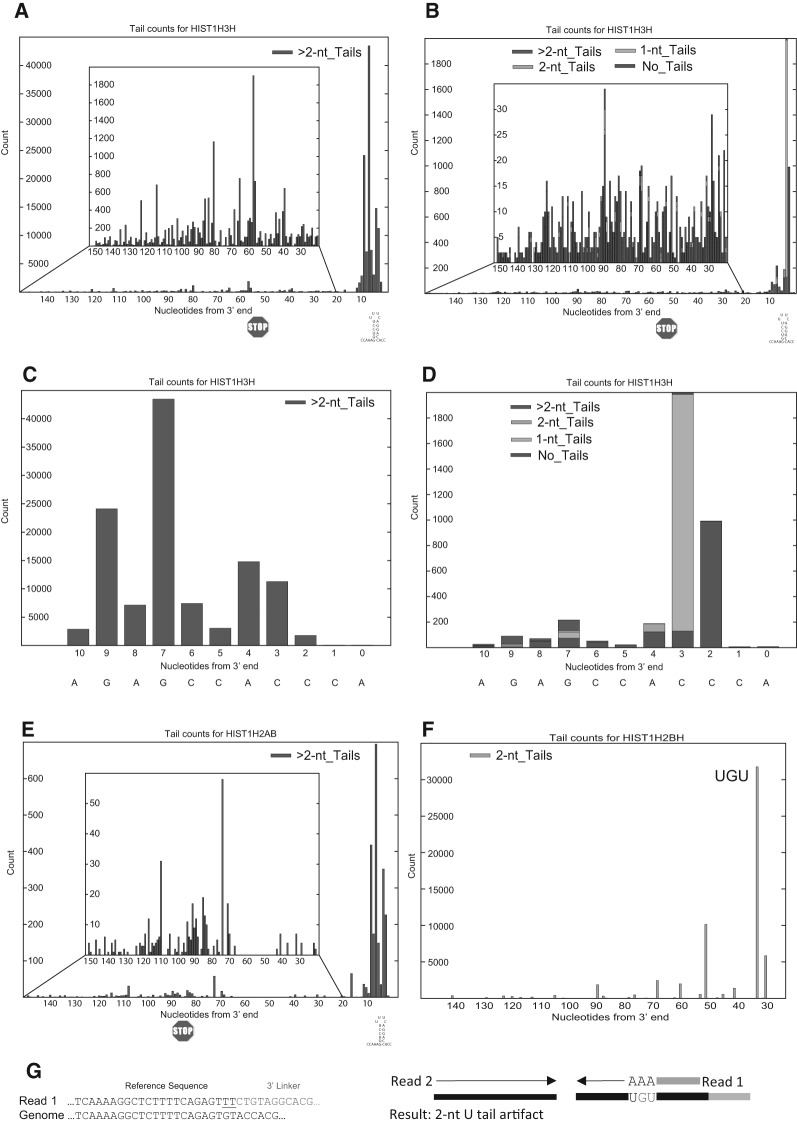
FIGURE 2.
Analysis of the 3′ end of histone mRNAs and degradation intermediates using a standard primer or an oligoadenylated primer for cDNA synthesis. (A) The 3′ ends of the HIST2H2AA3 mRNA were determined by high-throughput sequencing using EnD-Seq. The positions of the last templated nucleotides are indicated, and the diversity of lengths of nontemplated tails is indicated by the different colors in the histogram. These data are the results from four independent experiments with RNA from S-phase cells. The y-axis gives the number of reads at each position, and the x-axis is the position of the last templated nucleotide. The sequence below the figure indicates the sequence of the processed histone H2a mRNA formed in the nucleus which is quantitatively trimmed on cytoplasmic histone mRNA. (B) Sequence of the 3′ end of the histone mRNA after processing. (C) Sequences of the most abundant 3′ ends of cytoplasmic RNA without any nontemplated nucleotides. (D) Sequences of the most abundant 3′ ends ending in 1-nt or 2-nt nontemplated tails. (E) Distribution of the nontemplated nucleotides in the 1- nt (top) and 2-nt (bottom) tails. (F,G) The sequences of the 3′ end of the H2AA3 mRNA from exponentially growing HeLa cells treated with hydroxyurea for 20 min as identified using the standard EnD-Seq method (F), showing the major 1- and 2-nt tails at the 3′ end, and longer tails in the 3′ side of the stem. The y-axis gives the number of reads at each position. After priming cDNA with an oligo(A)3 tagged primer (G), the pattern of tails changes to reflect the distribution of molecules with tails of 3 nt or longer, and the number of these longer tails is dramatically increased since only oligouridylated histone mRNAs were sequenced. Note the pattern of tails in (G) is similar to the panel of >2-nt tails in (F). (H,I) The analysis of tails 5′ of the stem–loop from the same experiment in (F,G), with the 3′ ends of the RNAs determined by the standard EnD-Seq protocol (H) or the 3′ ends determined after priming cDNA with an oligo(A)3 tagged primer (I). The position of the stop codon is indicated. The y-axis is the total number of reads at each position. (J) AppEnD analysis of nontemplated U-tails detected by priming cDNA with the 3A-primer. Note that the tail is identified correctly. (K) AppEnD analysis of sequences resulting from adventitious priming at an oligo(U) stretch in the RNA when cDNA was primed with the 3A-primer. These sequences are not identified as nontemplated tails by AppEnD, since we required the oligo(U) tails to be at least 3 nt long.
Priming cDNA with a primer ending in 3 As enhances detection of oligo(U) tails
Degradation of replication-dependent-histone mRNAs is initiated by stopping DNA replication (Graves and Marzluff 1984), potentially allowing detection of degradation intermediates as the mRNA is rapidly degraded. To initiate histone mRNA degradation, longer oligo(U) tails are added to the 3′ end and the mRNA is degraded 3′–5′ (Mullen and Marzluff 2008; Hoefig et al. 2013; Slevin et al. 2014). Intermediates with oligo(U) tails accumulate in two regions of the transcript, in the stem on the 3′ side of the stem–loop (Hoefig et al. 2013; Slevin et al. 2014), and starting 15 nt 3′ of the stop codon, suggesting degradation pauses at these sites (Slevin et al. 2014). Since these intermediates represent a small percentage of the total histone mRNA molecules, they can be analyzed in depth only on the most abundant histone mRNAs.
To better detect these molecules, we primed reverse transcription with the 3A-primer, which ends in three adenosines, to enrich for molecules ending in at least three uridines. When the data generated by this approach is analyzed by AppEnD we obtain three distinct nontemplated populations. If there is a nontemplated tail ending in three U's or more the tail is detected and scored with its proper length and composition (Fig. 2J), even if there are substitutions between the three uridines at the end and the genomic sequence. If there is a stretch of U's encoded in the RNA which is primed adventitiously, then that sequence is not scored as a nontemplated tail (Fig. 2K), since it matches the genome sequence. We have observed mispriming at UGU or UCU in some contexts (likely when there is some additional homology with the primer sequence in the RNA) (see Fig. 3F). In this case the sequence obtained is scored as a 2U tail by AppEnD (Fig. 3G) and hence is easily identified as an artifact in the data, since we require a 3-nt nontemplated tail when we use the 3A-linker as a primer for cDNA. We observed that some of the reads on endogenous U-stretches or as a result of mispriming were very abundant, which reduces the degree of enrichment of the authentic oligo(U) tails, but does not interfere with the analysis.
