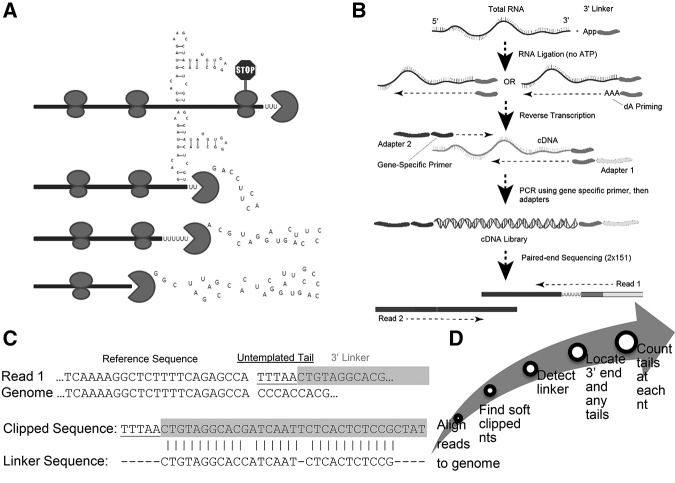
FIGURE 1.
EnD-Seq and AppEnD strategy. (A) Schematic of the 3′ end of a hypothetical RNA molecule, indicating potential intermediates in 3′–5′ degradation resulting from bound proteins or RNA secondary structure that might slow 3′–5′ exonuclease degradation. (B) EnD-seq sequencing strategy. We ligate a preadenylated 3′ linker onto the 3′ end to preserve the information at the 3′ end of RNAs in total cell RNA. We then prime cDNA synthesis using a primer antisense to the linker. Alternatively, we can prime cDNA with the antisense primer extended with a short sequence (e.g., three A's to enrich uridylated RNAs). Gene-specific primers can be used to enrich for transcripts of interest. (C) Example of sequence containing an untemplated tail and one containing a single U-tail obtained from the EnD-Seq data. Note that the linker sequence can be identified even if it contains sequencing errors. (D) Flow chart for the analysis of sequencing data by App-EnD.