FIGURE 3.
Priming with a primer ending in three A's enhances sensitivity and enriches oligouridylated RNAs. (A–D) The profile of sequences obtained from the HIST1H3H mRNA from the same sample analyzed in Fig. 2F–I obtained from cDNA primed with the oligo(dA) primer (A,C) or the standard linker (B,D) are shown. In A,B, the sequences 5′ of the stem–loop are shown in the inset, and in C,D, the sequences from the 3′ side of the stem–loop are shown. Since the HIST1H3H mRNA is expressed at about a fivefold lower level than the highly expressed histone mRNAs, there were very few sequences in the body of the mRNA obtained with the standard primer. (E) The profile of sequences from the HIST1H2AB mRNA from the same sample obtained from cDNA primed with the oligo(dA) primer. This histone mRNA is expressed at a very low level, and there were not enough sequences obtained with the standard primer to analyze this mRNA. (F) Artifacts obtained with the oligo(dA) primer from the HIST1H2BH mRNA. The major peak resulted from mispriming at a UGU sequence, resulting in sequences containing three U's at the 3′ end, which were identified as a 2-nt tail since one U is encoded in the genome. (G) AppEnD analysis of sequences misprimed at a UGU sequence when cDNA is primed with the 3A-primer. We observed a number of these examples, but the AppEnD pipeline removes them as authentic tails, since the nontemplated tail is only 2 nt.
We directly compared the same sample primed either with the standard primer or the 3A-primer, and sequenced the two samples in parallel. The RNA was prepared from cells 15 min after inhibition of DNA replication (20% degradation of histone mRNA in this experiment). With the standard primer we find that there is still a large amount of the mature histone mRNA present, but there is accumulation of tailed molecules on the 3′ side of the stem, and most of these tails were longer than 2 nt (Fig. 2F). In the same RNA sample primed with the 3A-primer, the degradation intermediates in the stem were the predominant RNA molecules detected, and the pattern of addition of these tails was similar to the pattern of tails >2 nt in the standard primer sample (Fig. 2G). We also detected longer tails added after the stem–loop. Note that we did not detect the 2-nt tails at the 3′ end of the mRNAs in the sample primed with the 3A-primer. Even if the 3A-primer initiates reverse transcription on a 2-nt tail, the resulting molecule would have been scored as a 3-nt tail. Thus we cannot detect tails shorter than 3 nt by this approach but we could analyze many more histone mRNAs in detail that were expressed at lower levels, as well as obtaining a >100-fold increase in the number of reads containing U-tails. We observed a wide variety of tail lengths since cDNA is primed 3 nt from the end of the ligated primer, resulting in the tail length being preserved.
We also mapped the 3′ ends of the histone H2AA3 mRNAs that ended 5′ of the stem–loop from the same two experiments, one primed with the standard primer (Fig. 2H) and the other with the 3A-primer (Fig. 2I). We detect the same pattern of degradation intermediates in each sample. While many of the molecules have no tails or 1- or 2-nt tails, tails >2 nt are found throughout the intermediates, and these were the only RNAs detected with the 3A primer. Thus we conclude that priming cDNA synthesis with the 3A-primer gives a similar pattern of degradation intermediates as priming with the standard primer but does not detect the very abundant mature length molecules with 1- or 2-nt tails. The enrichment of molecules with authentic U tails allowed a higher-resolution look at the spectrum of tailed intermediates, which are otherwise present in small numbers from many of the histone mRNAs. This result also suggests that we should be able to detect all the RNAs in the cell containing three or more nontemplated uridines using this strategy for cDNA synthesis.
To further demonstrate the ability of the 3A-primer to detect the oligouridylated intermediates, we analyzed two histone mRNAs that were present at low abundance. The HIST1H3H mRNA is expressed at moderate abundance, and the tailed intermediates in the 3′ side of the stem were detectable with the standard primer, but there were very few tailed intermediates detected in the body of the mRNA. With the 3A-primer the tailed intermediates in the body of the mRNA were readily detected (Fig. 3A), and their pattern was similar to those identified with the standard primer (Fig. 3B). When the tailed molecules in the 3′ side of the stem were analyzed (Fig. 3C,D) we observed a similar increase in the detection of the tailed degradation intermediates in the stem. When we analyzed a rare histone mRNA, HIST1H2AB, we could only detect significant number of tailed RNAs with the 3A-primer, but the distribution of the tails was similar to that of the other histone mRNAs (Fig. 3E). We did obtain a small number of sequences that are clearly artifacts with this approach due to internal priming at U-rich sequences, including at UGU or UCU sequences (Fig. 3F), which results in apparent 2U tails (Fig. 3G). When we primed with the 3A-primer we used a cut-off of nontemplated tails 3 nt or greater in AppEnd. Thus these artifactual “tails” were not scored by AppEnD.
Drosophila histone mRNAs also contain nontemplated nucleotides at the 3′ end
To investigate whether histone mRNA metabolism is conserved between human and Drosophila, we performed EnD-Seq on histone mRNAs from Drosophila embryos and ovaries. Drosophila histone mRNAs are cleaved 4 nt after the stem–loop, whereas vertebrate histone mRNAs are cleaved 5 nt after the stem–loop (Dominski et al. 2005). In both ovaries and embryos, the majority of the H2a and H3 histone mRNAs end in ACC (H2a) or AC (H3) 2 nt after the stem–loop, indicating that the mRNA is trimmed after processing as in human cells (Fig. 4A). There is no known ortholog of 3′hExo in Drosophila (Kupsco et al. 2006), and the enzyme that does the trimming is unclear. Both samples also had a substantial number of 1- to 2-nt nontemplated tails, resulting from further trimming of the mRNA followed by nucleotide addition, suggesting that a similar reaction can occur on histone mRNAs in Drosophila and mammalian cells to maintain the length of the mRNA. Surprisingly the tails in Drosophila ovaries were primarily A's while in embryos they were primarily U's.
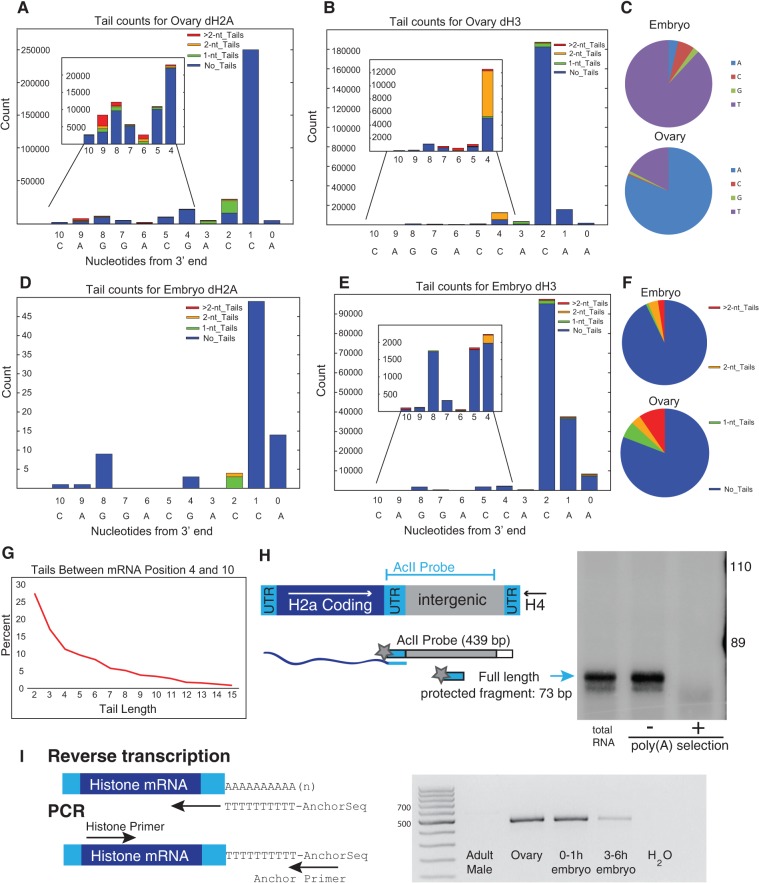
FIGURE 4.
Analysis of Drosophila histone mRNAs. (A,B) RNA from Drosophila ovary dH2a (A) and dH3 (B) was analyzed by EnD-Seq using a strategy to amplify the histone mRNAs selectively. Essentially all the 3′ ends mapped to the 3′ side of the stem in the stem–loop. The graphs represent the distribution of all recovered 3′ ends (with or without nontemplated tails) for each developmental stage. The inset shows the expansion of the distribution of 3′ ends from nucleotides −4 to −10 covering the 3′ side of the stem. (C) Pie charts showing nucleotide composition of single nucleotide tails in embryo (top) and ovary (bottom). (D,E) Distribution of 3′ ends and nontemplated tails in 3–6 h Drosophila embryos for (D) dH2a and (E) dH3. Note that the pattern of tail addition matches the ovary genes in (A) and (B) but the nucleotide composition is different. (F) Distribution of tail lengths for tails longer than 2 nt in the ovary RNA mapping between nucleotides 4 and 10 in the 3′ end of the stem. The tails >2 nt in the ovary were predominantly oligo(A). The tails of 1 or 2 nt in the embryo at nucleotides 4–10 were predominantly U's, and there were very few tails longer than 2 nt. (G) Chart showing the distribution of tail length between mRNA positions −4 and −10. Note that there are very few long tails. (H) S1 nuclease mapping of ovary histone mRNAs. Total RNA from ovaries was fractionated into poly(A+) and poly(A−) fractions on oligo(dT) cellulose. Total RNA, and equal proportions of the poly(A−) and poly(A+) RNA, were subjected to S1 nuclease mapping using the histone H2a gene labeled at the 3′ end of the AclI site as a probe. The S1 resistant fragments were resolved on a 6% polyacrylamine-7M urea gel and detected by autoradiography. A diagram of the S1 assay is shown on the left. The arrow indicates the fragment protected by the full-length mRNA. (I) RT-PCR analysis of the histone H2a mRNA. cDNA primed with oligo(dT) fused to an anchor primer was synthesized from 1 μg of total RNA from adult males, ovaries, 0–1 h embryos, and 3–6 h embryos, and then amplified using a primer near the 5′ end of the H2a mRNA. An amplicon of full-length H2a mRNA would be ∼500 nt long. Several amplicons were cloned and the majority had A-tails added in the 3′ end of the stem, consistent with the high-throughput sequencing data.
In addition to the mRNAs that ended after the stem–loop, a fraction of the RNAs from both the ovary and embryo ended in the 3′ side of the stem, and were similar to the major degradation intermediates found in mammalian cells. In the ovary, ∼8% of the histone ended in the 3′ side of the stem, and 10% of these contained nontemplated tails, ranging in length from 1 to 15 nt (Fig. 4B). These tails were predominantly oligo(A) tails. In the 4–6 h embryos, when the maternal histone mRNA has been degraded, and all the mRNA has been expressed zygotically, ∼4% of the RNAs ended in the 3′ side of the stem. In both the ovary and the embryo these shorter histone RNAs are similar to the major degradation intermediate found in mammalian cells. The mRNAs in the ovary likely represent degradation intermediates that were not completely degraded after the last cycle of DNA replication in the nurse cells, and are then deposited in the egg along with the other maternal histone mRNA synthesized after the last cycle of DNA replication in the nurse cells (Ambrosio and Schedl 1985; Ruddell and Jacobs-Lorena 1985). The shorter mRNAs in the embryo likely represent histone mRNAs that are undergoing degradation.
To confirm our sequencing results we directly assayed the RNAs with an S1 nuclease protection assay, using a probe that would be sensitive to small changes in length at the 3′ end. We also fractionated the RNA on oligo(dT) cellulose prior to the S1 nuclease assay. There is a previous report that some histone mRNAs in Drosophila ovaries are polyadenylated (Akhmanova et al. 1997). Akhmanova and coworkers cloned some of these polyadenylated mRNAs after RT-PCR, and many of the A-tails started at nucleotides within the 3′ side of the stem rather than 3′ of the stem–loop. The A-tails in their clones were at least the length of the oligo(dT) primer, so the actual A-tail length could not be determined. The percentage of histone mRNA that was polyadenylated also could not be determined.
The S1 nuclease assay detects the length of the mRNA that matches the probe (i.e., it does not detect the nontemplated tails). A fraction (5%–10%) of the total ovary histone mRNA was shorter than the processed histone mRNA, 5–10 nt shorter than the mature mRNAs (Fig. 4C). Very few of the RNAs bound to oligo(dT) cellulose, consistent with the sequencing data, which indicated the majority of the RNAs did not have A-tails and that >90% of the A tails that were present were <10 nt (Fig. 4B). Previously in our studies of the oligoadenylated histone mRNA present in Xenopus oocytes, we showed that oligo(dT) cellulose did not bind histone mRNAs containing short (<10 nt) A tails (Sánchez and Marzluff 2004), although these RNAs had been reported to be polyadenylated (Ruderman and Pardue 1978).
We used oligo(dT) to prime reverse transcription followed by RT-PCR to assay for “polyadenylated” histone mRNAs (Fig. 4C). These RNAs were readily detected in the ovary and 1-h embryo (which contains only histone mRNA synthesized during oogenesis) by RT-PCR, but only very small amounts of “polyadenylated” histone mRNA were detected in the RNA from the 3–6 h embryo, consistent with the sequencing results that showed the embryo contains predominantly U-tailed histone mRNA.
AppEnD detects genome-wide addition of nontemplated tails on short capped RNAs
We used gene-specific primers to apply EnD-Seq to histone genes, but the method can readily be extended to allow a genome-wide analysis of RNAs with nontemplated nucleotides at the 3′ end. In addition, AppEnD is also useful for detecting nontemplated tails in other types of sequencing data. A common approach to this problem is to strip homopolymers from raw reads before genomic read alignment (Henriques et al. 2013; Yao and Shi 2014). Such a prealignment read stripping approach has the shortcoming that it cannot distinguish between genomically encoded and nontemplated nucleotides. In addition, such an approach relies on knowing the sequence composition of the pattern to trim from the reads; consequently, the pattern must be sufficiently long that it is distinctive and must be known in advance.
In contrast to prealignment read stripping, AppEnD detects nontemplated tails by direct comparison with the reference genome during the read alignment process. This strategy provides three advantages: (1) It allows the detection of nontemplated additions without any assumptions about tail composition; (2) it detects tails as short as one nucleotide, and (3) it more effectively distinguishes nontemplated homopolymers from repeated genomic bases by direct comparison with the genome.
To illustrate the usefulness of this approach, we mapped the data set from cultured Drosophila cells of short capped RNAs resulting from stalling of RNA polymerase II immediately after initiation produced by Adelman and coworkers (Henriques et al. 2013). These RNAs were sequenced from the 3′ end. When the exosome was knocked down, a large increase in the number of small RNAs with short nontemplated tails was reported (Henriques et al. 2013). AppEnD successfully mapped these reads to the Drosophila genome and confirmed a sixfold to 10-fold increase in the number of oligo(A) tails after exosome knockdown (Fig. 5A). Since there was not an anchor primer used in this data set, but the Illumina primer was ligated directly onto the RNA, we could only unambiguously detect tailed molecules 3 nt or longer (Fig. 5F), because the Illumina primer sequence is removed by the instrument during data processing. Because the histone genes are present as tandemly repeated units (Lifton et al. 1978), reads are often not mapped to them in sequencing experiments. Using a Drosophila genome modified to contain a single histone repeat allowed us to identify and map the short capped RNAs expressed from the Drosophila histone genes (Fig. 5B). The tails in the exosome knockdown samples were almost exclusively oligo(A) tails with occasional substitutions of a U or C in the A-stretch, ranging up to 15 nt in length (Fig. 5C). We show the results for the tails that mapped to two Drosophila histone genes, histone H2B and histone H4. Since there are 100 copies of each histone gene and large amounts of histone mRNA produced in a growing cell, we obtained a large number of short capped RNA reads. For all five histone genes, the major pause site was ∼40 nt from the transcription start site (Fig. 5C, and data not shown). The paused tailed RNAs remaining after inhibition of transcription are generally not at the major pause site, suggesting that they may preferentially contain molecules that never reached the length of the major paused RNAs or were cleaved to give them a new 3′ end, as well as molecules that were released from the pause site prior to initiation of degradation.
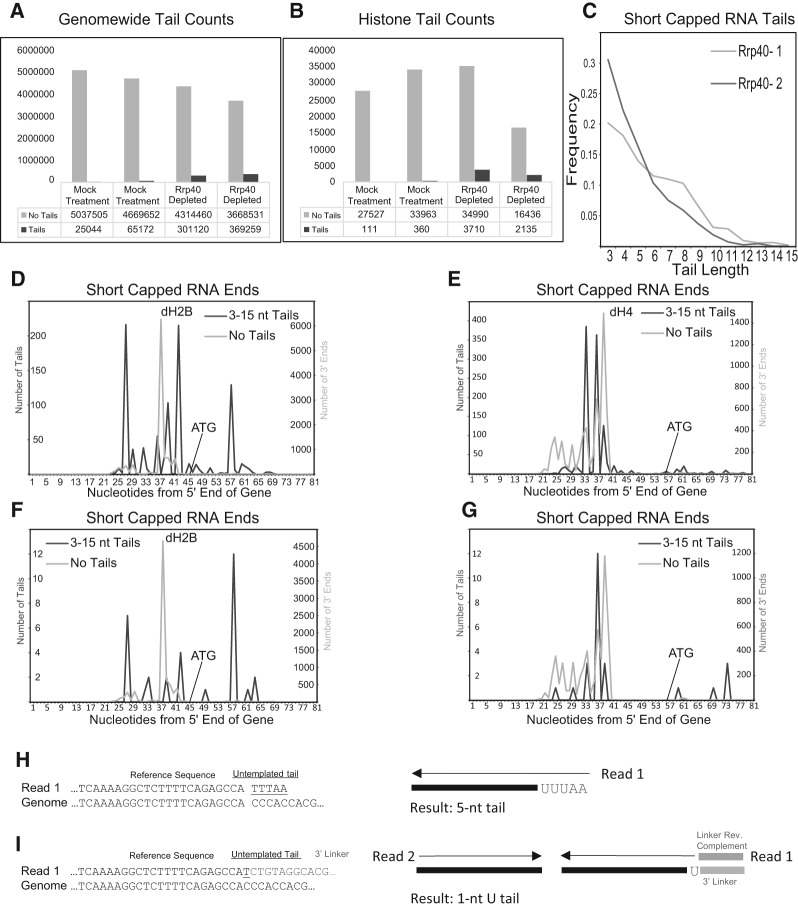
FIGURE 5.
AppEnD analysis of nontemplated tails on short capped transcripts. (A) We obtained the sequence data for paused capped RNAs from Drosophila tissue culture cells with and without exosome knockdown from Adelman and coworkers (Henriques et al. 2013), and analyzed the data using AppEnD. Note that the experimental results were generated by an approach different from EnD-Seq. These RNAs were sequenced from the 3′ end after ligation of the Illumina primer to the RNA, making it possible to reliably detect only tails 3 nt or longer. The number of reads that mapped with no tails (light gray) or tails (dark gray) are shown with or without knockdown of the exosome subunit Rrp40. (B) We obtained the reads on the tandemly repeated histone genes (not mapped in the initial paper) from the same data set by mapping the data to a Drosophila genome in which we placed a single histone repeat. (C) The length distribution of the tails 3 nt or longer, which were essentially all oligo(A) tails with occasional substitutions of a C or U in the A tail, is shown. (D,E) The distribution of sequences ending at the indicated nucleotide in the histone H2B (D) and histone H4 gene is shown in the sample with rrp40 knocked down. The light gray line is the untailed RNAs, and the dark gray line is the tailed RNAs. (F,G) Same as (D) and (E) but for the control sample (expressing Rrp40). Note that the patterns of tail additions match (D) and (E) well but there are far fewer tail additions. (H) Example of how AppEnD identifies and maps a short capped RNA read with a nontemplated tail. Very short tails (shorter than 3 nt) cannot be reliably be identified in this case because there is no 3′ linker marking the precise end of transcription. (I) Example showing how EnD-Seq with AppEnD can detect nontemplated additions as short as 1 nt due to the presence of the 3′ linker sequence in the read.
AppEnD can map alternative polyadenylation sites
Study of alternative polyadenylation requires the ability to accurately determine the position of the poly(A) tail on mRNAs. In both the PAS-Seq method (Shepard et al. 2011; Yao and Shi 2014) and the A-Seq method (Martin et al. 2012) cDNA is primed with an anchored oligo(dT) primer ending in a random dinucleotide, and the sequence obtained contains the 3′ UTR including the oligo(dT) primer and the anchor primer, allowing one to determine the site of adenylation at the nucleotide level. One challenge of the data analysis for these two methods is distinguishing priming at oligo(A) stretches encoded within the mRNA from authentic poly(A) tails. AppEnD automatically detects internally primed A-tails obtained from the PAS-Seq or A-Seq methods, keeping only the true poly(A) tails.
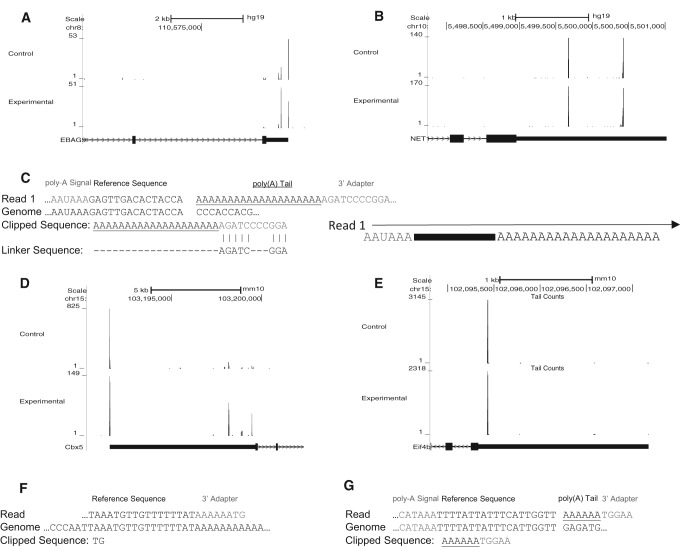
We applied AppEnD to two PAS-Seq data sets generated by sequencing from the 3′ UTR, through the oligo(A) tail from the primer, into the linker sequence (Fig. 6C). The results from two genes are shown, one, EBAG9, whose distribution of polyadenylation sites changed between the control and experimental conditions (Fig. 6A), and the other NET1, where the polyadenylation pattern remained constant (Fig. 6B). Figure 6C shows an example of a PAS-Seq read containing a poly(A) site. We also applied AppEnD to two A-Seq data sets from normal cells and cells with a polyadenylation factor knocked down, which were generated by sequencing from the 3′ UTR into the anchor primer. Examples of genes showing a change (Fig. 6D) and no change (Fig. 6E) between A-Seq experimental conditions are shown in Figure 6. Figure 6F,G show examples of a mispriming event and a true poly(A) site found from our analysis of the A-Seq data. We found that only 16% of the A-Seq reads contained authentic polyadenylation sites, 77% were misprimed, and 6% of the reads were uninformative since they did not get to the poly(A) tail. These numbers underscore the importance of filtering mispriming events, which make AppEnD useful for analyzing these types of data.
FIGURE 6.
Mapping alternative polyadenylation data with AppEnD. We analyzed PAS-Seq data (control and experimental sample) that was provided by Ami Ashar-Patel and Melissa Moore, as well as A-Seq data from Petar Grozdanov and Clint Macdonald. The PAS-Seq data were generated by sequencing from the 5′ end through the poly(A) tail (defined by the 20-nt dT primer used) into the anchor 3′ adapter incorporated as part of dT-priming for cDNA synthesis. The sites of poly(A) addition were mapped genome-wide. (A,B) The polyadenylation sites discovered by running AppEnD on PAS-Seq data. Poly(A) sites utilized by the EBAG9 gene (A) and the NET1 gene (B) are shown. The data from the control sample are at the top and the data for the experimental sample are at the bottom. Note that EBAG9 shows differential polyadenylation between control and experimental samples. (C) Example of PAS-Seq data. We required the length of the oligo(A) sequence that is nontemplated to be at least 4 nt to identify a polyadenylation site. Often the sequence of the 3′ adapter contained errors, presumably as a result of reading through the long oligo(A) stretch. (D,E) Poly(A) sites discovered using AppEnD on A-Seq data. The gene in (D) shows differential polyadenylation between control and experimental samples. (F,G) Examples of A-Seq reads containing (F) false positive poly(A) site due to adventitious priming and (G) a true poly(A) site.
DISCUSSION
Although the major 3′ ends of most RNAs, including rRNAs, tRNAs, mRNAs, miRNAs, and other structural RNAs have been defined, it has become apparent that there are a large number of RNA molecules whose 3′ ends can be altered either by addition of nontemplated nucleotides and/or by endonucleolytic or exonucleolytic cleavage. RNAs that have nontemplated nucleotides added onto the 3′ end are not readily detected or characterized by standard high-throughput sequencing methods, and identifying specific intermediates in mRNA degradation is hampered by their very low concentration among all RNA molecules in the cell. The combination of the EnD-Seq protocol and the AppEnD computational pipeline allows the identification of the 3′ end of RNA molecules, including identifying nontemplated nucleotides, in an unbiased manner. The AppEnD pipeline can also be used to analyze existing data sets where the information at the 3′ end of the RNA has been retained as part of the sequencing protocol.
The EnD-Seq protocol is similar to the protocols used for miRNA (Hafner et al. 2008) and pre-miRNA (Newman et al. 2011) analysis, and to the TAIL-Seq strategy recently reported by Narry Kim and coworkers (Chang et al. 2014). The similarity between these methods is that the information at the 3′ end of the RNA is preserved by efficient ligation of an activated linker in a relatively unbiased way onto the 3′ end (Hafner et al. 2011). The strength of the AppEnD computational pipeline is that it allows mapping of RNAs containing nontemplated nucleotides to the genome in a way that retains the information about the length and sequence of the nontemplated tail.
Additional insights into histone mRNA metabolism
Previously we used an earlier version of the methods described here to determine the pathway of 3′ to 5′ degradation of mammalian histone mRNA, detecting both intermediates that had no nontemplated tails and a wide variety of degradation intermediates with oligouridine tails (Slevin et al. 2014). Here we have extended these studies of histone mRNA to describe an additional feature of histone mRNA metabolism, the maintenance of the length of histone mRNAs by adding nontemplated nucleotides to the 3′ end. When we first directly sequenced the 3′ end of cytoplasmic histone mRNA by circular RT-PCR, we found that the 3′ end of histone mRNA is 2–3 nt shorter than the 3′ end formed in the nucleus during 3′ end formation (Mullen and Marzluff 2008). This result was confirmed by Heissmeyer and coworkers, who further showed that in cells with the 3′hExo knocked out the histone mRNAs end at the same site as the processed RNA formed in the nucleus, proving that 3′ hExo is responsible for trimming the 3′ end of histone mRNA (Hoefig et al. 2013). This finding is completely consistent with the properties of the 3′hExo/SLBP complex formed on the 3′ end of histone mRNA in vitro (Yang et al. 2009). Using the EnD-Seq approach, we were surprised to find that a substantial fraction of the 3′ end of every histone mRNA that we detected in mammalian S-phase cells (where histone mRNAs are stable) had lost an additional 1 or 2 nt from the 3′ end, but there were now either one or two nontemplated uridines at the 3′ end, restoring the normal length of cytoplasmic histone mRNA. We think this likely results from 3′hExo nibbling off an extra nucleotide or two, followed by addition of uridines by an unknown terminal uridyl transferase (TUTase). Since these modifications are abundant on histone mRNAs that are stable, and once histone mRNA degradation is initiated the same relative amounts of these two forms are maintained on the full-length RNAs that remain (Fig. 2A), we think that these are likely not degradation intermediates. In addition there are very few long tails observed in the nucleotides at the base of the stem, suggesting that normally initiation of degradation results in rapid degradation into the stem. Note that detection of these 1- to 2-nt tails was absolutely dependent on our technical and computational protocol. Adding an “anchor” primer to the 3′ end of the RNA unambiguously marks the 3′ end of the RNA, providing the resolution necessary to unambiguously detect single nucleotide tails, which could not have arisen from any PCR artifacts.
A major problem in detecting the nontemplated tails that are not present on mature RNAs is that they are often present in very low amounts, both because the degradation intermediates are present in low amounts, and the fraction of molecules of a particular length that are tailed is also low. In addition, in the case of the oligo(U) tails, there are relatively few long tails, probably because once the tails reach a certain length degradation of that molecule is triggered. One way to increase the detection of the tailed molecules is to select for tailed molecules, which requires knowing the tail sequence. We found that priming cDNA synthesis from the ligated RNA with the anchor primer ending in three additional A's greatly increased the number of oligouridine-tailed molecules that we detected. Most importantly, it did not change the pattern of tailed RNA molecules that were detected. The relatively abundant U-tailed molecules ending in the stem showed the same pattern with the 3A-primer, as was found when cDNA was primed with the anchor primer. In contrast, the molecules ending in one or two U's as a result of the reaction that restores the 3′ end were not detected with the 3A anchor primer.
As a result, we were able to obtain a large number of reads of low abundance tailed molecules both to give a clear description of the population of degradation intermediates on highly expressed histone mRNAs and to detect degradation intermediates on low-expressed histone mRNAs. The AppEnD computational pipeline is essential for rapid analysis of this data since it automatically removes sequences resulting from priming at internal U-stretches (those “tails” are identified as templated) and mispriming that results in tails shorter than 3 nt. We observed several very abundant species that resulted from mispriming (Fig. 3F) in some mRNAs. The disadvantage of this approach is that it requires at least a three uridine tail to make a definitive identification. However, this method likely will allow unambiguous detection of other oligouridylated RNAs which can then be further investigated.
Drosophila histone mRNAs are metabolized similarly to mammalian histone mRNAs
We applied the same approach to sequencing Drosophila histone mRNAs. In Drosophila ovaries and 0–1 h embryos (whose histone mRNAs are all maternal, synthesized in the oocyte) and the majority of the mRNAs ended 1 or 2 nt shorter than the nuclear processed Drosophila mRNA, similar to mammalian cells. There were also nontemplated tails of 1 or 2 nt on mRNAs that had been shortened by an additional 1 or 2 nt, which restored the length of the histone mRNA. Unlike in mammalian cells these tails were almost exclusive adenosine. Eight percent of the ovary histone mRNAs were shorter than the mature histone mRNAs ending in the 3′ side of the stem. Some of these contained oligo(A) tails up to 15 nt long. These are similar in structure to the major mammalian degradation intermediates. Many of these molecules did not have nontemplated tails. Most of these RNAs did not bind to oligo(dT) cellulose, although we could readily demonstrate the presence of adenylated histone mRNA by oligo(dT) priming cDNA from ovary or 0–1 h embryo RNA, which is all derived from the maternal mRNA provided during oogenesis.
In the embryo after destruction of the maternal mRNA and activation of zygotic histone mRNA synthesis, histone mRNAs are rapidly synthesized and degraded as cells continue rapidly dividing or initiate endocycles. In embryos most of the Drosophila histone mRNAs have U-tails rather than A-tails, both at the 3′ end as well as in the putative degradation intermediates partly into the stem. Our results suggest that the overall pathway of histone mRNA metabolism has been conserved between mammals and Drosophila, although in Drosophila there are developmental differences in the composition of the nontemplated nucleotides.
Application of AppEnD to other data sets
The Tail-Seq method recently reported by Narry Kim's group has a similar goal of identifying the 3′ ends of RNA molecules (Chang et al. 2014). It is similar to EnD-Seq (and methods used to sequence miRNAs) in that a linker is added on to the 3′ end prior to fragmenting the RNA and making the library using a primer complementary to the linker. Although the protocol can in principle identify any type of nontemplated tail addition, TAIL-seq is optimized for the analysis of poly(A) tail length. Since long homopolymer sequences (generally, longer than 30 nt) cannot be reliably read using Illumina sequencing technology, TAIL-seq relies upon a hidden Markov model to detect the transition from homopolymer runs to genomic DNA. It is not clear how accurately this approach can detect short homopolymer tails, and consequently in the TAIL-seq paper, they restricted analysis to tails 8 nt or longer, which included many oligo(A) tails.
We analyzed their TAIL-seq data (GSE51299) using AppEnD. This data set contains paired-end data in which read 1 starts at the 3′ end of the RNA and read 2 consists of 50 nt of sequence from 5′ end of the fragment. There is no linker sequence in the reads, since an Illumina linker was used and subsequently removed by the machine during the FASTQ creation process. AppEnD successfully identified both medium length (up to 30 nt) and short poly(A) tails, and some A tails that ended in nontemplated uridines. For poly(A) tails up to 30 nt, the precise site of polyadenylation could be identified. For long A tails this was not possible because the very long homopolymer stretches resulted in extremely low quality base calls in read 1. The TAIL-seq data set included 36 reads that mapped to the HIST2H2AA3 gene. Twelve of these ended at the trimmed 3′ end, 10 ended with an additional nt trimmed followed by a nontemplated uridine, and two ended with 2 nt trimmed and two nontemplated nucleotides. Although the number of reads are small, this is the same pattern we observed in our EnD-seq data, strongly suggesting that short U tails detected by each of the methods are not artifacts, and demonstrating that AppEnD can detect nontemplated short tails genome-wide.
The AppEnD method is applicable to any deep sequencing data set where the 3′ ends are sequenced. This includes small RNA data sets, such as miRNAs and pre-miRNAs (Newman et al. 2011), or the capped paused transcripts made by Pol II (Henriques et al. 2013). The data can be mapped genome-wide and does not require knowledge of the nontemplated nucleotides on the pre-miRNAs or miRNAs. One constraint is that for accurate mapping of 1- or 2-nt nontemplated tails, the data have to be generated using an anchor primer on the 3′ end to serve as the sequence that primes the cDNA. If that is not the case, we found we could not reliably map nontemplated nucleotides of <3 nt due to random heterogeneity at the end of many of the sequence reads. AppEnD is particularly applicable to the study of alternative polyadenylation, if the method used sequences the junction between the poly(A) tail and the mRNA. In such a case, AppEnD readily removes artifactual sequences resulting from internal priming at A-rich sequences.
In conclusion, EnD-Seq provides a platform for determining the 3′ end of RNA molecules together with any nontemplated nucleotides added to the transcript in a completely unbiased way, regardless of the length or composition of the nontemplated region. There are many potential applications of this platform for identifying novel cleavages and modifications of the 3′ ends of RNA molecules and for determining the details of RNA degradation proceeding in the 3′–5′ direction.
MATERIALS AND METHODS
RNA preparation
RNA samples are extracted from cultured cells using TRIzol. Prior to precipitation the aqueous phase containing RNA is extracted one time with an equal volume of chloroform, and the RNA was precipitated with ethanol. The RNA was recovered by centrifugation, resuspended in dH2O, and the samples treated with RQ1 DNase for 30 min. The DNase was heat inactivated for 20 min at 65°C, the sample extracted with phenol-chloroform, washed one time with chloroform, and precipitated with ethanol.
RNA 3′ end ligation
Preadenylated DNA linkers were made in-house as previously described (Hafner et al. 2008). We have used two different linkers successfully. The first linker was derived from Applied Biosystems SOLID system; the reverse SOLID sequence was preadenylated. Linker two was a modified version of NEB's universal miRNA cloning linker (Cat. #S1315S). We added 9 nt to the 3′ end of this linker to increase the Tm for the subsequent PCR reaction(s). We obtained similar results with both sets of linkers. Concatemerization of the preadenylated linker is prevented by blocking with 3′ end by addition of either a dideoxy nucleotide or an amine group. The amine blocker is equally effective and less expensive (Vigneault et al. 2008).
One and one-half micrograms of total RNA in a total volume of 10 µL was denatured for 10 min at 65°C and then cooled on ice for 2 min. Of note, 0.75 µg of preadenylated linker was added, followed by 200 units of truncated KQ T4 RNA ligase 2 (NEB Cat. # M0373S), using the buffer supplied by the manufacturer. Twenty units of RiboLock RNAse inhibitor was added to inhibit degradation. Ligation was carried out in the absence of ATP for 16 h at 16°C. After ligation the RNA was purified by phenol-chloroform extraction, followed by one extraction with chloroform and recovered by ethanol precipitation in the presence of glycoblue.
Reverse transcription
The ligated RNA was mixed with 0.4 mM linker complement (20 pmol) in water, heated to 65°C for 10 min and quickly cooled for 2 min on ice. The reaction was carried out in a final volume of 20 mL containing 2 mM of the 4 dNTPs, RT buffer (from a 10× RT buffer stock supplied by the manufacturer), 20 units RiboLock, 200 units Superscript III (Ambion), and 1 mM DTT for 1 h at 50°C.
RNA-sequencing primer design
We based the primer design on the Illumina TruSeq P5 and P7 adapters. These adapters were designed to hybridize by annealing the read one primer sequence (P5 arm) to the P7 flow cell sequence. To reduce PCR artifacts we replaced the read one primer with Illumina's V1.5 small RNA sequencing primer, and extended the primer on the 3′ end with a random 4 nt sequence to (1) increase library complexity and (2) in combination with bar codes, distinguish among sequences duplicated during PCR. Similarly for sequencing the histone mRNAs, Illumina's P7 adapter was modified by appending a histone gene-specific sequence at the 3′ end.
Multiple histone mRNAs of a particular class (i.e., H2A) were targeted by selecting a consensus sequence that was highly conserved in the coding region of each mRNA, and 200–300 nt from the 3′ end of the mRNA (Slevin et al. 2014). Initially we prepared libraries in a single round of PCR using 16 cycles. Later in order to multiplex 10 or more samples per MiSeq run we used additional indices, and the adapter/index addition was split into two limited rounds of PCR.
The first round primer set was universal to all samples: The ligated 3′ end was targeted using the reverse complement of the linker preceded by a random 4 nt sequence and part of the small RNA sequencing primer sequence (primer U5) while the 5′ end was targeted with a consensus histone sequence preceded by part of Illumina's read 2 primer (primer U7). During the second round the ligated/round 1 amplified end was targeted with the small RNA sequencing primer preceded by Illumina's P5 flow cell sequence (primer S5). In contrast the U7 amplified end was targeted with Illumina's read 2 primer followed by a one of Illumina's low-throughput indices (LTI) and the P7 Flow cell sequence (primer S7).
Library preparation
For the first round of PCR 14–16 cycles were performed with primers that target the linker and a gene specific sequence. PCR reactions were done with NEB Q5 polymerase (Cat. # M0491S). Removal of primer–dimers was done using an AMPure XP bead purification. Libraries were eluted in a volume of 20 µL of dH2O and then quantified using a Qubit Flourometer. During the second round of PCR 14–16 cycles were performed using ∼50 ng of the first round material (usually the entire reaction). During the second round the index and adapter sequences (P5 and P7) were added. Similar to the first round primer–dimer removal was accomplished using an AMPure XP bead purification. Libraries were eluted in a volume of 20 µL of dH2O and then quantified using a Qubit Flourometer.
Quality check
Libraries were initially checked using an Agilent Bioanalyzer. Secondary analysis was done with a conventional PCR using an aliquot of each library with a histone specific (P7 arm) and V1.5 (P5 arm) primer to check for the target insert and sequencing primer.
High-throughput sequencing
All high-throughput sequencing was done on an Illumina MiSEQ. The read one primer cocktail associated with the Illumina MiSeq does not contain the V1.5 small RNA sequencing primer. Therefore, 3 µL of a 100 µM stock of V1.5 was pipetted into the MiSeq cartridge well that contained the read one primer cocktail. Libraries were pooled and loaded at a final [C] of 8.5 pM. Due to the low complexity of our histone libraries we mixed in Illumina's PhiX control library. The histone/PhiX composition was 70% and 30%, respectively.
Computationally locating 3′ ends from EnD-Seq data
We used bowtie2 in local alignment mode (Langmead and Salzberg 2012) with default settings to map reads to either hg19 or dm3. A custom sequence including one copy of the histone repeat with the 5 histone genes (H1, H2A, H2B, H3, and H4) was added to the dm3 index, since the histone genes are not present in the dm3 assembly. Local alignment mode maximizes the alignment score of the whole read and will computationally remove (“soft clip”) portions of the beginning or end of a read that does not match the genome. We use this feature to detect the portion of EnD-Seq reads containing the 3′ ends of transcripts, including any nontemplated additions. Although spliced aligners are usually used for RNA-seq data, the histone genes are not generally spliced, so we chose to use bowtie2. A spliced aligner that performs soft clipping, such as Mapsplice (Wang et al. 2010) or Star (Dobin et al. 2013), could also be used.
Our EnD-seq sequencing strategy produces paired-end reads, although this is not essential, since sufficient information is present in the read that contains the 3′ end. Read 1 contains the reverse complement of the ligated linker followed by the reverse complement of any nontemplated additions, then the genomic portion of the transcript. Read 2 provides additional genomic context to aid in aligning read 1 but does not generally contain 3′ end information. We thus look for read 1 sequences whose alignments begin with a soft-clipped portion. To account for possible sequencing errors, we detect the linker within this soft-clipped portion by performing dynamic programming alignment to the known linker sequence using the Needleman–Wunsch algorithm. The remainder of the soft clipped portion of the read beyond the end of the linker as detected by this alignment represents a nontemplated addition. The end of the linker also indicates the precise position of the 3′ end of the RNA molecule being sequenced and thus provides important information that aids in the computational identification of 3′ nontemplated tails, and allows us to accurately determine nontemplated tails as short as 1 nt. Since the linker is at the beginning of read 1, this part of the read is generally of high quality. This represents a distinct advantage of EnD-seq over other sequencing strategies that either lack a 3′ linker or sequence it at the end of the read.
Mapping short capped RNAs and polyadenylation sites
We used AppEnD to map short capped RNAs (Henriques et al. 2013), PAS-Seq, and A-Seq data. This demonstrates the usefulness of the method for mapping other types of data than just EnD-seq. The protocol used to sequence short capped RNAs in this case produced single-end reads starting with the 3′ end of the RNA (Fig. 5H), since an Illumina linker was ligated onto the 3′ end of the RNA. These data sets were a single direction read from the 3′ end of the RNA. Unlike EnD-seq data, there is no linker present on the end of these short capped RNA reads, making it more difficult to distinguish short nontemplated additions from read errors. We therefore restricted analysis to nontemplated tails that were homopolymers at least 3 nt in length. We could not have reliably assigned reads that had shorter number of nontemplated bases or that had a mixed composition. This is in contrast to our ability to assign any nontemplated read regardless of length or composition with our EnD-Seq protocol.
The PAS-Seq protocol utilizes an anchored 20-nt dT primer ending in two random nucleotides to generate the cDNA, while the A-Seq strategy is similar but contains a 6-nt dT primer followed by a stem–loop and an additional 14 dTs. Short cDNA fragments were sequenced from the 5′ end of the mRNA producing a single end read with up to 20 nontemplated A’s (PAS-Seq) followed by the complement of the anchor on the dT primer (Fig. 6C) or six nontemplated A’s (A-Seq) followed by the sequencing adapter. In PAS-Seq, because the reads end with the sequencing adapter, the adapter sequence is generally of low quality, since it follows a long stretch of repeated A’s. Nevertheless, the presence of the adapter in the reads provides useful information that indicate how many nucleotides of the poly(A) tail were nontemplated, which helps distinguish authentic poly(A) tails from mispriming events. One of the challenges in analyzing PAS-Seq or A-Seq data is detecting false-positive polyadenylation sites due to mispriming events that can occur when the PAS-Seq primer anneals to stretches of repeated genomic A's. We detected such false positives by requiring that the 5 nt immediately preceding the soft-clipped portion of the read were not all A's. This shows the clear advantage of our method compared with a commonly used strategy in which reads are stripped of repeated A's before alignment to the genome; to such a strategy, mispriming events appear the same as true positive poly(A) sites, and must be identified in a separate computational step. However, by locating the precise position at which a read stops matching the genome, we are able to effectively detect misprimed reads.
S1 protection assay
Five micrograms of total RNA for each genotype was used for the S1 nuclease protection assay. The probe and method used have been previously described (Salzler et al. 2013). Briefly, the probe was generated by end labeling H2a DNA digested with AclI with α-32P-dCTP and Klenow Polymerase (NEB). After release from the TOPO TA vector (Invitrogen) by digestion with HindIII (NEB), the probe was gel purified and hybridized with the indicated RNA sample at either at 40°C overnight. Following digestion by S1 nuclease (Promega), protected DNA fragments were resolved on a 6% acrylamide-7M Urea gel and visualized by autoradiography.
RT-PCR
One microgram of total RNA was subject to DNAse treatment (Thermo) and hybridized with an anchor-oligo(dT) primer (5′-tttttttttttttttggaggttaggggaggttagg-3′). After reverse transcription (Superscript III, Invitrogen) cDNA was used as template for PCR with the H2a forward Primer (5′-ggccatgtctggacgtggaaaaggt-3′) and a reverse primer complementary to the anchor sequence (5′-cctaacctcccctaacctcc-3′). The products were resolved on a 2% agarose gel; the bands were excised and purified, subcloned (CloneJET PCR cloning kit, Thermo) and sequenced.
DATA DEPOSITION
The source code for AppEnD is available at https://code.google.com/p/append/ The data for Figure 2A–E are deposited at GEO (GSE54922); Figures 2F–I, 3, and 4 are GSE68471. The data in Figure 5 were from Henriques et al. 2013.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants GM29832 (W.F.M.), GM58921 (W.F.M. and R.J.D.), HG06272 and National Science Foundation (NSF) grant ABI/EF0850237 (J.F.P.), NSF Graduate Research fellowship DGE-1144081 (J.D.W.), and NIH Fellowship F32GM87059 (M.K.S.). We thank Ami Bashar and Melissa Moore for the PAS-Seq data; Petar Grozdanov and Clint Macdonald for the A-Seq data; and David Fargo and Karen Adelman for the short capped RNA data, and for helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.048785.114.
REFERENCES
- Akhmanova A, Miedema K, Kremer H, Hennig W. 1997. Two types of polyadenated mRNAs are synthesized from Drosophila replication-dependent histone genes. Eur J Biochem 244: 294–300. [DOI] [PubMed] [Google Scholar]
- Ambrosio L, Schedl P. 1985. Two discrete modes of histone gene expression during oogenesis in Drosophila melanogaster. Dev Biol 111: 220–231. [DOI] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Kaygun H, Marzluff WF. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol Cell 12: 295–305. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Marzluff WF. 2005. Differences and similarities between Drosophila and mammalian 3′ end processing of histone pre-mRNAs. RNA 11: 1835–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55. [DOI] [PubMed] [Google Scholar]
- Graves RA, Marzluff WF. 1984. Rapid reversible alterations in histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol 4: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, et al. 2011. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17: 1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. 2013. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 52: 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. 2013. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol 20: 73–81. [DOI] [PubMed] [Google Scholar]
- Hoque M, Li W, Tian B. 2014. Accurate mapping of cleavage and polyadenylation sites by 3′ region extraction and deep sequencing. Methods Mol Biol 1125: 119–129. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. 2006. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314: 1893. [DOI] [PubMed] [Google Scholar]
- Kupsco JM, Wu M-J, Marzluff WF, Thapar R, Duronio RJ. 2006. Genetic and biochemical characterization of Drosophila Snipper: a promiscuous member of the metazoan 3′hExo/ERI-1 family of 3′ to 5′ exonucleases. RNA 12: 2103–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton RP, Goldberg ML, Karp RW, Hogness DS. 1978. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harbor Symp Quant Biol 42: 1047–1051. [DOI] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep 1: 753–763. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. 2014. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510: 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48: 443–453. [DOI] [PubMed] [Google Scholar]
- Newman MA, Mani V, Hammond SM. 2011. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA 17: 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. 2009. 3′ uridylation precedes decapping in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddell A, Jacobs-Lorena M. 1985. Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc Natl Acad Sci 82: 3316–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman JV, Pardue ML. 1978. A portion of all major classes of histone messenger RNA in amphibian oocytes is polyadenylated. J Biol Chem 253: 2018–2025. [PubMed] [Google Scholar]
- Salzler HR, Tatomer DC, Malek PY, McDaniel SL, Orlando AN, Marzluff WF, Duronio RJ. 2013. A sequence in the Drosophila H3-H4 promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev Cell 24: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R, Marzluff WF. 2004. The oligoA tail on histone mRNA plays an active role in translational silencing of histone mRNA during Xenopus oogenesis. Mol Cell Biol 24: 2513–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl EC, Steitz JA. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J 13: 2432–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon JM, Bousquet-Antonelli C, Lange H, Gagliardi D. 2013. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res 41: 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM. 2004. Uridine addition after microRNA-directed cleavage. Science 306: 997. [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin MK, Meaux S, Welch JD, Bigler R, de Marval PLM, Su W, Prins JF, Rhoads RE, Marzluff WF. 2014. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol Cell 53: 1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, Darzynkiewicz E, Jemielity J, Marzluff WF, Rhoads RE. 2013. mRNAs containing the histone 3′ stem–loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA 19: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneault F, Sismour AM, Church GM. 2008. Efficient microRNA capture and bar-coding via enzymatic oligonucleotide adenylation. Nat Methods 5: 777–779. [DOI] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. 2010. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38: e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Purdy M, Marzluff WF, Dominski Z. 2006. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J Biol Chem 281: 30447–30454. [DOI] [PubMed] [Google Scholar]
- Yang XC, Sullivan KD, Marzluff WF, Dominski Z. 2009. Studies on the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol 29: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Shi Y. 2014. Global and quantitative profiling of polyadenylated RNAs using PAS-seq. Methods Mol Biol 1125: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]